ADVANCES IN ANALYSIS OF MICROBIAL METABOLIC FLUXES

ADVANCES IN ANALYSIS OF MICROBIAL METABOLIC FLUXES VIA 13C ISOTOPIC LABELING
Yinjie J.Tang,1,2,3,4Hector Garcia Martin,1Samuel Myers,4Sarah Rodriguez,5Edward E.K.Baidoo,2,3and Jay D.Keasling 1,2,3,4,6*1
Joint Bio-Energy Institute,Emeryville,CA 2
Virtual Institute for Microbial Stress and Survival,Berkeley,CA 3
Physical Biosciences Division,Lawrence Berkeley National Laboratory,Berkeley,CA 4
Department of Chemical Engineering,University of California,Berkeley,CA 5
Department of Molecular Cell Biology,University of California,Berkeley,CA 6
Department of Bioengineering,University of California,Berkeley,CA Received 8April 2008;received (revised)3July 2008;accepted 3July 2008
Published online in Wiley InterScience (https://www.360docs.net/doc/c88726505.html,)DOI 10.1002/mas.20191
Metabolic ?ux analysis via 13C labeling (13C MFA)quantita-tively tracks metabolic pathway activity and determines overall enzymatic function in cells.Three core techniques are necessary for 13C MFA:(1)a steady state cell culture in a de?ned medium with labeled-carbon substrates;(2)precise measurements of the labeling pattern of targeted metabolites;and (3)evaluation of the data sets obtained from mass spectrometry measurements with a computer model to calculate the metabolic ?uxes.In this review,we summarize recent advances in the 13C-?ux analysis technologies,including mini-bioreactor usage for tracer experiments,isotopomer analysis of metabolites via high resolution mass spectrometry (such as GC-MS,LC-MS,or FT-ICR),high performance and large-scale isotopomer modeling programs for ?ux analysis,and the integration of ?uxomics with other functional genomics studies.It will be shown that there is a signi?cant value for 13C-based metabolic ?ux analysis in many biological research ?elds.#2008Wiley Periodicals,Inc.,Mass Spec Rev 00:1–15,2008Keywords:steady state;mini-bioreactor;mass spectrometry;isotopomer modeling;functional genomics
I.INTRODUCTION
Microorganisms have evolved complex metabolic pathways that enable them to utilize various nutrients and survive in their local environment.To understand cell metabolism and its response to environmental and genetic changes,an array of genomic and
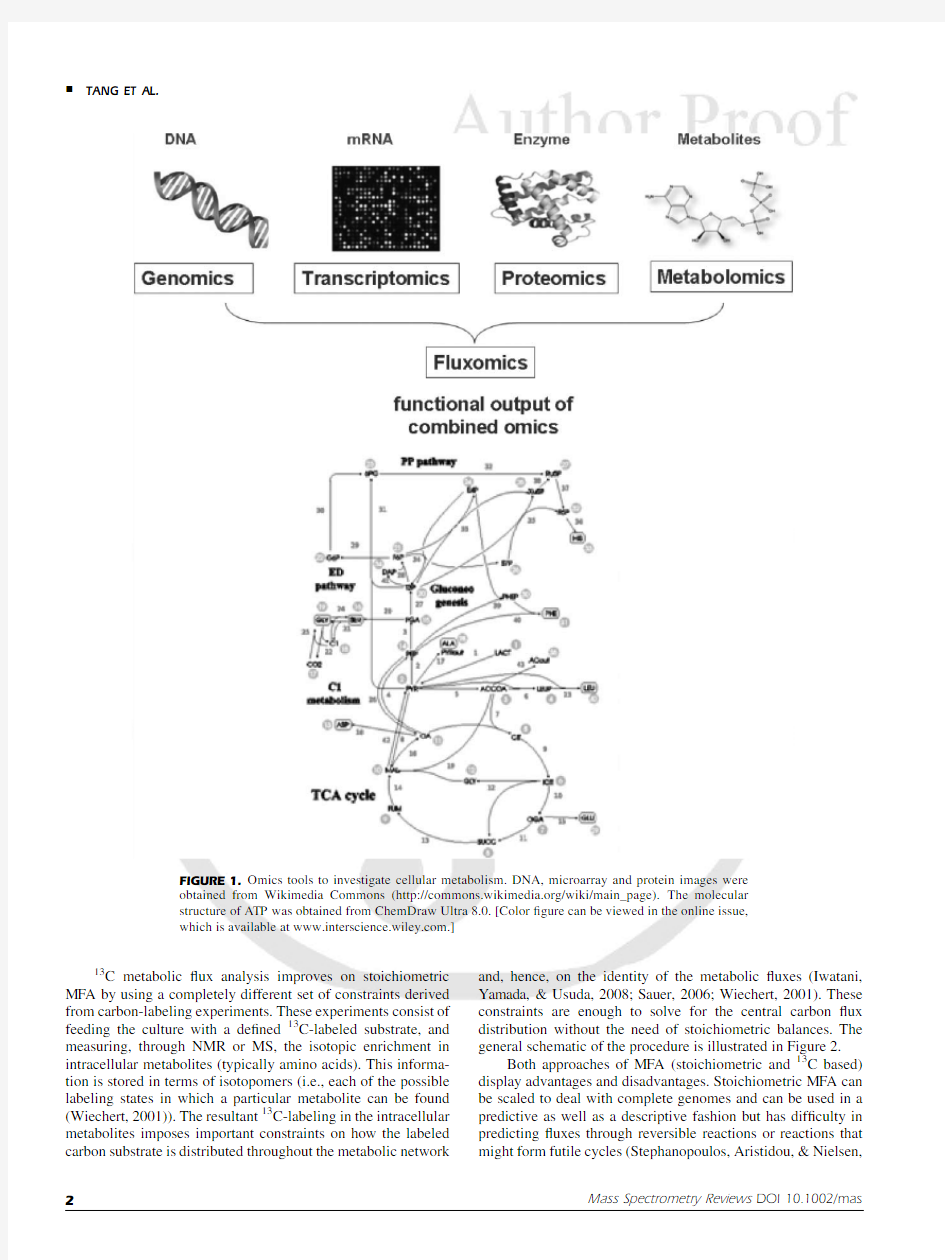
functional genomics tools are now available,including genomic and metagenomic sequencing (Alm et al.,2005;Tringe &Rubin,2005;Warnecke et al.,2007)and transcript,protein,and metabolite pro?ling (Sauer,2004;Wiechert,2001).However,the most physiologically relevant description of a cell’s metabolism remains the set of metabolic ?uxes,which represent the ?nal functional output of the interaction of all the molecular machinery studied by the other ‘‘omics’’?elds (Fig.1).Regulation of cellular processes might not always be re?ected in the gene annotation,transcript,or protein pro?les (Fong et al.,2006;Sauer,2004;Tang et al.,2007e).The transcription pro?le,moreover,might have little relationship to the ?nal ?ux pro?le of cells due to post-transcriptional regulation of protein synthesis and enzymes activities (Fong et al.,2006;Hua et al.,2007).The metabolic ?ux pro?le of a cell,however,re?ects the global reaction rates in the cellular metabolic network,and is a key determinant of cellular physiology (Sauer,2004).
A large number of comprehensive microbial ?ux studies have been performed with stoichiometric metabolic ?ux analyses (also referred to as ?ux balance analysis,FBA)(Varma &Palsson,1994).As its name implies,stoichiometric MFA uses the stoichiometry of the metabolic reactions (e.g.,global metabolite balances of cofactors such as ATP,NADH,and NADPH)in addition to a series of physical,chemical,and biological characteristics (extracellular ?uxes,thermodynamic direction-ality,enzymatic capacity,gene regulation,etc.)to constrain the feasible ?uxes for a given physiological condition.If the number of measured extracellular ?uxes equals the number of degrees of freedom it is possible to calculate the remaining ?uxes,but,typically,the number of constraints is much smaller than the number of reactions in the metabolic network (Vallino &Stephanopoulos,1993).The system is then underdetermined and it is necessary to postulate an objective function that one assumes the cell uses in its native ‘‘program’’(growth rate maximization,for example)to calculate a set of predicted ?uxes (Stephanopoulos,Aristidou,&Nielsen,1998).The general applicability of this optimization principle has been repeatedly called into question because cellular metabolism in several biological systems seems to display sub-optimal performance (Fischer &Sauer,2005;Schmidt et al.,1998;Schuetz,Kuepfer,&Sauer,2007).
MAS-637(20191)
Mass Spectrometry Reviews,2008,00,1–15#2008by Wiley Periodicals,Inc.
————
J.D.Keasling has a consulting relationship with and a ?nancial interest in Amyris and a ?nancial interest in LS9,both of which stand to bene?t from the commercialization of the results of this research.Y .J.Tang and H.G.Martin contributed equally to this study.
Contract grant sponsor:U.S.Department of Energy,Of?ce of Science,Of?ce of Biological and Environmental Research,Genomics (GTL Program)DE-AC02-05CH11231Q1.
Yinjie J.Tang’s present address is Department of Energy,Environ-mental and Chemical Engineering,Washington University,St.Louis,MO.
*Correspondence to:Jay D.Keasling,Joint Bio-Energy Institute,5885Hollis,Emeryville,CA 94608.E-mail:keasling@https://www.360docs.net/doc/c88726505.html,
13
C metabolic ?ux analysis improves on stoichiometric MFA by using a completely different set of constraints derived from carbon-labeling experiments.These experiments consist of feeding the culture with a de ?ned 13C-labeled substrate,and measuring,through NMR or MS,the isotopic enrichment in intracellular metabolites (typically amino acids).This informa-tion is stored in terms of isotopomers (i.e.,each of the possible labeling states in which a particular metabolite can be found (Wiechert,2001)).The resultant 13C-labeling in the intracellular metabolites imposes important constraints on how the labeled carbon substrate is distributed throughout the metabolic network
and,hence,on the identity of the metabolic ?uxes (Iwatani,Yamada,&Usuda,2008;Sauer,2006;Wiechert,2001).These constraints are enough to solve for the central carbon ?ux distribution without the need of stoichiometric balances.The general schematic of the procedure is illustrated in Figure 2.
Both approaches of MFA (stoichiometric and 13C based)display advantages and disadvantages.Stoichiometric MFA can be scaled to deal with complete genomes and can be used in a predictive as well as a descriptive fashion but has dif ?culty in predicting ?uxes through reversible reactions or reactions that might form futile cycles (Stephanopoulos,Aristidou,&
Nielsen,
FIGURE 1.Omics tools to investigate cellular metabolism.DNA,microarray and protein images were
obtained from Wikimedia Commons (https://www.360docs.net/doc/c88726505.html,/wiki/main_page).The molecular structure of ATP was obtained from ChemDraw Ultra 8.0.[Color ?gure can be viewed in the online issue,which is available at https://www.360docs.net/doc/c88726505.html,.]
&
TANG ET AL.
2
Mass Spectrometry Reviews DOI 10.1002/mas
1998;Wiechert &de Graaf,1997).On the other hand,13C MFA does not require the assumption of an optimization objective and usually provides more accurate ?ux estimations since it uses the (highly relevant)isotopomer data;however,13C MFA can only be used in a descriptive fashion (i.e.,it requires carbon labeling data),typically only tackles the central carbon metabolism,and is quite expensive to perform because of the high price of labeled feed.
The 14C-labeling experiments by Blum and Stein (1982)can be seen as the earliest direct precursor of 13C MFA.Since then,13
C-based ?ux methods have become a key technology to analyze metabolic networks and to provide support information for metabolic engineering applications (Wiechert,2001).In the last two decades,13C-based ?ux analysis has undergone signi ?cant developments that range from improvements in measuring the labeling patterns of targeted metabolites to computational algorithms for ?ux calculation.Such advancements have signi-?cantly extended the potential of 13C-based ?ux analysis for diverse applications in the ?elds of metabolic engineering,bioremediation,and biomedical research.This review summa-rizes the recent advances in the 13C-based ?uxomics ?eld for microbial systems and points out some possible future directions.
II.RECENT APPLICATION OF 13
C-BASED
FLUX ANALYSIS
Over the past decade,the high-throughput and high-content analysis of the cellular genome,transcriptome,proteome,and metabolome,commonly referred to as ‘‘omics,’’has been developed to investigate a variety of organisms.What has lagged well behind those omics studies,but might in fact be the most important indicator of cellular physiology,is the study of the cell
metabolic ?uxes (?uxome).A quick search of the PubMed database (https://www.360docs.net/doc/c88726505.html,)shows that the number of articles on ?ux analysis,speci ?cally 13C-based ?ux analysis to measure actual metabolic network,is several orders of magnitudes fewer than those for other omics studies (Table 1),in spite of the fact that ?ux pro ?les might provide a more accurate description of cell physiology (Sauer,2004,2006).This is due to a variety of reasons,including the high cost of labeled substrates,the requirement of specialized equipment (e.g.,MS or NMR)for determination of isotopic labeling and signi ?cant mathematical/statistical analysis (i.e.,isotopomer modeling of metabolism).Furthermore,this approach is not amenable to all biological systems,because many organisms cannot grow in a de ?ned minimal medium with labeled carbon substrates.Finally,13C ?ux analysis provides the information mostly regarding to ?ux distribution within central metabolic pathway.Precise determi-nation of all ?uxes through a large-scale metabolic network is still dif ?cult (Suthers et al.,2007).
During the last decade,measurement of metabolic ?uxes via 13
C-labeling has developed quickly across a diverse set of applications (summarized by Table 2),including:
*
Pathway bottleneck identi ?cation in industrial microorgan-isms with the ?nal objective of optimizing biomass and metabolite synthesis and,ultimately,providing guidelines for genetic engineering.Rational manipulation of cellular metabolism for product biosynthesis is the one of the main drivers for metabolic ?ux analysis.
*
Gene function validation in organisms and development of new insights into active pathways under speci ?c culture conditions.Every year,millions of dollars are spent on sequencing genomes of microorganisms and mammals.Whereas annotated and expressed genes may re ?ect the potential metabolism of a cell,?ux analysis provides
a
FIGURE 2.Protocol for 13C-based ?ux analysis.[Color ?gure can be viewed in the online issue,which is
available at https://www.360docs.net/doc/c88726505.html,.]
ADVANCES OF
13
C-BASED FLUX ANALYSIS
&
Mass Spectrometry Reviews DOI 10.1002/mas
3
valuable method to validate the proposed enzyme activities and the physiology of the cell.Flux analysis also reveals the responses of cellular metabolism to different growth con-ditions or environmental stresses.
*Drug-target search for a variety of diseases:pathogens or aberrant cells(e.g.,cancer cells)regulate speci?c metabolic pathways to bene?t their survival inside hosts.Studying the ?ux distributions in these cells’metabolic networks might reveal speci?c pathways that are essential for growth,and could be potential drug targets.
III.ADVANCES OF TECHNOLOGIES ASSOCIATED WITH13C METABOLIC FLUX PROFILING
Over the last few decades,a variety of methods associated with 13C?ux analysis have been developed.These methods include high-throughput cell culture experiments,accurate determina-tion of metabolites’labeling patterns by high-resolution mass spectrometry,and new algorithms for?ux calculation.
A.High Throughput13C-Labeled Cell Culture
13C-based?ux analysis is most easily done under metabolite and isotopomer steady state,which can be achieved by continuous feeding over three generation times(Tang et al.,2007c).Typical bioreactors with precise chemostat control often have a working volume over0.5L.Hence,experiments that use13C-labeled substrates can become very expensive,because the market price of labeled glucose or other carbon substrates is over$100/g (https://www.360docs.net/doc/c88726505.html,).One way to reduce the cost of labeled medium is to use unlabeled medium to achieve metabolic steady state,and then switch to an identical medium that contains labeled carbon substrates to obtain isotopic data.Because this approach might introduce signi?cant bias from the residual unlabeled carbon,it requires a computational method based on the standard wash-out kinetics of a chemostat culture in steady state to correct the measured isotopomer data(Dauner,Bailey,& Sauer,2001;Toya et al.,2007;Zhao&Shimizu,2003).On the other hand,high-resolution mass spectrometry technology can detect the metabolites at the level of nano-moles(i.e.,<5mg biomass is suf?cient to measure all proteinogenic amino acids),a large volume of expensive labeled culture is no longer necessary. To have a high-throughput and more economical method,shake ?asks or small-scale chemostat systems(<10mL)are often used for the purpose of reducing experiment costs(Nanchen et al., 2006).Shake?asks,for example,can be used for steady-state?ux analysis so long as the cells have maintained exponential growth for a suf?ciently long time(Sauer et al.,1999).Even in batch cultures as small as1mL,metabolic?uxes are directly comparable to those from cells grown in aerobic bioreactors (Fischer,Zamboni,&Sauer,2004).As such,deep-well micro-plates can be used to screen the intracellular?uxes of Escherichia coli and Saccharomyces cerevisiae(Cakar et al.,2005;Fischer, Zamboni,&Sauer,2004;Sauer,2004).Besides shake?asks and micro-plates,novel mini-bioreactors with a volume of1–10mL have been developed and are commercially available(Kostov et al.,2001;Maharbiz et al.,2004;Puskeiler et al.,2005;Tang et al.,2006).Micro-reactors not only control pH and temperature, but also automatically maintain the oxygen supply(a lack of which is a limitation often observed in shake?asks)and achieve stable dissolved oxygen levels throughout the entire growth period.These high-throughput cultivation systems have been shown to give reproducible results and extraordinary?exibility for tracer experiments under different growth conditions(Weiss et al.,2002;Yang,Wittmann,&Heinzle,2006).
Certain considerations need to be accounted for when choosing the position of the labeled carbon substrate and its composition in the cell culture medium.Generally,for fully
TABLE1.Publications with different omics tools from PubMed database
Omics Search key
words Total
Papers
Review papers Earliest paper
on PubMed
Genomics Genome sequence346,85919,459~1960s
Transcriptomics DNA microarray 28,2443,130 ~ 1995
Transcriptome32,3013,366~1982
Proteomics Protein analysis 1812,23289,892~1910s
Proteome10,3561,921~1995
Metabolomics Metabolite
analysis
31,581931~1950s
Fluxomics Metabolism+
Flux analysis
8,017388~1950s
13C + flux65126~1980s
Results as of March24th,2008.
&TANG ET AL.
4Mass Spectrometry Reviews DOI10.1002/mas
TABLE2.Examples of recent application of13C-based?ux analysis Authors Organisms Culture method/
Measurement
Findings Model Industrial Microorganisms
(Suthers et al. 2007)Escherichia coli Chemostat
culture; GC-MS
Use a large scale flux model to analyze
engineered amorphadiene producing strain.
(Fischer and Sauer 2003b)E. coli Chemostat
culture; GC-MS
Discover a novel metabolic cycle catalyzes
glucose oxidation and anaplerosis under
carbon limited conditions.
(Nanchen et al. 2006)E. coli Chemostat
culture;
GC-MS
Reveals the distribution of almost all major
fluxes varied nonlinearly with dilution rate
in chemostat culture.
(Dauner et al. 2001; Dauner et al. 2002; Fischer and Sauer 2005)Bacillus subtilis Chemostat or
shaking flask
culture; NMR or
GC-MS
Reveals the fluxes through central
metabolism when different carbon
substrates are used in carbon-limited
chemostat cultures. The robustness of
central metabolism was proposed.
(Blank et al. 2005a)Saccharomyces
cerevisiae
Mini-batch
(deep-well
plates) culture;
GC-MS
Reveals mechanistic principles of
metabolic network robustness to null
mutations in yeast, i.e., 75% network
reactions have redundancy through
duplicate genes.
Novel Microorganisms
(Tang et al. 2007c)Shewanella
oneidensis
Chemostat
culture; GC-MS
+ NMR
Reveal the regulation of central
metabolism under various oxygen
conditions.
(Tang et al. 2007a)Shewanella
oneidensis
Batch culture;
GC-MS
Reveal the effect of fullerene nano-
particles on cellular metabolism
(Risso et al. 2008; Tang et al. 2007b)Geobacter
metallireducens
Anaerobic batch
culture; GC-MS
Confirm a complete TCA cycle under Fe3+
reducing condition and found an unusual
isoleucine biosynthesis pathway.
(Tang et Desulfovibrio Anaerobic batch Reveal an incomplete central pathway and al. 2007e)vulgaris culture; GC-MS
+ FTICR
found the R-type citrate synthase.
(McKinlay et al. 2007)Actinobacillus
succinogenes
Anaerobic batch
culture; GC-MS
+ NMR; in vitro
enzyme assay
performed
Study carbon flux distributions and redox
balance for succinate production.
(Yang et al. 2002)Cyanobacterial
Synechocystis
Photosynthetic
bioreactor, GC-
MS + NMR
Calculate cyanobacterial central carbon
metabolism in both heterotrophic and
mixotrophic conditions
(Continued)
ADVANCES OF13C-BASED FLUX ANALYSIS&
Mass Spectrometry Reviews DOI10.1002/mas5
labeled carbon substrates(also called[U-13C]),a mixture of10–20%with unlabeled substrates is used for cell culture.This type of mixture has been proved to give more reliable?ux results if NMR is used for isotopomer analysis rather than GC-MS, because NMR can better determine the labeled carbon positions (Tang et al.,2007c).On the other hand,if a substrate with the proper position labeled is chosen,the GC-MS data of resulting metabolites can also give con?dent?ux results through most pathways.For example,the reactions of the pentose phosphate and Entner–Doudoroff pathways can be particularly well
TABLE2.(Continued)
(Fuhrer et al. 2005)Agrobacterium
tumefaciens,
Sinorhizobium
meliloti,
Rhodobacter
sphaeroides,
Zymomonas
mobilis,
Paracoccus
versutus,
Batch cultures
and GC-MS
Seven tested species show that Entner-
Doudoroff pathway and pyruvate bypass
were commonly used. All aerobes
exhibited fully respiratory metabolism
without significant overflow metabolism.
(Blank et al. 2005b)fourteen
Hemiascomycetous
yeasts
Shaking flask
and GC-MS
Findings include: compartmentation of
amino acid biosynthesis in most species
was identical to that in Saccharomyces
cerevisiae. The flux through the pentose
phosphate (PP) pathway was correlated to
the yield of biomass, but the operation of a
yet unidentified mechanism for NADPH
reoxidation in Pichia angusta is suggested.
Plant or mammalian cells
(Sriram et al. 2007)Catharanthus
roseus
Liquid medium
batch culture;
NMR
Fluxes analysis of plant hairy root system
quantifies the carbon flows through three-
compartments: plastid, cytosol and
mitochondrion.
(Allen et al. 2007)Soybeans(Glycine
max)
Grown in a
greenhouse with
labeled glucose
medium; GC-MS
Measurements of labeling of monomers
from starch, cell wall and protein glycans
estimate key carbon fluxes in the
compartmentalized flux network of plant
and MNR cells.
(Forbes et al. 2006)breast cancer cells roller bottles
with rich
medium ; NMR
The observed dependence of breast cancer
cells on pentose phosphate pathway
activity and glutamine consumption for
estradiol stimulated biosynthesis suggests
that these pathways may be targets for
estrogen-independent breast cancer
therapies.
(Yang et al. 2008a)mammary
carcinoma cells
Culture dishes
with rich
medium; NMR
and GC-MS
An integrated approach for the analysis of
metabolome and fluxomics to understand
fluxes through the key central metabolic
pathways and biosynthetic pathways of
fatty acids / amino acids in cancer cells.
(Meadows et al. 2008)human epithelial
breast cells
Culture dishes
with rich
medium; GC-MS
The unique metabolic characteristics of
cancerous breast cells are revealed by a
simple flux model based on isotopic
Enrichment in free metabolites
&TANG ET AL.
6Mass Spectrometry Reviews DOI10.1002/mas
differentiated using1st position labeled glucose but not with fully labeled glucose(Fischer,Zamboni,&Sauer,2004;Suthers et al., 2007).To achieve the most reliable metabolic?uxes computed based on the isotopomer distribution measured using NMR and GC-MS,mixture of singly labeled,doubly labeled,and fully labeled carbon substrates are recommended.One statistical analysis showed that the best feasible mixture of labeled carbon substrates for cultivation of Synechocystis sp.is70%unlabeled, 10%[U-13C]and20%[1,2-13C2]labeled glucose(Arauzo-Bravo&Shimizu,2003).Finally,the labeled carbon substrates purchased from commercial companies often contain a small amount of impure substrates(purity is<99%),which may adversely affect cell growth(Tang et al.,2007e).Hereby,if large amount of labeled carbon substrates is used in the culture, the purchased labeled substrates have to be further puri?ed to remove the potential toxic compounds.
B.Accurate Isotopomer Determination
Isotopomer analysis of metabolites allows branching(e.g.,the pentose phosphate pathway vs.glycolysis)and circular(e.g., TCA cycle)pathway?uxes to be determined,because the labeling pattern of metabolites in these pathways is very sensitive to the amount of?ux through them(Stephanopoulos,Aristidou, &Nielsen,1998).Proteinogenic amino acids are often used for isotopomer analysis because they acquire the labeling pattern of their central metabolic precursors and are abundant and stable. There are total20proteinogenic amino acids,but only16amino acids can be accessed after protein hydrolysis via6M HCl at high temperature(cysteine and tryptophan are degraded;glutamine and asparagines are converted to glutamate and aspartate respectively)(Daunder&Sauer,2000).The16amino acids can provide the isotopic labeling information of8crucial pre-cursor metabolites:pyruvate,acetyl-CoA,3-P-glycerate,phos-pho enol pyruvate,erythrose-4-P,oxaloacetate,2-oxo-glutarate, and ribose-5-P.Therefore,knowing the isotopomer distributions in these key metabolites provides enough constraints on the central carbon metabolic network model to con?dently calculate the?ux.
The experimental measurements of labeled carbon in amino acids or other metabolites can be done either by nuclear magnetic resonance(NMR)spectroscopy or by gas chromatography-mass spectrometry(GC-MS).NMR spectroscopy was a common technique for the early period of modern13C-?ux analysis,because it is sensitive to the position of the labeled carbon atoms in metabolites(de Graaf,2000;Malloy,Sherry,&Jeffrey, 1988;Sauer et al.,1997;Szyperski,1995,1998).However,the sensitivity of NMR is signi?cantly lower than that of GC-MS. GC-MS,currently the most popular technique,detects the mass distributions,which are the fractions of the total population of any particular molecule or molecular fragment that are unlabeled,singly labeled,doubly labeled,etc.(Christensen& Nielsen,1999;Daunder&Sauer,2000;Wittmann,2007). Although this type of data offers no information about the position of all labeled atoms,some of this information can be determined by fragmenting molecules to provide additional labeling information at certain carbon positions(mostly the carboxyl group)(Wahl,Dauner,&Wiechert,2004).
Analysis of amino acids or other metabolites via gas chromatography requires that the metabolites are derivatized, commonly with silylation reagents,to render the molecules volatile enough to enter the GC column(Fig.3).N-(tert-butyldimethylsilyl)-N-methyltri?uoroacetamide(TBDMS method) is most common derivatizing agent(Antoniewicz,Kelleher,& Stephanopoulos,2007a;Daunder&Sauer,2000;Wahl,Dauner, &Wiechert,2004).To measure the labeling in free metabolic acids,such as pyruvate or succinate,a more gentle and sensitive derivatization agent,N,O-bis-(trimetylsilyl)tri?uoroacetamide (BSTFA),has also been used(Meadows et al.,2008;Tang et al.,2007d)(Fig.3).The derivatization step introduces signi?cant amounts of naturally labeled isotopes(‘‘noise’’), including13C(1.13%),18O(0.20%),29Si(4.70%),and30Si (3.09%),such that the raw mass isotopomer spectrum must be corrected prior to calculation of metabolic?uxes(Daunder& Sauer,2000;Hellerstein&Neese,1999;Lee,Bergner,&Guo, 1992;van Winden et al.,2002;Wahl,Dauner,&Wiechert,2004; Wiechert&de Graaf,1996).Another disadvantage of GC-MS is that the accuracy of isotopomer measurement can be affected by the choice of the GC-MS spectrum integration algorithm,sample concentration,and overlapping fragments(Antoniewicz,Kel-leher,&Stephanopoulos,2007a).
High-resolution and highly sensitive mass spectrometers can be used to precisely measure the labeling pattern of amino acids and metabolites in central metabolic pathways(at concentrations as low as nM).Liquid chromatography-mass spectrometry/mass spectrometry(LC-MS/MS)has been used to determine intracellular free amino acids to pro?le metabolic?ux changes during fed-batch cultivation(Iwatani et al.,2007;No¨h et al.,2007).This approach,which involves the labeling of
free FIGURE3.The molecular structure and the bond fragmentation positions between two silyation-
derivatized amino acids.The dotted line represents the cracking position during ionization.
ADVANCES OF13C-BASED FLUX ANALYSIS& Mass Spectrometry Reviews DOI10.1002/mas7
metabolites with fast-turnover times,has great potential to investigate metabolism during various cell growth phases, because the phase dependent metabolism is dif?cult to accomplish using proteinogenic amino acids.In a related fashion, GC-MS,LC-MS,and NMR have been combined to extract maximal isotopomer information from amino acids.The information has been used to accurately determine the metab-olism in mitochondria and the cytosol in S.cerevisiae strains (Kleijn et al.,2007).Capillary electrophoresis time-of-?ight mass spectrometry(CE-TOF MS)has been used to measure isotopomers of thirteen unstable metabolites in central metab-olism,including some unstable phosphorylated molecules such as3-P-glycerate,phospho enol pyruvate,and ribose-5-P(Toya et al.,2007).Thus,CE-MS allows metabolic?ux analysis directly from metabolites without any measurement of amino acids.Recently,Fourier transform-ion cyclotron resonance mass spectrometry(FT-ICR MS),with direct infusion via electrospray ionization,was used to measure the metabolite isotopomer
distribution in a biomass hydrolysate of Desulfovibrio vulgaris Hildenborough to unveil an unusual citrate synthase activity (Pingitore et al.,2007).This method can determine the13C positions in the skeleton of the amino acid based on speci?c fragmentation patterns.
Measurement of labeling in central metabolites,other than proteinogenic amino acids,can signi?cantly extend the scope of 13C-based?ux analysis applications and improve the accuracy of
?ux determination.Because13C MFA derives?ux distributions from the labeling of amino acids produced by the labeled feed once it percolates through the metabolic network,it requires a minimal medium to avoid the introduction of a bias into the isotopomer measurements.However,many pure cultures are non-viable without nutrient supplements.When cells are grown in a rich medium that contains amino acids,only those proteinogenic amino acids synthesized from the central metab-olism(e.g.,alanine,aspartate,and glutamate)and not taken up from the medium can be used for?ux determination using standard methods(Christiansen,Christensen,&Nielsen,2002). To circumvent this problem,highly sensitive mass spectrometers can be used to directly obtain the isotopomer information from free central metabolites(e.g.,acids in the TCA cycle).Provided that the amino acids are not metabolized,use of isotopomers of central metabolic intermediates will allow calculation of metabolic?uxes even when amino acids are supplemented into the medium.Additionally,this strategy avoids the possible mistakes of acquiring the labeling pattern of metabolites (e.g.,amino acids)for organisms that are not well-known or where holes in genomic annotation are present(e.g.,the alternative isoleucine pathway in Geobacter spp.contain holes in the annotation that might introduce errors in metabolic?ux calculations)(Risso et al.,2008;Tang et al.,2007b).
C.High-Performance Flux Calculation Algorithms Metabolic?uxes cannot be measured directly but,rather,must be inferred through model based data evaluation from the knowl-edge of the reactions involved,the information contained in the amino acid/metabolite labeling,and the external?uxes(Fig.4). Hence,computational algorithms are a key component of 13C-based?ux analysis(Sauer,2006;Schmidt,Nielsen,& Villadsen,1999a;Schmidt et al.,1999b;Wiechert,2001). Because?nding analytical expressions for the internal?uxes as a function of carbon-labeling data is,for all practical cases, impossible,the determination of?uxes is usually achieved through a heuristic recursive procedure(Fig.5).The computa-tional bottlenecks for this procedure are:(1)how to best choose the new set of?uxes based on the past information on the error function e({v i})so as to minimize the number of steps necessary to reach its global minimum without getting trapped in the relative minima,and(2)calculating the amino acid/metabolite labeling pattern from the assumed set of?uxes{v i}.
Finding the input that optimizes a function(function optimization)has been an intensely studied challenge in numerical analysis.An array of tools(Floudas&
Pardalos, FIGURE4.Inputs for metabolic?ux analysis.The information on the metabolic reactions,amino acid/metabolite labeling and extracellular ?uxes is combined to produce the error function e(average difference between measured and computed labeling patterns).The predicted labeling can be computed following different methods(see main text) and then coupled to a chosen nonlinear solver(see main text)to solve for the?uxes following the iterative procedure in Figure
5.
FIGURE5.Recursive procedure to obtain?uxes from amino acid/ metabolite labeling information.A set of?uxes{v i}is initially chosen and the expected amino acid/metabolite labeling is calculated under the assumed?uxes{v i}.This computationally generated labeling is compared with the labeling obtained experimentally and the difference is quanti?ed as the error function e({v i}).A new set of?uxes{v i}is then chosen so as to try to decrease e({v i}).This procedure is repeated until the calculated labeling and the experimental data are within the experimental error.[Color?gure can be viewed in the online issue,which is available at https://www.360docs.net/doc/c88726505.html,.]
&TANG ET AL.
8Mass Spectrometry Reviews DOI10.1002/mas
1992;Goldberg,1989;Nocedal&Wright,1999;Press et al., 1992)is available to tackle the?rst bottleneck.Among those methods used in13C-based?ux analysis are global search algorithms such as simulated annealing and evolutionary algorithms(Dauner,Bailey,&Sauer,2001;Forbes,Clark,& Blanch,2001;Schmidt et al.,1999b;Zhao&Shimizu,2003);and local search algorithms such as the Levenberg–Marquardt method(Yang et al.,2008a;Zhao&Shimizu,2003),the Nelder–Mead method(Wiechert et al.,2001),Sequential Quadratic Programming(SQP)(Wiechert et al.,2001),or a hybrid SQP/Newton algorithm(Yang,Frick,&Heinzle,2008b). Although local search algorithms might easily become trapped in local minima,they are typically much faster than global search algorithms.Hence,they can be run many times with different initial points and have the best solution chosen,in the same amount of time that a single global search would be performed. Although no systematic comparison between methods has been published,the Levenberg–Marquardt method(Young et al., 2008)and a range-restricted evolutionary algorithm(Mo¨llney et al.,1999;No¨h et al.,2007;No¨h&Wiechert,2006)are used for the most computationally demanding?ux analysis:non-sta-tionary metabolic?ux analysis.
Development of computational methods for13C-based?ux analysis has,therefore,focused on speeding up the calculation of amino acid/metabolite labeling from an assumed set of?uxes. Several methods have been proposed,including the iterative averaging isotopomer method(Schmidt,Nielsen,&Villadsen, 1999a),the cumomer method(Wiechert et al.,1999),the Elementary Metabolic Unit(EMU)method(Antoniewicz, Kelleher,&Stephanopoulos,2007b),the isotopomer path tracing method(Forbes,Clark,&Blanch,2001),and the fractional labeling method(Riascos,Gombert,&Pinto,2005).In practice, the only methods under continuous development are the cumomer and EMU method.The cumomer method was developed as a more ef?cient strategy to solve metabolite labeling than the iterative averaging isotopomer method:by casting the problem in terms of the new concept of cumomers (cumulated isotopomers fractions),the isotopomer labeling can be obtained from the solution to a cascade of linear equations. The EMU method uses the knowledge of atomic transitions in the reactions network to identify a set of variables containing the minimum amount of information necessary to simulate isotopic labeling in the system.The models resulting from the use of this set of variables require signi?cantly fewer equations to be solved than for the cumomer case.The EMU method results in computation times that are claimed to be several orders of magnitude lower than for the cumomer method(Young et al., 2008)and,hence,the possibility of tracking other labeling atoms such as2H,15N,and18O in tracer experiments with multiple labeled substrates.Nonetheless,a recent study attributes similar improvements in speed for the cumomer method based on careful study of the labeling network topology(Weitzel,Wiechert,& No¨h,2007).Because the insights obtained form network topology are applicable to the EMU method,signi?cant increases in performance are expected in the future(Weitzel,Wiechert,& No¨h,2007).
Not every13C-based?ux analysis strategy follows the scheme shown in Figure5.There are other alternatives:one of them is to choose a set of amino acids with well-known precursors and to extract the relative ratio of?uxes that contribute to their labeling(Fischer&Sauer,2003a).This method has been used to perform the?rst large-scale experimental analysis of intracellular?ux distributions for137null mutants of B.subtilis (Fischer&Sauer,2005).Although this approach provides direct evidence for the relative magnitude of each?ux and has the great bene?t of using local network data around single metabolite nodes,it is restricted to10–15pre-selected pathways directly accessible from the isotopomer data(Sauer,2006).Another alternative involves using a nonlinear problem(NLP)solver to search simultaneously among the total number of con?gurations of?uxes and metabolite labeling patterns to match experimental data(Riascos,Gombert,&Pinto,2005;Vo&Palsson,2006). Whereas initial applications of this method were limited to central carbon metabolism reactions,more recent work(Suthers et al.,2007)makes use of much more comprehensive metabolic networks and can,hence,take into account global metabolite balances of cofactors(e.g.,ATP,NADH,and NADPH),which traditional13C MFA does not.These studies integrate13C MFA with FBA and display the bene?ts of both methods.
As with every measurement,it is desirable to assign a con?dence interval to?ux estimates.A traditional approach has been to?nd a general mapping from the experimental data(i.e., isotopomer labeling and extracellular?uxes)to the best?ux estimate,linearize that mapping around the actual measured experimental data,and map the con?dence intervals from the measured data on to the?ux estimates(Arauzo-Bravo& Shimizu,2003;Dauner,Bailey,&Sauer,2001;Schmidt et al., 1999b;Wiechert&de Graaf,1997;Wiechert et al.,1997). Recently,nonlinear statistical methods(Gallant,1987)have been applied to take into account the intrinsic nonlinear nature of?ux analysis(Antoniewicz,Kelleher,&Stephanopoulos,2006). Under this scenario,each individual?ux is increased from its estimated value until the objective function for the recalculated ?uxes reaches the maximum value allowed for a given con?dence value.This marks the upper con?dence limit.The crucial difference with the linear approach is that the?ux estimates are recalculated for each individual?ux increase.The lower con?dence limit is obtained by decreasing each independent?ux and following the same procedure.An alternative(but more computationally intensive)way to take into account the nonlinear nature of?ux calculation is to use a Monte–Carlo approach in which new experimental data is randomly generated within the measurement errors and new?ux estimates are calculated.By doing this a large enough number of times,the probability distribution of estimated?uxes provides the con?dence intervals and correlation between individual?uxes(Schmidt et al.,1999b; Zhao&Shimizu,2003).
D.Non-Stationary Flux Analysis
One of the traditional prerequisites for13C-based?ux analysis is that the system must be in a metabolic and isotopic steady-state; that is,?uxes and amino acid/metabolite labeling does not change in time.This requirement means that the length of the experiment should be signi?cantly longer than the inverse of the growth rate, 1/m(No¨h&Wiechert,2006;Wiechert&No¨h,2005);that requirement leads to impractically long experiments in the case
ADVANCES OF13C-BASED FLUX ANALYSIS&
Mass Spectrometry Reviews DOI10.1002/mas9
of low growth rates,such as in the production phase of typical
bioprocesses.A way to circumvent this problem is to use the
information of the labeling of intracellular free amino acids as
input(Iwatani et al.,2007;Kro¨mer et al.,2004).Intracellular
amino acids are continuously renewed,and represent the actual
?ux state of the cells,as modi?ed by protein turnover,
transamination,mRNA degradation and other effects(Grotkjaer
et al.,2004).Proteinogenic amino acids,on the other hand,are an
accumulation of intracellular amino acids throughout the entire
cultivation process and display much longer turnover rates.
Another strategy to overcome this dif?culty is to use intracellular
metabolite labeling and non-stationary?ux analysis.This type of 13C MFA assumes a changing metabolite labeling and tracks these changes to estimate?uxes.Cumomer-and EMU-based
algorithms have been developed,and their application allows for
the quanti?cation of time-resolved metabolite labeling patterns
and?ux pro?les(Antoniewicz et al.,2007c;No¨h,Wahl,&
Wiechert,2006;Young et al.,2008).Those developments extend 13C-?ux analysis to non-stationary conditions like batch and fed-batch fermentations,and reduce the cost and duration of labeling experiments.
IV.STRATEGIES FOR SYSTEMS-LEVEL
13C-BASED FLUX ANALYSIS
Metabolic?ux distributions provide new insights into how metabolism works.However,13C-based?ux analysis,so far,is mainly focused on central metabolic pathways and biomass synthesis pathways(e.g.,amino acids).Because each pathway could be controlled by several genes/enzymes,?ux analysis alone might be insuf?cient to reveal exact gene targets or regulatory mechanisms in a complicated biological system. Hence,13C-based?ux measurement has been integrated with other‘‘omics’’tools to understand global metabolism.
1.Genomics combined with?ux analysis:Most?ux-analysis
studies focus on central metabolism and neglect other?ux routes that could contribute to biomass growth and metabolite synthesis.Recently,13C-based genome-scale?ux models of S.cerevisiae and E.coli have been developed to identify annotated gene functions in more comprehensive metabolic networks(Blank,Kuepfer,&Sauer,2005a;Suthers et al.,2007).
For the E.coli case,for example,a reaction network that consisted of350?uxes and184metabolites in E.coli,including global metabolite balances on cofactors such as ATP,NADH, and NADPH(Suthers et al.,2007),was developed.This approach demonstrated possible key genes in an E.coli strain engineered to produce amorphadiene(a precursor to the anti-malarial drug artemisinin).Additionally,conventional genome-scale?ux-balance analysis(FBA)can determine intracellular ?uxes,but it requires choosing proper objective functions to accurately describe certain metabolic conditions.The13C-based ?ux analysis approach can be used to verify the FBA model and to provide useful metabolic regulation information.For instance, different objective functions to predict?uxes in the genome-scale FBA have been evaluated via isotopomer?ux models (Schuetz,Kuepfer,&Sauer,2007).The study showed that
unlimited growth on glucose in oxygen-or nitrate-respiring batch cultures is best described by nonlinear maximization of the ATP yield with minimal enzyme usage,whereas under nutrient scarcity in continuous cultures,linear maximization of the overall ATP or biomass yields achieved the highest predictive accuracy.
2.Transcriptomics and?ux analysis:Metabolic?uxes and global
mRNA transcript analyses have been used to study the?exibility of the metabolic network of E.coli to compensate for genetic perturbations(Fong et al.,2006).Only activation of latent pathways and?ux changes in some tricarboxylic acid cycle pathways were found to correlate with molecular changes at the transcriptional level,whereas?ux alterations in other central metabolic pathways were not connected to changes in the transcriptional network;those data suggest complex regulatory mechanisms at transcription and enzyme activity levels.Similar observations were reported from the study on E.coli strains adapted to growth on lactate and S.cerevisiae grown under a variety of carbon sources;that is,no clear qualitative corre-lations between most transcriptional expression and metabolic ?ux changes(Hua et al.,2007).
3.In vitro enzyme chemistry,proteomics,and?ux analysis:
13C-labeling experiments combined with measurements of enzyme activities and intracellular metabolite pro?les are often used to clarify the unknown pathways and support the results of the in vivo?ux measurement(Klapa,Aon,&Stephanopoulos, 2003;McKinlay et al.,2007;McKinlay&Vieille,2008;Sauer et al.,2004).For example,when the metabolism in a pykF mutant of E.coli was studied,information on intracellular metabolic?ux distributions,enzyme activities,and intracellular metabolite concentrations were integrated to quantitatively reveal the regulation of phospho enol pyruvate carboxylase, malic enzyme,phosphofructokinase,acetate formation,and the oxidative pentose phosphate(PP)pathway in the mutant(Al Zaid Siddiquee,Arauzo-Bravo,&Shimizu,2004).Proteomics tools have also been used to provide labeling constraints for?ux analysis of individual strains in microbial communities(Shaikh et al.,2008).This study shows that it is possible to analyze the isotopomer distribution of amino acids from targeted organism via highly expressed His-tagged green?uorescent protein (GFP).
Multiple‘‘omics’’analysis,including transcriptomics,proteomics, metabolomics,and?uxomics,are beginning to be integrated to monitor cellular physiology,and those‘‘omics’’studies create a new concept of functional genomics.The combination of those high-throughput tools allows for the systematic quanti?cation of the interactions of thousands of metabolic network components under genetic or environmental perturbations(Ishii et al.,2007;Kro¨mer et al.,2004).
V.CONCLUSIONS
We have strived to show that there is a signi?cant value for 13C-based metabolic?ux analysis in many?elds.A series of new techniques associated with13C-?ux analysis have recently emerged,including:high-throughput cultivation
&TANG ET AL.
10Mass Spectrometry Reviews DOI10.1002/mas
systems,high-sensitivity,high-resolution mass spectrometry for
isotopomer analysis of metabolites(such as GC-MS,LC-MS,
ESI-TOF,or FT-ICR),high-performance modeling programs
for isotopomer?ux analysis,and integrated?ux analysis with
other‘‘omics’’tools.These techniques extend the application of 13C-based?ux analysis to diverse applications from microbial to mammalian cells to(1)discover or validate gene functions
involved in central metabolic pathways;(2)understand the
in vivo metabolisms under different culture conditions;
(3)provide information of the bottleneck pathways for biomass
or metabolite synthesis in engineered microorganisms;(4)and
identify pathogen-speci?c metabolic pathways for drug targets. ACKNOWLEDGMENTS
We thank Dr.Dominic Desiderio,Dr.Paramvir Dehal,
Dr.Taek Soon Lee,and Dr.Yisheng Kang for useful comments on
the manuscripts.This work was funded in part of the Virtual
Institute for Microbial Stress and Survival(https://www.360docs.net/doc/c88726505.html,)
supported by the U.S.Department of Energy,Of?ce of Science,
Of?ce of Biological and Environmental Research,Genomics:
GTL Program through contract DE-AC02-05CH11231between
the Lawrence Berkeley National Laboratory and the US Depart-
ment of Energy and in part by the Joint BioEnergy Institute
(https://www.360docs.net/doc/c88726505.html,)supported in part by the U.S.Department
of Energy,Of?ce of Science,Of?ce of Biological and Environ-
mental Research,Genomics Program:GTL through contract
DE-AC02-05CH11231between Lawrence Berkeley National
Laboratory and the U.S.Department of Energy.
REFERENCES
Al Zaid Siddiquee K,Arauzo-Bravo MJ,Shimizu K.2004.Metabolic?ux analysis of pykF gene knockout Escherichia coli based on13C-labeling experiments together with measurements of enzyme activities and intracellular metabolite concentrations.Appl Microbiol Biotechnol 63:407–417.
Allen Q2DK,Shachar-Hill Y,Ohlrogge https://www.360docs.net/doc/c88726505.html,partment-speci?c labeling information in13C metabolic?ux analysis of plants.
Phytochemistry68(16–18):2197–2210.
Alm EJ,Huang KH,Price MN,Koche RP,Keller K,Dubchak IL,Arkin AP.
2005.The MicrobesOnline Web site for comparative genomics.
Genome Res15:1015–1022.
Antoniewicz MR,Kelleher JK,Stephanopoulos G.2006.Determination of con?dence intervals of metabolic?uxes estimated from stable isotope measurements.Metab Eng8(4):324–337.
Antoniewicz MR,Kelleher JK,Stephanopoulos G.2007a.Accurate assess-ment of amino acid mass isotopomer distributions for metabolic?ux analysis.Anal Chem79(19):7554–7559.
Antoniewicz MR,Kelleher JK,Stephanopoulos G.2007b.Elementary metabolite units(EMU):A novel framework for modeling isotopic distributions.Metab Eng9(1):68–86.
Antoniewicz MR,Kraynie DF,Laffend LA,Gonza′lez-Lergier J,Kelleher JK, Stephanopoulos G.2007c.Metabolic?ux analysis in a nonstationary system:Fed-batch fermentation of a high yielding strain of E.coli producing1,3-propanediol.Metab Eng9(3):277–292.Arauzo-Bravo MJ,Shimizu K.2003.An improved method for statistical analysis of metabolic?ux analysis using isotopomer mapping matrices with analytical expressions.J Biotechnol105:117–133.
Blank LM,Kuepfer L,Sauer https://www.360docs.net/doc/c88726505.html,rge-scale13C-?ux analysis reveals mechanistic principles of metabolic network robustness to null mutations in yeast.Genome Biol6(6):R49.
Blank LM,Lehmbeck F,Sauer U.2005b.Metabolic-?ux and network analysis in fourteen hemiascomycetous yeasts.Yeast Res5(6–
7):545–558.
Blum JJ,Stein RB.1982.In:Golberberger editor Q3.On the analysis of metabolic networks.New York:Plenum Press.pp99–124.
Cakar ZP,Seker UO,Tamerler C,Sonderegger M,Sauer U.2005.
Evolutionary engineering of multiple-stress resistant Saccharomyces cerevisiae.FEMS Yeast Res5(6–7):569–578.
Christensen B,Nielsen J.1999.Isotopomer analysis using GC-MS.Metab Eng1:282–290.
Christiansen T,Christensen B,Nielsen J.2002.Metabolic network analysis of Bacillus claussi on minimal and semirich medium using13C-labeled glucose.Metab Eng???Q4(4):159–
Daunder M,Sauer U.2000.GC-MS analysis of amino acids rapidly provides rich information for isotopomer balancing.Biotechnol Prog16:642–649.
Dauner M,Bailey JE,Sauer U.2001.Metabolic?ux analysis with a comprehensive isotopomer model in Bacillus subtilis.Biotechnol Bioeng76(2):144–156.
Dauner M,Sonderegger M,Hochuli M,Szyperski T,Wuthrich K,Hohmann HP,Sauer U,Bailey JE.2002.Intracellular carbon?uxes in ribo?avin-producing Bacillus subtilis during growth on two-carbon substrate mixtures.Appl Environ Microbiol68(4):1760–
de Graaf https://www.360docs.net/doc/c88726505.html,e of13C labelling and NMR spectroscopy in metabolic ?ux analysis.In:Barbotin JN,Portais JC,NMR biotechnol-ogy:Theory and applications.Norwich,UK:Horizon Q5Scienti?c Press.
Fischer E,Sauer U.2003a.Metabolic?ux pro?ling of Escherichia coli mutants in central carbon metabolism using GC-MS.Eur J Biochem 270(5):880–891.
Fischer E,Sauer U.2003b.A novel metabolic cycle catalyzes glucose oxidation and anaplerosis in hungry Escherichia coli.J Biol Chem 278(47):46446–46451.
Fischer E,Sauer https://www.360docs.net/doc/c88726505.html,rge-scale in vivo?ux analysis shows rigidity and suboptimal performance of Bacillus subtilis metabolism.Nat Genet 37(6):636–640.
Fischer E,Zamboni N,Sauer U.2004.High-throughput metabolic?ux analysis based on gas chromatography-mass spectrometry derived13C constraints.Anal Biochem325:308–316.
Floudas CA,Pardalos PM.1992.Recent advances in global optimization.
Princeton,N.J.:Princeton University Press.
Fong SS,Nanchen A,Palsson BO,Sauer https://www.360docs.net/doc/c88726505.html,tent pathway activation and increased pathway capacity enable Escherichia coli adaptation to loss of key metabolic enzymes.J Biol Chem281(12):8024–8033. Forbes NS,Clark DS,Blanch https://www.360docs.net/doc/c88726505.html,ing isotopomer path tracing to quantify metabolic?uxes in pathway models containing reversible reactions.Biotechnol Bioeng74:196–211.
Forbes NS,Meadows AL,Clark DS,Blanch HW.2006.Estradiol stimulates the biosynthetic pathways of breast cancer cells:Detection by metabolic ?ux analysis.Metab Eng8(6):639–652.
Fuhrer T,Fischer E,Sauer U.2005.Experimental identi?cation and quanti?cation of glucose metabolism in seven bacterial species.
J Bacteriol187(5):1581–1590.
Gallant AR.1987.Nonlinear statistical models.New York:Wiley. Goldberg DE.1989.Genetic algorithms in search,optimization,and machine learning reading.Massachusetts:Addison-Wesley.
ADVANCES OF13C-BASED FLUX ANALYSIS&
Mass Spectrometry Reviews DOI10.1002/mas11
Grotkjaer T,Akesson M,Christensen B,Gombert AK,Nielsen J.2004.
Impact of transamination reactions and protein turnover on labeling dynamics in(13)C-labeling experiments.Biotechnol Bioeng86(2): 209–216.
Hellerstein MK,Neese RA.1999.Mass isotopomer distribution analysis at eight years:Theoretical,analytic,and experimental considerations.Am J Physiol Endocrinol Metab276(6):E1146–E1170.
Hua Q,Joyce AR,Palsson BO,Fong SS.2007.Metabolic characterization of Escherichia coli adapted to growth on lactate.Appl Environ Microbiol 73(14):4639–4647.
Ishii N,Nakahigashi K,Baba T,Robert M,Soga T,Kanai A,Hirasawa TNM, Hirai K,Hoque A,Ho PY,Kakazu Y,Sugawara K,Igarashi S,Harada S, Masuda T,Sugiyama N,Togashi T,Hasegawa M,Takai Y,Yugi K, Arakawa K,Iwata N,Toya Y,Nakayama Y,Nishioka T,Shimizu K, Mori H,Tomita M.2007.Multiple high-throughput analyses monitor the response of E.coli to perturbations.Science316(5824):593–597. Iwatani S,Van Dien S,Shimbo K,Kubota K,Kageyama N,Iwahata D, Miyano H,Hirayama K,Usuda Y,Shimizu K,et al.2007.Determination of metabolic?ux changes during fed-batch cultivation from measure-ments of intracellular amino acids by LC-MS/MS.J Biotechnol128(1): 93–111.
Iwatani S,Yamada Y,Usuda Y.2008.Metabolic?ux analysis in biotechnology processes.Biotechnol Lett30(5):791–799.
Klapa MI,Aon JC,Stephanopoulos G.2003.Systematic quanti?cation of complex metabolic?ux networks using stable isotopes and mass spectrometry.Eur J Biochem270(17):3525–3542.
Kleijn RJ,Geertman JM,Nfor BK,Ras C,Schipper D,Pronk JT,Heijnen JJ, van Maris AJ,van Winden W A.2007.Metabolic?ux analysis of a glycerol-overproducing Saccharomyces cerevisiae strain based on GC-MS,LC-MS and NMR-derived13C-labelling data.FEMS Yeast Res 7(2):216–231.
Kostov Y,Harms P,Randers-Eichhorn L,Rao G.2001.Low-cost micro-bioreactor for high-throughput bioprocessing.Biotechnol Bioeng 72(3):346–352.
Kro¨mer JO,Sorgenfrei O,Klopprogge K,Heinzle E,Wittmann C.2004.
In-depth pro?ling of lysine-producing Corynebacterium glutamicum by combined analysis of the transcriptome,metabolome,and?uxome.
J Bacteriol186(6):1769–1784.
Lee WN,Bergner EA,Guo ZK.1992.Mass isotopomer pattern and precursorproduct relationship.Biol Mass Spectrom21:114–122. Maharbiz MM,Holtz WJ,Howe RT,Keasling JD.2004.Microbioreactor arrays with parametric control for high-throughput experimentation.
Biotechnol Bioeng86(4):485–490.
Malloy CR,Sherry AD,Jeffrey FMH.1988.Evaluation of carbon?ux and substrate selection through alternate pathways involving the citric acid cycle of the heart by13C NMR Spectroscopy.J Biol Chem263:6964–6971.
McKinlay JB,Shachar-Hill Y,Zeikus JG,Vieille C.2007.Determining Actinobacillus succinogenes metabolic pathways and?uxes by NMR and GC-MS analyses of13C-labeled metabolic product isotopomers.
Metab Eng9(2):177–192.
McKinlay JB,Vieille C.2008.13C-metabolic?ux analysis of Actinobacillus succinogenes fermentative metabolism at different NaHCO3and H2 concentrations.Metab Eng10(1):55–68.
Meadows AL,Kong B,Berdichevsky M,Roy S,Rosiva R,Blanch HW,Clark DS.2008.Metabolic and morphological differences between rapidly proliferating cancerous and normal breast epithelial cells.Biotechnol Prog24(2):334–341.
Mo¨llney M,Wiechert W,Kownatzki D,de Graaf AA.1999.Bidirectional reaction steps in metabolic networks:IV.Optimal design of isotopomer labeling experiments.Biotechnol Bioeng66(2):86–103.
Nanchen A,Schicker A,Sauer U.2006.Nonlinear dependency of intra-cellular?uxes on growth rate in miniaturized continuous cultures of Escherichia coli.Appl Environ Microbiol72(2):1164–1172.Nocedal J,Wright SJ.1999.Numerical optimization.New York:Springer. No¨h K,Gro¨nke K,Luo B,Takors R,Oldiges M,Wiechert W.2007.Metabolic ?ux analysis at ultra short time scale:Isotopically non-stationary 13C labeling experiments.J Biotechnol129(2):249–267.
No¨h K,Wahl A,Wiechert https://www.360docs.net/doc/c88726505.html,putational tools for isotopically instationary13C labeling experiments under metabolic steady state conditions.Metab Eng8(6):554–577.
No¨h K,Wiechert W.2006.Experimental design principles for isotopically instationary13C labeling experiments.Biotechnol Bioeng94(2):234–251.
Pingitore F,Tang Y,Kruppa GH,Keasling JD.2007.Analysis of amino acid isotopomers using FT-ICR MS.Anal Chem79(6):2483–2490. Press WH,Teukolsky SA,Vetterling WT,Flannery BP.1992.Numerical recipes in FORTRAN.Cambridge:Cambridge University Press.
pp387–448.
Puskeiler R,Kusterer A,John GT,Weuster-Botz D.2005.Miniature bioreactors for automated high-throughput bioprocess design(HTBD): Reproducibility of parallel fed-batch cultivations with Escherichia coli.
Biotechnol Appl Biochem42(3):227–235.
Riascos CAM,Gombert AK,Pinto JM.2005.A global optimization approach for metabolic?ux analysis based on labeling https://www.360docs.net/doc/c88726505.html,put Chem Eng29:447–458.
Risso C,Van Dien SJ,Orloff A,Lovley DR,Coppi MV.2008.Elucidation of an alternate isoleucine biosynthesis pathway in Geobacter sulfurredu-cens.J Bacteriol190(7):2266–2274.
Sauer U.2004.High-throughput phenomics:Experimental methods for mapping?uxomes.Curr Opin Biotechnol15:58–63.
Sauer U.2006.Metabolic networks in motion:13C-based?ux analysis.Mol Syst Biol2:62.
Sauer U,Canonaco F,Heri S,Perrenoud A,Fischer E.2004.The soluble and membrane-bound transhydrogenases UdhA and PntAB have divergent functions in NADPH metabolism of Escherichia coli.J Biol Chem 279(8):6613–6619.
Sauer U,Hatzimanikatis V,Bailey JE,Hochuli M,Szyperski T,Wuthrich K.
1997.Metabolic?uxes in ribo?avin-producing Bacillus subtilis.Nat Biotechnol15(5):448–452.
Sauer U,Lasko DR,Fiaux J,Hochuli M,Glaser R,Szyperski T,Wuthrich K, Bailey JE.1999.Metabolic?ux ratio analysis of genetic and environ-mental modulations of Escherichia coli central carbon metabolism.
J Bacteriol181(21):6679–6688.
Schmidt K,Marx A,de Graaf AA,Wiechert W,Sahm H,Nielsen J,Villadsen J.1998.13C tracer experiments and metabolite balancing for metabolic ?ux analysis:Comparing two approaches.Biotechnol Bioeng58(2–3): 254–257.
Schmidt K,Nielsen J,Villadsen J.1999a.Quantitative analysis of metabolic ?uxes in Escherichia coli,using two-dimensional NMR spectroscopy and complete isotopomer models.J Biotechnol71:175–189. Schmidt K,Norregaard LC,Pedersen B,Meissner A,Nielsen JQ.1999b.
Quanti?cation of intracellular metabolic?uxes from fractional enrich-ment and13C-13C coupling constraints on the isotopomer distribution in labeled biomass components.Metab Eng1:166–179.
Schuetz R,Kuepfer L,Sauer U.2007.Systematic evaluation of objective functions for predicting intracellular?uxes in Escherichia coli.Mol Syst Biol3:119.
Shaikh AS,Tang YJ,Mukhopadhyay A,Keasling JD.2008.Isotopomer distributions in amino acids from a highly expressed protein as a proxy for those from total protein.Anal Chem80(3):886–890.
Sriram G,Fulton DB,Shanks JV.2007.Flux quanti?cation in central carbon metabolism of Catharanthus roseus hairy roots by13C labeling and comprehensive bondomer balancing.Phytochemistry68(16–18): 2243–2257.
Stephanopoulos GN,Aristidou AA,Nielsen J.1998.Metabolic engineering principles and methodologies,V ol.75.San Diego:Academic Press.
pp120–130.
&TANG ET AL.
12Mass Spectrometry Reviews DOI10.1002/mas
Suthers PF,Burgard AP,Dasika MS,Nowroozi F,Van Dien S,Keasling JD,
Maranas CD.2007.Metabolic ?ux elucidation for large-scale models using 13C labeled isotopes.Metab Eng 9(5–6):387–405.
Szyperski T.1995.Biosynthetically directed fractional 13C labeling of
proteinogenic amino acids:An ef ?cient analytical too to investigate intermediary metabolism.Eur J Biochem 232:433–448.
Szyperski T.1998.13C-NMR,MS and metabolic ?ux balancing in
biotechnology research.Q Rev Biophys 31(1):41–88.
Tang YJ,Ashcroft M,Chen D,Min G,Kim C,Murkhejee B,Larabell C,
Keasling JD,Chen FF.2007a.Charge-associated effects of fullerene derivatives on microbial structural integrity and central metabolism.Nano Lett 7(3):754–760.
Tang YJ,Chakraborty R,Martin HG,Chu J,Hazen TC,Keasling JD.2007b.
Flux analysis of central metabolic pathways in Geobacter metalliredu-cens during reduction of soluble Fe(III)-NTA.Appl Environ Microbiol 73(12):3859–3864.
Tang YJ,Hwang JS,Wemmer D,Keasling JD.2007c.The Shewanella
oneidensis MR-1?uxome under various oxygen conditions.Appl Environ Microbiol 73(3):718–729.
Tang YJ,Laidlaw D,Gani K,Keasling JD.2006.Evaluation of the effects of
various culture conditions on Cr(VI)reduction by Shewanella oneidensis MR-1in a novel high-throughput mini-bioreactor.Bio-technol Bioeng 95(1):176–184.
Tang YJ,Meadows AL,Kirby J,Keasling JD.2007d.Anaerobic central
metabolic pathways in Shewanella oneidensis MR-1reinterpreted in the light of isotopic metabolite labeling.J Bacteriol 189(3):894–901.Tang YJ,Pingitore F,Mukhopadhyay A,Phan R,Hazen TC,Keasling JD.
2007e.Pathway con ?rmation and ?ux analysis of central metabolic pathways in Desulfovibrio vulgaris Hildenborough using GC-MS and FT-ICR mass spectrometry.J Bacteriol 189(3):940–949.
Toya Y ,Ishii N,Hirasawa T,Naba M,Hirai K,Sugawara K,Igarashi S,
Shimizu K,Tomita M,Soga T.2007.Direct measurement of isotopomer of intracellular metabolites using capillary electrophoresis time-of-?ight mass spectrometry for ef ?cient metabolic ?ux analysis.J Chromatogr A 1159:134–141.
Tringe SG,Rubin EM.2005.Metagenomics:DNA sequencing of environ-mental samples.Nat Rev Genet 6(11):805–814.
Vallino JJ,Stephanopoulos G.1993.Metabolic ?ux distribution in
Corynebacterium glutamicum during growth and lysine overproduc-tion.Biotechnol Bioeng 41:633–646.
van Winden WA,Wittmann C,Heinzle E,Heijnen JJ.2002.Correcting mass
isotopomer distributions for naturally occuring isotopes.Biotechnol Bioeng 80:477–479.
Varma A,Palsson BO.1994.Metabolic ?ux balancing:Basic concepts,
scienti ?c and practical use.Bio/Technology 12:994–998.
V o TD,Palsson BO.2006.Isotopomer analysis of myocardial substrate
metabolism:A systems biology approach.Biotechnol Bioeng 95(5):972–983.
Wahl SA,Dauner M,Wiechert W.2004.New tools for mass isotopomer data
evaluation in 13C ?ux analysis:Mass isotope correction,data consistency checking,and precursor relationships.Biotechnol Bioeng 85(3):259–268.
Warnecke F,Luginbu
¨hl P,Ivanova N,Ghassemian M,Richardson TH,Stege JT,Cayouette M,McHardy AC,Djordjevic G,Aboushadi N,et al.2007.Metagenomic and functional analysis of hindgut microbiota of a wood-feeding higher termite.Nature 450(7169):560–565.
Weiss S,John GT,Klimant I,Heinzle E.2002.Modeling of mixing in 96-well
microplates observed with ?uorescence indicators.Biotechnol Prog 18(4):821–830.
Weitzel M,Wiechert W,No
¨h K.2007.The topology of metabolic isotope labeling networks.BMC Bioinformatics 8:315.
Wiechert W.2001.13C metabolic ?ux analysis.Metab Eng 3:195–206.Wiechert W,de Graaf AA.1996.In vivo stationary ?ux analysis by 13C
labeling experiments.Adv Biochem Eng Biotechnol 54:109–154.Wiechert W,de Graaf AA.1997.Bidirectional reaction steps in metabolic
networks I.Modeling and simulation of carbon isotope labeling experiments.Biotechnol Bioeng 55:101–117.
Wiechert W,Mo
¨llney M,Isermann N,Wurzel M,de Graaf AA.1999.Bidirectional reaction steps in metabolic networks:III.Explicit solution and analysis of isotopomer labeling systems.Biotechnol Bioeng 66:69–85.
Wiechert W,Mo
¨llney M,Petersen S,de Graaf AA.2001.A universal framework for 13C metabolic ?ux analysis.Metab Eng 3:265–283.
Wiechert W,No
¨h K.2005.From stationary to instationary metabolic ?ux analysis.Adv Biochem Eng Biotechnol 92:145–172.
Wiechert W,Siefke C,de Graaf AA,Marx A.1997.Bidirectional reaction
steps in metabolic networks:II.Flux estimation and statistical analysis.Biotechnol Bioeng 55(1):118–135.
Wittmann C.2007.Fluxome analysis using GC-MS.Microb Cell Factories
6:6.
Yang C,Hua Q,Shimizu K.2002.Metabolic ?ux analysis in Synechocystis
using isotope distribution from 13C-labeled glucose.Metab Eng 4(3):202–216.
Yang C,Richardson AD,Osterman A,Smith JW.2008a.Pro ?ling of central
metabolism in human cancer cells by two-dimensional NMR,GC-MS analysis,and isotopomer modeling.Metabolomics 4:13–29.
Yang TH,Frick O,Heinzle E.2008b.Hybrid optimization for 13C metabolic
?ux analysis using systems parametrized by compacti ?cation.BMC Syst Biol 2:29.
Yang TH,Wittmann C,Heinzle E.2006.Respirometric 13C ?ux analysis,Part
I:Design,construction and validation of a novel multiple reactor system using on-line membrane inlet mass spectrometry.Metab Eng 85417–431.
Young JD,Walther JL,Antoniewicz MR,Yoo H,Stephanopoulos G.2008.An
elementary metabolite unit (EMU)based method of isotopically nonstationary ?ux analysis.Biotechnol Bioeng 993686–699.
Zhao J,Shimizu K.2003.Metabolic ?ux analysis of Escherichia coli K12
grown on 13C-labeled acetate and glucose using GC-MS and powerful ?ux calculation method.J Biotechnol 101:101–117.
Yinjie J.Tang received his Ph.D.in Chemical Engineering from University of Washington in 2004and did post-doctoral work at Lawrence Berkeley National Lab.He joined Washington University in St.Louis as an assistant professor in
2008.
ADVANCES OF
13
C-BASED FLUX ANALYSIS
&
Mass Spectrometry Reviews DOI 10.1002/mas 13
Hector Garcia Martin graduated from the Universidad del Pais Vasco (UPV/EHU)in 1999with a degree in Physics and received his Ph.D.in theoretical condensed matter physics in 2004from the University of Illinois at Urbana-Champaign.He then worked on metagenomics as a post-doctoral fellow at the DOE Joint Genome Institute under the supervision of Phil Hugenholtz,developing quantitative predictive models of microbial ecosystems.Since 2007he is a Project Computational Scientist at Lawrence Berkeley Lab and the Joint Bioenergy Institute,and works on using ?ux analysis for developing quantitative models for both pure cultures and microbial communities.
Samuel Myers received B.S.in Biochemistry from California Polytechnic University,San Luis Obispo in 2007.He was a staff research associate in Department of Chemical Engineering,University of California,Berkeley.He will begin his graduate career in the fall of 2008.
Sarah Rodriguez received B.S.in Biochemistry from University of Texas at Austin 2006.She is a Ph.D.candidate in the Department of Molecular and Cellular Biology,University of California,Berkeley.She was awarded an NSF Graduate Fellowship for 2006–2009.Her major research is engineered yeast to produce the anti-malarial drug artemisinin,and advanced isotopomer ?ux analysis of Mycobacterium tuberculosis .
Edward E.K.Baidoo was born in Prestea,Ghana,1974.Upon completing his B.Sc.(Hons)(1998)and M.Sc.(1999)at University of Wolverhampton,he received his Ph.D.(2003)from Shef ?eld Hallam University under the directions of Professor R.Malcolm Clench,Dr.Robert F.Smith and Dr.Lee W.Tetler.He is currently a post-doctoral fellow at Lawrence Berkeley National Laboratory (2004–2008)in the laboratory of Professor Jay D.Keasling.
Jay D.Keasling received his B.S.in Chemistry and Biology from the University of Nebraska in 1986;his Ph.D.in Chemical Engineering from the University of Michigan in 1991;and did post-doctoral work in Biochemistry at Stanford University from 1991to 1992.Keasling joined the Department of Chemical Engineering at the University of California,Berkeley as an assistant professor in 1992,where he is currently professor.Keasling is also a professor in the Department of Bioengineering at Berkeley,a Sr.Faculty Scientist and Director of the Physical Biosciences Division at the Lawrence Berkeley National Laboratory,Director of the Berkeley Center for Synthetic Biology,and Chief Executive Of ?cer of Joint Bioenergy Institute.Dr.Keasling ’s research focuses on engineering microorganisms for environmentally friendly synthesis of small molecules or degradation of environmental contaminants.Keasling ’s laboratory has engineered bacteria and yeast to produce polymers,a precursor to the anti-malarial drug artemisinin,and advanced biofuels and soil microorganisms to accumulate uranium and to degrade nerve
agents.
&
TANG ET AL.
14Mass Spectrometry Reviews DOI 10.1002/mas
ADVANCES OF13C-BASED FLUX ANALYSIS&
Q1:Please check the grant sponsors.
Q2:Please provide citation in the text for the
following references:Allen et al.,2007,
Blank et al.,2005b,Dauner et al.,2002,
Fischer and Sauer,2003b,Forbes et al.,
2006,Fuhrer et al.,2005,Sriram et al.,
2007,Tang et al.,2007a,Yang et al.,2002.
Q3:Please provide the chapter title.
Q4:Please provide the volume number.
Q5:Please provide the page range.
Mass Spectrometry Reviews DOI10.1002/mas15
111R I V E R S T R E E T,H O B O K E N,N J07030
E L E C T R O N I C P R O O
F C H E C K L I S T,M A S S S P E C T R O M E T R Y R E V I E W S
***IMMEDIATE RESPONSE REQUIRED***
Please follow these instructions to avoid delay of publication.
READ PROOFS CAREFULLY
?This will be your only chance to review these proofs.
?Please note that the volume and page numbers shown on the proofs are for position only.
ANSWER ALL QUERIES ON PROOFS (Queries for you to answer are attached as the last page of your proof.)?Mark all corrections directly on the proofs. Note that excessive author alterations may ultimately result in delay of publication and extra costs may be charged to you.
CHECK FIGURES AND TABLES CAREFULLY (Color figures will be sent under separate cover.)
?Check size, numbering, and orientation of figures.
?All images in the PDF are downsampled (reduced to lower resolution and file size) to facilitate Internet delivery.
These images will appear at higher resolution and sharpness in the printed article.
?Review figure legends to ensure that they are complete.
?Check all tables. Review layout, title, and footnotes.
COMPLETE REPRINT ORDER FORM
?Fill out the attached reprint order form. It is important to return the form even if you are not ordering reprints. You may, if you wish, pay for the reprints with a credit card. Reprints will be mailed only after your article appears in
print. This is the most opportune time to order reprints. If you wait until after your article comes off press, the
reprints will be considerably more expensive.
RETURN PROOFS
REPRINT ORDER FORM
CTA (If you have not already signed one)
RETURN WITHIN 48 HOURS OF RECEIPT VIA FAX TO Hattie Heavner AT 201-748-6052 QUESTIONS? Hattie Heavner, Production Editor
Phone: 201-748-6047
E-mail: hheavner@https://www.360docs.net/doc/c88726505.html,
Refer to journal acronym and article production number
(i.e., MAS 00-001 for Mass Spectrometry Reviews ms 00-001).
COPYRIGHT TRANSFER AGREEMENT
Re: Manuscript entitled____________________________________________________________________________________________________________________________________________________________________(the “Contribution”)for publication in ______________________________________________________________________(the “Journal”)published by Wiley Periodicals, Inc. (“Wiley”).Dear Contributor(s):
Thank you for submitting your Contribution for publication. In order to expedite the editing and publishing process and enable Wiley to disseminate your work to the fullest extent, we need to have this Copyright Transfer Agreement signed and returned to us as soon as possible. If the Contribution is not accepted for publication this Agreement shall be null and void.
A.COPYRIGHT
1.The Contributor assigns to Wiley, during the full term of copyright and any extensions or renewals of that term, all
copyright in and to the Contribution, including but not limited to the right to publish, republish, transmit, sell, distribute and otherwise use the Contribution and the material contained therein in electronic and print editions of the Journal and in derivative works throughout the world, in all languages and in all media of expression now known or later developed, and to license or permit others to do so.
2.Reproduction, posting, transmission or other distribution or use of the Contribution or any material contained
therein, in any medium as permitted hereunder, requires a citation to the Journal and an appropriate credit to Wiley as Publisher, suitable in form and content as follows: (Title of Article, Author, Journal Title and Volume/Issue Copyright ? [year] Wiley Periodicals, Inc. or copyright owner as specified in the Journal.)B.RETAINED RIGHTS
Notwithstanding the above, the Contributor or, if applicable, the Contributor’s Employer, retains all proprietary rights other than copyright, such as patent rights, in any process, procedure or article of manufacture described in the Contribution, and the right to make oral presentations of material from the Contribution.C.OTHER RIGHTS OF CONTRIBUTOR
Wiley grants back to the Contributor the following:
1.The right to share with colleagues print or electronic “preprints” of the unpublished Contribution, in form and
content as accepted by Wiley for publication in the Journal. Such preprints may be posted as electronic files on the Contributor’s own website for personal or professional use, or on the Contributor’s internal university or corporate networks/intranet, or secure external website at the Contributor’s institution, but not for commercial sale or for any systematic external distribution by a third party (e.g., a listserve or database connected to a public access server).Prior to publication, the Contributor must include the following notice on the preprint: “This is a preprint of an article accepted for publication in [Journal title] ? copyright (year) (copyright owner as specified in the Journal)”.After publication of the Contribution by Wiley, the preprint notice should be amended to read as follows: “This is a preprint of an article published in [include the complete citation information for the final version of the Contribution as published in the print edition of the Journal]”, and should provide an electronic link to the Journal’s WWW site,located at the following Wiley URL: https://www.360docs.net/doc/c88726505.html,/. The Contributor agrees not to update the preprint or replace it with the published version of the Contribution.2.The right, without charge, to photocopy or to transmit online or to download, print out and distribute to a colleague a
copy of the published Contribution in whole or in part, for the colleague’s personal or professional use, for the
111 River Street
Hoboken, NJ 07030, USA 201.748.6047
FAX 201.748.6052
Date:
To:
advancement of scholarly or scientific research or study, or for corporate informational purposes in accordance with Paragraph D.2 below.
3.The right to republish, without charge, in print format, all or part of the material from the published Contribution in
a book written or edited by the Contributor.
4.The right to use selected figures and tables, and selected text (up to 250 words, exclusive of the abstract) from the
Contribution, for the Contributor’s own teaching purposes, or for incorporation within another work by the
Contributor that is made part of an edited work published (in print or electronic format) by a third party, or for
presentation in electronic format on an internal computer network or external website of the Contributor or the
Contributor’s employer.
5.The right to include the Contribution in a compilation for classroom use (course packs) to be distributed to students
at the Contributor’s institution free of charge or to be stored in electronic format in datarooms for access by students at the Contributor’s institution as part of their course work (sometimes called “electronic reserve rooms”) and for in-house training programs at the Contributor’s employer.
D.CONTRIBUTIONS OWNED BY EMPLOYER
1.If the Contribution was written by the Contributor in the course of the Contributor’s employment (as a “work-made-
for-hire” in the course of employment), the Contribution is owned by the company/employer which must sign this Agreement (in addition to the Contributor’s signature), in the space provided below. In such case, the
company/employer hereby assigns to Wiley, during the full term of copyright, all copyright in and to the
Contribution for the full term of copyright throughout the world as specified in paragraph A above.
2.In addition to the rights specified as retained in paragraph B above and the rights granted back to the Contributor
pursuant to paragraph C above, Wiley hereby grants back, without charge, to such company/employer, its
subsidiaries and divisions, the right to make copies of and distribute the published Contribution internally in print
format or electronically on the Company’s internal network. Upon payment of Wiley’s reprint fee, the institution may distribute (but not resell) print copies of the published Contribution externally. Although copies so made shall not be available for individual re-sale, they may be included by the company/employer as part of an information package included with software or other products offered for sale or license. Posting of the published Contribution by the
institution on a public access website may only be done with Wiley’s written permission, and payment of any
applicable fee(s).
https://www.360docs.net/doc/c88726505.html,ERNMENT CONTRACTS
In the case of a Contribution prepared under U.S. Government contract or grant, the U.S. Government may reproduce, without charge, all or portions of the Contribution and may authorize others to do so, for official U.S. Government purposes only, if the U.S. Government contract or grant so requires. (U.S. Government Employees: see note at end.)
F.COPYRIGHT NOTICE
The Contributor and the company/employer agree that any and all copies of the Contribution or any part thereof
distributed or posted by them in print or electronic format as permitted herein will include the notice of copyright as stipulated in the Journal and a full citation to the Journal as published by Wiley.
G.CONTRIBUTOR’S REPRESENTATIONS
The Contributor represents that the Contribution is the Contributor’s original work. If the Contribution was prepared jointly, the Contributor agrees to inform the co-Contributors of the terms of this Agreement and to obtain their signature to this Agreement or their written permission to sign on their behalf. The Contribution is submitted only to this Journal and has not been published before, except for “preprints” as permitted above. (If excerpts from copyrighted works owned by third parties are included, the Contributor will obtain written permission from the copyright owners for all uses as set forth in Wiley’s permissions form or in the Journal’s Instructions for Contributors, and show credit to the sources in the Contribution.) The Contributor also warrants that the Contribution contains no libelous or unlawful statements, does not infringe upon the rights (including without limitation the copyright, patent or trademark rights) or the privacy of others, or contain material or instructions that might cause harm or injury.
CHECK ONE:
[____] Contributor-owned work
________________________________________________________ Type or print name and title
______________________________________ _________________ Co-contributor’s signature Date ________________________________________________________ Type or print name and title
ATTACHED ADDITIONAL SIGNATURE PAGE AS NECESSARY
[____] Company/Institution-owned work
(made-for-hire in the course of employment)
______________________________________ _________________ Authorized signature of Employer Date
[____] U.S. Government work
[____] U.K. Government work (Crown Copyright)______________________________________ _________________ Contributor’s signature Date ______________________________________ _________________
Company or Institution (Employer-for-Hire) Date
111 R IVER S TREET, H OBOKEN, NJ 07030 Telephone Number:?Facsimile Number:
To:Hattie Heavner
Company:
Phone: 201-748-6047
Fax: 201-748-6052
From:
Date:
Pages including this cover
page:
Message:
Re:
英语中的比较级与最高级 详解
比较级与最高级 1.as...as 与(not) as(so)...as as...as...句型中,as的词性 第一个as是副词,用在形容词和副词的原级前,常译为“同样地”。第二个as是连词,连接与前面句子结构相同的一个句子(相同部分常省略),可译为“同..... He is as tall as his brother is (tall) . (后面的as 为连词) 只有在否定句中,第一个as才可换为so 改错: He is so tall as his brother.(X) 2.在比较状语从句中,主句和从句的句式结构一般是相同的 与as...as 句式中第二个as一样,than 也是连词。as和than这两个连词后面的从句的结构与前面的句子大部分情况下结构是相同的,相同部分可以省略。 He picked more apples than she did. 完整的表达为: He picked more apples than she picked apples. 后而的picked apples和前面相同,用did 替代。 He walked as slowly as she did.完整表达为: He walked as slowly as she walked slowly. she后面walked slowly与前面相同,用did替代。
3.谓语的替代 在as和than 引导的比较状语从句中,由于句式同前面 主句相同,为避免重复,常把主句中出现而从句中又出现的动词用do的适当形式来代替。 John speaks German as fluently as Mary does. 4.前后的比较对象应一致 不管后面连词是than 还是as,前后的比较对象应一致。The weather of Beijing is colder than Guangzhou. x than前面比较对象是“天气”,than 后面比较对象是“广州”,不能相比较。应改为: The weather of Bejing is colder than that of Guangzhou. 再如: His handwriting is as good as me. 应改为: His handwriting is as good as mine. 5.可以修饰比较级的词 常用来修饰比较级的词或短语有: Much,even,far,a little,a lot,a bit,by far,rather,any,still,a great deal等。 by far的用法: 用于强调,意为“...得多”“最最...”“显然”等,可修饰形容词或副词的比较级和最高级,通常置于其后,但是若比较级或最高级前有冠词,则可置于其前或其后。
人教版(新目标)初中英语形容词与副词的比较级与最高级
人教版(新目标)初中英语形容词与副词的比较级与最高级 (一)规则变化: 1.绝大多数的单音节和少数双音节词,加词尾-er ,-est tall—taller—tallest 2.以不发音的e结尾的单音节词和少数以-le结尾的双音节词只加-r,-st nice—nicer—nicest , able—abler—ablest 3.以一个辅音字母结尾的重读闭音节词或少数双音节词,双写结尾的辅音字母,再加-er,-est big—bigger—biggest 4.以辅音字母加y结尾的双音节词,改y为i再加-er,-est easy—easier—easiest 5.少数以-er,-ow结尾的双音节词末尾加-er,-est clever—cleverer—cleverest, narrow—narrower—narrowest 6.其他双音节词和多音节词,在前面加more,most来构成比较级和最高级 easily—more easily—most easily (二)不规则变化 常见的有: good / well—better—best ; bad (ly)/ ill—worse—worst ; old—older/elder—oldest/eldest many / much—more—most ; little—less—least ; far—farther/further—farthest/furthest
用法: 1.原级比较:as + adj./adv. +as(否定为not so/as + adj./adv. +as)当as… as中间有名字时,采用as + adj. + a + n.或as + many / much + n. This is as good an example as the other is . I can carry as much paper as you can. 表示倍数的词或其他程度副词做修饰语时放在as的前面 This room is twice as big as that one. 倍数+as+adj.+as = 倍数+the +n.+of Your room is twice as larger as mine. = Your room is twice the size of mine. 2.比较级+ than 比较级前可加程度状语much, still, even, far, a lot, a little, three years. five times,20%等 He is three years older than I (am). 表示“(两个中)较……的那个”时,比较级前常加the(后面有名字时前面才能加冠词) He is the taller of the two brothers. / He is taller than his two brothers. Which is larger, Canada or Australia? / Which is the larger country, Canada or Australia? 可用比较级形式表示最高级概念,关键是要用或或否定词等把一事物(或人)与其他同类事物(或人)相分离 He is taller than any other boy / anybody else.
英语中的比较级和最高级
大多数形容词有三种形式,原级,比较级和最高级, 以表示形容词说明的性质在程度上的不同。 形容词的原级: 形容词的原级形式就是词典中出现的形容词的原形。例如: poor tall great glad bad 形容词的比较级和最高级: 形容词的比较级和最高级形式是在形容词的原级形式的基础上变化的。分为规则变化和不规则变化。 规则变化如下: 1) 单音节形容词的比较级和最高级形式是在词尾加 -er 和 -est 构成。 great (原级) (比较级) (最高级) 2) 以 -e 结尾的单音节形容词的比较级和最高级是在词尾加 -r 和 -st 构成。wide (原级) (比较级) (最高级) 3)少数以-y, -er, -ow, -ble结尾的双音节形容词的比较级和最高级是在词尾加 -er 和 -est 构成。 clever(原级) (比较级) (最高级) 4) 以 -y 结尾,但 -y 前是辅音字母的形容词的比较级和最高级是把 -y 去掉,加上 -ier 和-est 构成. happy (原形) (比较级) (最高级) 5) 以一个辅音字母结尾其前面的元音字母发短元音的形容词的比较级和最高级是双写该辅音字母然后再加 -er和-est。 big (原级) (比较级) (最高级) 6) 双音节和多音节形容词的比较级和最高级需用more 和 most 加在形容词前面来构成。 beautiful (原级) (比较级) (比较级) difficult (原级) (最高级) (最高级) 常用的不规则变化的形容词的比较级和最高级: 原级------比较级------最高级 good------better------best many------more------most much------more------most bad------worse------worst far------farther, further------farthest, furthest 形容词前如加 less 和 least 则表示"较不"和"最不 形容词比较级的用法: 形容词的比较级用于两个人或事物的比较,其结构形式如下: 主语+谓语(系动词)+ 形容词比较级+than+ 对比成分。也就是, 含有形容词比较级的主句+than+从句。注意从句常常省去意义上和主句相同的部分, 而只剩下对比的成分。
英语比较级和最高级的用法归纳
英语比较级和最高级的用法归纳 在学习英语过程中,会遇到很多的语法问题,比如比较级和最高级的用法,对于 这些语法你能够掌握吗?下面是小编整理的英语比较级和最高级的用法,欢迎阅读! 英语比较级和最高级的用法 一、形容词、副词的比较级和最高级的构成规则 1.一般单音节词和少数以-er,-ow结尾的双音节词,比较级在后面加-er,最高级 在后面加-est; (1)单音节词 如:small→smaller→smallest short→shorter→shortest tall→taller→tallest great→greater→greatest (2)双音节词 如:clever→cleverer→cleverest narrow→narrower→narrowest 2.以不发音e结尾的单音节词,比较在原级后加-r,最高级在原级后加-st; 如:large→larger→largest nice→nicer→nicest able→abler→ablest 3.在重读闭音节(即:辅音+元音+辅音)中,先双写末尾的辅音字母,比较级加-er,最高级加-est; 如:big→bigger→biggest hot→hotter→hottest fat→fatter→fattest 4.以“辅音字母+y”结尾的双音节词,把y改为i,比较级加-er,最高级加-est; 如:easy→easier→easiest heavy→heavier→heaviest busy→busier→busiest happy→happier→happiest 5.其他双音节词和多音节词,比较级在前面加more,最高级在前面加most; 如:bea utiful→more beautiful→most beautiful different→more different→most different easily→more easily→most easily 注意:(1)形容词最高级前通常必须用定冠词 the,副词最高级前可不用。 例句: The Sahara is the biggest desert in the world. (2) 形容词most前面没有the,不表示最高级的含义,只表示"非常"。 It is a most important problem. =It is a very important problem.
英语比较级和最高级的用法
More than的用法 A. “More than+名词”表示“不仅仅是” 1)Modern science is more than a large amount of information. 2)Jason is more than a lecturer; he is a writer, too. 3) We need more than material wealth to build our country.建设我们国家,不仅仅需要物质财富. B. “More than+数词”含“以上”或“不止”之意,如: 4)I have known David for more than 20 years. 5)Let's carry out the test with more than the sample copy. 6) More than one person has made this suggestion. 不止一人提过这个建议. C. “More than+形容词”等于“很”或“非常”的意思,如: 7)In doing scientific experiments, one must be more than careful with the instruments. 8)I assure you I am more than glad to help you. D. more than + (that)从句,其基本意义是“超过(=over)”,但可译成“简直不”“远非”.难以,完全不能(其后通常连用情态动词can) 9) That is more than I can understand . 那非我所能懂的. 10) That is more than I can tell. 那事我实在不明白。 11) The heat there was more than he could stand. 那儿的炎热程度是他所不能忍受的 此外,“more than”也在一些惯用语中出现,如: more...than 的用法 1. 比……多,比……更 He has more books than me. 他的书比我多。 He is more careful than the others. 他比其他人更仔细。 2. 与其……不如 He is more lucky than clever. 与其说他聪明,不如说他幸运。 He is more (a)scholar than (a)teacher. 与其说他是位教师,不如说他是位学者。 注:该句型主要用于同一个人或物在两个不同性质或特征等方面的比较,其中的比较级必须用加more 的形式,不能用加词尾-er 的形式。 No more than/not more than 1. no more than 的意思是“仅仅”“只有”“最多不超过”,强调少。如: --This test takes no more than thirty minutes. 这个测验只要30分钟。 --The pub was no more than half full. 该酒吧的上座率最多不超过五成。-For thirty years,he had done no more than he (had)needed to. 30年来,他只干了他需要干的工作。 2. not more than 为more than (多于)的否定式,其意为“不多于”“不超过”。如:Not more than 10 guests came to her birthday party. 来参加她的生日宴会的客人不超过十人。 比较: She has no more than three hats. 她只有3顶帽子。(太少了) She has not more than three hats. 她至多有3顶帽子。(也许不到3顶帽子) I have no more than five yuan in my pocket. 我口袋里的钱最多不过5元。(言其少) I have not more than five yuan in my pocket. 我口袋里的钱不多于5元。(也许不到5元) more than, less than 的用法 1. (指数量)不到,不足 It’s less than half an hour’s drive from here. 开车到那里不到半个钟头。 In less than an hour he finished the work. 没要上一个小时,他就完成了工作。 2. 比……(小)少 She eats less than she should. 她吃得比她应该吃的少。 Half the group felt they spent less than average. 半数人觉得他们的花费低于平均水平。 more…than,/no more than/not more than (1)Mr.Li is ________ a professor; he is also a famous scientist. (2)As I had ________ five dollars with me, I couldn’t afford the new jacket then. (3)He had to work at the age of ________ twelve. (4)There were ________ ten chairs in the room.However, the number of the children is twelve. (5)If you tel l your father what you’ve done, he’ll be ________ angry. (6)-What did you think of this novel? -I was disappointed to find it ________ interesting ________ that one. 倍数表达法 1. “倍数+形容词(或副词)的比较级+than+从句”表示“A比B大(长、高、宽等)多少倍” This rope is twice longer than that one.这根绳是那根绳的三倍(比那根绳长两倍)。The car runs twice faster than that truck.这辆小车的速度比那辆卡车快两倍(是那辆卡车的三倍)。 2. “倍数+as+形容词或副词的原级+as+从句”表示“A正好是B的多少倍”。
初中英语比较级和最高级讲解与练习
初中英语比较级和最高级讲解与练习 形容词比较级和最高级 一.绝大多数形容词有三种形式,原级,比较级和最高级, 以表示形容词说明的性质在程度上的不同。 1. 形容词的原级: 形容词的原级形式就是词典中出现的形容词的原形。例如: poor tall great glad bad 2. 形容词的比较级和最高级: 形容词的比较级和最高级形式是在形容词的原级形式的基 础上变化的。分为规则变化和不规则变化。 二.形容词比较级和最高级规则变化如下: 1) 单音节形容词的比较级和最高级形式是在词尾加-er 和-est 构成。 great (原级) greater(比较级) greatest(最高级) 2) 以-e 结尾的单音节形容词的比较级和最高级是在词尾加-r 和-st 构成。 wide (原级) wider (比较级) widest (最高级) 3) 少数以-y, -er, -ow, -ble结尾的双音节形容词的比较级和最高级是在词尾加 -er 和-est构成。 clever(原级) cleverer(比较级) cleverest(最高级), slow(原级) slower(比较级) slowest (最高级) 4) 以-y 结尾,但-y 前是辅音字母的形容词的比较级和最高级是把-y 去掉,加上-ier 和-est 构成. happy (原形) happier (比较级) happiest (最高级) 5) 以一个辅音字母结尾其前面的元音字母发短元音的形容词的比较级和最高级是双写该 辅音字母然后再加-er和-est。 原形比较级最高级原形比较级最高级 big bigger biggest hot hotter hottest red redder reddest thin thinner thinnest 6) 双音节和多音节形容词的比较级和最高级需用more 和most 加在形容词前面来构 成。 原形比较级最高级 careful careful more careful most careful difficult more difficult most difficult delicious more delicious most delicious 7)常用的不规则变化的形容词的比较级和最高级: 原级比较级最高级 good better best 好的 well better best 身体好的 bad worse worst 坏的 ill worse worst 病的 many more most 许多 much more most 许多 few less least 少数几个 little less least 少数一点儿 (little littler littlest 小的) far further furthest 远(指更进一步,深度。亦可指更远) far farther farthest 远(指更远,路程)
英语比较级和最高级
形容词比较级和最高级的形式 一、形容词比较级和最高级的构成 形容词的比较级和最高级变化形式规则如下 构成法原级比较级最高级 ①一般单音节词末尾加 er 和 est strong stronger strongest ②单音节词如果以 e结尾,只加 r 和 st strange stranger strangest ③闭音节单音节词如末尾只有一个辅音字母, 须先双写这个辅音字母,再加 er和 est sad big hot sadder bigger hotter saddest biggest hottest ④少数以 y, er(或 ure), ow, ble结尾的双音节词, 末尾加 er和 est(以 y结尾的词,如 y前是辅音字母, 把y变成i,再加 er和 est,以 e结尾的词仍 只加 r和 st) angry Clever Narrow Noble angrier Cleverer narrower nobler angriest cleverest narrowest noblest ⑤其他双音节和多音节词都在前面加单词more和most different more different most different 1) The most high 〔A〕mountain in 〔B〕the world is Mount Everest,which is situated 〔C〕in Nepal and is twenty nine thousand one hundred and fourty one feet high 〔D〕 . 2) This house is spaciouser 〔A〕than that 〔B〕white 〔C〕one I bought in Rapid City,South Dakota 〔D〕last year. 3) Research in the social 〔A〕sciences often proves difficulter 〔B〕than similar 〔C〕work in the physical 〔D〕sciences. 二、形容词比较级或最高级的特殊形式:
高中英语的比较级和最高级用法总结
比较级和最高级 1.在形容词词尾加上―er‖ ―est‖ 构成比较级、最高级: bright(明亮的)—brighter—brightest broad(广阔的)—broader—broadest cheap(便宜的)—cheaper—cheapest clean(干净的)—cleaner—cleanest clever(聪明的)—cleverer—cleverest cold(寒冷的)—colder—coldest cool(凉的)—cooler—coolest dark(黑暗的)—darker—darkest dear(贵的)—dearer—dearest deep(深的)—deeper—deepest fast(迅速的)—faster—fastest few(少的)—fewer—fewest great(伟大的)—greater—greatest hard(困难的,硬的)—harder—hardest high(高的)—higher—highest kind(善良的)—kinder—kindest light(轻的)—lighter—lightest long(长的)—longer—longest loud(响亮的)—louder—loudest low(低的)—lower—lowest near(近的)—nearer—nearest new(新的)—newer—newest poor(穷的)—poorer—poorest quick(快的)—quicker—quickest quiet(安静的)—quieter—quietest rich(富裕的)—richer—richest short(短的)—shorter—shortest slow(慢的)—slower—slowest small(小的)—smaller—smallest smart(聪明的)—smarter—smartest soft(柔软的)—softer—softest strong(强壮的)—stronger—strongest sweet(甜的)—sweeter—sweetest tall(高的)-taller-tallest thick(厚的)—thicker—thickest warm(温暖的)—warmer—warmest weak(弱的)—weaker—weakest young(年轻的)—younger—youngest 2.双写最后一个字母,再加上―er‖ ―est‖构成比较级、最高级: big(大的)—bigger—biggest fat(胖的)—fatter—fattest hot(热的)—hotter—hottest red(红的)—redder—reddest sad(伤心的)—sadder—saddest thin(瘦的)—thinner—thinnest wet(湿的)—wetter—wettest mad(疯的)—madder—maddest 3.以不发音的字母e结尾的形容词,加上―r‖ ―st‖ 构成比较级、最高级:able(能干的)—abler—ablest brave(勇敢的)—braver—bravest close(接近的)—closer—closest fine(好的,完美的)—finer—finest large(巨大的)—larger—largest late(迟的)—later—latest nice(好的)—nicer—nicest ripe(成熟的)—riper—ripest
(完整版)初中英语比较级和最高级的用法
英语语法---比较级和最高级的用法 在英语中通常用下列方式表示的词:在形容词或副词前加more(如 more natural,more clearly )或加后缀 -er(newer,sooner )。典型的是指形容词或副词所表示的质、量或关系的增加。英语句子中,将比较两个主体的方法叫做“比较句型”。其中,像“A比B更……”的表达方式称为比较级;而“A最……”的表达方式则称为最高级。组成句子的方式是将形容词或副词变化成比较级或最高级的形态。 一、形容词、副词的比较级和最高级的构成规则 1.一般单音节词和少数以-er,-ow结尾的双音节词,比较级在后面加-er,最高级在后面加-est; (1)单音节词 如:small→smaller→smallest short→shorter→shortest tall→taller→tallest great→greater→greatest (2)双音节词 如:clever→cleverer→cleverest narrow→narrower→narrowest 2.以不发音e结尾的单音节词,比较在原级后加-r,最高级在原级后加-st; 如:large→larger→largest nice→nicer→nicest able→abler→ablest 3.在重读闭音节(即:辅音+元音+辅音)中,先双写末尾的辅音字母,比较级加-er,最高级加-est; 如:big→bigger→biggest hot→hotter→hottest fat→fatter→fattest 4.以“辅音字母+y”结尾的双音节词,把y改为i,比较级加-er,最高级加-est; 如:easy→easier→easiest heavy→heavier→heaviest busy→busier→busiest happy→happier→happiest 5.其他双音节词和多音节词,比较级在前面加more,最高级在前面加most; 如:beautiful→more beautiful→most beautiful different→more different→most different easily→more easily→most easily
初中英语形容词比较级和最高级讲解与练习
初中英语形容词比较级和最高级讲解与练习 形容词比较级和最高级 绝大多数形容词有三种形式,原级,比较级和最高级, 以表示形容词说明的性质在程度上的不同。 形容词的原级: 形容词的原级形式就是词典中出现的形容词的原形。 例如: poor tall great glad bad 形容词的比较级和最高级: 形容词的比较级和最高级形式是在形容词的原级形式的基础上变化的。分为规则变化和不规则变化。 规则变化如下: 1) 单音节形容词的比较级和最高级形式是在词尾加 -er 和 -est 构成。 great (原级) (比较级) (最高级) 2) 以 -e 结尾的单音节形容词的比较级和最高级是在词尾加 -r 和 -st 构成。 wide (原级) (比较级) (最高级) 3)少数以-y, -er, -ow, -ble结尾的双音节形容词的比较级和最高级是在词尾加-er 和-est 构成。 clever(原级) (比较级) (最高级) 4) 以 -y 结尾,但 -y 前是辅音字母的形容词的比较级和最高级是把 -y 去掉,加上 -ier 和-est 构成. happy (原形) (比较级) (最高级) 5) 以一个辅音字母结尾其前面的元音字母发短元音的形容词的比较级和最高级是双写该辅音 字母然后再加-er和-est。 big (原级) (比较级) (最高级) 6) 双音节和多音节形容词的比较级和最高级需用more 和 most 加在形容词前面来构成。 beautiful (原级) (比较级) (比较级) difficult (原级) (最高级) (最高级) 常用的不规则变化的形容词的比较级和最高级: 原级------比较级------最高级 good------better------best many------more------most much------more------most bad------worse------worst far------farther, further------farthest, furthest 形容词前如加 less 和 least 则表示"较不"和"最不"
(完整)初中英语比较级和最高级
◇下列形容词和副词没有比较级和最高 (即表示“最高程度”或“绝对状态”的形容词和副词没有比较级和最高级) empty, wrong, perfect, unique, extreme, excellent, favourite, true, right, correct, extremely ... 形容词副词比较级最高级使用注意事项 ◇比较应在同类事物之间进行。 误:Your English is better than me. 正:Your English is better than mine. ◇比较级前可以有一个表示程度的状语,最常见的三大修饰词是:a little, much, even。 以下单词也可用来修饰:any, far, still, a lot, yet, rather。 My sister is a little taller than me. Their house is much larger than ours. 另外,名词短语也可修饰比较级,说明程度。 I’m three years older than he. 特别提醒:very, quite, too不可修饰比较级。 ◇避免重复使用比较级。 误:He is more kinder to small animals than I. 正:He is much kinder to small animals than I. 误:He is more cleverer than his brother. 正:He is cleverer than his brother. ◇比较要符合逻辑,在同一范围内比较时,避免将主语含在比较对象中,这时需使用other来排除自身。 误:China is larger that any country in Asia. 正:China is larger than any other country in Asia. 误:John studies harder than any student in his class. 正:John studies harder than any other student in his class. 正:John studies harder than any of the other students in his class. 正:John studies harder than anyone else in his class. ◇比较要遵循前后一致的原则,注意前后呼应。 The population of Shanghai is larger than that of Beijing. It is easier to make a plan than to carry it out. ◇序数词通常只修饰最高级。 Africa is the second largest continent. The Yellow River is the second longest river in China. This is the third most popular song of Michael Jackson. ◇为避免重复,我们通常用that, those, one, ones代替前面出现的名词。that 代替可数名词单数和不可数名词,those代替可数名词复数。one既可指人又可指物,只能 代替可数名词。 The weather in China is different from that in America. The book on the table is more interesting than that(或the one)on the desk. A box made of steel is stronger than one made of wood. 误:In winter, the weather of Beijing is colder than it of Shanghai. 正:In winter, the weather of Beijing is colder than that of Shanghai. ◇“否定词 + 比较级”相当于最高级。
英语比较级和最高级
英语比较级和最高级
一、比较级和最高级的讲解 变化规则 1.一般单音节词和少数以-er,-ow结尾的双音节词,比较级在后面加-er,最高级在后面加-est; (1)单音节词 如:small→smaller→smallest short→shorter→shortest tall→taller→tallest great→greater→greatest (2)双音节词 如:clever→cleverer→cleverest n arrow→narrower→narrowest 2.以不发音e结尾的单音节词,比较在原级后加-r,最高级在原级后加-st; 如:large→larger→largest nice→nicer→nicest able→abler→ablest 3.在重读闭音节(即:辅音+元音+辅音) 中,先双写末尾的辅音字母,比较级加-er,最高级加-est;
如:big→bigger→biggest hot→hotter→hottest fat→fatter→fattest 4.以“辅音字母+y”结尾的双音节词,把y 改为i,比较级加-er,最高级加-est; 如:easy→easier→easiest heavy→heavier→heaviest busy→busier→busiest happy→happier→happiest 5.其他双音节词和多音节词,比较级在前面加more,最高级在前面加most; 如:beautiful→more beautiful→most beautiful different→more different→most d ifferent easily→more easily→most easily 注意: (1)形容词最高级前通常必须用定冠词the,副词最高级前可不用。 例句:The Sahara is the biggest desert in the world. (2)形容词most前面没有the,不表示最高级的含义,只表示"非常"。
英语比较级与最高级的解释和种类
形容词的比较级和最高级: 绝大多数形容词有三种形式,原级,比较级和最高级, 以表示形容词说明的性质在程度上的不同。 形容词的原级: 形容词的原级形式就是词典中出现的形容词的原形。例如: poor tall great glad bad 形容词的比较级和最高级: 形容词的比较级和最高级形式是在形容词的原级形式的基础上变化的。分为规则变化和不规则变化。 规则变化如下: 1) 单音节形容词的比较级和最高级形式是在词尾加-er 和-est 构成。 great (原级) greater(比较级) greatest(最高级) 2) 以-e 结尾的单音节形容词的比较级和最高级是在词尾加-r 和-st 构成。 3)少数以-y, -er, -ow, -ble结尾的双音节形容词的比较级和最高级是在词尾加-er 和-est 构成。 clever(原级) cleverer(比较级) cleverest(最高级) 4) 以-y 结尾,但-y 前是辅音字母的形容词的比较级和最高级是把-y 去掉,加上-ier 和-est 构成. happy (原形) happier (比较级) happiest (最高级) 5) 以一个辅音字母结尾其前面的元音字母发短元音的形容词的比较级和最高级是双写该辅音字母然后再加-er和-est。 big (原级) bigger (比较级) biggest (最高级) ) 双音节和多音节形容词的比较级和最高级需用more 和most 加在形容词前面来构成。beautiful (原级)? difficult (原级) more beautiful (比较级) more difficult (比较级) 常用的不规则变化的形容词的比较级和最高级: 原级比较级最高级 good better best many more most much more most little less least ill worse worst far farther(further) farthest(furthest) 形容词前如加less 和lest 则表示"较不"和"最不" important 重要 less important 较不重要 lest important 最不重要 形容词比较级的用法: 形容词的比较级用于两个人或事物的比较,其结构形式如下: 主语+谓语(系动词)+ 形容词比较级+than+ 对比成分。也就是, 含有形容词比较级的主句+than+从句。注意从句常常省去意义上和主句相同的部分, 而只剩下对比的成分。 It is warmer today than it was yesterday. 今天的天气比昨天暖和。 This picture is more beautiful than that one.
英语单词的比较级和最高级
little:形容词比较级littler/less/lesser 形容词最高级littlest/least 副词比较级less 副词最高级least far:形容词比较级farther/further 形容词最高级farthest/furthest 副词比较级farther/further 副词最高级farthest/furthest well:形容词比较级better 形容词最高级best 副词比较级better 副词最高级best ill:形容词比较级worse 形容词最高级worst 副词比较级worse 副词最高级worst many:形容词比较级more 形容词最高级most bad:形容词比较级worse 形容词最高级worst good:形容词比较级better 形容词最高级:best old:形容词比较级:older/elder 形容词最高级:oldest/eldest good:比较级better 最高级the best hot:比较级hotter 最高级the hottest heavy:比较级heavier 最高级the heaviest fine:比较级finer 最高级the finest exciting:比较级more exciting 最高级the most exciting bad:比较级worse 最高级the worst creative:比较级more creative 最高级the most creative boring:比较级more boring 最高级the most boring far:比较级farther/further 最高级the farthest/the furthest near:比较级nearer
英语比较级和最高级的用法
英语比较级和最高级的用法 一、形容词、副词的比较级和最高级的构成规则 1.一般单音节词和少数以-er,-ow结尾的双音节词,比较级在后面加-er,最高级在后面加-est; (1)单音节词 如:small→smaller→smallest short→shorter→shortest tall→taller→tallest great→greater→greatest (2)双音节词 如:clever→cleverer→cleverest narrow→narrower→narrowest 2.以不发音e结尾的单音节词,比较在原级后加-r,最高级在原级后加-st; 如:large→larger→largest nice→nicer→nicest able→abler→ablest 3.在重读闭音节(即:辅音+元音+辅音)中,先双写末尾的辅音字母,比较级加-er,最高级加-est; 如:big→bigger→biggest hot→hotter→hottest fat→fatter→fattest 4.以“辅音字母+y”结尾的双音节词,把y改为i,比较级加-er,最高级加-est; 如:easy→easier→easiest heavy→heavier→heaviest busy→busier→busiest happy→happier→happiest 5.其他双音节词和多音节词,比较级在前面加more,最高级在前面加most; 如:beautiful→more beautiful→most beautiful different→more different→most different easi ly→more easily→most easily 注意:(1)形容词最高级前通常必须用定冠词the,副词最高级前可不用。 例句:The Sahara is the biggest desert in the world. (2) 形容词most前面没有the,不表示最高级的含义,只表示"非常"。 It is a most important problem. =It is a very important problem. 6.有少数形容词、副词的比较级和最高级是不规则的,必须熟记。 如:good→better→best well→better→best bad→worse→worst ill→worse→worst
