CRISPR技术综述

Leading Edge
Perspective The Heroes of CRISPR
Eric https://www.360docs.net/doc/233303519.html,nder1,2,3,*
1Broad Institute of MIT and Harvard,415Main Street,Cambridge,MA02142,USA
2Department of Biology,Massachusetts Institute of Technology,Cambridge,MA02139,USA
3Department of Systems Biology,Harvard Medical School,Boston,MA02115,USA
*Correspondence:lander@https://www.360docs.net/doc/233303519.html,
https://www.360docs.net/doc/233303519.html,/10.1016/j.cell.2015.12.041
Three years ago,scientists reported that CRISPR technology can enable precise and ef?cient genome editing in living eukaryotic cells.Since then,the method has taken the scienti?c community by storm,with thousands of labs using it for applications from biomedicine to agriculture.Yet,the preceding20-year journey—the discovery of a strange microbial repeat sequence;its recognition as an adaptive immune system;its biological characterization;and its repurposing for genome en-gineering—remains little known.This Perspective aims to?ll in this backstory—the history of ideas and the stories of pioneers—and draw lessons about the remarkable ecosystem underlying scien-ti?c discovery.
Introduction
It’s hard to recall a revolution that has swept biology more swiftly than CRISPR.Just3years ago,scientists reported that the CRISPR system—an adaptive immune system used by mi-crobes to defend themselves against invading viruses by recording and targeting their DNA sequences—could be repur-posed into a simple and reliable technique for editing,in living cells,the genomes of mammals and other organisms.CRISPR was soon adapted for a vast range of applications—creating complex animal models of human-inherited diseases and can-cers;performing genome-wide screens in human cells to pinpoint the genes underlying biological processes;turning spe-ci?c genes on or off;and genetically modifying plants—and is being used in thousands of labs worldwide.The prospect that CRISPR might be used to modify the human germline has stim-ulated international debate.
If there are molecular biologists left who have not heard of CRISPR,I have not met them.Yet,if you ask scientists how this revolution came to pass,they often have no idea.The immunolo-gist Sir Peter Medawar observed,‘‘The history of science bores most scientists stiff’’(Medawar,1968).Indeed,scientists focus relentlessly on the future.Once a fact is?rmly established, the circuitous path that led to its discovery is seen as a distraction.
Yet,the human stories behind scienti?c advances can teach us a lot about the miraculous ecosystem that drives biomedical progress—about the roles of serendipity and planning,of pure curiosity and practical application,of hypothesis-free and hy-pothesis-driven science,of individuals and teams,and of fresh perspectives and deep expertise.Such understanding is impor-tant for government agencies and foundations that together invest,in the U.S.alone,more than$40billion in biomedical research.It is also important for a general public who often imag-ines scientists as lone geniuses cloistered in laboratories.And, for trainees,it is especially valuable to have a realistic picture of scienti?c careers,as both guide and inspiration.
Over the past several months,I have sought to understand the 20-year backstory behind CRISPR,including the history of ideas and the stories of individuals.This Perspective is based on pub-lished papers,personal interviews,and other materials—including rejection letters from journals.At the end,I try to distill some general lessons.(As background,Figure1provides a brief overview of a type II CRISPR system,the variety that has been repurposed for genome engineering.)
Most of all,the Perspective describes an inspiring ensemble of a dozen or so scientists who—with their collaborators and other contributors whose stories are not elaborated here—discovered the CRISPR system,unraveled its molecular mechanisms,and repurposed it as a powerful tool for biological research and biomedicine.Together,they are the Heroes of CRISPR. Discovery of CRISPR
The story starts in the Mediterranean port of Santa Pola on Spain’s Costa Blanca,where the beautiful coast and vast salt marshes have for centuries attracted vacationers,?amingoes, and commercial salt producers.(The geography of the story is shown in Figure2.)Francisco Mojica,who grew up nearby,fre-quented those beaches,and it was no surprise that,when he began his doctoral studies in1989at the University of Alicante, just up the coast,he joined a laboratory working on Haloferax mediterranei,an archaeal microbe with extreme salt tolerance that had been isolated from Santa Pola’s marshes.His advisor had found that the salt concentration of the growth medium ap-peared to affect the way in which restriction enzymes cut the mi-crobe’s genome,and Mojica set out to characterize the altered fragments.In the?rst DNA fragment he examined,Mojica found a curious structure—multiple copies of a near-perfect,roughly palindromic,repeated sequence of30bases,separated by spacers of roughly36bases—that did not resemble any family of repeats known in microbes(Mojica et al.,1993).
The28-year-old graduate student was captivated and devoted the next decade of his career to unraveling the
mystery.
18Cell164,January14,2016a2016Elsevier Inc.
He soon discovered similar repeats in the closely related H.volcanii ,as well as in more distant halophilic https://www.360docs.net/doc/233303519.html,b-ing through the scienti?c literature,he also spotted a connection with eubacteria:a paper by a Japanese group (Ishino et al.,1987)had mentioned a repeat sequence in Escherichia coli that had a similar structure,although no sequence similarity,to the Halo-ferax repeats.These authors had made little of the observation,but Mojica realized that the presence of such similar structures in such distant microbes must signal an important function in pro-karyotes.He wrote up a paper reporting this new class of repeats (Mojica et al.,1995)before heading off for a short post-doctoral stint at Oxford.
Mojica returned home to take up a faculty position at the Uni-versity of Alicante.Because the school had hardly any start-up funds or lab space,he turned to bioinformatics to investigate the strange repeats,which he dubbed short regularly spaced re-peats (SRSRs);the name would later be changed,at his sugges-tion,to clustered regularly interspaced palindromic repeats (CRISPR)(Jansen et al.,2002;Mojica and Garrett,2012
).
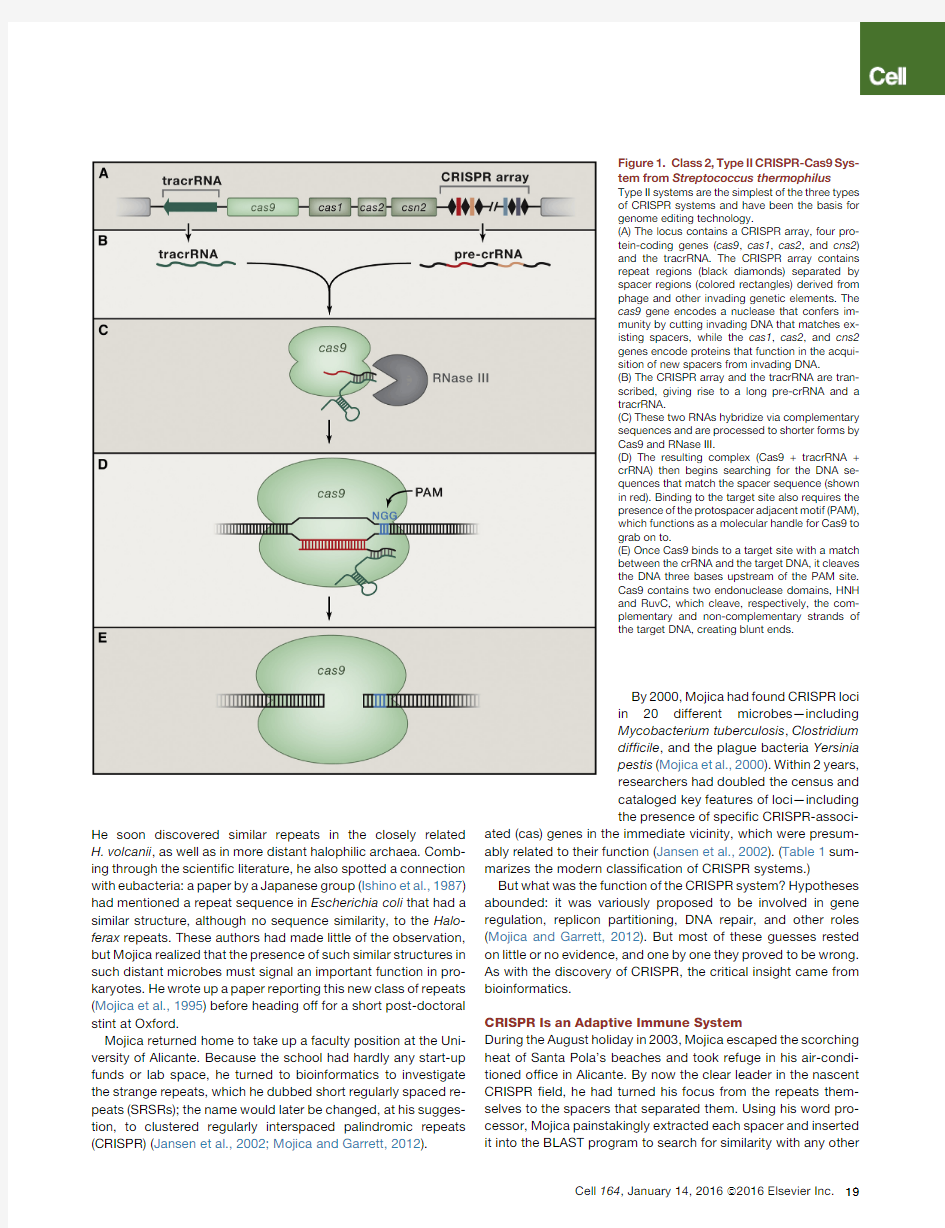
Figure 1.Class 2,Type II CRISPR-Cas9Sys-tem from Streptococcus thermophilus
Type II systems are the simplest of the three types of CRISPR systems and have been the basis for genome editing technology.
(A)The locus contains a CRISPR array,four pro-tein-coding genes (cas9,cas1,cas2,and cns2)and the tracrRNA.The CRISPR array contains repeat regions (black diamonds)separated by spacer regions (colored rectangles)derived from phage and other invading genetic elements.The cas9gene encodes a nuclease that confers im-munity by cutting invading DNA that matches ex-isting spacers,while the cas1,cas2,and cns2genes encode proteins that function in the acqui-sition of new spacers from invading DNA.
(B)The CRISPR array and the tracrRNA are tran-scribed,giving rise to a long pre-crRNA and a tracrRNA.
(C)These two RNAs hybridize via complementary sequences and are processed to shorter forms by Cas9and RNase III.
(D)The resulting complex (Cas9+tracrRNA +crRNA)then begins searching for the DNA se-quences that match the spacer sequence (shown in red).Binding to the target site also requires the presence of the protospacer adjacent motif (PAM),which functions as a molecular handle for Cas9to grab on to.
(E)Once Cas9binds to a target site with a match between the crRNA and the target DNA,it cleaves the DNA three bases upstream of the PAM site.Cas9contains two endonuclease domains,HNH and RuvC,which cleave,respectively,the com-plementary and non-complementary strands of the target DNA,creating blunt ends.
By 2000,Mojica had found CRISPR loci in 20different microbes—including Mycobacterium tuberculosis ,Clostridium dif?cile ,and the plague bacteria Yersinia pestis (Mojica et al.,2000).Within 2years,researchers had doubled the census and cataloged key features of loci—including the presence of speci?c CRISPR-associ-ated (cas)genes in the immediate vicinity,which were presum-ably related to their function (Jansen et al.,2002).(Table 1sum-marizes the modern classi?cation of CRISPR systems.)
But what was the function of the CRISPR system?Hypotheses abounded:it was variously proposed to be involved in gene regulation,replicon partitioning,DNA repair,and other roles (Mojica and Garrett,2012).But most of these guesses rested on little or no evidence,and one by one they proved to be wrong.As with the discovery of CRISPR,the critical insight came from bioinformatics.
CRISPR Is an Adaptive Immune System
During the August holiday in 2003,Mojica escaped the scorching heat of Santa Pola’s beaches and took refuge in his air-condi-tioned of?ce in Alicante.By now the clear leader in the nascent CRISPR ?eld,he had turned his focus from the repeats them-selves to the spacers that separated https://www.360docs.net/doc/233303519.html,ing his word pro-cessor,Mojica painstakingly extracted each spacer and inserted it into the BLAST program to search for similarity with any other
Cell 164,January 14,2016a2016Elsevier Inc.19
known DNA sequence.He had tried this exercise before without success,but the DNA sequence databases were continually ex-panding and this time he struck gold.In a CRISPR locus that he had recently sequenced from an E.coli strain,one of the spacers matched the sequence of a P1phage that infected many E.coli strains.However,the particular strain carrying the spacer was known to be resistant to P1infection.By the end of the week,he had slogged through 4,500spacers.Of 88spacers with sim-ilarity to known sequences,two-thirds matched viruses or con-jugative plasmids related to the microbe carrying the spacer.Mojica realized that CRISPR loci must encode the instructions for an adaptive immune system that protected microbes against speci?c infections.
Mojica went out to celebrate with colleagues over cognac and returned the next morning to draft a paper.So began an 18-month odyssey of frustration.Recognizing the importance of the discovery,Mojica sent the paper to Nature .In November 2003,the journal rejected the paper without seeking external re-view;inexplicably,the editor claimed the key idea was already known.In January 2004,the Proceedings of the National Acad-emy of Sciences decided that the paper lacked suf?cient ‘‘nov-elty and importance’’to justify sending it out to review.Molecular Microbiology and Nucleic Acid Research rejected the paper in turn.By now desperate and afraid of being scooped,Mojica
sent the paper to Journal of Molecular Evolution .After 12more months of review and revision,the paper reporting CRISPR’s likely function ?nally appeared on February 1,2005(Mojica et al.,2005).
At about the same time,CRISPR was the focus of attention in another,rather unlikely,venue:a unit of the French Ministry of Defense,some 30miles south of Paris.Gilles Vergnaud,a human geneticist trained at the Institut Pasteur,had received
doctoral and post-doctoral support from the Direction Ge
′ne ′rale de l’Armement.When he completed his studies in 1987,he joined the government agency to set up its ?rst molecular biology lab.For the next 10years,Vergnaud continued his work on human genetics.But when intelligence reports in the late 1990s raised concerns that Saddam Hussein’s regime in Iraq was developing biological weapons,the Ministry of Defense asked Vergnaud in 1997to shift his group’s focus to forensic microbiology—developing methods to trace the source of pathogens based on subtle genetic differences among strains.Establishing a joint lab with the nearby Institute
of Genetics and Microbiology at Universite
′Paris-Sud,he set out to use tandem-repeat polymorphisms—which were the workhorse of forensic DNA ?ngerprinting in humans—to char-acterize strains of the bacteria responsible for anthrax and
plague.
Figure 2.The Twenty-Year Story of CRISPR Unfolded across Twelve Cities in Nine Countries
For each ‘‘chapter’’in the CRISPR ‘‘story,’’the map shows the sites where the primary work occurred and the ?rst submission dates of the papers.Green circles refer to the early discovery of the CRISPR system and its function;red to the genetic,molecular biological,and biochemical characterization;and blue to the ?nal step of biological engineering to enable genome editing.
20Cell 164,January 14,2016a2016Elsevier Inc.
The French Defense Ministry had access to a unique trove of 61Y.pestis samples from a plague outbreak in Vietnam in 1964–1966.Vergnaud found that these closely related isolates were identical at their tandem-repeat loci—with a sole exception of a site that his colleague Christine Pourcel discovered was the CRISPR locus.The strains occasionally differed by the presence of new spacers,which were invariably acquired in a polarized fashion at the ‘‘front’’end of the CRISPR locus (Pourcel et al.,2005).Strikingly,many of the new spacers corresponded to a prophage present in the Y.pestis genome.The authors pro-posed that the CRISPR locus serves in a defense mecha-nism—as they put it,poetically,‘‘CRISPRs may represent a memory of ‘past genetic aggressions.’’’Vergnaud’s efforts to publish their ?ndings met the same resistance as Mojica’s.The paper was rejected from the Proceedings of the National Acad-emy of Sciences ,Journal of Bacteriology,Nucleic Acids Research ,and Genome Research ,before being published in Microbiology on March 1,2005.
Finally,a third researcher—Alexander Bolotin,a Russian e ′mi-gre
′who was a microbiologist at the French National Institute for Agricultural Research—also published a paper describing the extrachromosomal origin of CRISPR,in Microbiology in September 2005(Bolotin et al.,2005).His report was actually submitted a month after Mojica’s February 2005paper had already appeared—because his submission to another journal had been rejected.Notably,Bolotin was the ?rst to speculate how CRISPR conferred immunity—proposing that transcripts from the CRISPR locus worked by anti-sense RNA inhibition of phage gene expression.Although reasonable,the guess would prove to be wrong.
Experimental Evidence that CRISPR Confers Adaptive Immunity and Employs a Nuclease
Like Mojica,Philippe Horvath could hardly have chosen a thesis topic that was more local or less sexy.As a Ph.D.student at the University of Strasbourg,he concentrated on the genetics of a lactic-acid bacteria used in the production of sauerkraut—the central ingredient in the Alsatian specialty choucroute garnie .Given his interest in food science,Horvath skipped doing post-doctoral research and in late 2000joined Rhodia Food,a maker
of bacterial starter cultures located in Dange
′-Saint-Romain in western France,to set up its ?rst molecular biology lab.The company was later acquired by the Danish ?rm Danisco,which was itself acquired by DuPont in 2011.
Rhodia Food was interested in Horvath’s microbiological skills because other lactic-acid bacteria,such as Streptococcus ther-mophilus ,are used to make dairy products,such as yogurt and cheese.Horvath’s mission included developing DNA-based methods for precise identi?cation of bacterial strains and over-coming the frequent phage infections that plagued industrial cul-tures used in dairy fermentation.Understanding how certain S.thermophilus strains protect themselves from phage attack was thus of both scienti?c interest and economic importance.After learning about CRISPR at a Dutch conference on lactic-acid bacteria in late 2002,Horvath began using it to genotype his strains.By late 2004,he noticed a clear correlation between spacers and phage resistance—as would be reported just a few months later by Mojica and Vergnaud.In 2005,Horvath and colleagues—including Rodolphe Barrangou,a newly minted Ph.D.at Danisco USA,and Sylvain Moineau,a distinguished
phage biologist at Universite
′Laval in Que ′bec City—set out to directly test the hypothesis that CRISPR was an adaptive im-mune system.Notably,Moineau had also been an industrial sci-entist.He had earned his Ph.D in Food Sciences at Laval,also studying lactic-acid bacteria,and had worked at Unilever Corpo-ration before returning to academia at Laval;he had been collab-orating with Rhodia Food since 2000.
Using a well-characterized phage-sensitive S.thermophilus strain and two bacteriophages,these investigators performed genetic selections to isolate phage-resistant bacteria.Rather than harboring classical resistance mutations (such as in a cell-surface receptor required for phage entry),the
resistant
Table 1.Classi?cation and Examples of CRISPR Systems gle protein effectors.These classes are subdivided into ?ve types (Makarova et al.,2015),with type IV remaining a putative type within Class 1.Although only Class 2systems have been adapted for genome engineering,the results described in this review emerged from studying a diversity of CRISPR-Cas systems.(Type III-B systems are not discussed but represent an unusual system that targets RNA rather than DNA [Hale et al.,2009].)
Cell 164,January 14,2016a2016Elsevier Inc.21
strains had acquired phage-derived sequences at their CRISPR loci(Barrangou et al.,2007).Moreover,the insertion of multiple spacers correlated with increased resistance.They had seen ac-quired immunity in action.
They also studied the role of two of the cas genes:cas7and cas9.Bacteria required cas7in order to gain resistance,but those carrying a phage-derived spacer did not need the gene to remain resistant—suggesting that Cas7was involved in generating new spacers and repeats,but not in immunity itself. In contrast,cas9—whose sequence contained two types of nuclease motifs(HNH and RuvC)and whose product thus pre-sumably cut nucleic acids(Bolotin et al.,2005;Makarova et al., 2006)—was necessary for phage resistance;the Cas9protein was an active component of the bacterial immune system. (Warning:In the early CRISPR literature,the now-famous cas9 gene was called cas5or csn1.)
Finally,they found that rare phage isolates that overcame CRISPR-based immunity carried single-base changes in their genomes that altered the sequence corresponding to the spacers.Immunity thus depended on a precise DNA sequence match between spacer and target.
Programming CRISPR
John van der Oost,who got his Ph.D.from the Free University of Amsterdam in1989,originally set out to solve the world’s clean-energy needs by using cyanobacteria to produce biofuels. He studied metabolic pathways in bacteria,working in Helsinki and Heidelberg before returning to Amsterdam.In1995,Wage-ningen University offered him a permanent position—but with a catch:they wanted him to expand a group working on extremo-phile microbes.van der Oost,who had once heard a talk while in Germany about Sulfolobus solfataricus,which thrives in the hot springs of Yellowstone National Park,was game to investigate the evolutionary differences in the metabolic pathways of these strange microbes.He began to collaborate with Eugene Koonin—an expert in microbial evolution and computational biology at the National Center for Biotechnology Information (NCBI)at the National Institutes of Health.Koonin had begun working on classifying and analyzing CRISPR systems,and on a visit in2005,he introduced van der Oost to the then-obscure ?eld of CRISPR(Makarova et al.,2006).
van der Oost had just received a major grant from the Dutch National Science Foundation.In addition to working on the prob-lem described in his proposal,he decided to use some of the funding to study CRISPR.(In his report to the agency5years later,he underscored the value of the agency’s policy of allowing researchers the freedom to shift their scienti?c plans.)
He and his colleagues inserted an E.coli CRISPR system into another E.coli strain that lacked its own endogenous system. This allowed them to biochemically characterize a complex of ?ve Cas proteins,termed Cascade(Brouns et al.,2008).
(E.coli has the more complex Class1,type I CRISPR system, in which the functions of Cas9are instead performed by the Cascade complex,together with the nuclease Cas3.See Table1.)
By knocking out each component individually,they showed that Cascade is required for cleaving a long precursor RNA,tran-scribed from the CRISPR locus,into61-nucleotide-long CRISPR RNAs(crRNAs).They cloned and sequenced a set of crRNAs that co-puri?ed with the Cascade complex and found that all started with the last eight bases of the repeat sequence,followed by the complete spacer and the beginning of the next repeat region.This?nding supported earlier suggestions that the palin-dromic nature of the repeats would lead to secondary structure formation in the crRNA(Sorek et al.,2008).
To prove that the crRNA sequences are responsible for CRISPR-based resistance,they set out to create the?rst arti?cial CRISPR arrays—programming CRISPR to target four essential genes in lambda(l)phage.As they predicted,the strains car-rying the new CRISPR sequence showed resistance to phage l.It was the?rst case of directly programming CRISPR-based immunity—a?u shot for bacteria.
The results also hinted that the target of CRISPR was not RNA (as Bolotin had proposed)but,rather,DNA.The authors had de-signed two versions of the CRISPR array—one in the anti-sense direction(complementary to both the mRNA and coding strand of the DNA locus)and one in the sense direction(complementary only to the other DNA strand).Although the spacers varied in their ef?cacy,the fact that the sense version worked strongly suggested that the target was not mRNA.Still,the evidence was indirect.With the journal editors urging caution about draw-ing a?rm conclusion,van der Oost’s paper in Science offered the notion that CRISPR targets DNA as a‘‘hypothesis.’’
CRISPR Targets DNA
Luciano Marraf?ni was?nishing his Ph.D.,working on Staphylo-coccus,at the University of Chicago,when he learned about CRISPR from Malcolm Casadaban,a faculty member in the department who was a world authority on phage genetics.Casa-daban had immediately seen the importance of discovery in 2005that CRISPR was likely to be an adaptive immune system and talked about CRISPR to everyone who would listen.Like many in the phage community,Marraf?ni was convinced that CRISPR could not work by RNA interference because this mech-anism would be too inef?cient to overcome the explosive growth that occurs upon phage infection.Instead,he reasoned,CRISPR must cut DNA—functioning,in effect,like a restriction enzyme. Marraf?ni was eager to pursue post-doctoral work in one of the handful of groups in the world studying CRISPR,but his wife had a good job as a translator in the Cook County,Illinois criminal courts and he felt he should stay in Chicago.He persuaded Erik Sontheimer,a biochemist at Northwestern University who had been working on RNA splicing and RNA interference,to let him join his lab to work on CRISPR.
Even before moving to Northwestern,Marraf?ni started working on CRISPR even as he completed his graduate work—exploring whether the Staphylococcus CRISPR system could block plasmid conjugation.He noticed that a strain of Staphylococcus epidermidis had a spacer that matches a region of the nickase(nes)gene encoded on plasmids from antibiotic-resistant Staphylococcus aureus.He showed that these plas-mids cannot be transferred to S.epidermidis but that disrupting either the nes sequence in the plasmid or the matching spacer sequences in the CRISPR locus in the genome abolishes inter-ference(Marraf?ni and Sontheimer,2008).Clearly,CRISPR blocked the plasmids,just as it blocked viruses.
22Cell164,January14,2016a2016Elsevier Inc.
Marraf?ni and Sontheimer thought brie?y about trying to reconstitute the CRISPR system in vitro to demonstrate that it cuts DNA.But the S.epidermidis system was too compli-cated—it had nine cas genes—and it was still too poorly charac-terized.Instead,they turned to molecular biology.Cleverly,they modi?ed the nes gene in the plasmid targeted by the CRISPR system—inserting a self-splicing intron in the middle of its sequence.If CRISPR targeted mRNA,the change would not affect interference because the intronic sequence would be spliced out.If CRISPR targeted DNA,the insertion would abolish interference because the spacer would no longer match.The results were clear:the target of CRISPR was DNA.
Marraf?ni and Sontheimer recognized that CRISPR was essentially a programmable restriction enzyme.Their paper was the?rst to explicitly predict that CRISPR might be repur-posed for genome editing in heterologous systems.‘‘From a practical standpoint,’’they declared,‘‘the ability to direct the speci?c addressable destruction of DNA that contains any given 24-to48-nucleotide target sequence could have considerable functional utility,especially if the system can function outside of its native bacterial or archaeal context.’’They even?led a pat-ent application including the use of CRISPR to cut or correct genomic loci in eukaryotic cells,but it lacked suf?cient experi-mental demonstration and they eventually abandoned it(Son-theimer and Marraf?ni,2008).
Cas9Is Guided by crRNAs and Creates Double-Stranded Breaks in DNA
Following the seminal study in2007con?rming that CRISPR is an adaptive immune system(Barrangou et al.,2007),Sylvain Moineau continued to collaborate with Danisco to understand the mechanism by which CRISPR cleaves DNA.
The problem was that CRISPR was normally so ef?cient that Moineau and his colleagues could not readily observe how invading DNA was destroyed.However,they caught a lucky break while studying plasmid interference in S.thermophilus. The investigators found a handful of bacterial strains in which CRISPR conferred only partial protection against plasmid transformation by electroporation.In one such inef?cient strain, they could see linearized plasmids persisting inside the cells. Somehow,the process of plasmid interference had been slowed down enough to observe the direct products of CRISPR’s action (Garneau et al.,2010).
This strain allowed them to dissect the process of cutting. Consistent with their earlier results(Barrangou et al.,2007), they showed that the cutting of the plasmid depended on the Cas9nuclease.When they sequenced the linearized plas-mids,they found a single precise blunt-end cleavage event 3nucleotides upstream of the proto-spacer adjacent motif (PAM)sequence,a key sequence feature whose function they had characterized in earlier papers(Deveau et al.,2008;Horvath et al.,2008).Expanding their analysis,they showed that viral DNA is also cut in precisely the same position relative to the PAM sequence.Moreover,the number of distinct spacers matching a target corresponded to the number of cuts observed. Their results showed de?nitively that Cas9’s nuclease activity cut DNA at precise positions encoded by the speci?c sequence of the crRNAs.Discovery of tracrRNA
Despite intense study of the CRISPR-Cas9system,one addi-tional piece of the puzzle was missing—a small RNA that would come to be called trans-activating CRISPR RNA(tracrRNA).In fact,the discoverers,Emmanuelle Charpentier and Jo¨rg Vogel, were not speci?cally looking to study the CRISPR system;they were simply trying to identify microbial RNAs.
Charpentier had earned her Ph.D.in microbiology from Pasteur Institute in1995and did post-doctoral work in New York for6years before starting her own lab at the University of Vienna in2002and Umea?,Sweden in2008.After discovering an unusual RNA that controls virulence in Streptococcus pyo-genes(Mangold et al.,2004),she became interested in identi-fying additional regulatory RNAs in microbes.She used bio-informatics programs to scan intergenic regions in S.pyogenes for structures,suggesting that they might encode non-coding RNAs.She had found several candidate regions—including one near the CRISPR locus—but they were hard to follow up without direct information about the RNAs themselves.
The solution appeared when Charpentier met Vogel at the2007 meeting of RNA Society in Madison,Wisconsin.Trained as a microbiologist in Germany,Vogel had begun focusing on?nding RNAs in pathogens during his postdoctoral work in Uppsala and Jerusalem and had continued this work when he started his own group in2004at the Max Planck Institute for Infection Biology in Berlin.(Five years later,he would move to Wu¨rzburg to lead a research center on infectious disease.)With the recent advent of‘‘next-generation sequencing’’technology,Vogel realized that massively parallel sequencing would make it possible to pro-duce comprehensive catalogs of any microbial transcriptome.He had just applied the approach to Helicobacter pylori,the bacte-rium responsible for stomach ulcers(Sharma et al.,2010),and was working on various other bugs.Charpentier and Vogel decided to turn the shotgun on S.pyogenes as well.
The approach yielded a striking result:the third-most abun-dant class of transcript—after only ribosomal RNA and transfer RNA—was a novel small RNA that was transcribed from a sequence immediately adjacent to the CRISPR locus(in the region that had caught Charpentier’s attention)and had25 bases of near-perfect complementary to the CRISPR repeats. The complementarity suggested that this tracrRNA and the precursor of the crRNAs hybridized together and were pro-cessed into mature products by RNaseIII cleavage.Genetic deletion experiments con?rmed this notion,showing that tracrRNA was essential for processing crRNAs and thus for CRISPR function(Deltcheva et al.,2011).
Later studies would reveal that tracrRNA also has another key role.Subsequent biochemical studies showed that tracrRNA was not only involved in processing crRNA but was also essential for the Cas9nuclease complex to cleave DNA(Jinek et al.,2012; Siksnys et al.,2012).
Reconstituting CRISPR in a Distant Organism
Virginijus Siksnys grew up in Soviet-era Lithuania and graduated from Vilnius University before leaving home in the early1980s to get a Ph.D.at Moscow State University,where he studied enzyme kinetics.When he returned home to Vilnius,he joined the Institute of Applied Enzymology to study the then-hot?eld Cell164,January14,2016a2016Elsevier Inc.23
of restriction enzymes.After two decades,though,he was bored with characterizing restriction enzymes.Horvath,Barrangou, and Moineau’s2007paper re-ignited his fascination with bacte-rial barriers to foreign DNA.As a chemist,he felt that he would only understand CRISPR if he could reconstitute it in vitro.
His?rst step was to test whether he had all of the necessary components.He and his collaborators set out to see whether the CRISPR system from S.thermophilus could be reconstituted in fully functional form in a very distant microbe,E.coli.To their delight,they found that transferring the entire CRISPR locus was suf?cient to cause targeted interference against both plasmid and bacteriophage DNA(Sapranauskas et al.,2011).Using their heterologous system,they also proved that Cas9is the only pro-tein required for interference and that its RuvC-and HNH-nuclease domains(Bolotin et al.,2005;Makarova et al.,2006) are each essential for interference.
The?eld had reached a critical milestone:the necessary and suf?cient components of the CRISPR-Cas9interference system—the Cas9nuclease,crRNA,and tracrRNA—were now known.The system had been completely dissected based on elegant bioinformatics,genetics,and molecular biology.It was now time to turn to precise biochemical experiments to try to con?rm and extend the results in a test tube.
Studying CRISPR In Vitro
Using their heterologous expression system in E.coli,Siksnys and his colleagues puri?ed the S.thermophilus Cas9-crRNA complex by using a streptavidin tag on Cas9and studied its activity in a test tube(Gasiunas et al.,2012).They showed that the complex could cleave a DNA target in vitro,creating a double-stranded break precisely3nucleotides from the PAM sequence—matching the in vivo observations of Moineau and colleagues.Most dramatically,they demonstrated that they could reprogram Cas9with custom-designed spacers in the CRISPR array to cut a target site of their choosing in vitro.By mutating the catalytic residues of the HNH-and RuvC-nuclease domains,they also proved that the former cleaves the strand complementary to the crRNA while the latter cleaves the oppo-site strand.And,they showed that the crRNA could be trimmed down to just20nucleotides and still achieve ef?cient cleavage. Finally,Siksnys showed that the system could also be reconsti-tuted in a second way—by combining puri?ed His-tagged Cas9, in-vitro-transcribed tracrRNA and crRNA,and RNase III—and that both RNAs were essential for Cas9to cut DNA.(They would ultimately drop the second reconstitution from their revised paper,but they reported all of the work in their published U.S. patent application?led in March2012[Siksnys et al.,2012]). Around the same time,Charpentier had begun biochemical characterization of CRISPR with a colleague in Vienna.When she lectured about tracrRNA at an American Society for Micro-biology meeting in Puerto Rico in March2011,she met Jennifer Doudna—a world-renowned structural biologist and RNA expert at the University of California,Berkeley.After growing up in Hawaii,Doudna had received her Ph.D.at Harvard,working with Jack Szostak to re-engineer an RNA self-splicing intron into a ribozyme capable of copying an RNA template,and had then done postdoctoral work with Tom Cech at the University of Colorado,where she had solved crystal structures of ribo-zymes.In her own lab(?rst at Yale in1994and then at Berkeley starting in2002),she characterized RNA-protein complexes underlying diverse phenomena,such as internal ribosome entry sites and processing of microRNAs.She had been using crystal-lography and cryo-electron microscopy to solve structures of components of the Cascade complex of type I CRISPR systems, the more complex systems used in microbes such as E.coli. The two scientists decided to join forces.They used recombi-nant Cas9(from S.pyogenes expressed in E.coli)and crRNA and tracrRNA that had been transcribed in vitro(Jinek et al., 2012).Like Siksnys,they showed that Cas9could cut puri?ed DNA in vitro,that it could be programmed with custom-designed crRNAs,that the two nuclease domains cut opposite strands, and that both crRNA and tracrRNA were required for Cas9to function.In addition,they showed that the two RNAs could function in vitro when fused into a single-guide RNA(sgRNA). The concept of sgRNAs would become widely used in genome editing,after modi?cations by others to make it work ef?ciently in vivo.
Siksnys submitted his paper to Cell on April6,2012.Six days later,the journal rejected the paper without external review.(In hindsight,Cell’s editor agrees the paper turned out to be very important.)Siksnys condensed the manuscript and sent it on May21to the Proceedings of the National Academy of Sciences, which published it online on September 4.Charpentier and Doudna’s paper fared better.Submitted to Science2months after Siksnys’s on June8,it sailed through review and appeared online on June28.
Both groups clearly recognized the potential for biotech-nology,with Siksnys declaring that‘‘these?ndings pave the way for engineering of universal programmable RNA-guided DNA endonucleases,’’and Charpentier and Doudna noting ‘‘the potential to exploit the system for RNA-programmable genome editing.’’(A few years later,Doudna would call the world’s attention to the important societal issues raised by the prospect of editing the human germline.)
Genome Editing in Mammalian Cells
When scientists in the late1980s devised a way to alter mamma-lian genomes in living cells,it transformed biomedical research—including making it possible to insert DNA at a speci?c location in mouse embryonic stem cells and then produce mice carrying the genetic modi?cation(reviewed in Capecchi,2005).While revolu-tionary,the process was inef?cient—requiring selection and screening to identify the one-in-a-million cells in which homolo-gous recombination had swapped a gene with a modi?ed version supplied by the experimenter.In the mid-1990s, mammalian biologists—building on observations by yeast ge-neticists—found that introducing a double-stranded break at a genomic locus(using a‘‘meganuclease,’’an endonuclease with an extremely rare recognition site)dramatically increased the frequency of homologous recombination,as well as small deletions caused by non-homologous end joining(reviewed in Haber,2000and Jasin and Rothstein,2013).The secret to ef?-cient genome editing,they realized,was to?nd a reliable method to produce a double-stranded break at any desired location. The?rst general strategy was to use zinc-?nger nucleases (ZFNs)—fusion proteins composed of a zinc-?nger DNA-binding
24Cell164,January14,2016a2016Elsevier Inc.
domain and a DNA-cleavage domain,taken from a restriction enzyme,that bind and cut a genomic locus(Bibikova et al., 2001).Scientists soon demonstrated the use of ZFNs for site-speci?c gene editing by homologous recombination in the fruit ?y and mouse(Bibikova et al.,2003;Porteus and Baltimore, 2003).By2005,a group at Sangamo Biosciences reported suc-cessful correction of a mutation in the gene for severe combined immune syndrome in a human cell line(Urnov et al.,2005). However,fashioning ZFNs that reliably recognize speci?c sites proved to be slow and?nicky.A better solution emerged after two groups described in late2009a remarkable class of transcription-activating proteins called TALEs,from the plant pathogen Xanthomonas(Boch et al.,2009;Moscou and Bogdanove,2009),which use a precise code of modular domains to target speci?c DNA sequences.Still,the approach entailed considerable work,requiring a new protein for each target.
Since Marraf?ni and Sontheimer’s2008paper showing that CRISPR was a programmable restriction enzyme,researchers had grasped that CRISPR might provide a powerful tool for cutting,and thereby editing,speci?c genomic loci—if it could be made to work in mammalian cells.But this was a big‘‘if.’’In contrast to microbes,mammalian cells have very different internal environments and their genomes are1,000-fold larger, reside in nuclei,and are embedded in an elaborate chromatin structure.Attempts to transfer other simple microbial systems, such as self-splicing group II introns,had failed,and efforts to use nucleic acids to target genomic loci had been problematic. Could CRISPR be re-engineered to become a robust system for editing the human genome?As late as September2012, experts were skeptical(Barrangou2012;Carroll,2012).
Feng Zhang moved at age11from Shijiazhuang,China to Des Moines,Iowa.He got hooked on molecular biology at a Saturday enrichment course and,by age16,was working20hours a week in a local gene-therapy lab.As a Harvard undergraduate,he became interested in the brain when a classmate was stricken by severe depression,and he later pursued a Ph.D.in chemistry at Stanford with neurobiologist and psychiatrist Karl Deisseroth, where they(together with Edward Boyden)developed optoge-netics—a revolutionary technique whereby neurons carrying a microbial light-dependent channel protein can be triggered to ?re by light pulses.As an independent investigator in Boston (?rst as a Junior Fellow at Harvard and then at MIT’s Department of Brain and Cognitive Sciences and the Broad Institute),Zhang aimed to further expand the molecular toolbox for studying neurobiology.After developing a way to use light to activate gene expression(by coupling a DNA-binding domain and a transcription-activation domain to two plant proteins that bind each other in the presence of light),he began searching for a general way to program transcription factors.When TALEs were deciphered,Zhang,with his collaborators Paola Arlotta and George Church(and,independently,a group from Sangamo BioSciences),successfully repurposed them for mammals—making it possible to activate,repress,or edit genes with preci-sion(Zhang et al.,2011;Miller et al.,2011).Still,he remained on the lookout for a better approach.
In February2011,Zhang heard a talk about CRISPR from Michael Gilmore,a Harvard microbiologist,and was instantly captivated.He?ew the next day to a scienti?c meeting in Miami but remained holed up in his hotel room digesting the entire CRISPR literature.When he returned,he set out to create a version of S.thermophilus Cas9for use in human cells(with optimized codons and a nuclear-localization signal).By April 2011,he had found that,by expressing Cas9and an engineered CRISPR RNA targeting a plasmid carrying a luciferase gene,he could decrease luminescence levels in human embryonic kidney cells.Still,the effect was modest.
Zhang spent the next year optimizing the system.He explored ways to increase the proportion of Cas9that went to the nucleus. When he discovered that S.thermophilus Cas9was unevenly distributed within the nucleus(it clumped in the nucleolus), he tested alternatives and found that S.pyogenes Cas9was much better distributed.He found that mammalian cells,though lacking microbial RNaseIII,could still process crRNA,albeit differently than in bacteria.He tested various isoforms of tracrRNA to identify one that was stable in human cells.
By mid-2012,he had a robust three-component system con-sisting of Cas9from either S.pyogenes or S.thermophilus, tracrRNA,and a CRISPR array.Targeting16sites in the human and mouse genomes,he showed that it was possible to mutate genes with high ef?ciency and accuracy—causing deletions via non-homologous end-joining and inserting new sequences via homologous recombination with a repair template.Moreover, multiple genes could be edited simultaneously by programming the CRISPR arrays with spacers matching each.When Char-pentier and Doudna’s paper appeared in early summer,he also tested a two-component system with the short sgRNA fusion described in their in vitro study.The fusion turned out to work poorly in vivo,cutting only a minority of loci with low ef?-ciency,but he found that a full-length fusion that restored a crit-ical30hairpin solved the problem(Cong et al.,2013;Zhang, 2012).(Zhang would soon go on to show that CRISPR was even more versatile:it could be used to create complex mouse models of inherited diseases and somatic cancer in weeks and to perform genome-wide screening to?nd the essential genes in a biological process—and it could be made more accurate by decreasing‘‘off-target’’cutting.He and Koonin,the computa-tional biologist who had worked with van der Oost,would also ?nd new Class2CRISPR systems,including one with a nuclease that cuts differently than Cas9and requires only crRNA without tracrRNA[Zetsche et al.,2015]).Zhang submitted a paper re-porting mammalian genome editing on October5,2012,which appeared in Science on January3,2013(Cong et al.,2013);it would become the most cited paper in the?eld,with his reagents being distributed by the non-pro?t organization Addgene in response to more than25,000requests over the next3years. About a month later,on October26,George Church,a bril-liant—and colorful—senior Harvard professor with deep exper-tise in genomics and synthetic biology who had collaborated with Zhang,submitted a paper on genome editing in human cells.Since his time as a graduate student with DNA-sequencing pioneer Walter Gilbert at Harvard in the late1970s,Church had focused on developing powerful technologies for‘‘reading’’and‘‘writing’’genomes at large scale—as well as stirring societal debate with provocative proposals,such as to use synthetic biology to revive wooly mammoths and Neanderthals.Aware Cell164,January14,2016a2016Elsevier Inc.25
of Zhang’s efforts and stimulated by Charpentier and Doudna’s paper,Church set out to test crRNA-tracrRNA fusions in mammalian cells.Like Zhang,he found that short fusions were inef?cient in vivo but that full-length fusions worked well.He tar-geted seven sites and demonstrated both non-homologous end-joining and homologous recombination.His paper appeared back-to-back with Zhang’s(Mali et al.,2013).(Church and others would soon use CRISPR to create improved‘‘gene drives’’—synthetic genes able to spread rapidly through natural popula-tions—raising excitement about applications like?ghting malaria-carrying mosquitos and worries about disrupting eco-systems.He would also seek to facilitate pig-to-human trans-plants by using CRISPR to inactivate retroviruses in the porcine genome.)
By late summer2012—with the in vitro studies gaining atten-tion and news of successful in vivo genome editing spreading before publication—several other groups were racing to perform proof of principle experiments demonstrating genome cleavage, albeit not editing.Doudna,with assistance from Church,submit-ted a paper demonstrating low-level cutting at one genomic site (Pandika2014;Jinek et al.,2013).Jin-Soo Kim,a professor at Ko-rea’s Seoul National University who had worked on genome edit-ing with ZFNs and TALEs,showed cutting at two sites(Cho et al., 2013).In both cases,the cleavage was inef?cient because the sgRNAs lacked the critical30hairpin of tracrRNA.Keith Joung, a Harvard professor who had also been a leader in using ZFNs and TALEs for genome editing,went https://www.360docs.net/doc/233303519.html,ing the full-length sgRNA structure provided by his collaborator Church,Joung es-tablished through experiments in zebra?sh that CRISPR could be used to ef?ciently produce deletions in the germline(Hwang et al., 2013).These short papers,submitted in late2012and accepted soon after Zhang’s and Church’s papers were published in early January2013,appeared online in late January.
CRISPR Goes Viral
In early2013,Google searches for‘‘CRISPR’’began to skyrocket—a trend that has continued unabated.Within a year,investigators had reported the use of CRISPR-based genome editing in many organisms—including yeast,nematode, fruit?y,zebra?sh,mouse,and monkey.Scienti?c and commer-cial interest in potential applications in human therapeutics and commercial agriculture began to heat up—as did social con-cerns about the prospect that the technology could be used to produce designer babies.
The early pioneers of CRISPR continued to push the frontiers, but they were no longer alone.Scientists around the world poured in—a new cadre of heroes who further elucidated the biology of CRISPR,improved and extended the technology for genome editing,and applied it to a vast range of biological problems.It is impossible within the bounds of this Perspective to do justice to these contributions;the reader is referred to recent reviews(Barrangou and Marraf?ni,2014;Hsu et al., 2014;van der Oost et al.,2014;Sander and Joung,2014;Jiang and Marraf?ni,2015;Sternberg and Doudna,2015;Wright et al., 2016).
The once-obscure microbial system—discovered20years earlier in a Spanish salt marsh—was now the focus of special issues of scienti?c journals,headlines in the New York Times,biotech start-ups,and international ethics summits(Travis, 2015).CRISPR had arrived.
The Lessons of CRISPR
The story of CRISPR is rich with lessons about the human ecosystem that produces scienti?c advances,with relevance to funding agencies,the general public,and aspiring researchers. The most important is that medical breakthroughs often emerge from completely unpredictable origins.The early heroes of CRISPR were not on a quest to edit the human genome—or even to study human disease.Their motivations were a mix of per-sonal curiosity(to understand bizarre repeat sequences in salt-tolerant microbes),military exigency(to defend against biological warfare),and industrial application(to improve yogurt production). The history also illustrates the growing role in biology of ‘‘hypothesis-free’’discovery based on big data.The discovery of the CRISPR loci,their biological function,and the tracrRNA all emerged not from wet-bench experiments but from open-ended bioinformatic exploration of large-scale,often public, genomic datasets.‘‘Hypothesis-driven’’science of course re-mains essential,but the21st century will see an increasing part-nership between these two approaches.
It is instructive that so many of the Heroes of CRISPR did their seminal work near the very start of their scienti?c careers (including Mojica,Horvath,Marraf?ni,Charpentier,Vogel,and Zhang)—in several cases,before the age of30.With youth often comes a willingness to take risks—on uncharted directions and seemingly obscure questions—and a drive to succeed.It’s an important reminder at a time that the median age for?rst grants from the NIH has crept up to42.
Notably,too,many did their landmark work in places that some might regard as off the beaten path of science(Alicante, Spain;France’s Ministry of Defense;Danisco’s corporate labs;and Vilnius,Lithuania).And,their seminal papers were often rejected by leading journals—appearing only after considerable delay and in less prominent venues.These ob-servations may not be a coincidence:the settings may have afforded greater freedom to pursue less trendy topics but less support about how to overcome skepticism by journals and reviewers.
Finally,the narrative underscores that scienti?c breakthroughs are rarely eureka moments.They are typically ensemble acts, played out over a decade or more,in which the cast becomes part of something greater than what any one of them could do alone.It’s a wonderful lesson for the general public,as well as for a young person contemplating a life in science. REFERENCES
Barrangou,R.(2012).RNA-mediated programmable DNA cleavage.Nat.Bio-technol.30,836–838.
Barrangou,R.,and Marraf?ni,L.A.(2014).CRISPR-Cas systems:Prokaryotes upgrade to adaptive immunity.Mol.Cell54,234–244.
Barrangou,R.,Fremaux,C.,Deveau,H.,Richards,M.,Boyaval,P.,Moineau, S.,Romero,D.A.,and Horvath,P.(2007).CRISPR provides acquired resis-tance against viruses in prokaryotes.Science315,1709–1712.
Bibikova,M.,Carroll,D.,Segal,D.J.,Trautman,J.K.,Smith,J.,Kim,Y.G.,and Chandrasegaran,S.(2001).Stimulation of homologous recombination through targeted cleavage by chimeric nucleases.Mol.Cell.Biol.21,289–297.
26Cell164,January14,2016a2016Elsevier Inc.
Bibikova,M.,Beumer,K.,Trautman,J.K.,and Carroll,D.(2003).Enhancing gene targeting with designed zinc?nger nucleases.Science300,764.
Boch,J.,Scholze,H.,Schornack,S.,Landgraf,A.,Hahn,S.,Kay,S.,Lahaye, T.,Nickstadt,A.,and Bonas,U.(2009).Breaking the code of DNA binding speci?city of TAL-type III effectors.Science326,1509–1512.
Bolotin,A.,Quinquis,B.,Sorokin,A.,and Ehrlich,S.D.(2005).Clustered regu-larly interspaced short palindrome repeats(CRISPRs)have spacers of extra-chromosomal origin.Microbiology151,2551–2561.
Brouns,S.J.J.,Jore,M.M.,Lundgren,M.,Westra,E.R.,Slijkhuis,R.J.H., Snijders,A.P.L.,Dickman,M.J.,Makarova,K.S.,Koonin,E.V.,and van der Oost,J.(2008).Small CRISPR RNAs guide antiviral defense in prokaryotes. Science321,960–964.
Capecchi,M.R.(2005).Gene targeting in mice:functional analysis of the mammalian genome for the twenty-?rst century.Nat.Rev.Genet.6, 507–512.
Carroll,D.(2012).A CRISPR approach to gene targeting.Mol.Ther.20,1658–1660.
Cho,S.W.,Kim,S.,Kim,J.M.,and Kim,J.-S.(2013).Targeted genome engineering in human cells with the Cas9RNA-guided endonuclease.Nat. Biotechnol.31,230–232.
Cong,L.,Ran,F.A.,Cox,D.,Lin,S.,Barretto,R.,Habib,N.,Hsu,P.D.,Wu,X., Jiang,W.,Marraf?ni,L.A.,and Zhang,F.(2013).Multiplex genome engineering using CRISPR/Cas systems.Science339,819–823.
Deltcheva,E.,Chylinski,K.,Sharma,C.M.,Gonzales,K.,Chao,Y.,Pirzada, Z.A.,Eckert,M.R.,Vogel,J.,and Charpentier,E.(2011).CRISPR RNA matura-tion by trans-encoded small RNA and host factor RNase III.Nature471, 602–607.
Deveau,H.,Barrangou,R.,Garneau,J.E.,Labonte′,J.,Fremaux,C.,Boyaval, P.,Romero,D.A.,Horvath,P.,and Moineau,S.(2008).Phage response to CRISPR-encoded resistance in Streptococcus thermophilus.J.Bacteriol. 190,1390–1400.
Garneau,J.E.,Dupuis,M.-E`.,Villion,M.,Romero, D.A.,Barrangou,R., Boyaval,P.,Fremaux,C.,Horvath,P.,Magada′n,A.H.,and Moineau,S. (2010).The CRISPR/Cas bacterial immune system cleaves bacteriophage and plasmid DNA.Nature468,67–71.
Gasiunas,G.,Barrangou,R.,Horvath,P.,and Siksnys,V.(2012).Cas9-crRNA ribonucleoprotein complex mediates speci?c DNA cleavage for adaptive immunity in https://www.360docs.net/doc/233303519.html,A109,E2579–E2586.
Haber,J.E.(2000).Lucky breaks:analysis of recombination in Saccharo-myces.Mutat.Res.451,53–69.
Hale,C.R.,Zhao,P.,Olson,S.,Duff,M.O.,Graveley,B.R.,Wells,L.,Terns, R.M.,and Terns,M.P.(2009).RNA-guided RNA cleavage by a CRISPR RNA-Cas protein complex.Cell139,945–956.
Horvath,P.,Romero,D.A.,Cou?te′-Monvoisin,A.-C.,Richards,M.,Deveau,H., Moineau,S.,Boyaval,P.,Fremaux,C.,and Barrangou,R.(2008).Diversity, activity,and evolution of CRISPR loci in Streptococcus thermophilus. J.Bacteriol.190,1401–1412.
Hsu,P.D.,Lander,E.S.,and Zhang,F.(2014).Development and applications of CRISPR-Cas9for genome engineering.Cell157,1262–1278.
Hwang,W.Y.,Fu,Y.,Reyon,D.,Maeder,M.L.,Tsai,S.Q.,Sander,J.D., Peterson,R.T.,Yeh,J.R.,and Joung,J.K.(2013).Ef?cient genome editing in zebra?sh using a CRISPR-Cas system.Nat.Biotechnol.31,227–229.
Ishino,Y.,Shinagawa,H.,Makino,K.,Amemura,M.,and Nakata,A.(1987). Nucleotide sequence of the iap gene,responsible for alkaline phosphatase isozyme conversion in Escherichia coli,and identi?cation of the gene product. J.Bacteriol.169,5429–5433.
Jansen,R.,Embden,J.D.A.V.,Gaastra,W.,and Schouls,L.M.(2002).Identi?-cation of genes that are associated with DNA repeats in prokaryotes.Mol. Microbiol.43,1565–1575.
Jasin,M.,and Rothstein,R.(2013).Repair of strand breaks by homologous recombination.Cold Spring Harb.Perspect.Biol.5,a012740.Jiang,W.,and Marraf?ni,L.A.(2015).CRISPR-Cas:New tools for genetic manipulations from bacterial immunity systems.Annu.Rev.Microbiol.69, 209–228.
Jinek,M.,Chylinski,K.,Fonfara,I.,Hauer,M.,Doudna,J.A.,and Charpentier, E.(2012).A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity.Science337,816–821.
Jinek,M.,East,A.,Cheng,A.,Lin,S.,Ma,E.,and Doudna,J.(2013).RNA-pro-grammed genome editing in human cells.eLife2,e00471.
Makarova,K.S.,Grishin,N.V.,Shabalina,S.A.,Wolf,Y.I.,and Koonin,E.V. (2006).A putative RNA-interference-based immune system in prokaryotes: computational analysis of the predicted enzymatic machinery,functional analogies with eukaryotic RNAi,and hypothetical mechanisms of action. Biol.Direct1,7.
Makarova,K.S.,Wolf,Y.I.,Alkhnbashi,O.S.,Costa,F.,Shah,S.A.,Saunders, S.J.,Barrangou,R.,Brouns,S.J.J.,Charpentier,E.,Haft,D.H.,et al.(2015).An updated evolutionary classi?cation of CRISPR-Cas systems.Nat.Rev.Micro-biol.13,722–736.
Mali,P.,Yang,L.,Esvelt,K.M.,Aach,J.,Guell,M.,DiCarlo,J.E.,Norville,J.E., and Church,G.M.(2013).RNA-guided human genome engineering via Cas9. Science339,823–826.
Mangold,M.,Siller,M.,Roppenser,B.,Vlaminckx,B.J.M.,Penfound,T.A., Klein,R.,Novak,R.,Novick,R.P.,and Charpentier,E.(2004).Synthesis of group A streptococcal virulence factors is controlled by a regulatory RNA molecule.Mol.Microbiol.53,1515–1527.
Marraf?ni,L.A.,and Sontheimer, E.J.(2008).CRISPR interference limits horizontal gene transfer in staphylococci by targeting DNA.Science322, 1843–1845.
Medawar,P.(1968).Lucky Jim(New York Review of Books),March28,1968.
Miller,J.C.,Tan,S.,Qiao,G.,Barlow,K.A.,Wang,J.,Xia,D.F.,Meng,X., Paschon,D.E.,Leung,E.,Hinkley,S.J.,et al.(2011).A TALE nuclease architec-ture for ef?cient genome editing.Nat.Biotechnol.29,143–148.
Mojica,F.J.M.,and Garrett,R.A.(2012).Discovery and Seminal Developments in the CRISPR Field.In CRISPR-Cas Systems,R.Barrangou and J.van der Oost,eds.(Berlin,Heidelberg:Springer Berlin Heidelberg),pp.1–31.
Mojica,F.J.M.,Juez,G.,and Rodr?′guez-Valera,F.(1993).Transcription at different salinities of Haloferax mediterranei sequences adjacent to partially modi?ed PstI sites.Mol.Microbiol.9,613–621.
Mojica,F.J.M.,Ferrer,C.,Juez,G.,and Rodr?′guez-Valera,F.(1995).Long stretches of short tandem repeats are present in the largest replicons of the Archaea Haloferax mediterranei and Haloferax volcanii and could be involved in replicon partitioning.Mol.Microbiol.17,85–93.
Mojica,F.J.M.,D?′ez-Villasen?or,C.,Soria,E.,and Juez,G.(2000).Biological signi?cance of a family of regularly spaced repeats in the genomes of Archaea, Bacteria and mitochondria.Mol.Microbiol.36,244–246.
Mojica,F.J.M.,D?′ez-Villasen?or,C.,Garc?′a-Mart?′nez,J.,and Soria,E.(2005). Intervening sequences of regularly spaced prokaryotic repeats derive from foreign genetic elements.J.Mol.Evol.60,174–182.
Moscou,M.J.,and Bogdanove,A.J.(2009).A simple cipher governs DNA recognition by TAL effectors.Science326,1501–1501.
Pandika,M.(2014)Jennifer Doudna,CRISPR Code Killer,https://www.360docs.net/doc/233303519.html,,January 7,2014.
Porteus,M.H.,and Baltimore,D.(2003).Chimeric nucleases stimulate gene targeting in human cells.Science300,763.
Pourcel,C.,Salvignol,G.,and Vergnaud,G.(2005).CRISPR elements in Yersinia pestis acquire new repeats by preferential uptake of bacteriophage DNA,and provide additional tools for evolutionary studies.Microbiology 151,653–663.
Sander,J.D.,and Joung,J.K.(2014).CRISPR-Cas systems for editing,regu-lating and targeting genomes.Nat.Biotechnol.32,347–355.
Cell164,January14,2016a2016Elsevier Inc.27
Sapranauskas,R.,Gasiunas,G.,Fremaux,C.,Barrangou,R.,Horvath,P.,and Siksnys,V.(2011).The Streptococcus thermophilus CRISPR/Cas system pro-vides immunity in Escherichia coli.Nucleic Acids Res.39,9275–9282. Sharma,C.M.,Hoffmann,S.,Darfeuille,F.,Reignier,J.,Findeiss,S.,Sittka,A., Chabas,S.,Reiche,K.,Hackermu¨ller,J.,Reinhardt,R.,et al.(2010).The primary transcriptome of the major human pathogen Helicobacter pylori. Nature464,250–255.
Siksnys,V.,Gasiunas,G.,and Karvelis,T.(2012).RNA-directed DNA cleavage by the Cas9-crRNA complex from CRISPR3/Cas immune system of Strepto-coccus thermophilus.U.S.Provisional Patent Application61/613,373,?led March20,2012;later published as US2015/0045546(pending). Sontheimer,E.,and Marraf?ni,L.(2008).Target DNA interference with crRNA. U.S.Provisional Patent Application61/009,317,?led September23,2008; later published as US2010/0076057(abandoned).
Sorek,R.,Kunin,V.,and Hugenholtz,P.(2008).CRISPR–a widespread system that provides acquired resistance against phages in bacteria and archaea.Nat. Rev.Microbiol.6,181–186.
Sternberg,S.H.,and Doudna,J.A.(2015).Expanding the Biologist’s Toolkit with CRISPR-Cas9.Mol.Cell58,568–574.
Travis,J.(2015).GENETIC ENGINEERING.Germline editing dominates DNA summit.Science350,1299–1300.Urnov,F.D.,Miller,J.C.,Lee,Y.-L.,Beausejour,C.M.,Rock,J.M.,Augustus, S.,Jamieson, A.C.,Porteusm,M.H.,Gregory,P.D.,and Holmes,M.C. (2005).Highly ef?cient endogenous human gene correction using designed zinc-?nger nucleases.Nature435,646–651.
van der Oost,J.,Westra,E.R.,Jackson,R.N.,and Wiedenheft,B.(2014).Un-ravelling the structural and mechanistic basis of CRISPR-Cas systems.Nat. Rev.Microbiol.12,479–492.
Wright,A.V.,James,K.,Nun?ez,J.K.,and Doudna,J.A.(2016).Biology and ap-plications of CRISPR systems:Harnessing nature’s toolbox for genome engi-neering.Cell164,this issue,29–44.
Zetsche,B.,Gootenberg,J.S.,Abudayyeh,O.O.,Slaymaker,I.M.,Makarova,K.S., Essletzbichler,P.,Volz,S.E.,Joung,J.,van der Oost,J.,Regev,A.,et al.(2015). Cpf1Is a Single RNA-Guided Endonuclease of a Class2CRISPR-Cas System.Cell.
Zhang,F.,Cong,Le,Lodato,S.,Kosuri,S.,Church,G.M.,and Arlotta,P. (2011).Ef?cient construction of sequence-speci?c TAL effectors for modu-lating mammalian transcription.Nat.Biotechnol.29,149–153.
Zhang,F.(2012).Systems Methods and Compositions for Sequence Manipu-lation.U.S.Provisional Patent Application61/736,527,?led December12, 2012;later published as US008697359B1(awarded).
28Cell164,January14,2016a2016Elsevier Inc.
光纤通信技术的发展历史
论文题目:光纤通信技术发展历史 姓名:谢新云 学号:0932002231 专业班级:通信技术(2) 院系:电子通信工程学院 指导老师:彭霞 完成时间:2011年10月22日
概论 目前,在实际运用中相当有前途的一种通信技术之一,即光纤通信技术已成为现代化通信非常重要的支柱。作为全球新一代信息技术革命的重要标志之一,光纤通信技术已经变为当今信息社会中各种多样且复杂的信息的主要传输媒介,并深刻的、广泛的改变了信息网架构的整体面貌,以现代信息社会最坚实的通信基础的身份,向世人展现了其无限美好的发展前景。 自上世纪光纤通信技术在全球问世以来,整个的信息通讯领域发生了本质的、革命性的变革,光纤通信技术以光波作为信息传输的载体,以光纤硬件作为信息传输媒介,因为信息传输频带比较宽,所以它的主要特点是:通信达到了高速率和大容量,且损耗低、体积小、重量轻,还有抗电磁干扰和不易串音等一系列优点,从而备受通信领域专业人士青睐,发展也异常迅猛。 光纤通信不仅可以应用在通信的主干线路中,也可以在电力通信控制系统中发挥作用,进行工业监测、控制,现在在军事上也被广泛应用,基于各领域对信息量的需求不断增长,光纤通信技术的应用发展趋势也备受关注。一条完整的光纤链路除受光纤本身质量影响外,还取决于光纤链路现场的施工工艺和环境。 本文针对光纤通信技术的发展及趋势展开研究,分别介绍了光纤通信技术的发展历史和现状,以及光纤通信技术的发展趋势,对一些先进的光纤通信技术进行了介绍。 关键字:光纤通信技术,发展历史,现状,发展趋势
目录 概论 (1) 目录 (2) 第一章光纤通信技术的形成 (3) 1.1早期的光通信 (3) 1.2 现在光纤通信技术的形成 (3) 1.2.1 光纤通信器件的发展 (3) 1.2.2 光纤 (5) 第二章光纤通信技术的现状 (8) 2.1 光纤光缆 (8) 2.2 光电子器件 (8) 2.3光纤通信系统 (14) 第三章我国光纤通信技术的发展 (15) 参考文献 (16)
光纤通信技术概述解析
3.3 光纤通信技术 一、光纤通信系统概述及基本结构 光纤通信系统是以光纤为传输媒介, 光波为载波的通信系统。主要由光发送机、光纤光缆、中继器和光接收机组成, 其基本结构原理如图所示。 系统中还包含了一些互联和光信号处理部件, 如光纤连接器、隔离器、光开关等。图中电端机和光端机均包括发送和接收两部分, 两者合起来构成发送器和接收器。其中发送光端机是将电信号变换成光信号,接收光端机则是将光信号转换成电信号。 1、发送器 发送器由发送光端机和电端机构成, 其核心是一个光源。光源的主要功能就是将一个信息信号从电子格式转换为光格式。今天的光纤通信系统采用发光二极管或激光二极管作为光源。两者都是小型的半导体
设备, 可以有效地将电信号转换为光信号。LD 输出的光功率较大, 谱线窄, 一般适合长距离、大容量的通信系统, 但其寿命较短, 价格高; LED 光源发出的光功率较小, 光谱线较宽, 调制速率较低, 输出线性好, 寿命长, 成本低, 适用于短距离和中小容量的系统。它们需要与电源相连并且需要调制电路。 2、光纤 光纤通信系统中的传输介质是光纤。光纤通信系统中发送器端的光信息信号就是通过光纤传送到接收器端的。实际上, 同任何其他通信链路一样, 光纤提供发送器和接收器间的连接。同时, 光纤对光信号进行传导, 就像铜线和同轴线传导电信号一样。它大概和人的头发的粗细相同, 为了保护非常脆弱的光纤, 使其不受恶劣的外部环境和机械的损害, 通常将光纤封装在特定的结构中。裸露的光纤包上保护膜后封装到其他几层中, 所有这些就构成了光纤光缆。 3、接收器 接收器由接收光端机和电端机构成。接收光端机的主要部分包括光检测器、放大器、均衡器、判决器、自动增益控制电路和时钟电路。其中光检测器是接收光端机的核心, 光检测器的主要功能就是把光信息信号转换回电信号( 光电流) 。光纤通信系统中的光检测器主要有PIN 二极管、雪崩光电二极管( APD) 。APD 比PIN 更灵敏, 而且对外部放大功能要求更低。A PD 的缺点是具有相对较长的渡越时间以及由于雪崩放大造成的附加内部噪声。 4、光中继器
通信工程专业综述
通行工程专业综述 通信是通过某种媒体进行的信息传递。古代,人们通过驿站、飞鸽传书、烽火报警等方式进行信息传递。今天,随着科学水平的飞速发展,相继出现了无线电,固话,手机,互联网甚至可视电话等各种通信方式。 1.培养目标:本专业主要培养从事通信工程及计算机网络系统的研究、制造、开发和培养目标:培养目标 应用的高级人才。毕业后可从事无线通信、电视、大规模集成电路、智能仪器及应用电子技术领域的研究,设计和通信工程的研究、设计、技术引进和技术开发工作。近年来的毕业生集中在通信系统、高科技开发公司、科研院所、设计单位、金融系统、民航、铁路及政府和大专院校等。 2.主修学科:电路分析、低频电子线路、脉冲与数字电路、高频电子线路、电主修学科学科:磁场理论、信号与系统、微机原理及应用、单片机技术、微波技术与天线、通讯原理、程控交换技术、移动通讯、计算机网络通讯、光纤通讯等。毕业生应掌握电子技术、通讯技术和计算机技术的基本理论与设计方法及程控交换技术、光纤通讯、 1 移动通讯和计算机网络通讯的基本原理及应用方法,具有各类通讯系统的设计、研究及开发的工作能力。 通信工程专业课程关系图 高等数学高级语言程序设计电路分析 2 3.专业特点:专业特点:专业特点 (1)发展速度快;(2)应用范围广;(3)涉及知识领域广;(4)交叉学科领域广。 4.毕业生应获得以下几方面的知识和能力:毕业生应获得以下几方面的知识和能力:毕业生应获得以下几方面的知识和能力 1.掌握通信领域内的基本理论和基本知识; 2.掌握光波、无线、多媒体等通信技术; 3.掌握通信系统和通信网的分析与设计方法; 4.具有设计、开发、调测、应用通信系统和通信网的基本能力; 5.了解通信系统和通信网建设的基本方针、政策和法规; 6.掌握文献检索、资料查询的基本方法,具有一定的科学研究和实际工作能力。 5.发展前景:面向新的世纪,通信工程专业将会迎来其发展的广阔天地。随发展前景:发展前景 着通信技术应用的日趋广泛,上至太空,下至海底,无不活跃着这一专业的技术人才。现在中国已经加入 WTO,这势必会给中国信息产业的发展带来更大的发展空间。而通信工程专业优秀人才的短缺成为我国参与国际间竞争的一个十分不利的因素。因此,在未来若干年,我国势必会更加重视本专业人才的培养,更加重视通信工程专业的教育,提高教育水平。三、目前通信专业的分类——选择通信的理由目前通信专业的分类——选择通信的理由—— 通信工程专业主要为研究信号的产生、信息的传输、交换和处理,以及在计算机通信、数字通信、卫星通信、光纤通信、蜂窝通信、个人通信、平流层通信、多媒体技术、信息高速公路、数字程控交换等方面的理论和工程应用问题。 1、对数据通信的兴趣通信的兴趣、对数据通信数据通信可以说已经深入到社会生活的各个领域,电子邮件,浏览网页,在线电影都可以归结为数据通信。数据通
数据库技术的发展史
数据库技术的发展史 数据库技术的发展,已经成为先进信息技术的重要组成部分,是现代计算机信息系统和计算机应用系统的基础和核心。数据库技术最初产生于20世纪60年代中期,到今天近几十年的历史,其发展速度之快,使用X围之广是其它技术所远不及的。 先介绍一下数据模型的概念:数据模型是数据库系统的核心和基础。数据模型的发展经历了格式化数据模型(包括层状数据模型和网状数据模型)、关系数据模型两个阶段,正在走向面向对象的数据模型等非传统数据模型的阶段。 层状数据模型每个节点间是一对多的父子之间的联系,比如一个父亲三个儿子;中心下的几个部门,部门里的人。网状数据模型中允许任意两个节点间有多种联系,层次模型实际上是网状模型的一个特例;如同学生选课,一个学生可以选修多门课程,某一课程也可被多名学生选修。关系数据模型,职工,比如我(编号,XX,性别,所属部门,籍贯),我和马薇,X晖,陈曙光等就组成了一X关系模型的数据表。 根据数据模型的发展,数据库技术可以相应地划分为三个阶段:第一代的网状、层次数据库系统;第二代的关系数据库系统;第三代的以面向对象模型为主要特征的数据库系统。
第一代数据库的代表是1969年IBM公司研制的层次模型的数据库管理系统IMS和70年代美国数据库系统语言协商CODASYL下属数据库任务组DBTG提议的网状模型。层次数据库的数据模型是有根的定向有序树,网状模型对应的是有向图。这两种数据库奠定了现代数据库发展的基础。这两种数据库具有如下共同点: 1.支持三级模式(外模式、模式、内模式),模式之间具有转换(或成为映射)功能,保证了数据库系统具有数据与程序的物理独立性和一定的逻辑独立性; 2.用存取路径来表示数据之间的联系; 3.有独立的数据定义语言; 4.导航式的数据操纵语言。 网状数据库 最早出现的是网状DBMS。网状模型中以记录为数据的存储单位。记录包含若干数据项。网状数据库的数据项可以是多值的和复合的数据。每个记录有一个惟一地标识它的内部标识符,称为码(DatabaseKey,DBK),它在一个记录存入数据库时由DBMS自动赋予。DBK可以看作记录的逻辑地址,可作记录的替身,或用于寻找记录。网状数据库是导航式(Navigation)数据库,用户在操作数据库时不但说明要做什么,还要说明怎么做。例如在查找语句中不但要说明查找的对象,而且要规定存取路径。
数据库仓库综述
数据库仓库综述 摘要:自从计算机出现后,计算机对数据的管理经历了程序管理、文件管理和数据库管理三个阶段。数据库是数据管理的高级阶段,是数据管理最有效的手段,是现代计算机信息系统和计算机应用系统的基础和核心。本文介绍了数据库的定义、发展历史及各代数据库所采用的数据模型、各代数据库的优缺点。结合当今应用需求和新技术对数据库发展趋势、应用前景作了展望。关键词:数据模型;关系数据库;面向对象数据库 1 引言 数据库技术是20世纪60年代初开始发展起来的一门数据管理自动化的综合性新技术,它是应数据管理任务的需要而产生的,是数据管理最有效的手段。数据库就是为了一定的目的,在计算机系统中与特定的结构组织、存储和应用相关联的数据集合。在数据库出现之前,计算机管理数据经过了程序管理和文件管理两个阶段。数据库是数据管理的高级阶段,它与传统的数据管理相比有许多明显的差别,其中主要的有两点:一是使数据独立于应用程序而集中管理,实现了数据共享,减少了数据冗余,提高了数据的效益;二是在数据间建立了联系,从而能反映出现实世界中信息的联系。 数据库的应用领域相当广泛,从一般事务处理到各种专门化数据的存
储与管理。它的出现极大地促进了计算机应用的发展,数据库技术已经成为先进信息技术的重要组成部分,是现代计算机信息系统和计算机应用系统的基础和核心[1]。目前基于数据库技术的计算机应用已成为计算机应用的主流。 2 数据库发展的历史 数据模型是数据库系统的核心和基础。数据模型是数据库系统中关于数据和联系的逻辑组织的形式表示,通常由数据结构、数据操作和完整性约束三部分组成。数据结构是所研究的对象类型的集合,在数据库系统中通常按照数据结构的类型来命名数据模型。传统的数据模型有层次模型、网状模型和关系模型,现在随着面向对象技术的发展,数据库模型也有基于面向对象的数据模型。数据操作是指对数据库中各种对象的实例允许执行的操作的集合。数据库主要有检索和更新两大类操作。数据的约束条件是完整性规则的集合。保证数据的完整性是对数据库的一个很重要的要求。所谓完整性就是数据的正确性、有效性和相容性。每一个具体的数据库都是由一个相应的数据模型来定义的。按照数据模型的进展,数据库技术可以相应地分为三个发展阶段:第一代的网状、层次数据库系统;第二代的关系数据库系统;第三代的以面向对象模型为主要特征的数据库系统。目前关系数据库系统已经逐渐淘汰了网状数据库和层次数据库,成为当今最为流行的商用数据库系统。211 第一代数据库系统———层次和网状数据库系统第一代数据库系统的数据模型为层次模型和网状模型。层次模型是将数据组织成有向有序的树结构。层次模型由处于不同层次的各个结点组成。
水下光通信-综述
水下光通信-综述
水下光通信 综述 一、水下光通信的国内外研究现状 光通信起源最早可追溯到19 世纪70 年代,当时Alexander Graham Bell 提出采用可见光为媒介进行通信,但是当时既不能产生一个有用的光载波,也不能将光从一个地方传到另外一个地方。因此直到1960 年激光器的发明,光通信才有了突破性的发展,但研究领域基本上集中在光纤通信和不可见光无线通信领域。由于海水对光的强吸收特性,水下光通信技术一直没有得到重视。直到1963 年,Dimtley 等人在研究光波在海洋中的传播特性时, 发现海水在450- 550 纳米波段内蓝绿光的衰减比其它光波段的衰减要小很多, 证实在海洋中亦存在一个类似于大气中存在的透光窗口。这一物理现象的发现为解决长期水下目标探测、通 信等难题提供了基础。 水下光学通信技术研究前期主要集中在军
事领域,长期以来一直是水下潜艇通信中的关键技术。美国海军从1977 年提出卫星与潜艇间通信的可行性后, 就与美国国防研究远景规划局开始执行联合战略激光通信计划。从1980 年起, 以几乎每两年一次的频率, 进行了迄今为止共6 次海上大型蓝绿激光对潜通信试验, 这些试验包括成功进行的12 千米高空对水下300 米深海的潜艇的单工激光通信试验, 以及在更高的天空、长续航时间的模拟无人驾驶飞机与以正常下潜深度和航速航行的潜艇间的双工激光通信可行性试验, 证实了蓝绿激光通信能在天气不正常、大暴雨、海水浑浊等恶劣条件下正常进行。1983 年底, 前苏联在黑海舰队的主要基地塞瓦斯托波尔附近也进行了把蓝色激光束发送到空间轨道反射镜后再转发到水下弹道潜艇的激光通信试验。 澳大利亚国立大学信息科学与工程研究学院的研究小组开发了一种低成本、小体积、结构简单的光学通信系统,选用LuxeonⅢLED 的蓝(460nm)、青(490nm)、绿(520nm)光,接收器电路采用对蓝青绿三种光灵敏度很高的SLD—70BG2A 光电二极管,这套系统在兼顾速
通信工程专业综述 0905076016
通信工程专业综述 前言: 时间飞逝,眨眼之间,大学的时光已经过去一半,我们将要迎来了大三时期的专业选择。记得刚入校时,老师就给我们上过专业的相关知识,两年的时间里系里也分阶段给我们上了专业导论课,我们渐渐对本系的三个专业有了一定的了解。我们系共分电子信息工程、自动化和通信工程三个专业,学生大一时按大类入学,大三第一学期面临专业选择。在今天即将面对专业的选择时刻,经过平时对各个专业的学习、了解以及导论课老师们介绍,再综合自己的各方面因素,我毅然决定选择通信工程专业。随着各种新的技术日新月异,层出不穷。下面我就个人而言分别介绍通信各个领域的发展现状及前景。 一、三个专业主干课程的了解 电子信息工程的主干课程电路理论系列课程、计算机技术系列课程、信息理论与编码、信号与系统、数字信号处理、电磁场理论、自动控制原理、感测技术等。主要偏理论。自动化的主干课程有电路原理、电子技术基础、计算机原理及应用、计算机软件技术基础、过程工程基础、电机与电力拖动基础、电力电子技术、自动控制理论、信号与系统分析等。通信工程的主干课程和电子信息有相似之处,有电路理论与应用的系列课程、计算机技术系列课程、信号与系统、电磁场理论、数字系统与逻辑设计、数字信号处理、通信原理等。小结:结合主干课程的开设情况,根据自己的兴趣特点,我还是比较适合通信工小结程这个专业的。 二、通信工程简介 通信工程专业是信息科学技术发展迅速并极具活力的一个领域,尤其是数字移动通信、光纤通信、Internet网络通信使人们在传递信息和获得信息方面达到了前所未有的便捷程度。通信工程具有极广阔的发展前景,也是人才严重短缺的专业之一。本专业学习通信技术、通信系统和通信网等方面的知识,能在通信领域中从事研究、设计、制造、运营及在国民经济各部门和国防工业中从事开发、应用通信技术与设备。 三、通信工程专业的分析 通信是通过某种媒体进行的信息传递。古代,人们通过驿站、飞鸽传书、烽火报警等方式进行信息传递。今天,随着科学水平的飞速发展,相继出现了无线电、固话、手机、互联
数据库技术的发展(一)
数据库技术的发展(一) (总分:15.00,做题时间:90分钟) 一、{{B}}选择题{{/B}}(总题数:5,分数:5.00) 1.采用扩展关系数据模型的方法建立的数据库系统,称做 ______。 (分数:1.00) A.对象-关系数据库系统√ B.扩展关系数据库系统 C.拓展关系数据库系统 D.以上都不正确 解析: 2.下列哪一种结构是支持并行数据库系统最好的结构? ______。 (分数:1.00) A.共享内存 B.共享磁盘 C.无共享√ D.层次模式 解析: 3.下面属于并行数据库系统目标的是 ______。Ⅰ.高性能Ⅱ.高可用性Ⅲ.高扩充性 (分数:1.00) A.Ⅰ和Ⅱ B.Ⅱ和Ⅲ C.Ⅰ和Ⅲ D.Ⅰ、Ⅱ和Ⅲ√ 解析: 4.下列属于粗粒度并行机特点的是 ______。 (分数:1.00) A.拥有大量的处理器 B.共享一个主存√ C.单个事务运行得更快 D.数据库一般将一个查询分配到多个处理器上 解析: 5.操作型数据和分析型数据具有不同的特征,下列哪一个是操作型数据的特征? ______。 (分数:1.00) A.可更新的√ B.历史的(包括过去数据) C.支持管理决策的 D.面向主题的 解析: 二、{{B}}填空题{{/B}}(总题数:5,分数:10.00) 6.在客户机/服务器工作模式中,客户机可以使用{{U}} 【1】 {{/U}}向数据库服务器发送查询命令。(分数:2.00) 填空项1:__________________ (正确答案:结构化查询语言/SQL) 解析: 7.分布式数据库系统与集中式数据库系统最大的区别是分布式数据库中的数据{{U}} 【2】 {{/U}} 存储在多个场地。 (分数:2.00)
光通信中的重要技术及发展趋势
光通信中的重要技术及发展趋势 [摘要] 随着信息化社会的到来,通信技术也得到了日新月异的发展。在过去的几年中,人们对传输速率的要求越来越高,使用高速率数据传输的用户数量每年都在递增,而光通信技术在过去几年中也有了长足的发展,光纤通信凭借其传输高速率的数据,成为广域通信网的骨干网络,如今在广域通信网中绝大部分是通过光纤传输的。本文主要讨论在光通信中的主要技术以及未来光通信的几个发展趋势。 [关键词] 光通信光接入光交换全光网无线光通信 随着用户对接入带宽要求的日益增加以及三网融合后对数字高清信号的传送,对运营商接入侧及骨干核心传输有了更高的要求,而光通信在其中起了举足轻重的作用,光通信技术的发展决定了电信业的未来方向,近几年,不论在接入层以及核心层,光通信技术都有了长足的发展。 1.在接入层: 1.1无源光网络(PON) 无源光网络主要用于解决宽带最终用户接入终端局的问题,由于这种接入技术使得接入网的局端(OLT)与用户(ONU)之间只需光纤、光分路器等光无源器件,不需租用机房和配备电源,因此被称为无源光网络。无源光网络以其容量大、传输距离长、较低成本、全业务支持等优势成为热门技术。目前已经逐步商用化的无源光网络主要有TDM-PON(APON、EPON、GPON)和WDM-PON。 无论是核心网、传输网还是接入网,其发展的首要因素就是业务,是终端用户的需求。从业务发展现状来看,高带宽的消耗业务逐步涌现,带宽提速成为迫切需求,而PON以其容量大、传输距离长、较低成本、全业务支持等优势成为宽带接入的热点,它在提供业务组合的同时,实现了高可靠性和高性能,已经成为了下一代光接入网的发展方向。 1.2无线光通信技术 从光纤骨干网到用户之间的”最后一英里”,如果铺设光缆,不仅花费大而且耗时;许多无线通信技术可以解决”最后一英里”的问题,但是这些技术需要向无线电管理委员会申请频率执照,不仅要使用户支付大量的频率占用费,而且申请也要花费数月的时间。无线光通信因为无需频率申请,机型小方便架设,能够简单的解决最后一英里的问题,为宽带接入的快速部署提供一种灵活的解决方案。 无线光通信系统是以大气作为传输媒质来进行光信号的传送的。只要在收发两个端机之间存在无遮挡的视距路径和足够的光发射功率,就可以进行通信。一个无线光通信系统包括三个基本部分:发射机、信道和接收机。在点对点传输的
光纤通信设备概述
光纤通信设备概述 1.走进通信机房 通信机房,无论大小,走进去看到的是: 一排排的机柜,里面装有各种各样的设备,大部分机柜是19英寸宽,有2米高,也有2.2米高的. 地板,下面往往是走线槽, 上面也许有走线槽(地槽和顶槽2选1). 网管系统:用计算机管理通信设备. 电源系统
2.从电话机到机房的线路 家里的电话机通过双绞线连接到楼道里的电话分线盒,然后用50对或100对的音频电缆, 连到了小区附近的电缆交接箱,再用更大对数的电缆接到电话局里的音频配线架,也叫总配线架,就是112机房,在音频配线架上,每个电话机都对应有1对电话线接点,并且一般都配有防雷击的音频保安器,电话线在电话局内部还用电缆连到了交换机.或PCM30设备。 3.112机房的总配线架,也叫MDF,还叫VDF 4.电话交换机 交换机可以分为3部分,一是用户电路,负责为用户馈电,发铃流,发送忙音,拨号音,记录用户话机所拨的号码,同时将模拟的电话语音变成数字信号;二叫绳路,也就是交换系统,负责电话的交换接续;三是中继器,分入局中继器和出局中继器,中继器的接口是数字信号是2.048Mb/s的速率,叫E1口。 5.PCM30设备 电话机到电话局,如果距离近(2公里),可以用电缆直接连接,如果距离远,就必须用光纤 连接光纤通信中传输的信号是数字信号,而电话机使用的是模拟信号,因此必须要变换
PCM30设备就是将模拟信号变成数字信号的设备,它将30路电话,变成1路E1接口的数字信号。 6.同轴电缆与同轴头 7.数字配线架DDF 无论是交换机的中继器接口,还是PCM30的数字口,都是E1口,要用同轴电缆接到光端机,为了方便电缆的检修,和调换电路,就要使用数字配线架(DDF)设备.DDF就是一块装有同轴 头的面板,同轴电缆上的同轴头,接到DDF的同轴头上。 8.光传输设备(光端机) 将多路E1接口的数字信号变成1路光信号的设备叫光端机,来自交换机,或PCM30设备的数字信号E1信号,靠同轴电缆经过DDF接到光端机。光端机的输出就是激光了光端机的光接口有2根光纤,1根是发光的,另1个是收光的。 9.光缆线路器材 光缆每2公里就要有1个接头,2根光缆的接续是在光纤接续盒里完成。1条完整的光缆的两个终端是通信机房里的光缆终端盒,它将光缆里的很细的光纤与尾纤相连,尾纤是单根的,有外套,有牙签那样粗,一般是黄色的,尾纤带有1个光接头,可以通过法兰盘跟另1根尾纤相连,尾纤线束,是多根尾纤做在一起的,但是比单根尾纤细一点。 10.其他设备1 电源和电池:通信机房为了保证供电,一直采用电池作为停电后的供电,电池是直流的,所以电源设备就是将交流220V的交流电,变成-48V的直流电。电源列头柜:通信机房里有很多设备,光通信的,交换机,载波机,微波等,这些设备都要用到-48V的电源,列头柜就是将总电源通过保险然后再分配到各个通信机柜的设备。 11.其他设备2 接口变换器,传输设备的接口是E1口,在通信领域是标准的但是计算机领域的标准跟通信不同,随着计算机通信的发展,两者的接口越来越多,计算机通常采用以太网接口,和V35接口,因此他们跟E1口的变换器,就经常要用到。以太网光纤收发器,计算机的局域网已经趋向于以太网,而用光纤组网是越来越多,这就要用到光纤收发器。
4G通信技术综述
4G通信技术综述 近年来,第三代通信技术在全世界范围内,受到人们的青睐,并得到了广泛的应用。在此背景下,我国通信领域应当与时俱进,顺应潮流,积极发展我国通信技术,重视第四代通信技术的研究和开发工作。4G与3G相比较,具有安全性高、智能人性化以及传输速度快等优势,能够更好地满足人们互联通信的需求。文章简单地介绍了第四代移动通信的基本概念以及相关的技术,阐述了4G相关技术的主要用途,希望能够起到抛砖引玉的作用,为相关工作的发展提供一些建议。 标签:通信技术;发展趋势;综述 1 第四代移动通信系统的基本概念 1.1 4G的基本定义 4G通信技术又被人们称之为宽带接入和分布网络,其不仅非对称数据传输能力能够超过2Mbit/s,数传率高达100Mps。同时还兼具自动切换功能,能够实现不同速率之间的自动切换,从而为用户提供多种多样的服务,并达到兼容的目的。此外,4G通信技术实现了我国通信技术史上的首次三维图像的高质量传输,从而为移动用户提供更高质量的移动服务。其中包括4大部分,一是移动宽带系统;二是宽带无线固定接入;三是宽带无线局域网以及互操作的广播网络。在这些无线平台以及跨越不同频带的网络中,4G通信技术一方面能够为用户提供优质的无线服务,以及信息通信和数据采集外,另一方面还具有定时定位、远程控制等全方面的功能。 1.2 4G的研发背景 当前我国3G通信技术的发展已经十分成熟,与2G通信技术相比,3G通信技术灵活性更佳,速度更快,同时也进一步满足了人们对多媒体的需求。然而3G通信技术在发展过程中,为人们带来全新的通信感受的同时,也受到了一定的局限性,主要表现在:3G通信技术通信速率低下,灵活性不足,以及无法真正实现不同频段、不同业务环节之间的无缝漫游,并无法为用户提供动态范围内多速率的业务。这些都在一定程度上制约了我国通信技术的发展,基于这些压力,许多公司为了更好的发展通信事业,已经开始研究第四代概念通信系统。该系统与3G通信相比,一方面应当全面提高运行速率和灵活性,并配备更好的兼容性,从而有效的实现任何人在任何地点以任何形式接入网络。另一方面为了将核心基础设施通过开放式接口共享給不同的网络运营商以及服务供应商,应当建立一个为客户服务的开放性平台,该平台具有比传统封闭环境下的网络安全性能更高的优势。 1.3 4G移动系统网络的特点
密文数据库检索技术综述
密文数据库检索技术综述 摘要 关键词 1 引言 2 相关技术 3 研究分类 3.1 数值型数据 2002年,Hakan等人首次提出了在数据库即服务(Database as a service, DaaS)1模型下,针对加密数据执行SQL查询的方法2。其核心思想是:提出了一种过滤技术(桶划分技术)缩小解密范围,从而快速查询加密数据。并基于桶划分技术提出了一种对关系数据库进行加密和存储的模型,在此模型上存储数据时,除了对关系表中的记录采用常规加密外,还给每个属性值增加一个桶号,桶号表示明文数据值位于某段区间内。在该模型中,数据拥有者(即用户)对数据库进行加密后将数据库密文保存在服务提供商处,只有数据拥有者能够解密。用户提交查询指令后,服务器端无需对密文解密即可进行粗粒度的查询,得到包含查询结果的一个候选结果集合,然后将该候选结果集合返回给用户,用户解密该候选结果集合并对明文进行计算即可得到最终的查询结果。 该方法返回一个比正确结果集合更大一些的集合,其中可能包含一些并不匹配查询条件的密文元组,因此需要再对这个结果集合进行解密和过滤处理,才能得到最终的查询结果。此外,该方法仅通过值域分区的方式建立数据库值索引,容易造成数据库信息泄漏。数据库通常采用哈希技术分区的方式,这种方式的分区数量越多,检索性能越好,但同时会造成更多的数据冗余。当每个分区中的数据记录较多时,检索效率会受到较大影响。 2003年,Damiani等人提出基于索引的密文检索方法3。与桶划分方法不同,该方法将数据进行元组级的加密,因此能够进行元组级的检索。该方法不按数值的顺序分类,增加了安全性。其缺点是不能实现范围搜索。Damiani又使用B-tree 编码方式,这种方法可以实现范围检索,但是每次进行检索时需要检索的次数等
数据库新技术的发展综述
数据库新技术的发展综述
数据库技术的现状 及发展趋势 院系:数学科学学院 学号:20121014401 姓名:徐高扬 班级:统计122
数据库技术的现状与发展趋势 关键词:数据库;面向对象数据库;演绎面向对象数据库;数据仓库; 数据挖掘;发展;主流数据库新技术 1、引言 自从计算机问世以后,就有了处理数据、管理数据的需求,由此,计算机技术新的研究分支数据库技术应运而生。随着计算机应用领域的不断拓展和多媒体技术的发展,数据库已是计算机科学技术中发展最快、应用最广泛的重要分支之一。从20世纪60年代末开始,数据库系统已从第一代层次数据库、网状数据库,第二代的关系数据库系统,发展到第三代以面向对象模型为主要特征的数据库系统。关系数据库理论和技术在70~80年代得到长足的发展和广泛而有效地应用,80年代,关系数据库成为应用的主流,几乎所有新推出的数据库管理系统(DataBaseManagementSystem,DBMS)产品都是关系型的,他在计算机数据管理的发展史上是一个重要的里程碑,这种数据库具有数据结构化、最低冗余度、较高的程序与数据独立性、易于扩充、
易于编制应用程序等优点,目前较大的信息系统都是建立在关系数据库系统理论设计之上的。但是,这些数据库系统包括层次数据库、网状数据库和关系数据库,不论其模型和技术上有何差别,却主要是面向和支持商业和事务处理应用领域 的数据管理。然而,随着用户应用需求的提高、硬件技术的发展和InternetIntranet提供的丰 富多彩的多媒体交流方式,促进了数据库技术与网络通信技术、人工智能技术、面向对象程序设计技术、并行计算技术等相互渗透,互相结合, 成为当前数据库技术发展的主要特征,形成了数据库新技术。目前,数据库技术已相当成熟,被广泛应用于各行各业中,成为现代信息技术的重要组成部分,是现代计算机信息系统和计算机应用系统的基础和核心。 2、数据库技术的现状及发展趋势 1980年以前,数据库技术的发展,主要体现在数据库的模型设计上。进入90年代后,计算机领域中其它新兴技术的发展对数据库技术产生 了重大影响。数据库技术与网络通信技术、人工智能技术、多媒体技术等相互渗透,相互结合,使数据库技术的新内容层出不穷。数据库的许多
光纤通信综述报告
光纤通信综述报告 摘要:光纤通信技术(optical fiber communications)从光通信中脱颖而出,已成为现代通信的主要支柱之一,在现代电信网中起着举足轻重的作用。光纤通信作为一门新兴技术,其近年来发展速度之快、应用面之广是通信史上罕见的,也是世界新技术革命的重要标志和未来信息社会中各种信息的主要传送工具。 关键词:光纤通信新技术新器件新材料 仅在过去5年中,光纤技术领域取得了大量突破性进展,其中包括10Gbit/s网络的构建和单根光纤上每秒太比特容量的成功演示。不久前,业内成功演示了40Gbit/s和80Gbit/s网络。这些演示进一步突出了对速度更高、容量更大的网络的需求和期望。 一、光纤通信的发展史 世界光纤通信发展史 光纤的发明,引起了通信技术的一场革命,是构成21世纪即将到来的信息社会的一大要素。 1966年出生在中国上海的英籍华人高锟,发表论文《光频介质纤维表面波导》,提出用石英玻璃纤维(光纤)传送光信号来进行通信,可实现长距离、大容量通信。 于1970年损失为20db/km的光纤研制出来了。据说康宁公司花费3000万美元,得到30米光纤样品,认为非常值得。这一突破,引起整个通信界的震动,世界发达国家开始投入巨大力量研究光纤通信。1976年,美国贝尔实验室在亚特兰大到华盛顿间建立了世界第一条实用化的光纤通信线路,速率为45Mb/s,采用的是多模光纤,光源用的是发光管LED,波长是0.85微米的红外光。在上世纪70年代末,大容量的单模光纤和长寿命的半导体激光器研制成功。光纤通信系统开始显示出长距离、大容量无比的优越性。 按理论计算:就光纤通信常用波长1.3微米和1.55微米波长窗口的容量至少有25000GHz。自然会想到采用多波长的波分复用技术WDM (WavelengthDivisionMultiplex)。1996年WDM技术取得突破,贝尔实验室发展了WDM技术,美国MCI公司在1997年开通了商用的WDM线路。光纤通信系统的速率从单波长的2.5Gb/s和10Gb/s爆炸性地发展到多波长的Tb/s(1Tb/s=1000Gb/s)传输。当今实验室光系统速率已达10Tb/s,几乎是用之不尽的,所以它的前景辉煌。 中国光纤通信发展史 1973年,世界光纤通信尚未实用。邮电部武汉邮电科学研究院(当时是武汉邮电学院)就开始研究光纤通信。由于武汉邮电科学研究院采用了石英光纤、半导体激光器和编码制式通信机正确的技术路线,使我国在发展光纤通信技术上少走了不少弯路,从而使我国光纤通信在高新技术中与发达国家有较小的差距。 我国研究开发光纤通信正处于十年动乱时期,处于封闭状态。国外技术基本无
面向对象数据库技术的研究综述
面向对象数据库技术的研究综述 摘要:本文在提出传统数据库技术的不足及新应用领域需求的同时,介绍了面向对象数据库的特征与功能,并探讨了该技术面l临的一些问题;最后还对这一新技术的前景进行了展望。 关键词:面向对象;数据库技术;面向对象数据库 面向对象的思想首先出现在程序设计方法中。这一思想指导下产生的面向对象技术是一种按照人们对现实世界习惯的认识论思维方式来研究和模拟客观世界的方法学。它将现实世界中的任何事物均视为“对象”.将客观世界看成是由许多不同种类的对象构成。不同对象之间的相互联系和相互作用就构成了完整的客观世界。面向对象方法学所引入的对象、方法、消息、类、实例、继承性、封装性等一系列重要概念和良好机制为人们认识和模拟客观世界分析、设计和实现大型复杂系统奠定了良好的科学技术基础。 随着研究的不断深入和发展。面向对象技术已大大地超出了程序设计语言的范围。并渗透和应用到了诸多复杂的工程领域。并给软件工程、信息系统、工业设计与制造等带来了深远的影响。如面向对象的软件工程、面向对象的信息管理系统、面向对象的操作系统、面向对象的数据库系统、面
向对象的专家系统、面向对象的开发工具和面向对象的用户界面等的出现。其中,面向对象的数据库系统已成为当今数据库领域研究和发展的主要方向之一。 数据库技术与面向对象技术相结合已成为当前数据库技术研究、应用和发展的一个重要方向。将面向对象技术应用到数据库系统中。使数据库管理系统能够支持面向对象数据模型和数据库模式。这对于提高数据库系统模拟和操纵客观世界的能力,扩大数据库应用领域具有重要的意义:将面向对象技术应用到数据库的集成开发环境中。使数据库应用开发工具能够支持面向对象的开发方法井提供相应的开发手段,这对于提高应用软件的开发质量和扩大软件的应用推广是十分重要的。纵观数据库系统的发展,面向对象(00)技术的诞生为数据库的发展带来了希望。尽管目前面向对象数据库技术的实际发展与关系数据库系统相比,它的理论研究和形式化、标准化等方面还不完备和成熟。但是。从面向对象技术的前景和应用来看,面向对象数据库系统将代表着新一代数据库系统的发展方向。 一、新应用领域的需求及面向对象数据库的发展 从80年代以来,数据库技术在商业领域的巨大成功激发了其它领域对数据库技术需求的迅速增长。这些新的领域包括:CAD/CAM、CIM、CASE、OIS(办公信息系统)、GlS (地理信息系统)、知识库系统和实时系统等。新的应用领
键值对数据库综述
键值对数据库综述与典型KV数据库介绍 一、键值数据库概述 键值数据库是一种非关系数据库,它使用简单的键值方法来存储数据。键值数据库将数据存储为键值对集合,其中键作为唯一标识符。键和值都可以是从简单对象到复杂复合对象的任何内容。键值数据库是高度可分区的,并且允许以其他类型的数据库无法实现的规模进行水平扩展。 Key-Value 键值对数据模型实际上是一个映射,即key是查找每条数据地址的唯一关键字,value是该数据实际存储的内容。例如键值对:(“”,“张三”),其key:“”是该数据的唯一入口,而value:“张三”是该数据实际存储的内容.Key-Value 数据模型典型的是采用哈希函数实现关键字到值的映射,查询时,基于key 的hash值直接定位到数据所在的点,实现快速查询,并支持大数据量和高并发查询。 二、基本原理 从API的角度来看,键值数据库是最简单的NoSQL数据库。客户端可以根据键查询值,设置键所对应的值,或从数据库中删除键。“值”只是数据库存储的一块数据而已,它并不关心也无需知道其中的内容;应用程序负责理解所存数据的含义。由于键值数据库总是通过主键访问,所以它们一般性能较高,且易于扩展。基本上所有的编程语言都带有应用在内存中的键值对存储。C++STL的映射容器(map container)和Java的HashMap以及Python的字典类型都是键值对存储。键值对存储通常都有如下接口: -Get( key ): 获取之前存储于某标示符“key”之下的一些数据,或者“key”下没有数据时报错。 -Set( key, value ): 将“value”存储到存储空间中某标示符“key”下,使得我们可以通过调用相同的“key”来访问它。如果“key”下已经有了一些数据,旧的数据将被替换。 -Delete( key ): 删除存储在“key”下的数据。 三、基本特性 键值数据库具有以下几个特性:
光通信的历史及其发展现状
光通信的历史、现状、发展趋势 06007235 方云龙光通信的历史: 原始形式的光通信是通过中国古代的“烽火台”报警,欧洲人用旗语传送信息。1880年,美国人贝尔(Bell)发明了用光波作载波传送话音的“光电话”。贝尔光电话是现代光通信的雏型。 1960年,美国人梅曼(Maiman)发明了第一台红宝石激光器,给光通信带来了新的希望。激光器的发明和应用,使沉睡了80年的光通信进入一个崭新的阶段。 1966年,英籍华裔学者高锟(C.K.Kao)和霍克哈姆(C.A.Hockham)发表了关于传输介质新概念的论文,指出了利用光纤(Optical Fiber)进行信息传输的可能性和技术途径,奠定了现代光通信——光纤通信的基础。通过“原材料的提纯制造出适合于长距离通信使用的低损耗光纤”这一发展方向。 1970年,美国康宁(Corning)公司研制成功损耗20dB/km的石英光纤。把光纤通信的研究开发推向一个新阶段。 1973 年,美国贝尔(Bell)实验室的光纤损耗降低到2.5dB/km。1974 年降低到1.1dB/km。 1976 年,日本电报电话(NTT)公司将光纤损耗降低到0.47 dB/km(波长1.2μm)。在以后的10 年中,波长为1.55 μm的光纤损耗:1979 年是0.20 dB/km,1984年是0.157 dB/km,1986 年是0.154 dB/km,接近了光纤最低损耗的理论极限。 1970年,美国贝尔实验室、日本电气公司(NEC)和前苏联先后,研制成功室温下连续振荡的镓铝砷(GaAlAs)双异质结半导体激光器(短波长)。虽然寿命只有几个小时,但它为半导体激光器的发展奠定了基础。1977 年,贝尔实验室研制的半导体激光器寿命达到10万小时。1979年美国电报电话(AT&T)公司和日本电报电话公司研制成功发射波长为1.55 μm的连续振荡半导体激光器。 1976 年,美国在亚特兰大(Atlanta)进行了世界上第一个实用光纤通信系统的现场试验。1980 年,美国标准化FT - 3光纤通信系统投入商业应用。 1976 年和1978 年,日本先后进行了速率为34 Mb/s的突变型多模光纤通信系统,以及速率为100 Mb/s的渐变型多模光纤通信系统的试验。1983年敷设了纵贯日本南北的光缆长途干线。 随后,由美、日、英、法发起的第一条横跨大西洋TAT-8海底光缆通信系统于1988年建成。第一条横跨太平洋TPC-3/HAW-4 海底光缆通信系统于1989年建成。从此,海底光缆通信系统的建设得到了全面展开,促进了全球通信网的发展。 现状: 目前国内光纤光缆的生产能力过剩,供大于求。特种光纤如FTTH(光纤到户)用光纤仍需进口,但总量不大,国内生产光纤光缆价格与国际市场没有差别,成本无法再降,已经是零利润,在国际市场没有太强竞争力,出口量很小。二十年来的光技术的两个主要发展,WDM(Wavelength Division Multiplexing:波分复用)和PON(Passive Optical Network:无源光纤网络),这两个已经相对比较成熟。 今天,40Gbps的光通信系统得到广泛商用。作为新一代光网络的领军技术,40G商用大门的开启,满足日益增长的带宽需求同时,还为ROADM、先进光调制技术、超强EFC等新技术的应用赢得了市场发展空间,并为全光网的演进、升级创造了条件。不过,这只是40Gbps的一个开始,要承担起未来传输主力的重任,40G还需要很多路要走。现在对40Gbps,乃至更高速率的100Gbps而言,光学硬件的发展是关键,同时还必须与其他光通讯技术协同发展,包括复杂的调制技术、信号处理技术、并行接口、主动追踪和补偿技术,这些条件
数据库技术的现状及其发展趋势
数据库技术的现状及其发展趋势 (班级:041011 姓名:罗英学号:04101001) 一数据库技术的基本概述 数据库技术是信息系统的一个核心技术。是一种计算机辅助管理数据的方法,它研究如何组织和存储数据,如何高效地获取和处理数据。是通过研究数据库的结构、存储、设计、管理以及应用的基本理论和实现方法,并利用这些理论来实现对数据库中的数据进行处理、分析和理解的技术。即:数据库技术是研究、管理和应用数据库的一门软件科学。 数据库技术是现代信息科学与技术的重要组成部分,是计算机数据处理与信息管理系统的核心。数据库技术研究和解决了计算机信息处理过程中大量数据有效地组织和存储的问题,在数据库系统中减少数据存储冗余、实现数据共享、保障数据安全以及高效地检索数据和处理数据。 数据库技术研究和管理的对象是数据,所以数据库技术所涉及的具体内容主要包括:通过对数据的统一组织和管理,按照指定的结构建立相应的数据库和数据仓库;利用数据库管理系统和数据挖掘系统设计出能够实现对数据库中的数据进行添加、修改、删除、处理、分析、理解、报表和打印等多种功能的数据管理和数据挖掘应用系统;并利用应用管理系统最终实现对数据的处理、分析和理解。 数据库技术涉及到许多基本概念,主要包括:信息,数据,数据处理,数据库,数据库管理系统以及数据库系统等。 数据库技术是现代信息科学与技术的重要组成部分,是计算机数据处理与信息管理系统的核心。数据库技术研究和解决了计算机信息处理过程中大量数据有效地组织和存储的问题,在数据库系统中减少数据存储冗余、实现数据共享、保障数据安全以及高效地检索数据和处理数据。数据库技术的根本目标是要解决数据的共享问题。 二数据库技术发展历史 数据模型是数据库技术的核心和基础,因此,对数据库系统发展阶段的划分应该以数据模型的发展演变作为主要依据和标志。按照数据模型的发展演变过程,数据库技术从开始到现在短短的30年中,主要经历了三个发展阶段:第一代是网状和层次数据库系统,第二代是关系数据库系统,第三代是以面向对象数据模型为主要特征的数据库系统。数据库技术与网络通信技术、人工技能技术面向对象程序设计技术、并行计算技术等相互渗透、有机结合,成为当代数据库技术发展的重要特征。 第一代数据库系统 第一代数据库系统是20世纪70年代研制的层次和网状数据库系统。层次数据库系统的典型代表是1969年IBM公司研制出的层次模型的数据库管理技术IMS。20世纪60年代末70年代初,美国数据库系统语言协会
