Protein and cell final edition
R EVIEW
Role of tegument proteins in herpesvirus assembly and egress
Haitao Guo1,2,Sheng Shen1,2,Lili Wang1,2,Hongyu Deng1?
1CAS Key Laboratory of Infection and Immunity,Institute of Biophysics,Chinese Academy of Sciences,Beijing100101,China 2Graduate School of the Chinese Academy of Sciences,Beijing100080,China
?Correspondence:rrain6@https://www.360docs.net/doc/457826919.html,
Received October4,2010Accepted November4,2010
ABSTRACT
Morphogenesis and maturation of viral particles is an essential step of viral replication.An infectious herpes-viral particle has a multilayered architecture,and con-tains a large DNA genome,a capsid shell,a tegument and an envelope spiked with glycoproteins.Unique to herpesviruses,tegument is a structure that occupies the space between the nucleocapsid and the envelope and contains many virus encoded proteins called tegu-ment proteins.Historically the tegument has been described as an amorphous structure,but increasing evidence supports the notion that there is an ordered addition of tegument during virion assembly,which is consistent with the important roles of tegument proteins in the assembly and egress of herpesviral particles.In this review we?rst give an overview of the herpesvirus assembly and egress process.We then discuss the roles of selected tegument proteins in each step of the process,i.e.,primary envelopment,deenvelopment, secondary envelopment and transport of viral particles. We also suggest key issues that should be addressed in the near future.
KEYWORDS herpesvirus,tegument,assembly, egress,transport
INTRODUCTION
The family Herpesviridae consists of three subfamilies including Alphaherpesvirinae,Betaherpesvirinae and Gam-maherpesvirinae whose members infect a broad range of animal species including mammals,birds and reptiles (Davison et al.,2009).Until now,more than130herpes-viruses have been identi?ed,among which there are eight human pathogens,including the Alphaherpesvirinae mem-bers herpes simplex virus type1(HSV-1),herpes simplex virus type2(HSV-2)and varicella zoster virus(VZV),the Betaherpesvirinae members human cytomegalovirus (HCMV),human herpesvirus type6(HHV-6)and human herpesvirus type7(HHV-7),and the Gammaherpesvirinae members Epstein-Barr virus(EBV)and Kaposi’s sarcoma-associated herpesvirus(KSHV).HSV most commonly cause mucocutaneous infections,resulting in recurrent orolabial or genital lesions(Roizman et al.,2007).HCMV infection is responsible for approximately8%of infectious mononucleo-sis cases and is also associated with in?ammatory and proliferative diseases(S?derberg-Nauclér,2006;Steininger, 2007).EBV and KSHV are associated with several malig-nancies,such as Burkitt’s lymphoma and Kaposi’s sarcoma (Chang et al.,1994;Kutok and Wang,2006).
All members of the Herpesviridae family comprise two distinct stages in their life cycle:lytic replication and latency (Sunil-Chandra et al.,1992;Decker et al.,1996a,b;Dupin et al.,1999;Fla?o et al.,2000).Members of the Alphaherpesvir-inae subfamily establish latency in neurons,members of the Betaherpesvirinae subfamily establish latency in a range of nonneuronal cells,whereas members of Gammaherpesvir-inae subfamily members establish latency mainly in B and T https://www.360docs.net/doc/457826919.html,tency provides the viruses with advantages to escape host immune surveillance so as to establish lifelong persistent infection and thus contributes to transformation and development of malignancies.However,it is through lytic replication that viruses propagate and transmit among hosts to maintain viral reservoirs.Both viral latency and lytic replication play important roles in herpesvirus pathogenesis.
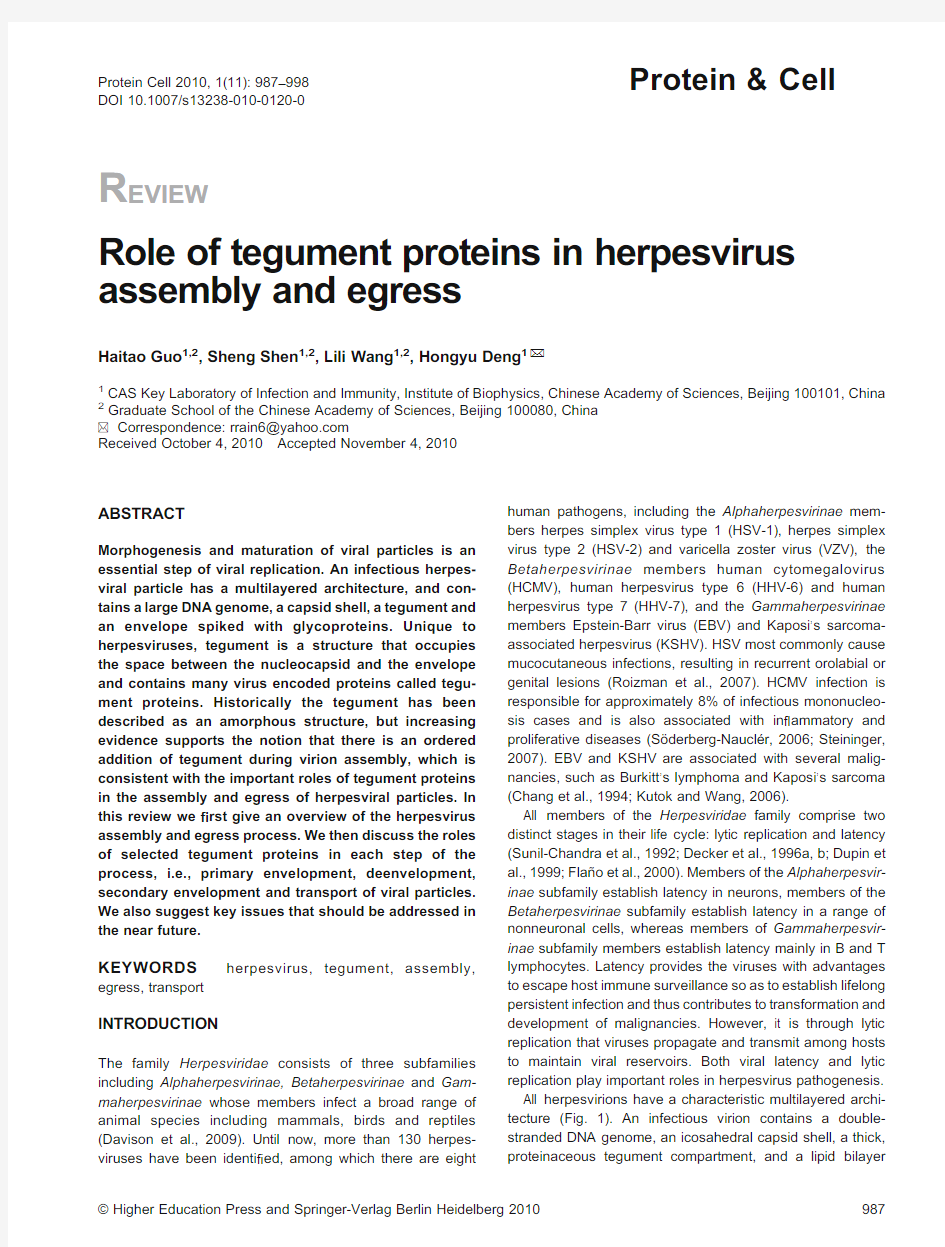
All herpesvirions have a characteristic multilayered archi-tecture(Fig.1).An infectious virion contains a double-stranded DNA genome,an icosahedral capsid shell,a thick, proteinaceous tegument compartment,and a lipid bilayer
Protein Cell2010,1(11):987–998
DOI10.1007/s13238-010-0120-0
Protein&Cell
envelope spiked with glycoproteins (Rixon,1993;Roizman and Pellett,2001;Liu and Zhou,2006;Dai et al.,2008).As a unique structure of herpesviruses,the tegument plays important roles in multiple aspects of the viral life cycle,including translocation of nucleocapsids into the nucleus as well as virion assembly and egress (Subak-Sharpe and Dargan,1998;Fuchs et al.,2002a).As components that are carried into a host cell during viral infection,tegument proteins can also transactivate the expression of viral immediate-early genes and cellular genes as well as modulate,host signal transduction and innate immunity (Bresnahan and Shenk,2000;Castillo and Kowalik,2002;Ishov et al.,2002;Zhu et al.,2002).
Virion assembly and egress is an essential stage of herpesviral lytic replication,hence contributes indirectly to herpesvirus pathogenesis.In this review we summarize the growing range of functions attributed to tegument proteins during the course of herpesvirus assembly and egress.We mainly focus on tegument proteins from the Alphaherpesvir-inae subfamily,in particular HSV-1and the related porcine pathogen pseudorabies virus (PrV),since they are the most extensively studied.Meanwhile,some well studied tegument proteins from the Betaherpesvirinae and Gammaherpesvir-inae subfamilies are also discussed.
OVERVIEW OF HERPESVIRUS ASSEMBLY AND EGRESS
During herpesvirus lytic infection,after attachment and penetration,capsids are transported to the nucleus via interaction with microtubules,docking at the nuclear pore where the viral genome is released into the nucleus (D?hner et al.,2002;Wolfstein et al.,2006),where transcription of viral immediate-early,early and late genes and genome replication take place.Then virion assembly and egress proceed from the nucleus to the cytoplasm.Several models have been proposed to explain this process,among which the most widely accepted is the envelopment-deenvelopment-reenvelopment model:capsid formation and viral DNA packaging in the nucleus,primary envelopment at the inner nuclear membrane and deenvelopment at the outer nuclear membrane,association with tegument proteins and second-ary envelopment (i.e.,reenvelopment)in the cytoplasm,and budding of complete virions into Golgi body-derived compart-ments for egress via the cellular secretory pathway (Metten-leiter,2002,2004).In each stage of virion assembly and egress,tegument proteins are found to play important roles.There are also lines of evidence suggesting direct exit from the nucleus of unenveloped nucleocapsids via enlarged nuclear pores or transport of perinuclear enveloped nucleo-capsids through a continuum of outer nuclear,rough ER and Golgi membranes (Wild et al.,2002;Leuzinger et al.,2005;Wild et al.,2005,2009).In this review,the functional roles of tegument proteins will be discussed in the context of the envelopment-deenvelopment-reenvelopment model of virion assembly.
TEGUMENT PROTEINS
Composition of tegument in all three Herpesviridae subfami-lies has been studied.At least 26,38or 17virus-encoded tegument components are recruited into an HSV-1,HCMV or EBV virion,respectively (Johannsen et al.,2004;Varnum et al.,2004;Zhu et al.,2005;Loret et al.,2008).Some of these proteins,such as pUL36,pUL47,pUL48and pUL49,are classi ?ed as major,structurally signi ?cant components (Heine et al.,1974),whereas some,such as vhs and the protein kinase pUL13,are minor but nonetheless important compo-nents (Schek and Bachenheimer,1985;Overton et al.,1992).Some tegument proteins are conserved in all herpesvirus members (Table 1),and their functions are progressively unrevealed.
As an indispensable part of mature virion,tegument plays important roles in herpesviral particle production.Much effort has been placed on elucidating the molecular interactions between various tegument proteins with the hope of under-standing the mechanisms by which this complex virus is assembled.Tegument addition is thought to originate in the nucleus,then more tegument proteins are added to capsids in the cytoplasm following nuclear egress and ?nally at trans-Golgi network (TGN)-derived membranes during maturation budding (Mettenleiter,2006).The complexity of virion assembly route clearly raises the issue that different tegu-ment proteins may be incorporated into herpesviral particles at different stages of egress and that the composition of virions in the perinuclear space may not be the same as that of the mature extracellular viruses.For example,the UL31and UL34gene products of PrV are present in viral particles in the perinuclear space but are absent from virions at later stages of assembly and mature virions (Fuchs et al.,2002c).In contrast,the pUL36,pUL37,pUL46,pUL47,pUL48
and
Figure 1.Diagram of herpesvirion structure.The virion particle is approximately 200nm in diameter.A linear double-stranded DNA genome is packaged within an icosahedral capsid to form the nucleocapsid.The capsid is embedded in a matrix known as the tegument layer,which contains many virus coded proteins.The tegument is itself surrounded by the envelope,a lipid membrane containing several viral glycopro-teins.
Haitao Guo et al.
Protein &Cell
pUL49tegument proteins of PrV are found in only cytoplasmic enveloped virions and mature PrV particles and are therefore presumably not recruited into the virion during primary envelopment(Fuchs et al.,2002a;Klupp et al.,2002;Kopp et al.,2002).The PrV pUS3protein kinase,on the other hand, is a component of both primary and mature enveloped virions (Granzow et al.,2004).However,some researchers showed con?icting data on the related HSV-1homologs,in which HSV tegument proteins pUL36,pUL37,pUL48and pUL49have been found associated with intranuclear capsids or existing in primary enveloped virions(Mossman et al.,2000;Naldinho-Souto et al.,2006;Bucks et al.,2007;Padula et al.,2009). These results indicate that the sequence of tegument protein addition and the regions in which capsid acquires the tegument are very complicated.The inner tegument proteins may interact with the capsid while the outer tegument proteins may be recruited by interacting with the cytoplasmic tails of viral glycoproteins.Several molecular interactions have been implicated in linking the viral capsid,tegument and membrane during the envelopment process.Examples include pUL25(capsid)-pUL36(tegument),pUL49(tegument)-gD(mem-brane),pUL48(tegument)-gH(membrane),and pUL11 (membrane)-pUL16(tegument/capsid)interactions(McNab et al.,1998;Mettenleiter,2002,2004;Coller et al.,2007). These interactions will be elaborated in the following sections. PRIMARY ENVELOPMENT AND DEENVELOPMENT
After capsid assembly,the nucleocapsids will bud at the inner nuclear membrane(NM).There,the viruses encounter the nuclear lamina,a rigid network of lamin proteins lining the inner NM,which is a major obstacle for budding of the nucleocapsids.Herpesviruses disrupt the nuclear lamina in order to bud into the perinuclear space and acquire the inner NM as the envelope,the so called primary envelopment (Scott and O'Hare,2001;Muranyi et al.,2002;Leach et al., 2007;Mou et al.,2007;Mou et al.,2008).HSV-1and PrV disrupt the lamina through two viral proteins,pUL31and pUL34,which form a complex colocalized to the inner NM (Reynolds et al.,2002;Reynolds et al.,2004;Simpson-Holley et al.,2004;Simpson-Holley et al.,2005;Bjerke and Roller, 2006).Research suggested that pUL31and pUL34promote viral nuclear egress by several mechanisms,including(1) affecting maturation of viral replication intermediates so that capsids assemble adjacent to the nuclear envelope,making it convenient for primary egress(Simpson-Holley et al.,2004, 2005),(2)causing displacement of and conformational changes in lamins A/C and B(Reynolds et al.,2004; Simpson-Holley et al.,2004;Bjerke and Roller,2006),and (3)mislocalizing the membrane lamin receptors,such as the lamin B receptor and emerin,which tether lamins to the inner NM(Scott and O'Hare,2001;Leach et al.,2007;Morris et al., 2007;Mou et al.,2008).Amazingly,expression of PrV pUL31 and pUL34in RK-13cells resulted in formation of vesicles in the perinuclear space,indicating that they may compose the essential budding machinery(Klupp et al.,2007).
pUL31and pUL34are conserved in all three Herpesviridae subfamilies.In EBV,the pUL34positional homolog BFRF1 and the pUL31positional homolog BFLF2also interact with each other.HSV-1UL34or EBV BFRF1mutant viruses showed a phenotype of nuclear retention(Lake and Hutt-Fletcher,2004;Farina et al.,2005;Gonnella et al.,2005; Calderwood et al.,2007).EBV BFLF2functions in both viral DNA packaging and primary envelopment,as deletion of BFLF2led to sequestration of nucleocapsids in the nucleus and production of more empty capsids without viral genomes (Granato et al.,2008).
Alphaherpesviruses also express a viral serine/threonine protein kinase,pUS3,that phosphorylates lamins and lamin receptor emerin,which are thought to be involved in maintaining nuclear integrity(Leach et al.,2007;Morris et al.,2007;Mou et al.,2008).In addition,both HSV-1and PrV pUL31and pUL34have been shown to be phosphorylated in a pUS3-dependent manner,and all three proteins have been found to be incorporated into primary virions(Purves et al.,
Table1Conservation of herpesvirus tegument proteins
HSV-1tegu-ment proteins nomenclature of HSV-1tegument homologs HCMV EBV KSHV
UL7UL103BBRF2ORF42 UL11UL99BBLF1ORF38 UL13UL97BGLF4ORF36 UL14UL95BGLF3ORF34 UL16UL94BGLF2ORF33 UL21UL87BcRF1ORF24 UL23NA BXLF2ORF21 UL36UL48BPLF1ORF64 UL37UL47BOLF1ORF63 UL51UL71BSRF1ORF55 UL41NA NA NA UL46NA NA NA UL47NA NA NA UL48NA NA NA UL49NA NA NA UL50NA NA NA UL55NA NA NA US2NA NA NA US3NA NA NA US10NA NA NA US11NA NA NA RL1(ICP34.5)NA NA NA RL2(ICP0)NA NA NA RS1(ICP4)NA NA NA NA:not applicable;HSV:herpes simplex virus.
1991;Granzow et al.,2004;Ryckman and Roller,2004;Kato et al.,2005;Mou et al.,2007,2009).pUS3also regulates the localization of the pUL31/pUL34complex,as deletion of pUS3disrupts their localization from a smooth nuclear rim distribution to distinct foci(Wagenaar et al.,1995;Klupp et al., 2001a;Reynolds et al.,2002;Ryckman and Roller,2004). These observations indicate that pUS3might function in virion primary envelopment.
The other tegument protein kinase,pUL13,may also play a role in regulating the localization of pUL31and pUL34in HSV-1either directly or indirectly through phosphorylation of pUS3 (Kato et al.,2006,2008),indicating its participating in primary envelopment.The HCMV homolog of HSV-1pUL13,pUL97, which is also a kinase,is incorporated into virions(Michel et al.,1996).pUL97has been reported to play an important role in virion nuclear egress,as a marked decrease in the number of mature capsids was observed in the cytoplasm of cells infected with UL97null mutant compared to the wild type virus (Wolf et al.,2001;Krosky et al.,2003).Deletion of HCMV UL97causes an aggregation of tegument and other structural proteins in nuclear inclusion,suggesting that pUL97kinase is required for the normal function of tegument proteins in the nucleus and may function in initiation of tegumentation or primary envelopment(Prichard et al.,2005).Although it is not clear where pUL97is added onto the nucleocapsid,interac-tion between pUL97and the HCMV major tegument protein pp65is required for the incorporation of pUL97into viral particles(Kamil and Coen,2007;Chevillotte et al.,2009).
HSV-1tegument proteins pUL36,pUL37,pUL48and pUL49have been found to be associated with intranuclear capsids or exist in primary enveloped virions,though their speci?c functions in nuclear egress are not clear yet(Moss-man et al.,2000;Naldinho-Souto et al.,2006;Bucks et al., 2007;Padula et al.,2009).pUL36contains a capsid binding domain and interacts with a minor capsid protein pUL25 previously found to participate in DNA encapsidation(McNab et al.,1998;Coller et al.,2007).Such an interaction is required for assembly of pUL36onto capsid,indicating that pUL36can bind to capsid directly.This result is consistent with those from electron microscope studies,which suggest presence of HSV-1pUL36in the inner most layer of the tegument,attached to the vertices of the capsid(McNabb and Courtney,1992;Zhou et al.,1999).pUL36is conserved throughout the Herpesviridae.It forms a complex with pUL37 as part of the capsid-associated inner tegument.Deletion of either of the genes encoding pUL36or pUL37blocks further tegumentation of the capsid in the cytoplasm(Desai,2000; Desai et al.,2001;Klupp et al.,2001b;Fuchs et al.,2004; Leege et al.,2009;Roberts et al.,2009).By interacting with pUL48,pUL36is a major factor for recruiting pUL48onto capsids(Ko et al.,2010).However,in PrV,pUL36and pUL48 only exist in mature virions,and are undetectable from nuclear or primary enveloped virions,indicating that these two tegument proteins are acquired in the cytoplasm (Naldinho-Souto et al.,2006;M?hl et al.,2009).Deletion of pUL48does not affect the capsid assembly and nuclear egress of PrV(Fuchs et al.,2002a).These con?icting results suggest that pUL36and pUL48of HSV-1and PrV may function differently.
Deenvelopment is a step of herpesvirus nuclear egress whose details are poorly understood.pUL48might play a role in this process for HSV-1,as large numbers of enveloped particles accumulate between the inner and outer NM in the absence of pUL48(Mossman et al.,2000).In addition,HSV-1 gB and gH/gL have redundant or overlapping roles in fusion with the outer NM(Farnsworth et al.,2007b).Consistent with this notion,EBV,KSHV and PrV gB null mutants all exhibit defects in nuclear egress(Peeters et al.,1992;Lee and Longnecker,1997;Krishnan et al.,2005).In the primary enveloped virions,pUS3can phosphorylate the gB cytoplas-mic domain and this modi?cation is important for gB-mediated fusion of the virion envelope and the outer NM(Wisner et al., 2009).Thus,pUS3plays important roles in both primary envelopment and deenvelopment.
SECONDARY ENVELOPMENT
Most tegument proteins are recruited onto the nucleocapsid in the cytoplasm.The addition of outer tegument is accompa-nied with virion secondary envelopment.During this process, several molecular interactions,such as pUL11(membrane)-pUL16(tegument),pUL48(tegument)-gH(membrane)and pUL49(tegument)-gE(membrane),have been implicated in linking the viral capsid,tegument and membrane(Mettenlei-ter,2002,2004).pUL16is conserved throughout the Herpesviridae,and plays a pivotal role in virion morphogen-esis in the cytoplasm.pUL16binds to the capsid by interacting with pUL21which is also a tegument protein associated with capsid in both the nucleus and the cytoplasm (de Wind et al.,1992;Takakuwa et al.,2001;Klupp et al., 2005).pUL16has another binding partner,named pUL11, which is a membrane-bound tegument protein(Loomis et al., 2003).Two distinct binding sites exist in pUL16for pUL11and pUL21,as covalent modi?cation of its free cysteines with N-ethylmaleimide blocks its binding to pUL11but not to pUL21 (Yeh et al.,2008;Harper et al.,2010).This observation suggests that pUL16may interact with pUL21and pUL11in sequence and both interactions contribute to ef?cient incorporation of pUL16into virions(Meckes et al.,2010). The pUL11tegument protein is also conserved among all herpesviruses,and each homolog contains amino acid motifs that allow covalent modi?cations with two fatty acids, myristate and palmitate(MacLean et al.,1989;Loomis et al.,2001).These two modi?cations allow pUL11accumu-lating at TGN-derived vesicles in the absence of other viral proteins(Loomis et al.,2001).In addition,there exist leucine-isoleucine and acidic cluster motifs,responsible for recovering pUL11from plasma membrane to the internal
Haitao Guo et al.
Protein&Cell
membrane.This might enable the recruitment of other membrane proteins(virus or host encoded)from the plasma membrane for secondary envelopment occurring around the TGN-vesicles(Loomis et al.,2001).
Similar to the conservation of pUL16and pUL11,their interaction is also conserved among all herpesviruses.In HCMV,the pUL11homolog pUL99has been found to interact with the pUL16homolog pUL94(Liu et al.,2009).Consistent with the previously described model of capsid transport and budding,herpesviruses lacking pUL16,pUL21or pUL11(or their homologs)have defects in virus egress,resulting in decreased amounts of extracellular viruses produced and the accumulation of non-enveloped capsids in the cytoplasm (MacLean et al.,1989,1992;Baines and Roizman,1992; Baines et al.,1994;Kopp et al.,2003;Schimmer and Neubauer,2003;Silva et al.,2003;Britt et al.,2004;Jones and Lee,2004;Silva et al.,2005;Seo and Britt,2006;Guo et al.,2009).
In murine gammaherpesvirus-68(MHV-68),the homolog of pUL16is ORF33.Our laboratory has con?rmed that ORF33 indeed encodes a tegument protein for MHV-68.Through constructing and analyzing an ORF33null mutant,we demonstrated that ORF33is not required for viral genome replication or gene expression,but is essential for virion morphogenesis and egress.More interestingly,ORF33null mutation caused a partial retention of nucleocapsids in the nucleus,and maturation of virions was arrested at a cytoplasmic stage of partially tegumented nucleocapsids, indicating that ORF33participates in both primary envelop-ment of nucleocapsids and morphogenesis of virions in the cytoplasm(Guo et al.,2009).We also found that ORF33 localizes in the nucleus during early infection and interacts with another nuclear tegument protein(Guo et al.,unpub-lished data).We hypothesize that such interaction may facilitate the nuclear budding or egress of nucleocapsids, although such an interaction is relatively redundant.
A number of interactions have been identi?ed involving HSV-1pUL48and inner tegument proteins,outer tegument proteins or the cytoplasmic tails of glycoproteins,supporting the concept that pUL48contributes to linking capsid and envelope during virion formation.For instance,pUL48has been shown to interact with the outer tegument proteins pUL41,pUL46,pUL47and pUL49,through a combination of yeast two-hybrid,in vitr o pull-down and co-immunoprecipita-tion studies(Smibert et al.,1994;Elliott et al.,1995;Vittone et al.,2005).HSV-2pUL48has also been shown to copurify and colocalize with pUL46during the course of infection(Kato et al.,2000).HSV-1pUL46is associated with cellular membrane independent of the presence of other viral proteins.Surprisingly,membrane association within the cell does not translate to association of this protein with the viral envelope,as pUL46exhibits a strong af?nity for the capsid of puri?ed viral particles(Murphy et al.,2008).However,deletion of pUL46in HSV-1and PrV does not block virion assembly (Zhang et al.,1991;Kopp et al.,2002).Although deletion of pUL47causes an approximately10-fold decrease in viral titer, pUL47is not essential for virion assembly in the cytoplasm (Kopp et al.,2002).HSV-1pUL49interacts with pUL48,but this interaction is not required for incorporation of pUL49into viral particles(O’Regan et al.,2007).Similar to pUL46, deletion of pUL49in HSV-1does not have a signi?cant effect on virus assembly(Elliott et al.,2005).In the case of PrV, deletion of any of pUL46,pUL47,pUL48or pUL49does not prevent incorporation of the remaining three proteins into the tegument(Fuchs et al.,2002a;Michael et al.,2006). Furthermore,an increased incorporation of cellular actin into virions was observed when pUL46,pUL47or pUL49was deleted,suggesting that actin may?ll the void left by the absence of these tegument proteins(del Rio et al.,2005; Michael et al.,2006).These observations indicate that redundancy exists in tegument assembly.In addition to tegument proteins mentioned above,HSV-1pUL48also interacts with the cytoplasmic tails of HSV-1glyoprotein gB, gD and gH,as identi?ed by pull-down,co-immunoprecipita-tion and chemical cross-linking assays(Zhu and Courtney, 1994;Gross et al.,2003;Kamen et al.,2005).However,the role of these interactions in HSV-1assembly requires further investigation.
pUL49,besides an interaction with pUL48,also interacts with a number of viral envelope proteins.Several studies have shown that pUL49interacts with the cytoplasmic tails of HSV-1gE,gD and envelope protein pUS9(Chi et al.,2005; Farnsworth et al.,2007a;O’Regan et al.,2007).In PrV, interactions of pUL49with the cytoplasmic tail of both gE and gM were identi?ed(Fuchs et al.,2002b).Furthermore,for both HSV-1and PrV,gE or gM is suf?cient for recruitment of pUL49into virions through a direct interaction(Fuchs et al., 2002b;Michael et al.,2006;O’Regan et al.,2007;Stylianou et al.,2009).
Other tegument proteins promoting secondary envelop-ment have also been identi?ed.ORF52is a tegument protein conserved in gammaherpesviruses.The MHV-68ORF52 localizes in TGN-derived vesicles in both transfection and viral infection.Furthermore,ORF52was found to speci?cally function in virus budding into the vesicles in the cytoplasm (Bortz et al.,2007).The crystal structure of ORF52has been determined at2.1?resolution,revealing a dimeric associa-tion of this protein.The self-association was con?rmed by co-immunoprecipitation and?uorescence resonance energy transfer experiments.Functional complementation assay demonstrated that both the N-terminalα-helix and the conserved Arg95residue are critical for ORF52function (Benach et al.,2007).The detailed mechanism responsible for involvement of ORF52in secondary envelopment is currently under investigation in our laboratory.
In HCMV,the tegument protein pUL32has been reported to be important in cytoplasmic maturation of virions,espe-cially in virus egress(Tandon and Mocarski,2008).pUL32
interacts with Bicaudal D1(BicD1),a protein thought to play a role in traf?cking within the secretory pathway(Indran et al., 2010).This interaction helps pUL32to traf?c to the virus assembly compartment,suggesting a primary contribution of this protein in the morphogenesis and/or cytoplasmic trans-port of progeny virion particles to sites of virion secondary envelopment.
TRANSPORT AND RELEASE
How do nucleocapsids or partially tegumented nucleocapsids transport to the site where secondary envelopment takes place?Microtubule has been found to be involved in transporting non-enveloped viral particles.Several tegument proteins have been identi?ed as candidates linking deenve-loped nucleocapsids to the anterograde cellular microtubule-dependent molecular motor kinesin.In alphaherpesviruses, pUS11was found to interact with both kinesin-1and the kinesin-related protein PAT-1(Diefenbach et al.,2002; Benboudjema et al.,2003).An association between pUL21 and microtubules was also detected(Takakuwa et al.,2001), raising the possibility that pUL21could be involved with capsid transport to the TGN-derived vesicles,where pUL16 interacts with pUL11,facilitating the budding process by linking capsids to the membrane.In gammaherpesviruses, an interaction between KSHV tegument protein ORF45and kinesin-2,determined by yeast two-hybrid and co-immunoprecipitation assays,has been reported(Sathish et al.,2009).Disrupting the interaction between KSHV ORF45and kinesin-2by a dominant negative mutant of ORF45leads to a decreased?nal production of mature viruses.This result supports the idea that the association between a viral tegument protein and a cellular molecular motor is important for the intracellular movement of deenve-loped nucleocapsids.The HSV-1pUL36is a multifunctional large tegument protein.As part of the inner tegument,pUL36 is also important for the transport of enveloped virions.A potential mechanism was proposed that,during viral infection, pUL36resides on the surface of organelles which nucleo-capsids bud into,and pUL36would directly or indirectly recruit kinesin motors to the organelles to enable motility(Shanda and Wilson,2008).
CONCLUSIONS AND PERSPECTIVES
Through interacting with and manipulating the host micro-environment,tegument proteins are multifunctional during the complete cycle of herpesvirus lytic replication.In particular, they play key structural roles during virion primary envelop-ment,secondary envelopment and virion traf?cking by forming a bridge between capsid or capsid-associated proteins and membrane-associated viral proteins or
cellular
Figure2.Diagram summarizing the roles of selected tegument proteins in herpesvirus assembly and egress.Viral genomes are packaged into preformed capsids in the nucleus.Through primary envelopment at the inner nuclear membrane and deenvelopment at the outer nuclear membrane,the nucleocapsid is transported into the cytoplasm from the nucleus.Acquisition of tegument proteins onto the nucleocapsid is completed in the cytoplasm.The immature virion is transported via the microtubules into
a trans-Golgi derived vesicle containing viral glycoproteins.After transport of the vesicle to the cell surface,the vesicle and plasma
membrane fuse,resulting in the egress of a mature,enveloped virion.
Haitao Guo et al. Protein&Cell
molecular motors(Fig.2).The herpesvirion assembly and egress process is complex and dynamic,and so are the roles of tegument proteins.As a complex process,herpesvirion assembly and egress involves many protein–protein interac-tions with marked redundancy.Indeed,interactions between tegument proteins and other viral and cellular proteins are increasingly reported in vitro or in vitro.However,future research on tegument will require identifying herpesviral protein–protein interaction and herpesviral protein-cellular protein interaction maps during the course of infecting host cells and validating such interactions by viral genetics approach.This has been done to some extent with the Alphaherpesvirinae subfamily member VZV and the Gamma-herpesvirinae subfamily members KSHV and EBV(Uetz et al.,2006;Calderwood et al.,2007;Rozen et al.,2008).Such information will improve our understanding of the biology of herpesvirus and the roles of tegument proteins.More importantly,by examining the importance of each pair of protein–protein interaction,useful therapeutic targets may be identi?ed.The current antiviral strategy for treatment of herpesviruses employs inhibitors of viral DNA replication which have varying ef?cacies depending on the Herpesvir-inae subfamily being treated.Therefore,new targets for therapeutic interventions through different mechanisms are useful,especially when combined with the viral DNA replication inhibitors.
The herpesvirion assembly and egress is also a dynamic process,and dissecting the roles of tegument proteins in such a dynamic process calls for integration of different technical approaches.Recently,combinations of molecular tags visible in light and electron microscopes have become particularly advantageous in the analysis of dynamic events at the cellular level(Martin et al.,2005;Gaietta et al.,2006;Lanman et al., 2008).Engineering such tags into herpesvirus tegument proteins as well as capsid/envelope proteins will enable optical live cell imaging and correlated ultrastructural analysis by electron microscopy,and help provide high-resolution information for the dynamic process of herpesvirion assembly and egress.
ACKNOWLEDGEMENTS
We thank Dr.Z.Hong Zhou and members of the Deng′laboratory for helpful discussions.This work was supported by the“One Hundred Talents Program”from the Chinese Academy of Sciences and the National Protein Science Project(No.2006CB910901)from the Ministry of Science and Technology.
REFERENCES
Baines,J.D.,Koyama,A.H.,Huang,T.,and Roizman,B.(1994).The UL21gene products of herpes simplex virus1are dispensable for growth in cultured cells.J Virol68,2929–2936.
Baines,J.D.,and Roizman,B.(1992).The UL11gene of herpes simplex virus1encodes a function that facilitates nucleocapsid
envelopment and egress from cells.J Virol66,5168–5174. Benach,J.,Wang,L.,Chen,Y.,Ho,C.K.,Lee,S.,Seetharaman,J., Xiao,R.,Acton,T.B.,Montelione,G.T.,Deng,H.,et al.(2007).
Structural and functional studies of the abundant tegument protein ORF52from murine gammaherpesvirus68.J Biol Chem282, 31534–31541.
Benboudjema,L.,Mulvey,M.,Gao,Y.,Pimplikar,S.W.,and Mohr,I.
(2003).Association of the herpes simplex virus type1Us11gene product with the cellular kinesin light-chain-related protein PAT1 results in the redistribution of both polypeptides.J Virol77, 9192–9203.
Bjerke,S.L.,and Roller,R.J.(2006).Roles for herpes simplex virus type1UL34and US3proteins in disrupting the nuclear lamina during herpes simplex virus type1egress.Virology347,261–276. Bortz,E.,Wang,L.,Jia,Q.,Wu,T.T.,Whitelegge,J.P.,Deng,H., Zhou,Z.H.,and Sun,R.(2007).Murine gammaherpesvirus68 ORF52encodes a tegument protein required for virion morpho-genesis in the cytoplasm.J Virol81,10137–10150. Bresnahan,W.A.,and Shenk,T.E.(2000).UL82virion protein activates expression of immediate early viral genes in human cytomegalovirus-infected cells.Proc Natl Acad Sci U S A97, 14506–14511.
Britt,W.J.,Jarvis,M.,Seo,J.Y.,Drummond,D.,and Nelson,J.(2004).
Rapid genetic engineering of human cytomegalovirus by using a lambda phage linear recombination system:demonstration that pp28(UL99)is essential for production of infectious virus.J Virol 78,539–543.
Bucks,M.A.,O’Regan,K.J.,Murphy,M.A.,Wills,J.W.,and Courtney, R.J.(2007).Herpes simplex virus type1tegument proteins VP1/2 and UL37are associated with intranuclear capsids.Virology361, 316–324.
Calderwood,M.A.,Venkatesan,K.,Xing,L.,Chase,M.R.,Vazquez,
A.,Holthaus,A.M.,Ewence,A.E.,Li,N.,Hirozane-Kishikawa,T.,
Hill,D.E.,et al.(2007).Epstein-Barr virus and virus human protein interaction maps.Proc Natl Acad Sci U S A104,7606–7611. Castillo,J.P.,and Kowalik,T.F.(2002).Human cytomegalovirus immediate early proteins and cell growth control.Gene290,19–34. Chang,Y.,Cesarman, E.,Pessin,M.S.,Lee, F.,Culpepper,J., Knowles, D.M.,and Moore,P.S.(1994).Identi?cation of herpesvirus-like DNA sequences in AIDS-associated Kaposi’s sarcoma.Science266,1865–1869.
Chevillotte,M.,Landwehr,S.,Linta,L.,Frascaroli,G.,Lüske,A., Buser,C.,Mertens,T.,and von Einem,J.(2009).Major tegument protein pp65of human cytomegalovirus is required for the incorporation of pUL69and pUL97into the virus particle and for viral growth in macrophages.J Virol83,2480–2490.
Chi,J.H.,Harley,C.A.,Mukhopadhyay,A.,and Wilson,D.W.(2005).
The cytoplasmic tail of herpes simplex virus envelope glycoprotein
D binds to the tegument protein VP22and to capsids.J Gen Virol
86,253–261.
Coller,K.E.,Lee,J.I.,Ueda,A.,and Smith,G.A.(2007).The capsid and tegument of the alphaherpesviruses are linked by an interaction between the UL25and VP1/2proteins.J Virol81, 11790–11797.
Dai,W.,Jia,Q.,Bortz,E.,Shah,S.,Liu,J.,Atanasov,I.,Li,X.,Taylor, K.A.,Sun,R.,and Zhou,Z.H.(2008).Unique structures in a tumor herpesvirus revealed by cryo-electron tomography and micro-scopy.J Struct Biol161,428–438.
Davison,A.J.,Eberle,R.,Ehlers,B.,Hayward,G.S.,McGeoch,D.J., Minson,A.C.,Pellett,P.E.,Roizman,B.,Studdert,M.J.,and Thiry,
E.(2009).The order Herpesvirales.Arch Virol154,171–177.
de Wind,N.,Wagenaar,F.,Pol,J.,Kimman,T.,and Berns,A.(1992).
The pseudorabies virus homology of the herpes simplex virus UL21gene product is a capsid protein which is involved in capsid maturation.J Virol66,7096–7103.
Decker,L.L.,Klaman,L.D.,and Thorley-Lawson, D.A.(1996a).
Detection of the latent form of Epstein-Barr virus DNA in the peripheral blood of healthy individuals.J Virol70,3286–3289. Decker,L.L.,Shankar,P.,Khan,G.,Freeman,R.B.,Dezube,B.J., Lieberman,J.,and Thorley-Lawson,D.A.(1996b).The Kaposi sarcoma-associated herpesvirus(KSHV)is present as an intact latent genome in KS tissue but replicates in the peripheral blood mononuclear cells of KS patients.J Exp Med184,283–288.
del Rio,T.,DeCoste,C.J.,and Enquist,L.W.(2005).Actin is a component of the compensation mechanism in pseudorabies virus virions lacking the major tegument protein VP22.J Virol79, 8614–8619.
Desai,P.,Sexton,G.L.,McCaffery,J.M.,and Person,S.(2001).A null mutation in the gene encoding the herpes simplex virus type1 UL37polypeptide abrogates virus maturation.J Virol75, 10259–10271.
Desai,P.J.(2000).A null mutation in the UL36gene of herpes simplex virus type1results in accumulation of unenveloped DNA-?lled capsids in the cytoplasm of infected cells.J Virol74,11608–11618. Diefenbach,R.J.,Miranda-Saksena,M.,Diefenbach,E.,Holland,D.
J.,Boadle,R.A.,Armati,P.J.,and Cunningham, A.L.(2002).
Herpes simplex virus tegument protein US11interacts with conventional kinesin heavy chain.J Virol76,3282–3291.
D?hner,K.,Wolfstein,A.,Prank,U.,Echeverri,C.,Dujardin,D., Vallee,R.,and Sodeik,B.(2002).Function of dynein and dynactin in herpes simplex virus capsid transport.Mol Biol Cell13, 2795–2809.
Dupin,N.,Fisher,C.,Kellam,P.,Ariad,S.,Tulliez,M.,Franck,N.,van Marck,E.,Salmon,D.,Gorin,I.,Escande,J.P.,et al.(1999).
Distribution of human herpesvirus-8latently infected cells in Kaposi’s sarcoma,multicentric Castleman’s disease,and primary effusion lymphoma.Proc Natl Acad Sci U S A96,4546–4551. Elliott,G.,Hafezi,W.,Whiteley,A.,and Bernard,E.(2005).Deletion of the herpes simplex virus VP22-encoding gene(UL49)alters the expression,localization,and virion incorporation of ICP0.J Virol 79,9735–9745.
Elliott,G.,Mouzakitis,G.,and O’Hare,P.(1995).VP16interacts via its activation domain with VP22,a tegument protein of herpes simplex virus,and is relocated to a novel macromolecular assembly in coexpressing cells.J Virol69,7932–7941.
Farina,A.,Feederle,R.,Raffa,S.,Gonnella,R.,Santarelli,R.,Frati, L.,Angeloni,A.,Torrisi,M.R.,Faggioni,A.,and Delecluse,H.J.
(2005).BFRF1of Epstein-Barr virus is essential for ef?cient primary viral envelopment and egress.J Virol79,3703–3712. Farnsworth,A.,Wisner,T.W.,and Johnson,D.C.(2007a).Cytoplas-mic residues of herpes simplex virus glycoprotein gE required for secondary envelopment and binding of tegument proteins VP22 and UL11to gE and gD.J Virol81,319–331.
Farnsworth, A.,Wisner,T.W.,Webb,M.,Roller,R.,Cohen,G., Eisenberg,R.,and Johnson,D.C.(2007b).Herpes simplex virus glycoproteins gB and gH function in fusion between the virion
envelope and the outer nuclear membrane.Proc Natl Acad Sci U S A104,10187–10192.
Fla?o,E.,Husain,S.M.,Sample,J.T.,Woodland,D.L.,and Black-man,M.A.(2000).Latent murine gamma-herpesvirus infection is established in activated B cells,dendritic cells,and macrophages.
J Immunol165,1074–1081.
Fuchs,W.,Granzow,H.,Klupp,B.G.,Kopp,M.,and Mettenleiter,T.C.
(2002a).The UL48tegument protein of pseudorabies virus is critical for intracytoplasmic assembly of infectious virions.J Virol 76,6729–6742.
Fuchs,W.,Klupp,B.G.,Granzow,H.,Hengartner,C.,Brack,A., Mundt,A.,Enquist,L.W.,and Mettenleiter,T.C.(2002b).Physical interaction between envelope glycoproteins E and M of pseudora-bies virus and the major tegument protein UL49.J Virol76, 8208–8217.
Fuchs,W.,Klupp,B.G.,Granzow,H.,and Mettenleiter,T.C.(2004).
Essential function of the pseudorabies virus UL36gene product is independent of its interaction with the UL37protein.J Virol78, 11879–11889.
Fuchs,W.,Klupp,B.G.,Granzow,H.,Osterrieder,N.,and Mettenlei-ter,T.C.(2002c).The interacting UL31and UL34gene products of pseudorabies virus are involved in egress from the host-cell nucleus and represent components of primary enveloped but not mature virions.J Virol76,364–378.
Gaietta,G.M.,Giepmans,B.N.,Deerinck,T.J.,Smith,W.B.,Ngan,L., Llopis,J.,Adams,S.R.,Tsien,R.Y.,and Ellisman,M.H.(2006).
Golgi twins in late mitosis revealed by genetically encoded tags for live cell imaging and correlated electron microscopy.Proc Natl Acad Sci U S A103,17777–17782.
Gonnella,R.,Farina,A.,Santarelli,R.,Raffa,S.,Feederle,R.,Bei,R., Granato,M.,Modesti,A.,Frati,L.,Delecluse,H.J.,et al.(2005).
Characterization and intracellular localization of the Epstein-Barr virus protein BFLF2:interactions with BFRF1and with the nuclear lamina.J Virol79,3713–3727.
Granato,M.,Feederle,R.,Farina,A.,Gonnella,R.,Santarelli,R., Hub,B.,Faggioni,A.,and Delecluse,H.J.(2008).Deletion of Epstein-Barr virus BFLF2leads to impaired viral DNA packaging and primary egress as well as to the production of defective viral particles.J Virol82,4042–4051.
Granzow,H.,Klupp, B.G.,and Mettenleiter,T.C.(2004).The pseudorabies virus US3protein is a component of primary and of mature virions.J Virol78,1314–1323.
Gross,S.T.,Harley,C.A.,and Wilson,D.W.(2003).The cytoplasmic tail of Herpes simplex virus glycoprotein H binds to the tegument protein VP16in vitro and in vivo.Virology317,1–12.
Guo,H.,Wang,L.,Peng,L.,Zhou,Z.H.,and Deng,H.(2009).Open reading frame33of a gammaherpesvirus encodes a tegument protein essential for virion morphogenesis and egress.J Virol83, 10582–10595.
Harper,A.L.,Meckes,D.G.Jr,Marsh,J.A.,Ward,M.D.,Yeh,P.C., Baird,N.L.,Wilson,C.B.,Semmes,O.J.,and Wills,J.W.(2010).
Interaction domains of the UL16and UL21tegument proteins of herpes simplex virus.J Virol84,2963–2971.
Heine,J.W.,Honess,R.W.,Cassai,E.,and Roizman,B.(1974).
Proteins speci?ed by herpes simplex virus.XII.The virion polypeptides of type1strains.J Virol14,640–651.
Indran,S.V.,Ballestas,M.E.,and Britt,W.J.(2010).Bicaudal D1-dependent traf?cking of human cytomegalovirus tegument protein
Haitao Guo et al.
Protein&Cell
pp150in virus-infected cells.J Virol84,3162–3177.
Ishov, A.M.,Vladimirova,O.V.,and Maul,G.G.(2002).Daxx-mediated accumulation of human cytomegalovirus tegument protein pp71at ND10facilitates initiation of viral infection at these nuclear domains.J Virol76,7705–7712.
Johannsen, E.,Luftig,M.,Chase,M.R.,Weicksel,S.,Cahir-McFarland,E.,Illanes,D.,Sarracino,D.,and Kieff,E.(2004).
Proteins of puri?ed Epstein-Barr virus.Proc Natl Acad Sci U S A 101,16286–16291.
Jones,T.R.,and Lee,S.W.(2004).An acidic cluster of human cytomegalovirus UL99tegument protein is required for traf?cking and function.J Virol78,1488–1502.
Kamen,D.E.,Gross,S.T.,Girvin,M.E.,and Wilson,D.W.(2005).
Structural basis for the physiological temperature dependence of the association of VP16with the cytoplasmic tail of herpes simplex virus glycoprotein H.J Virol79,6134–6141.
Kamil,J.P.,and Coen,D.M.(2007).Human cytomegalovirus protein kinase UL97forms a complex with the tegument phosphoprotein pp65.J Virol81,10659–10668.
Kato,A.,Tanaka,M.,Yamamoto,M.,Asai,R.,Sata,T.,Nishiyama,Y., and Kawaguchi,Y.(2008).Identi?cation of a physiological phosphorylation site of the herpes simplex virus1-encoded protein kinase Us3which regulates its optimal catalytic activity in vitro and in?uences its function in infected cells.J Virol82,6172–6189. Kato,A.,Yamamoto,M.,Ohno,T.,Kodaira,H.,Nishiyama,Y.,and Kawaguchi,Y.(2005).Identi?cation of proteins phosphorylated directly by the Us3protein kinase encoded by herpes simplex virus
1.J Virol79,9325–9331.
Kato,A.,Yamamoto,M.,Ohno,T.,Tanaka,M.,Sata,T.,Nishiyama, Y.,and Kawaguchi,Y.(2006).Herpes simplex virus1-encoded protein kinase UL13phosphorylates viral Us3protein kinase and regulates nuclear localization of viral envelopment factors UL34 and UL31.J Virol80,1476–1486.
Kato,K.,Daikoku,T.,Goshima, F.,Kume,H.,Yamaki,K.,and Nishiyama,Y.(2000).Synthesis,subcellular localization and VP16 interaction of the herpes simplex virus type2UL46gene product.
Arch Virol145,2149–2162.
Klupp,B.G.,B?ttcher,S.,Granzow,H.,Kopp,M.,and Mettenleiter,T.
C.(2005).Complex formation between the UL16and UL21
tegument proteins of pseudorabies virus.J Virol79,1510–1522. Klupp,B.G.,Fuchs,W.,Granzow,H.,Nixdorf,R.,and Mettenleiter,T.
C.(2002).Pseudorabies virus UL36tegument protein physically
interacts with the UL37protein.J Virol76,3065–3071.
Klupp,B.G.,Granzow,H.,Fuchs,W.,Keil,G.M.,Finke,S.,and Mettenleiter,T.C.(2007).Vesicle formation from the nuclear membrane is induced by coexpression of two conserved herpes-virus proteins.Proc Natl Acad Sci U S A104,7241–7246. Klupp,B.G.,Granzow,H.,and Mettenleiter,T.C.(2001a).Effect of the pseudorabies virus US3protein on nuclear membrane localization of the UL34protein and virus egress from the nucleus.J Gen Virol 82,2363–2371.
Klupp,B.G.,Granzow,H.,Mundt,E.,and Mettenleiter,T.C.(2001b).
Pseudorabies virus UL37gene product is involved in secondary envelopment.J Virol75,8927–8936.
Ko,D.H.,Cunningham,A.L.,and Diefenbach,R.J.(2010).The major determinant for addition of tegument protein pUL48(VP16)to capsids in herpes simplex virus type1is the presence of the major tegument protein pUL36(VP1/2).J Virol84,1397–1405.Kopp,M.,Granzow,H.,Fuchs,W.,Klupp,B.G.,Mundt,E.,Karger,A., and Mettenleiter,T.C.(2003).The pseudorabies virus UL11protein is a virion component involved in secondary envelopment in the cytoplasm.J Virol77,5339–5351.
Kopp,M.,Klupp,B.G.,Granzow,H.,Fuchs,W.,and Mettenleiter,T.C.
(2002).Identi?cation and characterization of the pseudorabies virus tegument proteins UL46and UL47:role for UL47in virion morphogenesis in the cytoplasm.J Virol76,8820–8833. Krishnan,H.H.,Sharma-Walia,N.,Zeng,L.,Gao,S.J.,and Chandran, B.(2005).Envelope glycoprotein gB of Kaposi’s sarcoma-associated herpesvirus is essential for egress from infected cells.J Virol79,10952–10967.
Krosky,P.M.,Baek,M.C.,and Coen, D.M.(2003).The human cytomegalovirus UL97protein kinase,an antiviral drug target,is required at the stage of nuclear egress.J Virol77,905–914. Kutok,J.L.,and Wang,F.(2006).Spectrum of Epstein-Barr virus-associated diseases.Annu Rev Pathol1,375–404.
Lake,C.M.,and Hutt-Fletcher,L.M.(2004).The Epstein-Barr virus BFRF1and BFLF2proteins interact and coexpression alters their cellular localization.Virology320,99–106.
Lanman,J.,Crum,J.,Deerinck,T.J.,Gaietta,G.M.,Schneemann,A., Sosinsky,G.E.,Ellisman,M.H.,and Johnson,J.E.(2008).
Visualizing?ock house virus infection in Drosophila cells with correlated?uorescence and electron microscopy.J Struct Biol161, 439–446.
Leach,N.,Bjerke,S.L.,Christensen,D.K.,Bouchard,J.M.,Mou,F., Park,R.,Baines,J.,Haraguchi,T.,and Roller,R.J.(2007).Emerin is hyperphosphorylated and redistributed in herpes simplex virus type1-infected cells in a manner dependent on both UL34and US3.J Virol81,10792–10803.
Lee,S.K.,and Longnecker,R.(1997).The Epstein-Barr virus glycoprotein110carboxy-terminal tail domain is essential for lytic virus replication.J Virol71,4092–4097.
Leege,T.,Granzow,H.,Fuchs,W.,Klupp,B.G.,and Mettenleiter,T.C.
(2009).Phenotypic similarities and differences between UL37-deleted pseudorabies virus and herpes simplex virus type1.J Gen Virol90,1560–1568.
Leuzinger,H.,Ziegler,U.,Schraner,E.M.,Fraefel,C.,Glauser,D.L., Heid,I.,Ackermann,M.,Mueller,M.,and Wild,P.(2005).Herpes simplex virus1envelopment follows two diverse pathways.J Virol 79,13047–13059.
Liu, F.,and Zhou,Z.H.(2006).Comparative virion structures of human herpesviruses.In Human herpesviruses:biology,therapy, and immunoprophylaxis,G.C.-F.A.Arvin,P.Moore,et al.,ed.
(Cambridge,United Kingdom,Cambridge University Press),pp.
27–42.
Liu,Y.,Cui,Z.,Zhang,Z.,Wei,H.,Zhou,Y.,Wang,M.,and Zhang,X.
E.(2009).The tegument protein UL94of human cytomegalovirus
as a binding partner for tegument protein pp28identi?ed by intracellular imaging.Virology388,68–77.
Loomis,J.S.,Bowzard,J.B.,Courtney,R.J.,and Wills,J.W.(2001).
Intracellular traf?cking of the UL11tegument protein of herpes simplex virus type1.J Virol75,12209–12219.
Loomis,J.S.,Courtney,R.J.,and Wills,J.W.(2003).Binding partners for the UL11tegument protein of herpes simplex virus type1.J Virol77,11417–11424.
Loret,S.,Guay,G.,and Lippé,R.(2008).Comprehensive character-ization of extracellular herpes simplex virus type1virions.J Virol
82,8605–8618.
MacLean,C.A.,Clark,B.,and McGeoch,D.J.(1989).Gene UL11of herpes simplex virus type1encodes a virion protein which is myristylated.J Gen Virol70,3147–3157.
MacLean, C.A.,Dolan, A.,Jamieson, F.E.,and McGeoch, D.J.
(1992).The myristylated virion proteins of herpes simplex virus type1:investigation of their role in the virus life cycle.J Gen Virol 73,539–547.
Martin,B.R.,Giepmans,B.N.,Adams,S.R.,and Tsien,R.Y.(2005).
Mammalian cell-based optimization of the biarsenical-binding tetracysteine motif for improved?uorescence and af?nity.Nat Biotechnol23,1308–1314.
McNab,A.R.,Desai,P.,Person,S.,Roof,L.L.,Thomsen, D.R., Newcomb,W.W.,Brown,J.C.,and Homa,F.L.(1998).The product of the herpes simplex virus type1UL25gene is required for encapsidation but not for cleavage of replicated viral DNA.J Virol 72,1060–1070.
McNabb,D.S.,and Courtney,R.J.(1992).Characterization of the large tegument protein(ICP1/2)of herpes simplex virus type1.
Virology190,221–232.
Meckes, D.G.Jr,Marsh,J.A.,and Wills,J.W.(2010).Complex mechanisms for the packaging of the UL16tegument protein into herpes simplex virus.Virology398,208–213.
Mettenleiter,T.C.(2002).Herpesvirus assembly and egress.J Virol 76,1537–1547.
Mettenleiter,T.C.(2004).Budding events in herpesvirus morphogen-esis.Virus Res106,167–180.
Mettenleiter,T.C.(2006).Intriguing interplay between viral proteins during herpesvirus assembly or:the herpesvirus assembly puzzle.
Vet Microbiol113,163–169.
Michael,K.,Klupp,B.G.,Mettenleiter,T.C.,and Karger,A.(2006).
Composition of pseudorabies virus particles lacking tegument protein US3,UL47,or UL49or envelope glycoprotein E.J Virol80, 1332–1339.
Michel,D.,Pavi?,I.,Zimmermann,A.,Haupt,E.,Wunderlich,K., Heuschmid,M.,and Mertens,T.(1996).The UL97gene product of human cytomegalovirus is an early-late protein with a nuclear localization but is not a nucleoside kinase.J Virol70,6340–6346. M?hl,B.S.,B?ttcher,S.,Granzow,H.,Kuhn,J.,Klupp,B.G.,and Mettenleiter,T.C.(2009).Intracellular localization of the pseudora-bies virus large tegument protein pUL36.J Virol83,9641–9651. Morris,J.B.,Hofemeister,H.,and O’Hare,P.(2007).Herpes simplex virus infection induces phosphorylation and delocalization of emerin,a key inner nuclear membrane protein.J Virol81, 4429–4437.
Mossman,K.L.,Sherburne,R.,Lavery,C.,Duncan,J.,and Smiley,J.
R.(2000).Evidence that herpes simplex virus VP16is required for viral egress downstream of the initial envelopment event.J Virol 74,6287–6299.
Mou,F.,Forest,T.,and Baines,J.D.(2007).US3of herpes simplex virus type1encodes a promiscuous protein kinase that phosphor-ylates and alters localization of lamin A/C in infected cells.J Virol 81,6459–6470.
Mou,F.,Wills,E.,and Baines,J.D.(2009).Phosphorylation of the U(L)31protein of herpes simplex virus1by the U(S)3-encoded kinase regulates localization of the nuclear envelopment complex and egress of nucleocapsids.J Virol83,5181–5191.
Mou,F.,Wills,E.G.,Park,R.,and Baines,J.D.(2008).Effects of lamin
A/C,lamin B1,and viral US3kinase activity on viral infectivity, virion egress,and the targeting of herpes simplex virus U(L)34-encoded protein to the inner nuclear membrane.J Virol82, 8094–8104.
Muranyi,W.,Haas,J.,Wagner,M.,Krohne,G.,and Koszinowski,U.
H.(2002).Cytomegalovirus recruitment of cellular kinases to
dissolve the nuclear lamina.Science297,854–857.
Murphy,M.A.,Bucks,M.A.,O’Regan,K.J.,and Courtney,R.J.(2008).
The HSV-1tegument protein pUL46associates with cellular membranes and viral capsids.Virology376,279–289.
Naldinho-Souto,R.,Browne,H.,and Minson,T.(2006).Herpes simplex virus tegument protein VP16is a component of primary enveloped virions.J Virol80,2582–2584.
O’Regan,K.J.,Murphy,M.A.,Bucks,M.A.,Wills,J.W.,and Courtney, R.J.(2007).Incorporation of the herpes simplex virus type1 tegument protein VP22into the virus particle is independent of interaction with VP16.Virology369,263–280.
Overton,H.A.,McMillan,D.J.,Klavinskis,L.S.,Hope,L.,Ritchie,A.J., and Wong-kai-in,P.(1992).Herpes simplex virus type1gene UL13encodes a phosphoprotein that is a component of the virion.
Virology190,184–192.
Padula,M.E.,Sydnor,M.L.,and Wilson,D.W.(2009).Isolation and preliminary characterization of herpes simplex virus1primary enveloped virions from the perinuclear space.J Virol83, 4757–4765.
Peeters,B.,de Wind,N.,Hooisma,M.,Wagenaar,F.,Gielkens,A., and Moormann,R.(1992).Pseudorabies virus envelope glyco-proteins gp50and gII are essential for virus penetration,but only gII is involved in membrane fusion.J Virol66,894–905.
Prichard,M.N.,Britt,W.J.,Daily,S.L.,Hartline,C.B.,and Kern,E.R.
(2005).Human cytomegalovirus UL97Kinase is required for the normal intranuclear distribution of pp65and virion morphogenesis.
J Virol79,15494–15502.
Purves, F.C.,Spector, D.,and Roizman, B.(1991).The herpes simplex virus1protein kinase encoded by the US3gene mediates posttranslational modi?cation of the phosphoprotein encoded by the UL34gene.J Virol65,5757–5764.
Reynolds,A.E.,Liang,L.,and Baines,J.D.(2004).Conformational changes in the nuclear lamina induced by herpes simplex virus type1require genes U(L)31and U(L)34.J Virol78,5564–5575. Reynolds,A.E.,Wills,E.G.,Roller,R.J.,Ryckman,B.J.,and Baines, J.D.(2002).Ultrastructural localization of the herpes simplex virus type1UL31,UL34,and US3proteins suggests speci?c roles in primary envelopment and egress of nucleocapsids.J Virol76, 8939–8952.
Rixon,F.J.(1993).Structure and assembly of herpesviruses.Semin Virol4,135–144.
Roberts,A.P.,Abaitua,F.,O’Hare,P.,McNab,D.,Rixon,F.J.,and Pasdeloup,D.(2009).Differing roles of inner tegument proteins pUL36and pUL37during entry of herpes simplex virus type1.J Virol83,105–116.
Roizman,B.,Knipe,D.M.,and Whitely,R.J.(2007).Herpes Simplex Virus.In Fields Virology,D.M.Knipe,Howley,P.M.,ed.(Philadel-phia,Lippincott Williams&Wilkins),pp.2501–2601.
Roizman, B.,and Pellett,P.E.(2001).Herpesviridae:a brief introduction,In Fields virology,D.M.K.a.P.M.Howley,ed.(Phila-delphia,PA.,Lippincott-Raven Publishers),pp.2381–2398. Rozen,R.,Sathish,N.,Li,Y.,and Yuan,Y.(2008).Virion-wide protein
Haitao Guo et al.
Protein&Cell
interactions of Kaposi’s sarcoma-associated herpesvirus.J Virol 82,4742–4750.
Ryckman,B.J.,and Roller,R.J.(2004).Herpes simplex virus type1 primary envelopment:UL34protein modi?cation and the US3-UL34catalytic relationship.J Virol78,399–412.
Sathish,N.,Zhu,F.X.,Yuan,Y.,and Farzan,M.(2009).Kaposi’s sarcoma-associated herpesvirus ORF45interacts with kinesin-2 transporting viral capsid-tegument complexes along microtubules.
PLoS Pathog5,e1000332.
Schek,N.,and Bachenheimer,S.L.(1985).Degradation of cellular mRNAs induced by a virion-associated factor during herpes simplex virus infection of Vero cells.J Virol55,601–610. Schimmer,C.,and Neubauer,A.(2003).The equine herpesvirus1 UL11gene product localizes to the trans-golgi network and is involved in cell-to-cell spread.Virology308,23–36.
Scott, E.S.,and O’Hare,P.(2001).Fate of the inner nuclear membrane protein lamin B receptor and nuclear lamins in herpes simplex virus type1infection.J Virol75,8818–8830.
Seo,J.Y.,and Britt,W.J.(2006).Sequence requirements for localization of human cytomegalovirus tegument protein pp28to the virus assembly compartment and for assembly of infectious virus.J Virol80,5611–5626.
Shanda,S.K.,and Wilson,D.W.(2008).UL36p is required for ef?cient transport of membrane-associated herpes simplex virus type1 along microtubules.J Virol82,7388–7394.
Silva,M.C.,Schr?er,J.,and Shenk,T.(2005).Human cytomegalo-virus cell-to-cell spread in the absence of an essential assembly protein.Proc Natl Acad Sci U S A102,2081–2086.
Silva,M.C.,Yu,Q.C.,Enquist,L.,and Shenk,T.(2003).Human cytomegalovirus UL99-encoded pp28is required for the cytoplas-mic envelopment of tegument-associated capsids.J Virol77, 10594–10605.
Simpson-Holley,M.,Baines,J.,Roller,R.,and Knipe,D.M.(2004).
Herpes simplex virus1U(L)31and U(L)34gene products promote the late maturation of viral replication compartments to the nuclear periphery.J Virol78,5591–5600.
Simpson-Holley,M.,Colgrove,R.C.,Nalepa,G.,Harper,J.W.,and Knipe, D.M.(2005).Identi?cation and functional evaluation of cellular and viral factors involved in the alteration of nuclear architecture during herpes simplex virus1infection.J Virol79, 12840–12851.
Smibert,C.A.,Popova,B.,Xiao,P.,Capone,J.P.,and Smiley,J.R.
(1994).Herpes simplex virus VP16forms a complex with the virion host shutoff protein vhs.J Virol68,2339–2346.
S?derberg-Nauclér,C.(2006).Does cytomegalovirus play a causa-tive role in the development of various in?ammatory diseases and cancer?J Intern Med259,219–246.
Steininger,C.(2007).Clinical relevance of cytomegalovirus infection in patients with disorders of the immune system.Clin Microbiol Infect13,953–963.
Stylianou,J.,Maringer,K.,Cook,R.,Bernard,E.,and Elliott,G.
(2009).Virion incorporation of the herpes simplex virus type1 tegument protein VP22occurs via glycoprotein E-speci?c recruit-ment to the late secretory pathway.J Virol83,5204–5218. Subak-Sharpe,J.H.,and Dargan, D.J.(1998).HSV molecular biology:general aspects of herpes simplex virus molecular biology.
Virus Genes16,239–251.
Sunil-Chandra,N.P.,Efstathiou,S.,and Nash,A.A.(1992).Murine
gammaherpesvirus68establishes a latent infection in mouse B lymphocytes in vivo.J Gen Virol73,3275–3279.
Takakuwa,H.,Goshima,F.,Koshizuka,T.,Murata,T.,Daikoku,T., and Nishiyama,Y.(2001).Herpes simplex virus encodes a virion-associated protein which promotes long cellular processes in over-expressing cells.Genes Cells6,955–966.
Tandon,R.,and Mocarski, E.S.(2008).Control of cytoplasmic maturation events by cytomegalovirus tegument protein pp150.J Virol82,9433–9444.
Uetz,P.,Dong,Y.A.,Zeretzke,C.,Atzler,C.,Baiker,A.,Berger,B., Rajagopala,S.V.,Roupelieva,M.,Rose,D.,Fossum,E.,et al.
(2006).Herpesviral protein networks and their interaction with the human proteome.Science311,239–242.
Varnum,S.M.,Streblow,D.N.,Monroe,M.E.,Smith,P.,Auberry,K.J., Pasa-Tolic,L.,Wang,D.,Camp,D.G.2nd,Rodland,K.,Wiley,S., et al.(2004).Identi?cation of proteins in human cytomegalovirus (HCMV)particles:the HCMV proteome.J Virol78,10960–10966. Vittone,V.,Diefenbach,E.,Triffett,D.,Douglas,M.W.,Cunningham,
A.L.,and Diefenbach,R.J.(2005).Determination of interactions
between tegument proteins of herpes simplex virus type1.J Virol 79,9566–9571.
Wagenaar,F.,Pol,J.M.,Peeters,B.,Gielkens,A.L.,de Wind,N.,and Kimman,T.G.(1995).The US3-encoded protein kinase from pseudorabies virus affects egress of virions from the nucleus.J Gen Virol76,1851–1859.
Wild,P.,Engels,M.,Senn,C.,Tobler,K.,Ziegler,U.,Schraner,E.M., Loepfe,E.,Ackermann,M.,Mueller,M.,and Walther,P.(2005).
Impairment of nuclear pores in bovine herpesvirus1-infected MDBK cells.J Virol79,1071–1083.
Wild,P.,Schraner,E.M.,Cantieni,D.,Loepfe,E.,Walther,P.,Müller, M.,and Engels,M.(2002).The signi?cance of the Golgi complex in envelopment of bovine herpesvirus1(BHV-1)as revealed by cryobased electron microscopy.Micron33,327–337.
Wild,P.,Senn,C.,Manera,C.L.,Sutter,E.,Schraner,E.M.,Tobler,K., Ackermann,M.,Ziegler,U.,Lucas,M.S.,and Kaech,A.(2009).
Exploring the nuclear envelope of herpes simplex virus1-infected cells by high-resolution microscopy.J Virol83,408–419. Wisner,T.W.,Wright,C.C.,Kato,A.,Kawaguchi,Y.,Mou,F.,Baines, J.D.,Roller,R.J.,and Johnson,D.C.(2009).Herpesvirus gB-induced fusion between the virion envelope and outer nuclear membrane during virus egress is regulated by the viral US3kinase.
J Virol83,3115–3126.
Wolf, D.G.,Courcelle, C.T.,Prichard,M.N.,and Mocarski, E.S.
(2001).Distinct and separate roles for herpesvirus-conserved UL97kinase in cytomegalovirus DNA synthesis and encapsida-tion.Proc Natl Acad Sci U S A98,1895–1900.
Wolfstein,A.,Nagel,C.H.,Radtke,K.,D?hner,K.,Allan,V.J.,and Sodeik,B.(2006).The inner tegument promotes herpes simplex virus capsid motility along microtubules in vitro.Traf?c7, 227–237.
Yeh,P.C.,Meckes,D.G.Jr,and Wills,J.W.(2008).Analysis of the interaction between the UL11and UL16tegument proteins of herpes simplex virus.J Virol82,10693–10700.
Zhang,Y.,Sirko,D.A.,and McKnight,J.L.(1991).Role of herpes simplex virus type1UL46and UL47in alpha TIF-mediated transcriptional induction:characterization of three viral deletion mutants.J Virol65,829–841.
Zhou,Z.H.,Chen,D.H.,Jakana,J.,Rixon,F.J.,and Chiu,W.(1999).
Visualization of tegument-capsid interactions and DNA in intact herpes simplex virus type1virions.J Virol73,3210–3218. Zhu,F.X.,Chong,J.M.,Wu,L.,and Yuan,Y.(2005).Virion proteins of Kaposi’s sarcoma-associated herpesvirus.J Virol79,800–811. Zhu, F.X.,King,S.M.,Smith, E.J.,Levy, D.E.,and Yuan,Y.
(2002).A Kaposi’s sarcoma-associated herpesviral protein inhibits
virus-mediated induction of type I interferon by blocking IRF-7 phosphorylation and nuclear accumulation.Proc Natl Acad Sci U S A99,5573–5578.
Zhu,Q.,and Courtney,R.J.(1994).Chemical cross-linking of virion envelope and tegument proteins of herpes simplex virus type1.
Virology204,590–599.
Haitao Guo et al.
Protein&Cell
蛋白质组学研究方法选择及比较
蛋白质组学研究方法选择及比较 目前研究蛋白组学的主要方法有蛋白质芯片及质谱法,本文将从多方面对两种研究方法进行了解与比较; 蛋白质芯片(Protein Array) 将大量不同的蛋白质有序地排列、固定于固相载体表面,形成微阵列。利用蛋白质分子间特异性结合的原理,实现对生物蛋白质分子精准、快速、高通量的检测。 主要类型: ●夹心法芯片(Sandwich-based Array) ●标记法芯片(Label-based Array) ●定量芯片(Quantitative Array) ●半定量芯片(Semi-Quantitative Array) 质谱(Mass Spectrometry) 用电场和磁场将运动的离子按它们的质荷比分离后进行检测,测出离子准确质量并确定离子的化合物组成,即通过对样品离子质荷比的分析而实现对样品进行定性和定量的一种方法。 主要类型:
●二维电泳+质谱(2D/Mass Spectrometry, MS) ●表面增强激光解吸电离飞行时间质谱(Surface-enhanced laser desorption/ionization- time of flight, SELDI) ●同位素标记相对和绝对定量(Isobaric tags for relative and absolute quantitation, iTRAQ) Protein Array or Mass Spectrometry? 如何选择合适的研究方法?以下将从六个方面进行比较与推荐: 1.筛查蛋白组学表达差异 建议选择:RayBiotech(1000个因子的芯片)+质谱 a)不同的方法学有不同的特点:对于质谱,可以筛查到未知的蛋白,但是对于分子量大、 低丰度的蛋白质,质谱的灵敏度和准确性有一定的限制。 b)不同的方法能筛查到的目标不同:根据Proteome Analysis of Human Aqueous Humor 一文中报道,质谱筛查到的差异蛋白集中在小分子与代谢物。而用RayBiotech芯片筛查到的结果,多是集中在细胞因子、趋化、血管、生长等等。 c)质谱筛查到355个蛋白,而RayBiotech抗体芯片也筛查到328个蛋白,且用定量芯片 验证25个蛋白有差异,这些蛋白是质谱找不到的。目前RayBiotech夹心法抗体芯片已经可以检测到1000个蛋白,采用双抗夹心法,尤其是对于低丰度蛋白,有很好的灵敏度和特异性,很多的低丰度蛋白是抗体芯片可以检测出来,而质谱检测不到的,且样品不经过变性和前处理,保持天然状态的样品直接检测,对于蛋白的检测准确度高。 d)质谱的重复性一直是质谱工作者纠结的问题,不同操作者的结果,不同样品处理条件, 峰值的偏移等影响因素都会产生大的影响;RayBiotech的夹心法芯片重复性高。
基因组学与蛋白质组学
《基因组学与蛋白质组学》课程教学大纲 学时: 40 学分:2.5 理论学时: 40 实验学时:0 面向专业:生物科学、生物技 术课程代码:B7700005先开课程:生物化学、分子生物 学课程性质:必修/选修执笔人:朱新 产审定人: 第一部分:理论教学部分 一、课程的性质、目的和任务 《基因组学与蛋白质组学》是随着生物化学、分子生物学、结构生物学、晶体学和计算机技术等的迅猛发展而诞生的,是融合了生物信息学、计算机辅助设计等多学科而发展起来的新兴研究领域。是当今生命科学研究的热点与前沿领域。由于基因组学与蛋白质组学学科的边缘性,所以本课程在介绍基因组学与蛋白质组学基本基本技术和原理的同时,兼顾学科发展动向,讲授基因组与蛋白组学中的热点和最新进展,旨在使学生了解现代基因组学与蛋白质组学理论的新进展并为相关学科提供知识和技术。 二、课程的目的与教学要求 通过本课程的学习,使学生掌握基因组学与蛋白质组学的基本理论、基础知识、主要研究方法和技术以及生物信息学和现代生物技术在基因组学与蛋白质组学上的应用及典型研究实例,熟悉从事基因组学与蛋白质组学的重要方法和途
径。努力培养学生具有科学思维方式、启发学生科学思维能力和勇于探索,善于思考、分析问题的能力,激发学生的学习热情,并通过学习提高自学能力、独立思考能力以及科研实践能力,为将来从事蛋白质的研究奠定坚实的理论和实践基础。 三、教学内容与课时分配 第一篇基因组学
第一章绪论(1学时) 第一节基因组学的研究对象与任务; 第二节基因组学发展的历程; 第三节基因组学的分子基础; 第四节基因组学的应用前景。 本章重点: 1. 基因组学的概念及主要任务; 2. 基因组学的研究对象。 本章难点: 1.基因组学的应用及发展趋势; 2.基因组学与生物的遗传改良、人类健康及生物进化。建议教学方法:课堂讲授和讨论 思考题: 查阅有关资料,了解基因组学的应用发展。 第二章人类基因组计划(1学时) 第一节人类基因组计划的诞生; 第二节人类基因组研究的竞赛; 第三节人类基因组测序存在的缺口; 第四节人类基因组中的非编码成分; 第五节人类基因组的概观; 第六节人类基因组多样性计划。 本章重点: 1. 人类基因组的研究; 2. 人类基因组多样性。 本章难点: 人类基因组序列的诠释。 建议教学方法:课堂讲授和讨论 思考题:
基因组学和蛋白质组学对新药研发的影响
通过校园网进入数据库例如维普期刊数据库、CNKI、超星电子图书等。完成 A、任选一题,检索相关资料,截取检索过程图片,做成一个ppt文件(50分)。 B、写综述形式的学术论文(学术论文格式,字数不限,正文字体小四),做成word文件(50分)。要求:按照自己的思路组织成文件,严禁抄袭。 写明班级学号,打印纸质版交给老师。 1、对检索课题“磷酸对草莓生长和开花的影响”检索中文信息。提示:磷酸的化学物质名称是“Phosphonic acid ”普通商业名称是“ethephon”, 2、基因组学和蛋白质组学对新药研发的影响 3、红霉素衍生物的设计、合成与抗菌活性研究 4、HPLC法测定复方谷氨酰胺肠溶胶囊中L-谷氨酰胺的释放度 姓名:朱艳红 班级: 11生科师范 学号: 11223074 学科教师:张来军
基因组学和蛋白质组学对新药研发的影响琼州学院生物科学与技术学院 11生科师范2班朱艳红 11223074 摘要 20世纪末伴随着人类基因组计划的实施,相继产生了基因组学和蛋白质组学,基因组学和蛋白质组学的迅速发展,对药学科学产生着深远的影响。文章在简介蛋白质组学基本概念、核心技术的基础上,综述了基因组学和蛋白质组学对新药研发带来的影响。 关键词:基因组学;蛋白质组学;药物研发 The impact of genomics and proteomics on the research and development of innovative drug abstract With the implementation of the 20th century,Genomics and proteomics had emerged one after the other. Driven by Soaring development of the omits,pharmaceutical industry presents a new vision,all human life faces a promising future. On the basis of proteomics Introduction to basic concepts, core technology, reviewed the genomics and proteomics research on the impact of new drugs. Keywords:Genomics; proteomics; drug development
蛋白质结构预测在线软件
蛋白质预测在线分析常用软件推荐 蛋白质预测分析网址集锦 物理性质预测: Compute PI/MW http://expaxy.hcuge.ch/ch2d/pi-tool.html Peptidemasshttp://expaxy.hcuge.ch/sprot/peptide-mass.html TGREASE ftp://https://www.360docs.net/doc/457826919.html,/pub/fasta/ SAPS http://ulrec3.unil.ch/software/SAPS_form.html 基于组成的蛋白质识别预测 AACompIdent http://expaxy.hcuge.ch ... htmlAACompSim http://expaxy.hcuge.ch/ch2d/aacsim.html PROPSEARCH http://www.e mbl-heidelberg.de/prs.html 二级结构和折叠类预测 nnpredict https://www.360docs.net/doc/457826919.html,/~nomi/nnpredict Predictprotein http://www.embl-heidel ... protein/SOPMA http://www.ibcp.fr/predict.html SSPRED http://www.embl-heidel ... prd_info.html 特殊结构或结构预测 COILS http://ulrec3.unil.ch/ ... ILS_form.html MacStripe https://www.360docs.net/doc/457826919.html,/ ... acstripe.html 与核酸序列一样,蛋白质序列的检索往往是进行相关分析的第一步,由于数据库和网络技校术的发展,蛋白序列的检索是十分方便,将蛋白质序列数据库下载到本地检索和通过国际互联网进行检索均是可行的。 由NCBI检索蛋白质序列 可联网到:“http://www.ncbi.nlm.ni ... gi?db=protein”进行检索。 利用SRS系统从EMBL检索蛋白质序列 联网到:https://www.360docs.net/doc/457826919.html,/”,可利用EMBL的SRS系统进行蛋白质序列的检索。 通过EMAIL进行序列检索 当网络不是很畅通时或并不急于得到较多数量的蛋白质序列时,可采用EMAIL方式进行序列检索。 蛋白质基本性质分析 蛋白质序列的基本性质分析是蛋白质序列分析的基本方面,一般包括蛋白质的氨基酸组成,分子质量,等电点,亲水性,和疏水性、信号肽,跨膜区及结构功能域的分析等到。蛋白质的很多功能特征可直接由分析其序列而获得。例如,疏水性图谱可通知来预测跨膜螺旋。同时,也有很多短片段被细胞用来将目的蛋白质向特定细胞器进行转移的靶标(其中最典型的
基因组学(结构基因组学和功能基因组学).
问:基因组学、转录组学、蛋白质组学、结构基因组学、功能基因组学、比较基因组学研究有哪些特点? 答:人类基因组计划完成后生物科学进入了人类后基因组时代,即大规模开展基因组生物学功能研究和应用研究的时代。在这个时代,生命科学的主要研究对象是功能基因组学,包括结构基因组研究和蛋白质组研究等。以功能基因组学为代表的后基因组时代主要为利用基因组学提供的信息。 基因组研究应该包括两方面的内容:以全基因组测序为目标的结构基因组学(struc tural genomics和以基因功能鉴定为目标的功能基因组学(functional genomics。结构基因组学代表基因组分析的早期阶段,以建立生物体高分辨率遗传、物理和转录图谱为主。功能基因组学代表基因分析的新阶段,是利用结构基因组学提供的信息系统地研究基因功能,它以高通量、大规模实验方法以及统计与计算机分析为特征。 功能基因组学(functional genomics又往往被称为后基因组学(postgenomics,它利用结构基因组所提供的信息和产物,发展和应用新的实验手段,通过在基因组或系统水平上全面分析基因的功能,使得生物学研究从对单一基因或蛋白质的研究转向多个基因或蛋白质同时进行系统的研究。这是在基因组静态的碱基序列弄清楚之后转入基因组动态的生物学功能学研究。研究内容包括基因功能发现、基因表达分析及突变检测。 基因的功能包括:生物学功能,如作为蛋白质激酶对特异蛋白质进行磷酸化修饰;细胞学功能,如参与细胞间和细胞内信号传递途径;发育上功能,如参与形态建成等采用的手段包括经典的减法杂交,差示筛选,cDNA代表差异分析以及mRNA差异显示等,但这些技术不能对基因进行全面系统的分析。新的技术应运而生,包括基因表达的系统分析,cDNA微阵列,DNA芯片等。鉴定基因功能最有效的方法是观察基因表达被阻断或增加后在细胞和整体水平所产生的表型变异,因此需要建立模式生物体。 功能基因组学
蛋白质结构预测和序列分析软件
蛋白质结构预测和序列分析软件蛋白质数据库及蛋白质序列分析 第一节、蛋白质数据库介绍 一、蛋白质一级数据库 1、 SWISS-PROT 数据库 SWISS-PROT和PIR是国际上二个主要的蛋白质序列数据 库,目前这二个数据库在EMBL和GenBank数据库上均建 立了镜像 (mirror) 站点。 SWISS-PROT数据库包括了从EMBL翻译而来的蛋白质序 列,这些序列经过检验和注释。该数据库主要由日内瓦大 学医学生物化学系和欧洲生物信息学研究所(EBI)合作维 护。SWISS-PROT的序列数量呈直线增长。 2、TrEMBL数据库: SWISS-PROT的数据存在一个滞后问题,即 进行注释需要时间。一大批含有开放阅读 了解决这一问题,TrEMBL(Translated E 白质数据库,它包括了所有EMBL库中的 质序列数据源,但这势必导致其注释质量 3、PIR数据库: PIR数据库的数据最初是由美国国家生物医学研究基金 会(National Biomedical Research Foundation, NBRF) 收集的蛋白质序列,主要翻译自GenBank的DNA序列。 1988年,美国的NBRF、日本的JIPID(the Japanese International Protein Sequence Database日本国家蛋 白质信息数据库)、德国的MIPS(Munich Information Centre for Protein Sequences摹尼黑蛋白质序列信息 中心)合作,共同收集和维护PIR数据库。PIR根据注释 程度(质量)分为4个等级。 4、 ExPASy数据库: 目前,瑞士生物信息学研究所(Swiss I 质分析专家系统(Expert protein anal 据库。 网址:https://www.360docs.net/doc/457826919.html, 我国的北京大学生物信息中心(www.cbi.
质谱技术在蛋白质组学研究中的应用_甄艳
第35卷 第1期2011年1月 南京林业大学学报(自然科学版) J o u r n a l o f N a n j i n g F o r e s t r y U n i v e r s i t y (N a t u r a l S c i e n c e E d i t i o n ) V o l .35,N o .1 J a n .,2011 h t t p ://w w w .n l d x b .c o m [d o i :10.3969/j .i s s n .1000-2006.2011.01.024] 收稿日期:2009-12-31 修回日期:2010-10-26 基金项目:国家自然科学基金项目(31000287);江苏省高校自然科学基础研究项目(10K J B 220002) 作者简介:甄艳(1976—),副教授,博士。*施季森(通信作者),教授。E -m a i l :j s h i @n j f u .e d u .c n 。 引文格式:甄艳,施季森.质谱技术在蛋白质组学研究中的应用[J ].南京林业大学学报:自然科学版,2011,35(1):103-108. 质谱技术在蛋白质组学研究中的应用 甄 艳,施季森 * (南京林业大学,林木遗传与生物技术省部共建教育部重点实验室,江苏 南京 210037) 摘要:随着蛋白质组学研究的迅速发展,质谱技术已成为应用于蛋白质组学研究中的强有力工具和核心技术。质谱技术的先进性在于为蛋白质组学研究提供的通量和分子信息。笔者重点概述了基于质谱路线的蛋白质组学研究,介绍了基于质谱的定量蛋白质组学﹑翻译后修饰蛋白质组学、定向蛋白质组学、功能蛋白质组学以及基于串联质谱技术的蛋白质组学数据解析的研究 进展。 关键词:质谱;蛋白质组学;定量蛋白质组学;翻译后修饰;定向蛋白质组学;功能蛋白质组学中图分类号:Q 81 文献标志码:A 文章编号:1000-2006(2011)01-0103-06 A p p l i c a t i o n o f m a s s s p e c t r o m e t r y i n p r o t e o m i c s s t u d i e s Z H E NY a n ,S H I J i s e n * (K e y L a b o r a t o r y o f F o r e s t G e n e t i c s a n d B i o t e c h n o l o g y M i n i s t r y o f E d u c a t i o n , N a n j i n g F o r e s t r y U n i v e r s i t y ,N a n j i n g 210037,C h i n a ) A b s t r a c t :W i t ht h e r a p i d d e v e l o p m e n t o f p r o t e o m i c s ,m a s s s p e c t r o m e t r y i s m a t u r i n g t o b e a p o w e r f u l t o o l a n dc o r e t e c h -n o l o g y f o r p r o t e o m i c s s t u d i e s d u r i n g t h e r e c e n t y e a r s .T h e s u p e r i o r i t y o f m a s s s p e c t r o m e t r y l i e s i n p r o v i d i n g t h e t h r o u g h -p u t a n d t h e m o l e c u l a r i n f o r m a t i o n ,w h i c hn o o t h e r t e c h n o l o g y c a n b e m a t c h e di np r o t e o m i c s .I nt h i s r e v i e w ,w e m a d e a g l a n c e o n t h e o u t l i n e o f m a s s s p e c t r o m e t r y -b a s e d p r o t e o m i c s .A n dt h e nw e a d d r e s s e d o n t h e a d v a n c e s o f d a t a a n a l y s i s o f m a s s s p e c t r o m e t r y -b a s e dp r o t e o m i c s ,q u a n t i t a t i v em a s ss p e c t r o m e t r y -b a s e dp r o t e o m i c s ,p o s t -t r a n s l a t i o n a l m o d i f i c a t i o n s b a s e d m a s s s p e c t r o m e t r y ,t a r g e t e d p r o t e o m i c s a n df u n c t i o n a l p r o t e o m i c s b a s e d -m a s s s p e c t r o m e t r y . K e yw o r d s :m a s ss p e c t r o m e t r y ;p r o t e o m i c s ;q u a n t i t a t i v ep r o t e o m i c s ;p o s t -t r a n s l a t i o n m o d i f i c a t i o n ;t a r g e t e d p r o -t e o m i c s ;f u n c t i o n a l p r o t e o m i c s 蛋白质组学(P r o t e o m i c s )是从整体水平上研究细胞内蛋白质的组成、活动规律及蛋白质与蛋白质的相互作用,是功能基因组学时代一门新的学科。目前蛋白质组学的研究主要有两条路线:一是基于双向电泳的蛋白质组学;二是基于质谱的蛋白质组学,其中基于双向电泳的蛋白质组学研究路线最终也离不开质谱技术的应用。自20世纪80年代末,两种质谱软电离方式即电喷雾电离(e l e c t r o s p r a y i o n i z a t i o n ,E S I )和基质辅助激光解析离子化(m a -t r i x a s s i s t e d l a s e r d e s o r p t i o n i o n i z a t i o n ,M A L D I )的发明和发展解决了极性大、热不稳定蛋白质和多肽分 析的离子化和分子质量大的测定问题[1] ,蛋白质组学研究中常用的质谱分析仪包括离子阱(i o n t r a p ,I T ),飞行时间(t i m e o f f l i g h t ,T O F ),串联飞行时间(T O F -T O F ),四级杆/飞行时间(q u a d r u p o l e /T O F h y b r i d s ),离子阱/轨道阱(I T /o r b i t r a ph y b r i d ) 和离子阱/傅里叶变换串联质谱分析仪(I T /F o u r i e r t r a n s f o r m i o n c y c l o t r o nr e s o n a n c em a s s s p e c t r o m e t e r s h y b r i d s ,I T /F T M S ),这些质谱仪具有不同的灵敏度、分辨率、质量精确度和产生不同质量的M S /M S 谱[2] 。质谱作为蛋白质组学研究的一项强有力的工具日趋成熟,并作为样品制备及数据分析的信息学工具被广泛地应用。因此,有学者指出质谱技术 已在蛋白质组学研究中处于核心地位[3] 。目前在通量及所包含的分子信息内容上,基于质谱的蛋白质组学技术在细胞生物学研究中可以鉴定和量化
蛋白质结构预测在线软件
蛋白质预测分析网址集锦? 物理性质预测:? Compute PI/MW?? ?? SAPS?? 基于组成的蛋白质识别预测? AACompIdent???PROPSEARCH?? 二级结构和折叠类预测? nnpredict?? Predictprotein??? SSPRED?? 特殊结构或结构预测? COILS?? MacStripe?? 与核酸序列一样,蛋白质序列的检索往往是进行相关分析的第一步,由于数据库和网络技校术的发展,蛋白序列的检索是十分方便,将蛋白质序列数据库下载到本地检索和通过国际互联网进行检索均是可行的。? 由NCBI检索蛋白质序列? 可联网到:“”进行检索。? 利用SRS系统从EMBL检索蛋白质序列? 联网到:”,可利用EMBL的SRS系统进行蛋白质序列的检索。? 通过EMAIL进行序列检索?
当网络不是很畅通时或并不急于得到较多数量的蛋白质序列时,可采用EMAIL方式进行序列检索。? 蛋白质基本性质分析? 蛋白质序列的基本性质分析是蛋白质序列分析的基本方面,一般包括蛋白质的氨基酸组成,分子质量,等电点,亲水性,和疏水性、信号肽,跨膜区及结构功能域的分析等到。蛋白质的很多功能特征可直接由分析其序列而获得。例如,疏水性图谱可通知来预测跨膜螺旋。同时,也有很多短片段被细胞用来将目的蛋白质向特定细胞器进行转移的靶标(其中最典型的例子是在羧基端含有KDEL序列特征的蛋白质将被引向内质网。WEB中有很多此类资源用于帮助预测蛋白质的功能。? 疏水性分析? 位于ExPASy的ProtScale程序(?)可被用来计算蛋白质的疏水性图谱。该网站充许用户计算蛋白质的50余种不同属性,并为每一种氨基酸输出相应的分值。输入的数据可为蛋白质序列或SWISSPROT数据库的序列接受号。需要调整的只是计算窗口的大小(n)该参数用于估计每种氨基酸残基的平均显示尺度。? 进行蛋白质的亲/疏水性分析时,也可用一些windows下的软件如,bioedit,dnamana等。? 跨膜区分析? 有多种预测跨膜螺旋的方法,最简单的是直接,观察以20个氨基酸为单位的疏水性氨基酸残基的分布区域,但同时还有多种更加复杂的、精确的算法能够预测跨膜螺旋的具体位置和它们的膜向性。这些技术主要是基于对已知
蛋白质结构预测方法综述
蛋白质结构预测方法综述 卜东波陈翔王志勇 《计算机不能做什么?》是一本好书,其中文版序言也堪称佳构。在这篇十余页的短文中,马希文教授总结了使用计算机解决实际问题的三步曲,即首先进行形式化,将领域相关的实际问题抽象转化成一个数学问题;然后分析问题的可计算性;最后进行算法设计,分析算法的时间和空间复杂度,寻找最优算法。 蛋白质空间结构预测是很有生物学意义的问题,迄今亦有很多的工作。有意思的是,其中一些典型工作恰恰是上述三步曲的绝好示例,本文即沿着这一路线作一总结,介绍于后。 1 背景知识 生物细胞种有许多蛋白质(由20余种氨基酸所形成的长链),这些大分子对于完成生物功能是至关重要的。蛋白质的空间结构往往决定了其功能,因此,如何揭示蛋白质的结构是非常重要的工作。 生物学界常常将蛋白质的结构分为4个层次:一级结构,也就是组成蛋白质的氨基酸序列;二级结构,即骨架原子间的相互作用形成的局部结构,比如alpha螺旋,beta片层和loop区等;三级结构,即二级结构在更大范围内的堆积形成的空间结构;四级结构主要描述不同亚基之间的相互作用。 经过多年努力,结构测定的实验方法得到了很好的发展,比较常用的有核磁共振和X光晶体衍射两种。然而由于实验测定比较耗时和昂贵,对于某些不易结晶的蛋白质来说不适用。相比之下,测定蛋白质氨基酸序列则比较容易。因此如果能够从一级序列推断出空间结构则是非常有意义的工作。这也就是下面的蛋白质折叠问题: 1蛋白质折叠问题(Protein Folding Problem) 输入: 蛋白质的氨基酸序列
输出: 蛋白质的空间结构 蛋白质结构预测的可行性是有坚实依据的。因为一般而言,蛋白质的空间结构是由其一级结构确定的。生化实验表明:如果在体外无任何其他物质存在的条件下,使得蛋白质去折叠,然后复性,蛋白质将立刻重新折叠回原来的空间结构,整个过程在不到1秒种内即可完成。因此有理由认为对于大部分蛋白质而言,其空间结构信息已经完全蕴涵于氨基酸序列中。从物理学的角度讲,系统的稳定状态通常是能量最小的状态,这也是蛋白质预测工作的理论基础。 2 蛋白质结构预测方法 蛋白质结构预测的方法可以分为三种: 同源性(Homology )方法:这类方法的理论依据是如果两个蛋白质的序列比较相似,则其结构也有很大可能比较相似。有工作表明,如果序列相似性高于75%,则可以使用这种方法进行粗略的预测。这类方法的优点是准确度高,缺点是只能处理和模板库中蛋白质序列相似性较高的情况。 从头计算(Ab initio ) 方法:这类方法的依据是热力学理论,即求蛋白质能量最小的状态。生物学家和物理学家等认为从原理上讲这是影响蛋白质结构的本质因素。然而由于巨大的计算量,这种方法并不实用,目前只能计算几个氨基酸形成的结构。IBM 开发的Blue Gene 超级计算机,就是要解决这个问题。 穿线法(Threading )方法:由于Ab Initio 方法目前只有理论上的意义,Homology 方法受限于待求蛋白质必需和已知模板库中某个蛋白质有较高的序列相似性,对于其他大部分蛋白质来说,有必要寻求新的方法。Threading 就此应运而生。 以上三种方法中,Ab Initio 方法不依赖于已知结构,其余两种则需要已知结构的协助。通常将蛋白质序列和其真实三级结构组织成模板库,待预测三级结构的蛋白质序列,则称之为查询序列(query sequence)。 3 蛋白质结构预测的Threading 方法 Threading 方法有三个代表性的工作:Eisenburg 基于环境串的工作、Xu Ying 的Prospetor 和Xu Jinbo 、Li Ming 的RAPTOR 。 Threading 的方法:首先取出一条模版和查询序列作序列比对(Alignment),并将模版蛋白质与查询序列匹配上的残基的空间坐标赋给查询序列上相应的残基。比对的过程是在我们设计的一个能量函数指导下进行的。根据比对结果和得到的查询序列的空间坐标,通过我们设计的能量函数,得到一个能量值。将这个操作应用到所有的模版上,取能量值最低的那条模版产生的查询序列的空间坐标为我们的预测结果。 需要指出的是,此处的能量函数却不再是热力学意义上的能量函数。它实质上是概率的负对数,即 ,我们用统计意义上的能量来代替真实的分子能量,这两者有大致相同的形式。 p E log ?=如果沿着马希文教授的观点看上述工作 ,则更有意思:Eisenburg 指出如果仅仅停留在简单地使用每个原子的空间坐标(x,y,z)来形式化表示蛋白质空间结构,则难以进一步深入研究。Eisenburg 创造性地使用环境串表示结构,从而将结构预测问题转化成序列串和环境串之间的比对问题;其后,Xu Ying 作了进一步发展,将蛋白质序列表示成一系列核(core )组成的序列,Core 和Core 之间存在相互作用。因此结构就表示成Core 的空间坐标,以及Core 之间的相互作用。在这种表示方法的基础上,Xu Ying 开发了一种求最优匹配的动态规划算法,得到了很好的结果。但是由于其较高的复杂度,在Prospetor2上不得不作了一些简化;Xu Jinbo 和Li Ming 很漂亮地解决了这个问题,将求最优匹配的过程表示成一个整数规划问题,并且证明了一些常用
浅析功能基因组学和蛋白质组学的概念及应用
【摘要】基因组相对较稳定,而且各种细胞或生物体的基因组结构有许多基本相似的特征;蛋白质组是动态的,随内外界刺激而变化。对蛋白质组的研究可以使我们更容易接近对生命过程的认识。蛋白质组学是在细胞的整体蛋白质水平上进行研究、从蛋白质整体活动的角度来认识生命活动规律的一门新学科,简要介绍功能基因组学和蛋白质组学的科学背景、概念及其应用。 【关键词】基因组;功能基因组学;蛋白质组学; 一、基因组及基因组学的概念 基因组(genome)一词系由德国汉堡大学H.威克勒教授于1920年首创,用以表示真核生物从其亲代所继承的单套染色体,或称染色体组。更准确地说,基因组是指生物的整套染色体所含有的全部DNA序列。由于在真核细胞的线粒体和植物的叶绿体中也发现存在遗传物质,因此又将线粒体或叶绿体所携带的遗传物质称为线粒体基因组或叶绿体基因组。原核生物基因组则包括细胞内的染色体和质粒DNA。此外非独立生命形态的病毒颗粒也携带遗传物质,称为病毒基因组。所有生命都具有指令其生长与发育,维持其结构与功能所必需的遗传信息,本书中将生物所具有的携带遗传信息的遗传物质总和称为基因组。[1] 基因组学(genomic)一词系由T.罗德里克(T.Roderick)于1986年首创,用于概括涉及基因组作图、测序和整个基因组功能分析的遗传学学科分支,并已用来命名一个学术刊物Genomics。基因组学是伴随人类基因组计划的实施而形成的一个全新的生命科学领域。[1] 基因组学与传统遗传学其他学科的差别在于,基因组学是在全基因组范围研究基因的结构、组成、功能及其进化,因而涉及大范围高通量收集和分析有关基因组DNA的序列组成,染色体分子水平的结构特征,全基因组的基因数目、功能和分类,基因组水平的基因表达与调控以及不同物种之间基因组的进化关系。基因组学的研究方法、技术和路线有许多不同于传统遗传学的特点,各相关领域的研究仍处于迅速发展和不断完善的过程中。 基因组学的主要工具和方法包括:生物信息学,遗传分析,基因表达测量和基因功能鉴定。 二、功能基因组学的概念及应用
蛋白质结构预测网址
蛋白质结构预测网址 物理性质预测: Compute PI/MW Peptidemass TGREASE SAPS 基于组成的蛋白质识别预测 AACompIdent PROPSEARCH 二级结构和折叠类预测 nnpredict Predictprotein SSPRED 特殊结构或结构预测 COILS MacStripe 与核酸序列一样,蛋白质序列的检索往往是进行相关分析的第一步,由于数据库和网络技校术的发展,蛋白序列的检索是十分方便,将蛋白质序列数据库下载到本地检索和通过国际互联网进行检索均是可行的。 由NCBI检索蛋白质序列 可联网到:“”进行检索。 利用SRS系统从EMBL检索蛋白质序列 联网到:”,可利用EMBL的SRS系统进行蛋白质序列的检索。 通过EMAIL进行序列检索 当网络不是很畅通时或并不急于得到较多数量的蛋白质序列时,可采用EMAIL方式进行序列检索。 蛋白质基本性质分析 蛋白质序列的基本性质分析是蛋白质序列分析的基本方面,一般包括蛋白质的氨基酸组成,分子质量,等电点,亲水性,和疏水性、信号肽,跨膜区及结构功能域的分析等到。蛋白质的很多功能特征可直接由分析其序列而获得。例如,疏水性图谱可通知来预测跨膜螺旋。同时,也有很多短片段被细胞用来将目的蛋白质向特定细胞器进行转移的靶标(其中最典型的例子是在羧基端含有KDEL序列特征的蛋白质将被引向内质网。WEB中有很多此类资源用于帮助预测蛋白质的功能。 疏水性分析 位于ExPASy的ProtScale程序()可被用来计算蛋白质的疏水性图谱。该网站充许用户计算蛋白质的50余种不同属性,并为每一种氨基酸输出相应的分值。输入的数据可为蛋白质序列或SWISSPROT数据库的序列接受号。需要调整的只是计算窗口的大小(n)该参数用于估计每种氨基酸残基的平均显示尺度。 进行蛋白质的亲/疏水性分析时,也可用一些windows下的软件如, bioedit,dnamana等。 跨膜区分析 有多种预测跨膜螺旋的方法,最简单的是直接,观察以20个氨基酸为单位的疏水性氨基酸残基的分布区域,但同时还有多种更加复杂的、精确的算法能够预测跨膜螺旋的具体位置和它们的膜向性。这些技术主要是基于对已知跨膜螺旋的研究而得到的。自然存在的跨膜螺旋Tmbase 数据库,可通过匿名FTP获得(),参见表一
蛋白质结构预测
实习 5 :蛋白质结构预测 学号20090***** 姓名****** 专业年级生命生技**** 实验时间2012.6.21 提交报告时间2012.6.21 实验目的: 1.学会使用GOR和HNN方法预测蛋白质二级结构 2.学会使用SWISS-MODEL进行蛋白质高级结构预测 实验内容: 1.分别用GOR和HNN方法预测蛋白质序列的二级结构,并对比异同性。 2.利用SWISS-MODEL进行蛋白质的三级结构预测,并对预测结果进行解释。 作业: 1. 搜索一条你感兴趣的蛋白质序列,分别用GOR和HNN进行二级结构预测,解释预测结果,分析两个方法结果有何异同。 答:所选用蛋白质序列为>>gi|390408302|gb|AFL70986.1| gag protein, partial [Human immunodeficiency virus] (1)GOR预测结果: 图1 图1是每个氨基酸在序列中所处的状态,可以看出序列的二级结构预测结果为: 1到9位个氨基酸为无规卷曲,10到33位氨基酸为α螺旋,34到37位为β折叠,38到45位为无规卷曲,46到49位为α螺旋,50到53位为无规卷曲,54到65为α螺旋,66到72位为无规卷曲,73到95位为α螺旋,96到101位为无规卷曲,102到108为β折叠,109到115位为无规卷曲,117位为β折叠。 图2 图2为各种结构在序列中所占的比例,其中Alpha helix占53.85%,Extended strand占11.11%,Random coil占35.04%,无他二级结构。
图3 图3为各个氨基酸在序列中的状态以及二级结构在全序列中二级结构分布情况。 (2)HNN预测: 图4 图4是每个氨基酸在序列中所处的状态,可以看出序列的二级结构预测结果为: 1到6位个氨基酸为无规卷曲,7到34位氨基酸为α螺旋,35到37位为β折叠,38位为α螺旋,39到44位为无规卷曲,45到49位为α螺旋,50到55位为无规卷曲,56到65为α螺旋,66到71位为无规卷曲,72到83位为α螺旋,84到86位为无规卷曲,87到95位为α螺旋,96到102为无规卷曲,103到108位为β折叠,108到117位为无规卷曲。 图5 图5为各种结构在序列中所占的比例,其中Alpha helix占55.56%,Extended strand占7.69%,Random coil占36.75%,无他二级结构。
蛋白质组学及其研究方法与进展
蛋白质组学及其研究方法与进展 蛋白质是生命活动的体现者,基因的表达最后是通过蛋白质来体现的,在这个过程中,蛋白质起了连接基因与表现的功能。蛋白质是有氨基酸组成的,组成蛋白质的氨基酸的种类及排列顺序构成了蛋白质的一级结构,而在一级机构基础上的多肽链本身的折叠和盘绕方式构成了蛋白质的二级结构,考虑到多肽链上原子在空间的分布,由二级结构进一步形成了蛋白质的三级结构,对于有多个亚基的蛋白质还存在四级结构。 蛋白质的一级结构决定了高级结构,而高级结构则决定着蛋白质的生物学功能。如今对于蛋白质研究已经单独形成了一个活跃的生物学分支学科―――蛋白质组学,在蛋白质的研究中发挥着很重要的作用,下面将介绍蛋白质组学的一些基本内容及研究进展。 一.产生背景[1] 在20世纪中后期随着DNA双螺旋结构的提出和蛋白质空间结构的解析,生命科学研究进入了分子生物学时代,对遗传信息载体DNA和生命功能的体现者蛋白质的研究,成为了其主要内容。90年代初期启动的庞大的人类基因组计划.在经过各国科学家多年的努力下,已经取得了巨大的成就。10多种低等模式生物的基因组序列测定L三完成;第一个多细胞生物一线虫基因组的DNA全序列测定也在1998年年底完成;人类所有基因的部分序列测定(EST)已经完成;人类基因组的全序列测定有可能提前到2003年完成。生命科学已跨入了后基因组时代。在后基因组时代,研究重心将从揭示生命的所有遗传信息转移到在整体水平上对功能的研究。这种转向的第一个标志是产生了功能基因组学这一新学科,即从基因组整体水平上对基因的活动规律进行阐述。如在mRNA 水平上,通过DNA 芯片(DNA chips)和微阵列(Microarray)法等技术检测大量基因的表达模式,并取得了很好的进展。但是,mRNA的表达水平(包括mRNA的种类和含量)由于mRNA储存和翻译调控以及翻译后加工等的存在.并不能直接反映蛋白质的表达水平}蛋白质自身特有的活动规律,如蛋白质的修饰加工、转运定位结构形成、代谢、蛋白质与蛋白质及其他生物大分子的相互作用等.均无法从在基因组水平上的研究获知。因此,对生物功能的主要体现者或执行者一蛋白质的表达模式和功能模式的研究就成为生命科学发展的必然。在此背景下.80年代中期,国际上葫发了一门研究细胞内垒部蛋白质的组成及其活动规律的新兴学科- 蛋白质组学(Proteomic)。 蛋白质组(proteome)一词是马克.威尔金斯(Marc Wilkins)最先提出来的, 最早见诸于1995年7月的“Electrophoresis”杂志上它是指一个有机体的全部蛋白质组成及其活动方式。蛋白质组研究虽然尚处于初始阶段, 但已经取得了一些重要进展。当前蛋白质组学的主要内容是, 在建立和发展蛋白质组研究的技术方法的同时, 进行蛋白质组分析。对蛋白质组的分析工作大致有两个方面。一方面,通过二维凝胶电泳得到正常生理条件下的机体、组织或细胞的全部蛋白质的图谱, 相关数据将作为待检测机体、组织或细胞的二维参考图谱和数据库。一系列这样的二维参考图谱和数据库已经建立并且可通过联网检索。二维参考图谱
生态毒理基因组学和生态毒理蛋白质组学研究进展_戴家银
第26卷第3期2006年3月生 态 学 报ACTA EC OLOGI CA SI NICA Vol .26,No .3Mar .,2006生态毒理基因组学和生态毒理蛋白质组学研究进展 戴家银,王建设 (中国科学院动物研究所,北京 100080) 基金项目:中国科学院知识创新工程重要方向性资助项目(KSCX2-SW -128) 收稿日期:2005-08-30;修订日期:2005-12-05 作者简介:戴家银(1965~),男,安徽怀宁人,博士,研究员,主要从事生态毒理学和生物化学研究.E -mail :daijy @ioz .ac .cn Foundation item :The project was supported by the Innovation Project of Chines e Academy of Sciences (No .KSCX2-SW -128) Received date :2005-08-30;Accepted date :2005-12-05 Biography :DAI Jia -Yin ,Ph .D .,Professor ,mainly engaged in ecotoxicology and biochemis try .E -mail :daijy @ioz .ac .cn 摘要:将基因组学和蛋白质组学知识整合到生态毒理学中形成了生态毒理基因组学和生态毒理蛋白质组学。通过生态毒理基因组学和生态毒理蛋白质组学的研究能够在基因组和蛋白质组水平更深入理解毒物的作用机制,寻找更敏感、有效的生物标记物,形成潜在的强有力的生态风险评价工具。介绍了生态毒理基因组学和生态毒理蛋白质组学的研究进展,以及DNA 芯片技术和2D -凝胶电泳技术在持久性有毒污染物的生态毒理学研究中的应用。 关键词:生态毒理基因组学;生态毒理蛋白质组学;DNA 芯片技术;2D -凝胶电泳;持久性有机污染物 文章编号:1000-0933(2006)03-0930-05 中图分类号:X171 文献标识码:A Progress in ecotoxicogenomics and ecotoxicoproteomics DAI Jia -Yin ,WANG Jian -She (Institut e of Zoology ,C hines e Acade my of Sci ence s ,Beijing 100080,C hina )..Acta Ecologica Sinica ,2006,26(3): 930~934.Abstract :Ec otoxicogeno mics and ecotoxic oproteo mics are integration of genomics and proteomics into ec otoxicology .Ecotoxic ogenomics is defined as the study of gene and pr otein expr ession in non -target organisms that is impor tant in responses to environmental toxicant exposures .Ecotoxic ogenomic toolsmay provide us with a better mechanistic understanding of ec otoxicology ,and they are likely to provide a vital r ole in ecological risk assessment .Pr ogress in ec otoxicogenomics and ecotoxicoprote omics are discussed in this paper .DNA gene c hip and 2D -gel usually used in ecotoxicogeno mics and ecotoxicoproteomics ar e also e xpounded by exa mples . Key words :ec otoxicogeno mics ;ecotoxic oproteo mics ;D NA micr oarra y ;2D -gel ;persistent organic pollutants 随着生态学和环境科学的深入发展,生态毒理学已成为生态学和环境科学前沿研究领域,正从基因、蛋白质、器官和整体水平深入开展研究工作。 在人类基因组计划实施的短短几年间,以“组学(-omics )”构成的学科及其相关研究如雨后春笋般在生命科学界迅速蔓延、蓬勃发展。在环境科学领域中也出现了环境基因组学(environmental genomics )、毒理基因组学(toxicogenomics )等学科。Snape 等人[1~3]将基因组学知识整合到生态毒理学中,于2004年提出了“生态毒理基因组学(ecotoxicogenomics )”的概念,通过生态毒理基因组学研究确认一系列毒物效应基因,从而在基因组水平更深入理解毒物的作用机制,并在基因和蛋白质水平寻找更敏感、有效的生物标记物(biomarkers ),形成潜在的强有力的生态风险评价工具。 持久性有机污染物(Persistent Organic Pollutants ,POPs )是指能持久存在于环境中、通过食物链蓄积、逐级传递,经直接或间接途径进入人体的化学物质。POPs 具有致癌、致畸、致突变性、内分泌干扰等毒作用。POPs 对人体健康和生态环境带来的危害受到全社会的普遍关注,引起世界各国的决策者和科学家的高度重视,也成为环境科学和生态毒理学研究的热点课题之一[4,5]。我国已于2001年5月签署了控制12种P OPs 对人类健康
