Epigenetics interaction of DNA methylation and chromatin

Review
Epigenetics:interaction of DNA methylation and chromatin q
Mitsuyoshi Nakao *
Department of Tumor Genetics and Biology,Kumamoto University School of Medicine,2-2-1Honjo,Kumamoto 860-0811,Japan
Received 25June 2001;received in revised form 28August 2001;accepted 14September 2001
Received by A.J.van Wijnen
Abstract
Epigenetic regulation is the mechanism by which gene function is selectively activated or inactivated in the cells.It provides higher-ordered and more speci?ed genetic information,compared with the whole genome itself.Recently,a variety of regulatory proteins including DNA methyltransferases,methyl-CpG binding proteins,histone-modifying enzymes,chromatin remodeling factors,and their multimole-cular complexes have been identi?ed.These facilitate our understanding of the molecular basis for transcription,DNA replication,mutation and repair,DNA recombination,and chromosome dynamics,which are crucial for normal cell regulation.Abnormalities in the epigenetic states represent human disease phenotypes,especially developmental defects and tumorigenesis.Therefore,epigenetics will become the focus and a major target for emerging biological and medical discoveries.q 2001Elsevier Science B.V.All rights reserved.
Keywords :Epigenetics;DNA methylation;Chromatin;Post-translational modi?cation;Histone;Transcription
1.Introduction
The twenty-?rst century,heralded as the century of life science,is coming to a point of re?ecting signi?cant intel-lectual properties of the genome projects,databases,and modern science and technology upon the next generation of research.Whole genome sequences have been deter-mined for some living species including the human being;thus,we can now clearly recognize what kinds of genes exist in the genome,as if we know all kinds of cards for a metaphorical card game.However,it is essential to under-stand when,where and how these cards will be used.Somatic cells in an individual multicellular organism have basically identical genomes,but each of these cells has a distinct structure and function.This is due to the different uses of genes on the genome,that is,epigenetics.Epigenetic modi?cation of the genome may involve cytosine methyla-tion and chromatin,and therefore produce alterations in gene expression without any differences in DNA sequence.Looking back over the last few years,issues such as DNA
methylation,histone acetylation,chromatin and chromo-somes,transcriptional control and genome dynamics,which have been discussed separately,have turned out to be closely interrelated,as is discussed below.In order to understand cellular and biological phenomena such as development,aging and tumorigenesis,we need an overall view of epigenetics.In this special issue,I wish to provide recent information regarding biochemical aspects of the epigenetic system and its implication in human diseases.2.Concept of epigenetics
Dr Alan Wolffe de?ned the term epigenetics as “heritable changes in gene expression that occur without a change in DNA sequence”(Wolffe and Matzke,1999).The de?nition agrees with the central theme of this review that epigenetics is achieved by DNA methylation and chromatin.Wolffe further placed most emphasis on the transcriptional repres-sion mechanism,as seen in the subtitle ‘regulation through repression’.When we observe a single cell,inactivated genes are widely known to outnumber actively transcribed genes,suggesting that repression of unnecessary genes may be a basic rule of transcriptional control.Hence,epigenetics can be understood as an ingenious system to selectively utilize genome information,through activating or inactivat-ing functional genes.At a molecular level,DNA methyl-transferases,methyl-CpG binding proteins,histone-modifying enzymes,chromatin remodeling factors,tran-
Gene 278(2001)
25–31
q
For further information,a special number featuring articles on the epigenetics was published by Science,volume 293,10August 2001.
Abbreviations:DNMT,DNA methyltransferase;HAT,histone acetyl-transferase;HDAC,histone deacetylase;MBD,methylated DNA-binding domain;MeCP,methyl-CpG binding protein;NuRD,nucleosome-remo-deling histone deacetylase
*Tel.:181-96-373-5118;fax:181-96-373-5120.
E-mail address:mnakao@gpo.kumamoto-u.ac.jp (M.Nakao).
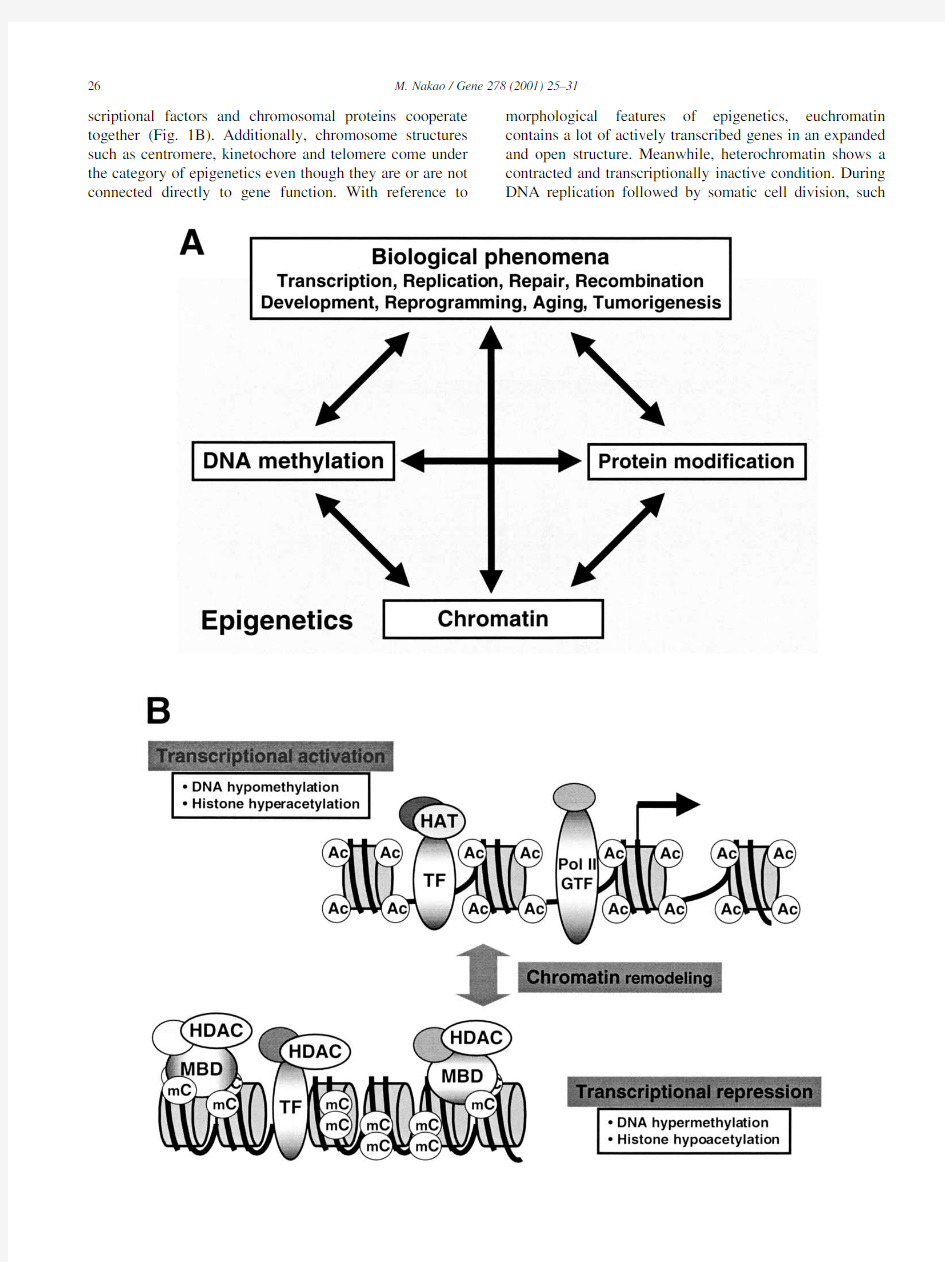
scriptional factors and chromosomal proteins cooperate together(Fig.1B).Additionally,chromosome structures such as centromere,kinetochore and telomere come under the category of epigenetics even though they are or are not connected directly to gene function.With reference to morphological features of epigenetics,euchromatin contains a lot of actively transcribed genes in an expanded and open structure.Meanwhile,heterochromatin shows a contracted and transcriptionally inactive condition.During DNA replication followed by somatic cell division,such
M.Nakao/Gene278(2001)25–31
26
epigenetic states in a parent cell are inherited identically by daughter cells,as DNA sequences are conserved during mitosis.Therefore,memories of epigenetic states,methyla-tion patterns and chromatin organization are maintained during the cell cycle(Rakyan et al.,2001).
Traditional depiction of gene expression shows that ‘DNA produces RNA produces protein’,the two steps being called transcription and translation.In such molecular conversion of DNA–RNA–protein,we occasionally regard the whole process except for the DNA sequence itself as epigenetics in a broad sense.In this case,it is an extensive concept that includes cytosine methylation and chromatin formation,RNA synthesis and degradation,polypeptide synthesis and post-translational modi?cation,and turnover of protein.Thus,the DNA sequence is a program of life,and gene function is inherently controlled at DNA–RNA–protein levels.However,there will be no doubt that tran-scription is the most important and substantial process of epigenetic regulation.
3.System of DNA methylation
3.1.DNA methylation
In vertebrate genomes,5-positioned carbon of cytosine in 50-CpG-30dinucleotide is usually modi?ed by a methyl group.Cytosine residue in complementary30-GpC-50that makes the base pairs is also methylated symmetrically,and these two methyl groups show a three-dimensional structure prominent in the major groove of the double-stranded DNA (Ohki et al.,2001).Approximately60–90%of all CpG sequences in the genome are methylated,while unmethy-lated CpG dinucleotides are mainly clustered in the CpG-rich sequence,termed CpG island,of the gene promoter region(Ng and Bird,1999).Normally,both core promoter and transcription start site are included within the CpG island,and gene expression is completely repressed when this region becomes hypermethylated.
DNA methylation,either reversibly or irreversibly,regu-lates genome functions through affecting gene transcription and chromatin formation(Bird and Wolffe,1999).During cell differentiation,for example muscle and T-cell differ-entiation,genome methylation may speci?cally change to activate or inactivate genes that affect the determination of cell fate,and cell type-speci?c gene expression is also controlled by methylation in the differentiated cells.Geno-mic methylation patterns in these somatic cells are gener-ally stable and heritable.However,genome-wide methylation patterns are fully reprogrammed in mamma-lian germ cells and in pre-implantation embryos.In geno-mic imprinting and X-chromosome inactivation,which are major epigenetic phenomena in mammals,methylation is believed to be indispensable.Abnormal methylation patterns in many cancers induce the inactivation of tumor suppressor genes and the instability of the whole genome. In addition,5-methyl-cytosine on the genome is sponta-neously converted to thymine in the deamination reaction. This is the main cause for generating gene mutations in hereditary diseases and cancers when the mismatched T-G is inherited in the cell division without correcting it by the repair system.Foreign genes introduced into the cells for gene therapy,research and industry purposes tend to be methylated and suppressed as a host defense mechanism. The DNA methylation system mentioned below will give important clues to reveal the molecular basis of these biological phenomena.
3.2.DNA methyltransferases and demethylase Mammalian DNA methyltransferases are classi?ed into two groups:maintenance DNA methyltransferase(DNMT1 or maintenance methylase)and de novo methylase(Bestor, 2000).Maintenance methylase is highly active to methylate a hemi-methylated DNA that is methylated in one strand and unmethylated in the other of double-stranded DNA.It provides the methylation pattern to the newly replicated daughter strand,based on the parent strand.In addition to the enzymatic activities,DNMT1was reported to repress transcription directly in cooperation with histone deacety-lases(HDAC).On the other hand,recently identi?ed de novo methylases(DNMT3a,DNMT3b)add a methyl group to unmethylated CpG base pairs,resulting in the crea-tion of a new hemi-methylated and then fully methylated CpG.For this reason,de novo methylation is considered to be implicated in cell growth and differentiation,and in altered methylation in tumorigenesis.Further,DNMT3b was found to be mutated in patients with ICF syndrome (immunode?ciency in association with centromere instabil-ity of chromosomes1,9and16,and facial anomalies) (Okano et al.,1999;Xu et al.,1999).
Demethylation activity still remains uncertain(Wolffe et al.,1999;Kress et al.,2001).There are two possible processes for removing a methyl group from methylated
M.Nakao/Gene278(2001)25–3127 Fig.1.Overview of epigenetics.(A)Epigenetics is an advanced biological system that selectively utilizes genomic information and is involved in various fundamental phenomena.Speci?cally,it puts emphasis on the regulation of gene expression,through DNA methylation,chromatin,and post-translational modi?cation of proteins such as histones.Arrows indicate possible functional interactions between them.DNA hypermethylation,histone hypoacetylation and inactive chromatin repress transcription.In contrast,a transcriptionally active condition may encourage DNA hypomethylation,histone hyperacetylation and active chromatin.Also,a particular chromatin structure may be required for establishing DNA methylation(see text).(B)DNA methylation and histone modi?cation play key roles in transcriptional control.The?gure shows transcriptional factors(TF),RNA polymerase(Pol II),general transcription factors (GTF),acetylated histone(Ac)and methylated cytosine(mC).Either HAT or HDAC recruited by TF and other DNA-binding proteins induce transcriptional activation or repression,respectively.Modi?cations(acetylation,phosphorylation,methylation)of histone tail domains are described in the text.Chromatin remodeling factors convert the chromatin to active(upper)and inactive(lower)states.
DNA.One is a passive mechanism whereby methylation is not maintained during the DNA replication,and the other is an active mechanism catalyzed by an as yet unidenti?ed DNA demethylase(s).Although MBD2noted below was reported to have demethylase activities,this result has not been reproduced by other research institutions.In addition, 5-methylcytosine DNA glycosylase is noted as a candidate for demethylase in vivo(Zhu et al.,2001).Demethylation of the genome may have great consequences for the regulation of transposons,imprinted genes,and genes on the inactive X-chromosome.At present,we need to focus on the funda-mental problems,that is,how DNA methylases and demethylation are functionally regulated,and how methyla-tion patterns are established,maintained and eliminated.
3.3.Methyl-CpG binding proteins
DNA methylation is known as an epigenetic mark identi-fying the template strand in DNA replication and the paren-tal origin in imprinted regions of the genome.It is also known that methylation alone hinders DNA-binding activ-ities of methylation-sensitive transcriptional factors includ-ing E2F,CREB,AP2,cMyc/Myn,NF-k B,cMyb,and ETS. In addition to methylated DNA,methyl-CpG binding proteins are theoretically required to inhibit transcription by methylation-insensitive transcriptional factors such as Sp1,CTF and YY1.Methyl-CpG binding proteins are deci-phering epigenetic methylation patterns and moreover mediate interactions between DNA methylation,histone deacetylation,and chromatin components.Currently,?ve family members with a conserved methylated DNA-binding domain(MBD)have been described.Among them,MeCP2, MBD1,MBD2and MBD3can be involved in methylation-mediated transcriptional repression,and MBD4has a DNA glycosylase activity for removing a thymine from T-G mismatch sites(Bird and Wolffe,1999;Ballestar and Wolffe,2001).The MeCP1complex,?rst noted by Dr Adrian Bird’s group,was reported to consist of MBD2 and other proteins,and some components may be different in cell types.The MeCP2gene locates on the X-chromo-some and was mutated in the Rett syndrome patients(Amir et al.,1999).This syndrome is the most frequent of the female neurodevelopmental disorders,with loss of speech, autism,ataxia,erratic hand movements and mental retarda-tion,being recognized from6–18months after birth.The MBD4gene is also altered in tumors with microsatellite instabilities.A recent gene knockout strategy in mice found that MBD3is required for embryonic development, whereas MBD2-de?cient mice are viable(Hendrich et al., 2001).MBD2-de?cient cells lacked MeCP1complex and can not ef?ciently repress exogenous methylated promoter. However,it is further necessary to analyze the inactivation of the endogenous methylated gene by MBD-containing proteins,and their functional interrelationship and redun-dancy in vivo.4.Chromatin conversion system
Genomic DNA is folded in the nucleus as a multimole-cular complex with proteins,called chromatin.Nucleosome is a fundamental unit of chromatin and consists of core histones bound to DNA.Dr David Allis and colleagues proposed the concept of‘histone code’whereby combina-tions of N-terminal modi?cations on histones,including acetylation,methylation,phosphorylation,ubiquitination and ADP-ribosylation,have an in?uence on gene expres-sion,DNA replication and chromatin-dependent processes (Strahl and Allis,2000).Phosphorylation at serine10of histone H3is important for chromosome condensation in mitosis and for an initial response to mitogens,and it is also suggested that this phosphorylation induces acetylation of neighboring lysine residues by histone acetylases. Recently,H3-speci?c methylase was identi?ed,and methy-lation at lysine9has been proved to inhibit phosphorylation at serine10by the Ipl-1/aurora kinase.The H3methylation at lysine9generates a binding site for heterochromatin-associated protein HP1(Jenuwein,2001).In contrast,H3 methylated at lysine4is speci?c to the euchromatic regions and correlates with H3acetylation.Thus,it can be assumed that the different modi?cations of one histone are function-ally related to each other.Further,Drosophila TAFII250,a major subunit of the basic transcription factor TFIID,was reported to be a ubiquitinating enzyme for histone H1,and mono-ubiquitination of H1promoted transcriptional activa-tion(Pham and Sauer,2000).A unique intranuclear struc-ture,PML body or nuclear dot10(ND10),widely modulates histone-modifying enzymes and transcriptional factors (Zhong et al.,2000).Interestingly,some molecules connected to this nuclear body are conjugated with the ubiquitin-like protein SUMO-1or sentrin.Thus,the details of these post-translational modi?cations are the subject of a number of studies.
Here,I focused on histone-modifying enzymes and chro-matin-remodeling complexes as the chromatin conversion systems.
4.1.Histone-modifying enzymes
Acetylation of histones H3and H4normally increases gene expression by promoting an open chromatin structure. Transcriptional co-activators such as CBP/p300and PCAF are intrinsic histone acetyltransferase(HAT)(Marmorstein and Roth,2001).Conversely,HDAC contribute to form transcriptional co-repressor complexes.Two complexes, SIN3and Mi2-NuRD(nucleosome-remodelling histone deacetylase),are known to take HDAC1/HDAC2as component molecules(Ahringer,2000;Knoep?er and Eisenman,1999;Wade,2001).As shown in Fig.2, HDAC1/HDAC2and RbAp46/RbAp48are core proteins common to these complexes.Sin3A/Sin3B,SAP30,and SAP18participate in the SIN3complex,while Mi2, MTA1/MTA2,and MBD3are speci?c subunits for the
M.Nakao/Gene278(2001)25–31 28
Mi2-NuRD complex.The SIN3complex has an effect on chromatin through interacting with sequence-speci?c tran-scription factors or co-repressors,including Mad-Max, nuclear hormone receptor and N-CoR/SMRT,and methyl-CpG binding proteins such as MeCP2and MBD2. On the other hand,MBD3,one of components in the Mi2-NuRD complex,has a MBD-like sequence but very weak binding af?nity to methylated DNA.NMR analysis suggested that some amino acids important for methyl-CpG binding of the MBD are substituted to another residue in MBD3(Ohki et al.,2001).It is postulated that the MBD3-MBD2interaction recruits the Mi2-NuRD complex to methylated DNA regions.Thus,DNA methylation and histone deacetylation are cooperatively involved in tran-scriptional repression.The SIN3and Mi2-NuRD complexes may be effective in long-term and short-term transcriptional repression,respectively,since the Mi2-NuRD has additional chromatin remodeling activity as seen below.It is of great interest how they share and split their functions in transcriptional repression.Until now,human HDACs have been roughly divided into three classes:HDAC1,HDAC2,HDAC3and HDAC8in class I,and HDAC4,HDAC5,HDAC6and HDAC7in class II.Based on the protein structure,classes I and II correspond to Rpd3and Hda1in yeast,respectively. Recently,Yeast Sir2has been reported to be a new class of HDAC,as a NAD-dependent HDAC enzyme(Guarente, 2000).
4.2.Chromatin remodeling and assembly factors Transcription,DNA replication,repair and recombina-tion are dynamically carried out at the chromatin level. Chromatin remodeling represents a change of nucleosome position and conformation,leading to chromatin assembly and disassembly.ATP-dependent chromatin remodeling complexes,especially the SWI/SNF and ISWI families, were initially found in yeast and Drosophila(Kingston and Narlikar,1999;Workman and Kingston,1998).As an ATPase subunit of chromatin remodeling complexes in human,BRG1and hBRM for SWI/SNF and hSNF2L/ hSNF2H for ISWI are well known(Fig.3).The hSWI/ SNF complex,which is a huge multimolecular structure including either BRG1or hBRM and tumor suppressor protein hSNF5/Ini-1,mainly activates gene transcription (though gene inactivation was also reported).These complexes are also associated with the cell cycle and the assembly of immunoglobulin and TCR genes by V(D)J recombination.The RSF heterodimer complex including hSNF2H is involved in the initiation of transcription.The above-mentioned Mi2-NuRD complex has been reported to convert active to inactive chromatin due to both ATPase of Mi2and HDAC activities(Fig.2).In addition,the HuCHRAC complex including hSNF2H and chromatin assembly factor hACF1is considered to associate with the replication and maintenance of heterochromatin.There is a possibility that topoisomerase II may be included in the HuCHRAC.As a chromatin assembly complex,the CAF1 complex has a function in the maintenance of chromatin coupled to DNA replication.In some interesting analyses of mutants in arabidopsis,mutations of chromatin remodel-ing factor termed DDM1were found to cause hypomethyla-tion of the genome(Mittelsten Scheid and Paszkowski, 2000).It is thus suggested that DDM1-mediated chromatin
M.Nakao/Gene278(2001)25–31
29
Fig.2.HDAC complexes.Both SIN3and Mi2-NuRD complexes contain HDAC1/HDAC2and RbAp46/RbAp48as core molecules.The SIN3complex additionally has Sin3A/Sin3B,SAP18,and SAP30,while the Mi2-NuRD complex contains Mi2,MTA1/MTA2,p66,and MBD3.The SIN3complex is recruited to methylated DNA by interaction with MBD2and MeCP2.The Mi2-NuRD complex seems to be localized in the methylated region by MBD3-MBD2interaction.Many other factors interacting with these complexes are known but not shown for simplicity.
formation may be a requisite for the maintenance of genome methylation.We expect that information on chromatin remodeling will rapidly increase in the near future.
5.Conclusions
The ‘shape ’of chromatin effectively supports the ‘language ’of genetic information.Epigenetics is a physio-logical system that enables ‘motion ’of the genome,leading to the conducting of all the biological activities of the genome.Replicated DNA is immediately methylated in somatic cells,and methyl-CpG binding proteins recognize the methylation patterns.Histone-modifying enzymes,chromatin remodeling factors,transcriptional factors and co-regulators,and chromosomal proteins such as Poly-comb-group enter chromatin complexes to establish and maintain the epigenetic states (Muller and Leutz,2001).Looking at the processes for gametogenesis or early embry-ogenesis,both DNA methylation and chromatin are dyna-mically reconstructed in the whole genome.In addition to such global roles,local epigenetics in a single chromosome,a chromosomal subdomain,or a speci ?c gene locus also
plays an important role.We will be able to understand the overall concept of epigenetics by investigating relationships between DNA methylation,chromatin,and post-transla-tional modi ?cation of histones and other proteins,and many biological phenomena (Fig.1A).It is of note that human diseases related to abnormal epigenetic conditions are dramatically increasing (Table 1),showing that the molecules and protein complexes discussed here can be appropriate targets for new medical therapies and drug discoveries.In fact,HDAC inhibitors are extensively proposed as a new anti-cancer drug (Marks et al.,2000).Epigenetics is responsible for clarifying mechanisms for selective utilization of genes on the genome.Accordingly,the universal signi ?cance of epigenetics will ensure it is one of the central ?elds of life science in the future.
Acknowledgements
Drs Naoyuki Fujita,Kazuhito Matsuzaki,Masahide Tojo,Sugiko Watanabe,Hideyuki Saya,and Masahiro Shirakawa are thanked for their help.I apologize that all works were not cited here due to space limitations.Our work is
M.Nakao /Gene 278(2001)25–31
30Fig.3.Chromatin remodeling and assembly factors.The hSWI/SNF complex,which is a large multimolecular structure containing either BRG1or hBRM and tumor repressor protein hSNF5/Ini-1,mainly activates gene transcription.The heterodimer RSF complex containing hSNF2H is involved in initiation of transcription.The CAF1complex is related to chromatin assembly in DNA replication.The HuCHRAC complex containing hSNF2H and hACF1maintains heterochromatin.The Mi2-NuRD complex represses transcription through Mi2and HDAC activities (Fig.2).BRG1,hBRM,hSNF2H,and Mi2show ATPase activities.Variant forms of these complexes may occur,but are not all indicated here.
supported by a Grant-in-Aid for Scienti?c Research on Priority Areas from the Ministry of Education,Science, Sports and Culture of Japan.When preparing this review, Dr Alan P.Wolffe tragically died.His contribution will have a great impact on the biochemical aspects of epigenetics research.
References
Ahringer,J.,2000.NuRD and SIN3histone deacetylase complexes in development.Trends Genet.16,351–356.
Amir,R.E.,Van den Veyver,I.B.,Wan,M.,Tran,C.Q.,Francke,U., Zoghbi,H.Y.,1999.Rett syndrome is caused by mutations in X-linked MECP2,encoding methyl-CpG-binding protein2.Nat.Genet.23,185–188.
Ballestar,E.,Wolffe,A.P.,2001.Methyl-CpG-binding proteins.Eur.J.
Biochem.268,1–6.
Bestor,T.H.,2000.The DNA methyltransferases of mammals.Hum.Mol.
Genet.9,2395–2402.Bird,A.P.,Wolffe,A.P.,1999.Methylation-induced repression–belts, braces,and chromatin.Cell99,451–454.
Guarente,L.,2000.Sir2links chromatin silencing,metabolism,and aging.
Genes Dev.14,1021–1026.
Hendrich,B.,Guy,J.,Ramsahoye,B.,Wilson,V.A.,Bird,A.,2001.
Closely related proteins MBD2play distinctive but interacting roles in mouse development.Genes Dev.15,710–723.
Jenuwein,T.,2001.Re-SET-ting heterochromatin by histone methyltrans-ferases.Trends Cell Biol.11,266–273.
Kingston,R.E.,Narlikar,G.J.,1999.ATP-dependent remodeling and acet-ylation as regulators of chromatin?uidity.Genes Dev.13,2339–2352. Knoep?er,P.S.,Eisenman,R.N.,1999.Sin meets NuRD and other tails of repression.Cell99,447–450.
Kress,C.,Thomassin,H.,Grange,T.,2001.Local DNA demethylation in vertebrates:how could it be performed and targeted?FEBS Lett.494, 135–140.
Marks,P.A.,Richon,V.M.,Rifkind,R.A.,2000.Histone deacetylase inhi-bitors:inducers of differentiation or apoptosis of transformed cells.J.
Natl.Cancer Inst.92,1210–1216.
Marmorstein,R.,Roth,S.Y.,2001.Histone acetyltransferases:function, structure,and catalysis.Curr.Opin.Genet.Dev.11,155–161. Mittelsten Scheid,O.,Paszkowski,J.,2000.Transcriptional gene silencing mutants.Plant Mol.Biol.43,235–241.
Muller,C.,Leutz,A.,2001.Chromatin remodeling in development and differentiation.Curr.Opin.Genet.Dev.11,167–174.
Ng,H.H.,Bird,A.,1999.DNA methylation and chromatin modi?cation.
Curr.Opin.Genet.Dev.9,158–163.
Ohki,I.,Shimotake,N.,Fujita,N.,Jee,J.,Ikegami,T.,Nakao,M.,Shir-akawa,M.,2001.Solution structure of the methyl-cpg binding domain of human MBD1in complex with methylated DNA.Cell105,487–497. Okano,M.,Bell,D.W.,Haber,D.A.,Li,E.,1999.DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development.Cell99,247–257.
Pham,A.D.,Sauer,F.,2000.Ubiquitin-activating/conjugating activity of TAFII250,a mediator of activation of gene expression in Drosophila.
Science289,2357–2360.
Rakyan,V.K.,Preis,J.,Morgan,H.D.,Whitelaw,E.,2001.The marks, mechanisms and memory of epigenetic states in mammals.Biochem.
J.356,1–10.
Strahl,B.D.,Allis,C.D.,2000.The language of covalent histone modi?ca-tions.Nature403,41–45.
Wade,P.A.,2001.Transcriptional control at regulatory checkpoints by histone deacetylases:molecular connections between cancer and chro-matin.Hum.Mol.Genet.10,693–698.
Wolffe,A.P.,Matzke,M.A.,1999.Epigenetics:regulation through repres-sion.Science286,481–486.
Wolffe,A.P.,Jones,P.L.,Wade,P.A.,1999.DNA demethylation.Proc.
https://www.360docs.net/doc/fc16840611.html,A96,5894–5896.
Workman,J.L.,Kingston,R.E.,1998.Alteration of nucleosome structure as
a mechanism of transcriptional regulation.Annu.Rev.Biochem.67,
545–579.
Xu,G.-L.,Bestor,T.H.,Bourc’his,D.,Hsieh,C.-L.,Tommerup,N.,Bugge, M.,Hulten,M.,Qu,X.,Russo,J.J.,Viegas-Pequignot,E.,1999.Chro-mosome instability and immunode?ciency syndrome caused by muta-tions in a DNA methyltransferase gene.Nature402,187–191. Zhong,S.,Salomoni,P.,Pandol?,P.P.,2000.The transcriptional role of PML and the nuclear body.Nat.Cell Biol.2,E85–E90.
Zhu,B.,Benjamin,D.,Zheng,Y.,Angliker,H.,Thiry,S.,Siegmann,M., Jost,J.-P.,2001.Overexpression of5-methylcytosine DNA glycosylase in human embryonic kidney cells EcR293demethylates the promoter of
a hormone-regulated reporter https://www.360docs.net/doc/fc16840611.html,A98,
5031–5036.
M.Nakao/Gene278(2001)25–3131 Table1
Epigenetics and human diseases
Gene/protein Disease
DNA methylation system
MeCP2Rett syndrome
MBD2Colon cancer antigen
MBD4Tumors with microsatellite
instability
DNMT3b ICF syndrome
Epigenetic regulation of genes
FMR-1Fragile X mental retardation
IGF2Wilms’tumor
Imprinted genes Prader–Willi&Angelman
syndromes,Beckwith–
Wiedemann syndrome
Tumor suppressor genes Many tumors
X-Inactivation center Functional disomy of X-linked
genes
Histone acetylation system
CBP Rubinstein–Taybi syndrome
p300Gastric cancer,colon cancer,
brain tumor
MOZ-CBP Acute myelocytic leukemia
MLL-CBP Leukemias
Histone modi?cation
Phosphorylation defect of
histone H3
Cof?n–Lowry syndrome
Chromatin remodeling system
Mi2Autoantibody in
dermatomyositis
MTA1Metastatic potential of cancer
hSNF5/Ini-1Rhabdoid tumor
BRG1Tumors
ATRX a-Thalassemia/mental
retardation syndrome,X-linked
Transcriptional control
PML-RAR a Acute promyelocytic leukemia
