Galanthamine_hydrobromide_DataSheet_MedChemExpress
上海氘代试剂类技术参数

上海氘代试剂类技术参数
上海氘代试剂技术参数:
(一)性能参数
1、CAS号:148143-11-8
2、分子式:C3H8
3、密度:0.68~0.70 g/cm3
4、沸点:35.9℃
5、相对折射率:1.393~1.405
6、折光系数:1.382
7、闪点:33.3℃
8、吸水量:≤0.1%
(二)应用参数
1、用作开关破壁剂,提供生物系统中化学反应所需的能量
2、用于改进药物工作性能,如提高溶解性,减少口服非特异性应激症状和刺激等
3、还可用于长期输注悬液的上滤作用
4、它可以促进某些反应的中间产物的分解,从而促进反应的完成
5、可作为生物医药行业中的溶剂和佐剂,以便改善药物的稳定性和作用
6、可用于类酮的分解代谢,以抑制某些有害物质的产生
7、还可用作酯交换剂,可催化合成一氧化碳(CO)及其他有机物
(三)安全参数
1、Skydrol敏感: 是
2、D.S.T.属性: 不活性
3、危险标志:警示性T
4、安全说明:该试剂应储存在阴凉、通风的地方,以避免发生表面挥发或污染、温度超出允许范围。
5、伤害处理:对眼睛有危害时,自动消毒并用干布轻轻地擦拭清洁;有皮肤损伤时,用温水清洗;有口部、鼻部损伤时,用水进行冲洗清洁。
6、防护措施:工作时需戴橡胶手套,穿防护衣,穿口罩,备有足够的空气净化用具。
7、运输条件:按照国家有关规定及企业标准进行运输。
8、主要毒性:该试剂毒性很低,可引起皮肤的刺激症状,注意防护。
右旋泛醇81-13-0

主要症状和影响,急性和迟发效应
4.3 及时的医疗处理和所需的特殊处理的说明和指示
如必要的话,戴自给式呼吸器去救火。
5 消防措施
5.1 灭火介质
火灾特征 无数据资料 灭火方法及灭火剂 碳氧化物,氮氧化物
5.2 源于此物质或混合物的特别的危害
使用个人防护设备。防止吸入蒸汽、气雾或气体。保证充分的通风。
9 理化特性
9.1 基本的理化特性的信息
a) 外观与性状
形状 : 无数据资料
颜色 : 无数据资料
b) 气味
无数据资料
c) 气味临界值
无数据资料
d) pH值
无数据资料
e) 熔点/凝固点
无数据资料
f) 起始沸点和沸程
118-120℃
g) 闪点
无数据资料
h) 蒸发速率
无数据资料
i) 可燃性(固体,气体) 无数据资料
11.1 毒理学影响的信息
急性毒性 半数致死剂量 (LD50) 经口 - 老鼠 - 15,000 mg/kg 亚 急性毒性 无数据资料 刺激性(总述) 无数据资料 皮肤腐蚀/刺激 皮肤-兔子-轻度的皮肤刺激-4h 严重眼损伤 / 眼刺激 眼睛-兔子-轻度的眼睛刺激 呼吸道或皮肤过敏 如果通过皮肤吸收可能是有害的。可能引起皮肤刺激。 生殖细胞诱变 无数据资料 致癌性 IARC:此产品中没有大于或等于0.1%含量的组分被IARC鉴别为可能的或肯定的人类致癌物。 生殖毒性 生殖 特异性靶器官系统毒性(一次接触) 化学物质毒性作用登记:无数据资料 特异性靶器官系统毒性(反复接触) 无数据资料 潜在的健康影响
化学品安全技术说明书
https://
1 化学品及企业标识
大肠杆菌高产L丙氨酸

APPLIED GENETICS AND MOLECULAR BIOTECHNOLOGYProduction of L-alanine by metabolically engineered Escherichia coliXueli Zhang&Kaemwich Jantama&J.C.Moore&K.T.Shanmugam&L.O.IngramReceived:23May2007/Revised:13August2007/Accepted:16August2007/Published online:15September2007 #Springer-Verlag2007Abstract Escherichia coli W was genetically engineered to produce L-alanine as the primary fermentation product from sugars by replacing the native D-lactate dehydroge-nase of E.coli SZ194with alanine dehydrogenase from Geobacillus stearothermophilus.As a result,the heterolo-gous alanine dehydrogenase gene was integrated under the regulation of the native D-lactate dehydrogenase(ldhA) promoter.This homologous promoter is growth-regulated and provides high levels of expression during anaerobic fermentation.Strain XZ111accumulated alanine as the primary product during glucose fermentation.The methyl-glyoxal synthase gene(mgsA)was deleted to eliminate low levels of lactate and improve growth,and the catabolic alanine racemase gene(dadX)was deleted to minimize conversion of L-alanine to D-alanine.In these strains,re-duced nicotinamide adenine dinucleotide oxidation during alanine biosynthesis is obligately linked to adenosine triphosphate production and cell growth.This linkage provided a basis for metabolic evolution where selection for improvements in growth coselected for increased glycolytic flux and alanine production.The resulting strain, XZ132,produced1,279mmol alanine from120g l−1 glucose within48h during batch fermentation in the mineral salts medium.The alanine yield was95%on a weight basis(g g−1glucose)with a chiral purity greater than99.5%L-alanine.Keywords Alanine.Fermentation.E.coli.Evolution. GlycolysisIntroductionWorldwide production of L-alanine has been estimated at 500tons per year(Ikeda2003).In pharmaceutical and veterinary applications,L-alanine is used with other L-amino acids as a pre-and postoperative nutrition therapy(Hols et al.1999).Alanine is also used as a food additive because of its sweet taste(Lee et al.2004).The use of L-alanine is limited in part by the current high cost.L-Alanine is pro-duced commercially by the enzymatic decarboxylation of L-aspartic acid using immobilized cells or cell suspensions of Pseudomonas dacunhae as a biocatalyst with a yield greater than90%(Shibatani et al.1979).The substrate for this enzymatic production process,L-aspartate,is usually pro-duced from fumarate by enzymatic catalysis with aspartate ammonia-lyase.Fumaric acid is produced primarily from petroleum,a nonrenewable feedstock.An efficient fermen-tative process with a renewable feedstock such as glucose offers the potential to reduce L-alanine cost and facilitate a broad expansion of the alanine market into other products.Alanine is a central intermediate(Fig.1)and an essential component of cellular proteins.Most microorganisms produce alanine only for biosynthesis using a glutamate–pyruvate transaminase(Hashimoto and Katsumata1998). Some organisms such as Arthrobacter oxydans(Hashimoto and Katsumata1993;Hashimoto and Katsumata1998; Hashimoto and Katsumata1999),Bacillus sphaericus (Ohashima and Soda1979),and Clostridium sp.P2Appl Microbiol Biotechnol(2007)77:355–366DOI10.1007/s00253-007-1170-yElectronic supplementary material The online version of this article (doi:10.1007/s00253-007-1170-y)contains supplementary material, which is available to authorized users.X.Zhang:J.C.Moore:K.T.Shanmugam:L.O.Ingram(*) Department of Microbiology and Cell Science,University of Florida,Box110700,Gainesville,FL32611,USAe-mail:ingram@K.JantamaDepartment of Chemical Engineering,University of Florida, Gainesville,FL32611,USA(Orlygsson et al.1995)produce alanine from pyruvate and ammonia using an reduced nicotinamide adenine dinucleo-tide (NADH)-linked alanine dehydrogenase (ALD).How-ever,fermentations are slow,and yields from the best natural producers are typically 60%or less because of coproduct formation (Hashimoto and Katsumata 1998;Table 1).Plasmid-borne genes encoding NADH-linked ALD have been tested as an approach to develop improved biocatalysts with varying degrees of success (Table 1).Engineered strains of Zymomonas mobilis CP4expressing the B.sphaericus alaD gene produced low levels of racemic alanine during the anaerobic fermentation of 5%glucose (Uhlenbusch et al.1991).A native chromosomal lactate dehydrogenase gene (ldhA )-deleted strain of Lactococcus lactis containing a mutation in alanine racemase was engineered in a similar fashion and produced 12.6g l −1L -alanine from 1.8%glucose (Hols et al.1999).An Escherichia coli aceF ldhA double mutant containing pTrc99A-alaD plasmid produced 32g l −1racemic alanine in 27h during a two-stage (aerobic and anaerobic)fermentation with a yield of 0.63g alanine g −1glucose (Lee et al.2004).With further gene deletions and process optimization,the racemic alanine titer wasincreasedFig.1Alanine pathway in recombinant E.coli .a Native and recom-binant fermentation pathways.The foreign gene,G.stearothermophilus alaD ,is shown in bold .G.stearothermophilus alaD coding region and transcriptional terminator were integrated into the native ldhA gene under transcriptional control of the ldhA promoter.Solid stars represent deletions of native genes in XZ132.Note that the native biosynthetic route for alanine production is omitted for simplicity.ackA Acetate kinase,adhE alcohol/aldehyde dehydrogenase,alaD alanine dehydro-genase (Geobacillus stearothermophilus XL-65-6),aldA aldehyde dehydrogenase A,aldB aldehyde dehydrogenase B,alr alanine race-mase 1,dadX alanine racemase 2,frd fumarate reductase,gloA glyoxalase I,gloB glyoxalase II,gloC glyoxalase III,ldhA D -lactate dehydrogenase,mdh malate dehydrogenase,mgsA methylglyoxal synthase,pflB pyruvate –formate lyase,ppc phosphoenolpyruvate carboxylase,pta phosphate acetyltransferase.b Coupling of ATP production and growth to NADH oxidation and L -alanine production.Glucose is metabolized to pyruvate,ATP,and NADH.Energy conserved in ATP is utilized for growth and homeostasis,regenerating ADP.NADH is oxidized by alanine formation allowing glycolysis and ATP production to continueT a b l e 1C o m p a r i s o n o f a l a n i n e -p r o d u c i n g s t r a i n sO r g a n i s m s M o d i f i e d p r o p e r t yM e d i a ,s u b s t r a t e a n d p r o c e s s c o n d i t i o n sT i m e (h )A l a n i n e (g l −1)Y i e l d (%)L -A l a n i n ep u r i t y (%)R e f e r e n c e E .c o l i X Z 132I n t e g r a t e d G .s t e a r o t h e r m o p h i l u s a l a D ;Δp f l ,Δa c k A ,Δa d h E ,Δl d h A ,Δm g s A ,Δd a d XM i n e r a l m e d i u m ,b a t c h ,g l u c o s e 120g l −148.0114.095>99T h i s s t u d yA r t h r o b a c t e r o x y d a n s H A P -1M i n e r a l m e d i u m ,t w o -s t a g e f e d -b a t c h ,g l u c o s e 150g l −1120825560.0H a s h i m o t o a n d K a t s u m a t a 1998A .o x y d a n s D A N 75A l a n i n e r a c e m a c e d e f i c i e n tM i n e r a l m e d i u m ,t w o -s t a g e f e d -b a t c h ,g l u c o s e 150g l −1,0.2g l −1D -a l a n i n e120775198H a s h i m o t o a n d K a t s u m a t a 1998E c o l i A L 1(p O B P 1)P l a s m i d w i t h A .o x y d a n s H A P -1a l a D M i n e r a l m e d i u m ,g l u c o s e 20g l −1,l i m i t e d o x y g e n40841N o t r e p o r t e dK a t s u m a t a a n d H a s h i m o t o 1996C o r y n e b a c t e r i u m g l u t a m i c u m A L 107(p O B P 107)P l a s m i d w i t h A .o x y d a n s H A P -1a l a DC o r n s t e e p l i q u o r ,g l u c o s e 200g l −1,4g l −1D L -a l a n i n e ,l i m i t e d o x y g e n 707136>99K a t s u m a t a a n d H a s h i m o t o 1996Z y m o m o n a s m o b i l i s C P 4(p Z Y 73)P l a s m i d w i t h B .s p h a e r i c u s I F O 3525a l a D M i n e r a l s a l t s m e d i u m ,s i m p l e b a t c h ,g l u c o s e 50g l −126816N o t r e p o r t e dU h l e n b u s c h e t a l .1991L a c t o c o c c u s l a c t i s N Z 3950(p N Z 2650)P l a s m i d w i t h B .s p h a e r i c u s I F O 3525a l a D Δl d h AR i c h m e d i u m (M 17),g l u c o s e 18g l −117137085–90H o l s e t a l .1999L .l a c t i s P H 3950(p N Z 2650)P l a s m i d w i t h B .s p h a e r i c u s I F O 3525a l a D Δl d h A ,Δa l rR i c h m e d i u m (M 17),g l u c o s e 18g l −1,0.2g l −1D -a l a n i n e 17N o t k n o w nN o t k n o w n >99H o l s e t a l .1999E .c o l i A L S 887(p T r c 99A -a l a D )P l a s m i d w i t h B .s p h a e r i c u s I F O 3525a l a D Δl d h A ,Δa c e FY e a s t e x t r a c t ,t w o -s t a g e b a t c h ,g l u c o s e 50g l −1,a e r o b i c a i r 1l m i n −1273263N o t r e p o r t e d L e e e t a l .2004E .c o l i A L S 929(p T r c 99A -a l a D )P l a s m i d w i t h B .s p h a e r i c u s I F O 3525a l a D Δp f l ,Δp p s ,Δp o x B ,Δl d h A ,Δa c e E FY e a s t e x t r a c t a n d c a s a m i n o a c i d s ,t w o -s t a g e b a t c h (a e r o b i c c e l l g r o w t h a n d a n a e r o b i c f e r m e n t a t i o n )223486N o t r e p o r t e d S m i t h e t a l .2006E .c o l i A L S 929(p T r c 99A -a l a D )P l a s m i d w i t h B .s p h a e r i c u s I F O 3525a l a D Δp f l ,Δp p s ,Δp o x B ,Δl d h A ,Δa c e E FY e a s t e x t r a c t a n d c a s a m i n o a c i d s ,t w o -s t a g e f e d -b a t c h (a e r o b i c c e l l g r o w t h a n d a n a e r o b i c f e r m e n t a t i o n )4888100N o t r e p o r t e d S m i t h e t a l .2006to88g l−1in a more complex process with yields ap-proaching the theoretical maximum(Smith et al.2006). However,this strain produced only racemic alanine,utilized multicopy plasmids requiring antibiotic selection,and required complex media with a complex multistage fermen-tation process(Smith et al.2006).In this study,we developed novel biocatalysts that pro-duce chirally pure L-alanine in batch fermentations without using plasmid-containing biocatalysts,antibiotics,or com-plex nutrients.The resulting strains are based on a deriva-tive of E.coli W(strain SZ194)that produces D-lactate (Zhou et al.2006b).The ldhA gene in SZ194was replaced with a single,chromosomally integrated copy of the ALD gene from the thermophile,Geobacillus stearothermophilus XL-65-6(formerly B.stearothermophilus;Lai and Ingram 1993).After additional deletions of alanine racemase (dadX)and methylglyoxal synthase(mgsA)and metabolic evolution,the resulting strain produced L-alanine at high titers(over1M)and yields in batch fermentations using the mineral salts medium.Materials and methodsStrains,plasmids,media,and growth conditionsThe strains and plasmids used in this study are listed in Table2.Strain SZ194was previously engineered from a derivative of E.coli W(ATCC9637)and served as a starting point for constructions(Zhou et al.2006b).G. stearothermophilus XL-65-6(Lai and Ingram1993)was used for cloning the ALD gene.During sequencing of chro-mosomal genes,we discovered a20-year-old error in culture labeling.Strain SZ194,the parent used to construct the alanine strains,is a derivative of E.coli W(ATCC9637). Other constructs for ethanol production and lactate produc-tion that have been reported previously as derivatives of E. coli B are now known to be derivates of E.coli W(ATCC 9637).Primers used in this study are listed in Table3.During strain construction,cultures were grown aerobi-cally at30,37,or39°C in Luria broth(10g l−1Difco tryptone,5g l−1Difco yeast extract,and5g l−1NaCl) containing2%(w/v)glucose or5%(w/v)arabinose. Ampicillin(50mg l−1),tetracycline(12.5mg l−1), kanamycin(50mg l−1),or chloramphenicol(40mg l−1) were added as needed.For initial tests of fermentative alanine production,strains were grown without antibiotics at37°C in NBS mineral salts medium(Causey et al.2004) supplemented with100mM ammonia sulfate,1mM betaine,and2%(w/v)glucose.Fermentation experiments (2–12%sugar)were carried out in NBS medium and AM1 medium(Martinez et al.2007).Broth was maintained at pH 7by the automatic addition of5M NH4OH.Genetic methodsStandard methods were used for genomic deoxyribonucleic acid(DNA)extraction(Qiagen,Valencia,CA),polymerase chain reaction(PCR)amplification(Stratagene,La Jolla CA,and Invitrogen,Carlsbad,CA),transformation,plas-mid extration(Qiagen),and restriction endonuclease diges-tion(New England Biolabs,Ipswich,MA).Methods for foreign gene(alaD)integration and for chromosomal gene (mgsA and dadX)deletion are described below.DNA sequencing was provided by the University of Florida Interdisciplinary Center for Biotechnology Research.The Biocyc and Metacyc databases(Karp et al.2005)were instrumental in the design and completion of these studies. Cloning the alanine dehydrogenase gene alaD from G. stearothermophilus XL-65-6and detection of the enzyme activityThe primers for amplifying alaD from G.stearothermophilus XL-65-6were designed based on the alaD sequence of G. stearothermophilus strain10.The forward primers(5′–3′GGAAAAA GGAGGAAAAAGTG ATGAAGATCGG CATT)included the ribosomal-binding region(bold)and the amino terminus(italicized).The reverse primer(5′–3′GAA GGAGTTGATCATTGTTTAACGAGAGAGG)was down-stream from the putative transcriptional terminator region (Table3).ALD was verified in clones using an activity stain (Kuroda et al.1990).E.coli TOP10F′harboring plasmids containing alaD was grown on Luria–Bertani(LB)plates at 37°C,then transferred to a Whatman7.0-cm filter paper. The filter was immersed in10mM potassium phosphate buffer(pH7.2)and incubated for20min at80°C for lysis of the cells and denaturation of the E.coli proteins.The dried filter paper was assayed in a reaction mixture containing50mM L-alanine,50mM Tris–HCl buffer (pH9.0),0.625mM NAD+,0.064mM phenazine metho-sulfate,and0.24mM nitro blue tetrazolium.The cells with ALD appeared as blue spots on the filter.Integration of alaD into E.coli SZ194The alaD gene was integrated into the chromosomal ldhA gene of SZ194.The fragment(Sma I–Kpn I,1.7kb)con-taining a tet gene flanked by two FRT sites was isolated from pLOI2065and cloned into pLOI4211between a unique Bam HI site(Klenow-treated)and Kpn I site to produce plasmid pLOI4213(6.0kb).In this plasmid,transcription of alaD and tet are oriented in the same direction.The Apa I(treated with T4DNA polymerase to produce a blunt end)–Kpn I fragment(2.2kb)containing alaD and tet was isolated from pLOI4213and cloned into pLOI2395Table2 E.coli strains and plasmids used in this studyRelevant characteristics Source or referenceStrainsSZ194plfB frd adhE ackA deletions Zhou et al.2006bXZ103-110SZ194,ldhA::FRT-tet-FRT::This studyG.stearothermophilus alaDXZ111XZ105,ldhA::G.stearothermophilus alaD This studyXZ112XZ111,metabolic evolution in NBS medium with2%glucose This studyXZ113XZ112,metabolic evolution in NBS medium with5%glucose This studyXZ115XZ113,metabolic evolution in NBS medium with8%glucose This studyXZ121XZ115,mgsA deletion This studyXZ123XZ121,metabolic evolution in NBS medium with8%glucose This studyXZ126XZ123,dadX deletion This studyXZ129XZ126,metabolic evolution in NBS medium with8%glucose This studyXZ130XZ129,metabolic evolution in AM1medium with8%glucose This studyXZ131XZ130,metabolic evolution in AM1medium with10%glucose This studyXZ132XZ131,metabolic evolution in AM1medium with12%glucose This studyPlasmidspCR2.1-TOPO bla kan;TOPO TA cloning vector InvitrogenDatsenko and Wanner2000pKD46Blaγβexo(Red recombinase),temperature conditionalpSC101repliconpFT-A Bla flp,temperature conditional pSC101replicon Posfai et al.1997pEL04cat-sacB targeting cassette Lee et al.2001;Thomason et al.2005 pLOI2224kan;R6K conditional integration vector Martinez-Morales et al.1999pLOI2065bla;FRT-tet-FRT cassette Zhou et al.2003bpLOI2395bla;ldhA franked by two Asc I site Zhou et al.2003apLOI3421 1.8kbp SmaI fragment containing aac Wood et al.2005pLOI4151bla cat;cat-sacB cassette This studyalaD integrationThis studypLOI4211bla kan alaD;alaD(PCR)from G.stearothermophilus XL-65-6cloned into pCR2.1-TOPO vectorpLOI4213bla kan;alaD-FRT-tet-FRT Kpn I-Sma I fragment(FRT-tet-FRT)This studyfrom pLOI2065cloned into Kpn I-BamH I(blunted)site of pLOI4211This studypLOI4214bla kan;ldhA’-alaD-FRT-tet-FRT-ldhA”Apa I(blunted)-Kpn I fragment(alaD-FRT-tet-FRT)from pLOI4213cloned into ldhA at Hinc II-Kpn Isites of pLOI2395This studypLOI4215kan;ldhA’-alaD-FRT-tet-FRT-ldhA”Asc I fragment(ldhA’-alaD-FRT-tet-FRT-‘ldhA)from pLOI4214cloned into Asc I sites of pLOI2224mgsA deletionThis studypLOI4228bla kan;yccT’-mgsA-helD’(PCR)from E.coli W clonedinto PCR2.1-TOPO vectorThis studypLOI4229cat-sacB cassette PCR amplified from pLOI4151(Eco RV digested)cloned into mgsA in pLOI4228This studypLOI4230PCR fragment amplified from pLOI4228(using mgsA-1/mgsA-2primers),kinase treated,and self-ligateddadX deletionThis studypLOI4216bla kan;dadA’-dadX-cvrA’(PCR)from E.coli W clonedinto PCR2.1-TOPO vectorpLOI4218cat-sacB cassette PCR amplified from pLOI4151(Eco RV digested)This studycloned into dadX in pLOI4216This studypLOI4220PCR fragment amplified from pLOI4216(using dadX-4/dadX-5primers),kinase treated,and self-ligated(Hinc II to Kpn I sites)to produce pLOI4214(6.5kb).In this plasmid,ldhA ,alaD ,and tet genes are transcribed in the same direction.The Asc I fragment (4.3kb)containing these three genes was isolated from pLOI4214and cloned into the R6K integration vector pLOI2224to produce pLOI4215(6.2kb).Plasmid pLOI4215contains resistance genes for both tetracycline and kanamycin (Fig.2).The Asc I fragment (4.3kb)containing ldhA ,alaD ,and tet genes was isolated from pLOI4215,further cut by Xmn I to eliminate any remaining uncut plasmid DNA,and electroporated into SZ194containing the Red recombinase plasmid pKD46(Datsenko and Wanner 2000).Integrants were selected for tetracycline resistance,confirmed by sensitivity to kanamycin and ampicillin and by PCR analysis using the primers of ldhA and its neighboring genes ydbH and hslJ (Table 3).Deletion of mgsA and dadX genesA modified method for deleting E.coli chromosomal genes was developed using two steps of homologous recom-bination (Thomason et al.2005).With this method,no antibiotic genes or scar sequences remain on the chromo-some after gene deletion.In the first recombination,part of the target gene was replaced by a DNA cassette containing a chloramphenicol resistance gene (cat )and levansucrase gene (sacB ).In the second recombination,the cat –sacBcassette was removed by selection for resistance to sucrose.Cells containing the sacB gene accumulate levan during incubation with sucrose and are killed.Surviving recombi-nants are highly enriched for loss of the cat –sacB cassette.A new cassette was constructed as a template to facilitate gene deletions.The cat –sacB region was amplified from pEL04(Lee et al.2001;Thomason et al.2005)by PCR using the JM catsacB up Nhe I and JM catsacB down Nhe I primers (Table 3),digested with Nhe I,and ligated into the corresponding site in pLOI3421to produced pLOI4151.The cat –sacB cassette was amplified by PCR using pLOI4151as a template with the cat -up2and sacB -down2primers (Eco RV site included in each primer),digested with Eco RV ,and used in subsequent ligations.The mgsA gene and neighboring 500-bp regions (yccT ′–mgsA –helD ′,1,435bp)were amplified using the mgsA -up and mgsA -down primers and cloned into the pCR 2.1-TOPO vector (Invitrogen)to produce plasmid pLOI4228.A 1,000-fold diluted plasmid preparation of this plasmid served as a template for inside-out amplification using the mgsA -1and mgsA -2primers (both within the mgsA gene and facing outward).The resulting 4,958-bp fragment containing the replicon was ligated to the Eco RV-digested cat –sacB cassette from pLOI4151to produce pLOI4229(Fig.3a).This 4,958-bp fragment was also used to construct a second plasmid,pLOI4230(Fig.3b),by phosphorylation and self-ligation.In pLOI4230,the central region of mgsA is deleted (yccT ′–mgsA ′–mgsA ″–helD ′).After digestion of pLOI4229and pLOI4230with Xmn I (within the vector),each served as a template for amplifica-tion using the mgsA -up and mgsA -down primers to produce linear DNA for integration step 1(yccT ′–mgsA ′–cat –sacB –mgsA ″–helD ′)and step II (yccT ′–mgsA ′–mgsA ″–helD ′),respectively.After electroporation of the step 1fragment into XZ115containing pKD46(Red recombinase)and 2h ofTable 3Primers used in this study Primers SequencealaD -forward GGAAAAAGGAGGAAAAAGTGATGAA GATCGGCATTalaD -reverse GAAGGAGTTGATCATTGTTTAACGA GAGAGGldhA -forward AGTACCTGCAACAGGTGAAC ldhA -reverse CAGGCGACGGAATACGTCAT ldhA -up (ydbH )CTGATAACGCAGTTGCTGGA ldhA -down (hslJ )TTCATTAAATCCGCCAGCTTJM catsacB up NheI TTAGCTAGCATGTGACGGAAGATC ACTTCGJM catsacB down NheI CCGCTAGCATCAAAGGGAAAACTGT CCATATcat -up2AGAGAGGATATCTGTGACGGAAGAT CACTTCGsacB -down2AGAGAGGATATCGAATTGATCCGGT GGATGACmgsA -up CAGCTCATCAACCAGGTCAA mgsA -down AAAAGCCGTCACGTTATTGG mgsA -1AGCGTTATCTCGCGGACCGT mgsA -2AAGTGCGAGTCGTCAGTTCC dadX -up AGGCTACTCGCTGACCATTC dadX -down GGTTGTCGGTGACCAGGTAG dadX -4TGGGCTATGAGTTGATGTGC dadX -5CTGTATCGGACGGGTCATCTFig.2Integration vector used for chromosomal insertion of G.stearothermophilus alaD into E.coli ldhA .Sequence encoding the N-terminal and C-terminal regions are designated ldhA ′and ldhA ″,respectivelyincubation at 30°C to allow expression and segregation,recombinants were selected for chloramphenicol (40mg l −1)and ampicillin (50mg l −1)resistance in Luria broth at 30°C (18h).Three clones were selected,grown in Luria broth containing ampicillin and 5%(w/v)arabinose (to induce expression of red recombinase),and prepared for electro-poration.After electroporation with the step 2fragment,cells were incubated at 30°C for 4h and then transferred into a 250-ml flask containing 100ml of modified LB (100mM 3-(N -morpholino)propanesulfonic acid [MOPS]buffer added and NaCl omitted)containing 10%sucrose.After overnight incubation (30°C),clones were selected on modified LB plates (no NaCl;100mM MOPS added)containing 6%sucrose (39°C,16h).Resulting clones were tested for loss of ampicillin and chloramphenicol resistance.Construction was confirmed by PCR using the mgsA-up/down primer set.A clone containing a deletion in the central region of mgsA was selected and designated XZ121.The dadX gene was deleted in a manner analogous to that used to delete the mgsA gene.Primers for dadX deletion are shown in Table 3,and the corresponding plasmids are shown in Table 2.FermentationNBS mineral salts medium (Causey et al.2004)with 1mM betaine (Zhou et al.2006a )was used in the initial fermentation (pH 7.0).Preinoculum was grown by inocu-lating three colonies into a 250ml flask (100ml NBS medium,2%glucose,and 100mM ammonium sulfate).After 16h (37°C,120rpm),this preinoculum was diluted into 500-ml fermentation fleakers containing 300ml NBS medium (2–8%glucose,100mM ammonium sulfate,and 1mM betaine)with 33mg cell dry weight (CDW)l −1.In early experiments,pH was maintained at 7.0by automat-ically adding 2M potassium hydroxide.In later experi-ments,5M ammonium hydroxide was used to maintain pH,and a low salt medium,AM1(Martinez et al.2007),was used to replace the NBS medium for fermentation (8–12%glucose).AM1medium contains much less salt and has been optimized for E.coli .Metabolic evolutionCells from pH-controlled fermentations were serially transferred at 24-h intervals to facilitate metabolic evolution through competitive,growth-based selection (Fig.1b).At the beginning,sequentially transferred cultures were inoc-ulated with an initial density of 33mg CDW l −1.As growth increased,the inoculum was changed to a 1:100dilution and subsequently to a 1:300dilution.Periodically,clones were isolated from these experiments,assigned new strain designations,and frozen for storage.AnalysesCell mass was estimated by measuring the optical density at anic acids and glucose concentrations were mea-sured by high-performance liquid chromatography (HPLC,Underwood et al.2002).Analysis of fermentation products by mass spectroscopy and amino acid analyzer were provided by the University of Florida Interdisciplinary Center for Bio-technology Research.Alanine was found to be the predominant product.The alanine concentration and isomeric purity were further measured by HPLC using the Chiralpak MA(+)chiral column (Chiral Technologies,West Chester,PA).ResultCloning of the alanine dehydrogenase geneALD is found in Bacillus (and Geobacillus )species where it plays a pivotal role in energy generation during sporulation (Ohashima and Soda 1979;Kuroda et al.1990).ALD from B.sphaericus IFO3525has beenwidelyFig.3Plasmids used to delete mgsA .Plasmid pLOI4229(a )was used to delete the mgsA gene and insert the cat-sacB cassette in the first recombina-tion step.Plasmid pLOI4230(b )was used to remove the cat-sacB cassette to create a deletion devoid of foreign sequence.Se-quence encoding the N-terminal and C-terminal regions are des-ignated mgsA ′and mgsA ″,respectivelyused with varying degrees of success to engineer alanine production in recombinant bacteria(Uhlenbusch et al. 1991;Hols et al.1999;Lee et al.2004;Smith et al.2006). Selection of the B.sphaericus IFO3525is presumed to be due in part to the high specific activity(Ohashima and Soda 1979).In contrast,we have selected a thermostable ALD from the thermophile,G.stearothermophilus XL-65-6, based on our prior experience in expressing genes from this organism in recombinant E.coli(Burchhardt and Ingram 1992;Lai and Ingram1993;Lai and Ingram1995).The ribosomal-binding region,coding region,and tran-scriptional terminator of alaD were amplified from G. stearothermophilus XL-65-6and sequenced(EF154460in GenBank).The deduced amino acid sequence was identical to that reported for Geobacillus kaustophilus HTA426and very similar to G.stearothermophilus strain10(99%iden-tity)and G.stearothermophilus strain IFO12550(94% identity).The nucleotide sequence(65%identity)and the deduced ALD amino acid sequence(74%identity)were quite different from the B.sphaericus IFO3525gene,the gene pre-viously used for alanine production in recombinant bacteria.Modification of E.coli W for homoalanine productionE.coli W strain SZ194(pflB frdBC adhE ackA)was previously constructed to produce only D-lactic acid.All major fermentation pathways except lactate have been blocked in this strain by gene deletions(Fig.1a).To convert this strain to the production of alanine,part of the native ldhA-coding region was replaced by a DNA fragment containing the ribosomal-binding region,coding region,and transcriptional terminator of alaD from G. stearothermophilus XL-65-6.The promoterless alaD was oriented in the same direction as ldhA to allow expression from the native ldhA promoter(Fig.2).After electroporation,approximately500colonies were recovered with tetracycline resistance and sensitivity to kana-mycin,consistent with a double-crossover event.These colo-nies were further examined by PCR using ldhA forward and reverse primer set(Table3).Only eight colonies of the500 tested were correct based on an analysis of PCR fragments. These eight colonies were further verified using primer sets for alaD,ldhA forward and alaD reverse,alaD forward and ldhA reverse,and ldhA outside primers(Table3)and de-signated XZ103,XZ104,XZ105,XZ106,XZ107,XZ108, XZ109,and XZ110,respectively.These eight strains were initially tested in15-ml screw-cap tubes containing NBS medium with2%glucose and100mM ammonium sulfate, which were filled to the brim.Strain XZ105appeared to grow faster than the other strains(37°C for48h)and was selected for further development.XZ105was transformed with pFT-A,which contains an inducible flippase(FLP)recombinase(Martinez-Morales et al.1999;Posfai et al.1997).The chromosomal FRT-flanked tet gene in XZ105was removed by inducing the FLP recombinase.After growing in39°C to eliminate the temperature-sensitive plasmid pFT-A,resulting strain was designated XZ111.Expression of G.stearothermophilus alaD in XZ111is transcriptionally regulated by the ldhA promoter,the same promoter that regulates the production of lactate dehydrogenase(dominant fermentation pathway) in native E.coli.pH-controlled batch fermentation for alanine production Alanine production by strain XZ111was tested in500-ml fermentation vessels containing300ml NBS medium, 20g l−1glucose,100mM ammonium sulfate,and1mM betaine.Broth pH was automatically controlled by adding 2N potassium hydroxide.After96h,181mM alanine was produced.The alanine yield from total glucose was 81%(g/g),and84%based on glucose that had been metabo-lized.The chiral purity of L-alanine was96.1%(Table4). Very low levels of other products(lactate,succinate,ace-tate,ethanol)were present,typically below1mM.This result demonstrated that the integrated G.stearothermophilus alaD gene as a single chromosomal copy under the control of the native ldhA promoter can provide sufficient levels of ALD to support E.coli growth from the production of alanine as the sole fermentation product.Metabolic evolution of strain XZ111Although XZ111could accumulate alanine as the primary product,incubation times were long,and volumetric productivity was limited.When using a high-glucose concentration(80g l−1),growth and alanine productivity were further reduced(Table4).In this strain,adenosine triphosphate(ATP)production and growth are tightly coupled to NADH oxidation and alanine production by ALD(Fig.1b).This coupling provided a basis for strain improvement by selecting for increased growth during serial cultivation,i.e.,metabolic evolution.Cells with increased growth because of spontaneous mutations will successively displace their parents while coselecting for increased alanine productivity.Serial transfers of XZ111were carried out at24-h intervals in NBS mineral salts medium with1mM betaine.Cultures were first transferred in the medium containing20g l−1 glucose,and the pH was controlled by automatically adding 2N potassium hydroxide.However,after ten transfers to strain XZ112,little improvement was observed(data not shown).Because ammonia is essential for alanine pro-duction,it was thought that ammonia may be limiting for fermentation.Two normals potassium hydroxide containing 1N ammonia carbonate and5N ammonia hydroxide alone。
盐酸奥洛他定片说明书

核准日期:2010年10月13日修改日期:2012年09月14日 2013年10月15日 2015年02月09日 2015年12月24日 2016年07月20日2017年01月16日 2019年12月20日 2020年01月03日 2020年08月17日 2021年06月28日 2022年01月06日盐酸奥洛他定片说明书请仔细阅读说明书并在医师指导下使用【药品名称】通用名称:盐酸奥洛他定片商品名称:阿洛刻英文名称:Olopatadine Hydrochloride Tablets汉语拼音:Yansuan Aoluotading Pian【成份】本品活性成份为盐酸奥洛他定。
化学名称:{11-[(1Z)-3-(二甲氨基) 亚丙基]-6, 11-二氢二苯并[b, e]恶庚英-2-基}乙酸盐酸盐化学结构式:分子式:C21H23NO3·HCl分子量:373.87辅料:乳糖水合物、结晶纤维素、交联羧甲基纤维素钠、聚乙烯醇(部分皂化物)、硬脂酸镁、羟丙基甲基纤维素、聚乙二醇6000、氧化钛、黄色氧化铁、氧化铁、巴西棕榈蜡【性状】本品为淡粉色的薄膜衣片,除去包衣后显白色。
【适应症】过敏性鼻炎、荨麻疹、瘙痒性皮肤病(湿疹、皮炎、痒疹、皮肤瘙痒症、寻常性银屑病、渗出性多形红斑)【规格】5mg【用法用量】本品为口服片剂。
成人用量通常为1日2次,1次5mg,早晨和晚上睡前各服1次。
根据年龄及症状适当增减。
【不良反应】在批准前临床试验、药物使用-结果调查及长期使用专项调查的9620例患者中,有1056例(发生率11.0%)发生不良反应共计1402件。
主要不良反应为嗜睡674件(7.0%),ALT (GPT)上升68件(0.7%),倦怠感53件(0.6%),AST (GOT)上升46件(0.5%),口渴36件(0.4%)等。
(在日本再审查结束时)1)有临床意义的不良反应暴发性肝炎、肝功能损害、黄疸(发生率不明):有可能发生暴发性肝炎、伴随AST (GOT)、ALT (GPT)、γ-GTP、LDH、Al-P上升等的肝功能损害、黄疸,故应注意观察。
盐酸拉维达韦片说明书
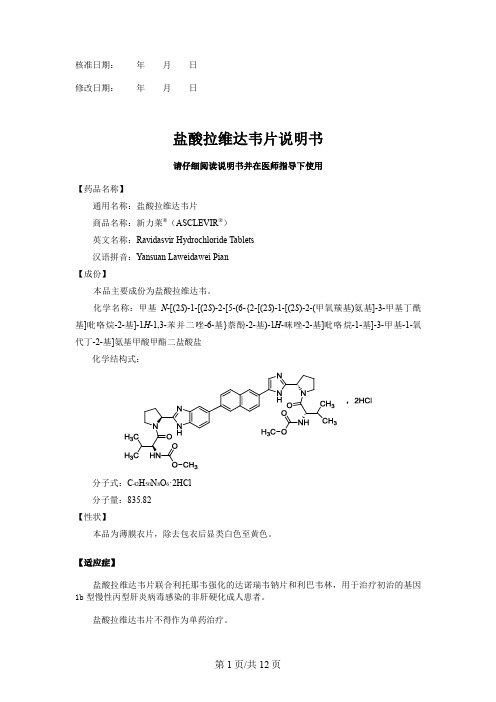
核准日期:年月日修改日期:年月日盐酸拉维达韦片说明书请仔细阅读说明书并在医师指导下使用【药品名称】通用名称:盐酸拉维达韦片商品名称:新力莱®(ASCLEVIR®)英文名称:Ravidasvir Hydrochloride Tablets汉语拼音:Yansuan Laweidawei Pian【成份】本品主要成份为盐酸拉维达韦。
化学名称:甲基N-[(2S)-1-[(2S)-2-[5-(6-{2-[(2S)-1-[(2S)-2-(甲氧羰基)氨基]-3-甲基丁酰基]吡咯烷-2-基]-1H-1,3-苯并二唑-6-基}萘酚-2-基)-1H-咪唑-2-基]吡咯烷-1-基]-3-甲基-1-氧代丁-2-基]氨基甲酸甲酯二盐酸盐化学结构式:分子式:C42H50N8O6·2HCl分子量:835.82【性状】本品为薄膜衣片,除去包衣后显类白色至黄色。
【适应症】盐酸拉维达韦片联合利托那韦强化的达诺瑞韦钠片和利巴韦林,用于治疗初治的基因1b型慢性丙型肝炎病毒感染的非肝硬化成人患者。
盐酸拉维达韦片不得作为单药治疗。
【规格】0.2g(以C42H50N8O6计)【用法用量】推荐剂量本品口服,可空腹或与食物同服。
本品用法用量:每次200mg,每日1次,连续12周。
服用本品时须同时应用达诺瑞韦钠片、利托那韦和利巴韦林。
推荐达诺瑞韦钠片用法用量:口服,每次100mg,每日2次;连续12周(详见达诺瑞韦钠片说明书)。
推荐利托那韦(RTV)用法用量:口服,每次100mg,每日2次,连续12周(详见利托那韦片说明书)。
推荐利巴韦林用法用量:利巴韦林的剂量根据体重确定。
如体重<75kg,每次500mg,每日2次;如体重≥75kg,每次600mg,每日2次;连续12周(详见利巴韦林制剂说明书)。
剂量调整、暂停给药和停止治疗不建议调整盐酸拉维达韦片的剂量,并应避免暂停给药。
但如果因不良反应需要暂停给药联合治疗方案中的任何一种药物,则不得单独应用盐酸拉维达韦片治疗。
盐酸苯海拉明(溶出数据库)

【盐酸苯海拉明】
日文名:塩酸ジフェンヒドラミン
结构式:
(ジフェンヒドラミン塩酸塩)
英文名:Diphenhydramine Hydrochloride
解离常数(25℃):pKa = 8.6(针对叔氨基、采用滴定法测定)
在各溶出介质中的溶解度(37℃):pH1.2:1.0g/ml以上pH4.0:1.0g/ml以上
pH 6.8:1.0g/ml以上水:1.0g/ml以上
在各溶出介质中的稳定性:
水:未测定。
在各pH值溶出介质中:在酸性溶出介质中缓慢降解。
光:缓慢降解。
《四条标准溶出曲线》
溶出度试验条件:桨板法/50转、溶出介质中不添加表面活性剂。
< 10mg规格片剂(A型)>
< 10mg规格片剂(B型)>
《质量标准》
取本品,照溶出度测定法(桨板法),以水900ml为溶出介质,转速为每分钟50转,依法操作,经30分钟时,取溶液适量,弃去至少10ml初滤液,取续滤液作为供试品溶液。
另精密称取经105℃干燥3小时的对照品22mg,置100ml量瓶中,加水溶解并稀释至刻度,摇匀,精密量取5ml,置100ml量瓶中,加水稀释至刻度,摇匀,作为对照品溶液。
取上述两种溶液照紫外-可见分光光度法,在220nm的波长处测定吸光度,计算出每片溶出量,限度为标示量的75%,应符合规定。
阿拓莫兰(还原型谷胱甘肽片)说明书

阿拓莫兰(还原型谷胱甘肽片)说明书之杨若古兰创作【阿拓莫兰药品名称】通用名:还原型谷胱甘肽片商品名:阿拓莫兰英文名:Reduced glutathione Tablets汉语拼音:Huanyuanxing Guguanggantai Pian【阿拓莫兰成份】阿拓莫兰次要成份为还原型谷胱甘肽,其化学名为N(NLγ谷氨酰基L半胱氨酰基)甘氨酸.【阿拓莫兰性状】阿拓莫兰为薄膜衣片,除去包衣后显白色.【阿拓莫兰产品介绍】阿拓莫兰的次要成分是还原型谷胱甘肽(GSH),GSH是哺乳动物细胞内含量丰富的低分子量多肽,由谷氨酸、半胱氨酸和甘氨酸残基构成,对人体具有十分次要的生理功能:解酒精毒性、呵护肝脏;解药物毒性(抗肿瘤药、抗结核药、中枢神经药物、对乙酰氨基酚等中毒),防止抗癌药物的副感化,预防和医治放射线损害;对抗自在基,抗氧化;提高机体免疫力.人体在很多形态下都可以使细胞内GSH生物合成能力降低,含量降低,特别是在病理形态下.外源性GSH的弥补,可以预防、减轻、中断、组织细胞的损伤,改变病理生理过程.药理毒理研讨标明:阿拓莫兰的次要成分(还原型谷胱甘肽)是含有巯基(SH)的三肽类化合物,在人体内具有活化氧化还原零碎,激活SH酶、解毒感化等次要生理活性.介入体内三羧酸轮回和糖代谢,促进体内发生高能量,起到辅酶感化.还原型谷胱甘肽是甘油醛磷酸脱氢酶的辅基,又是乙二醛酶及磷酸丙糖脱氢酶的辅酶.还原型谷胱甘肽能激活体内SH 酶等,促进碳水化合物、脂肪及蛋白质的代谢,以调节细胞膜的代谢过程.还原型谷胱甘肽介入多种外源性、内源性有毒物资结合生成减毒物资.【阿拓莫兰适应症】1)肝损伤:病毒性肝病,药物性肝病,中毒性肝损伤,脂肪肝,肝硬化等;2)肾损伤:急性药物性肾损伤,尿毒症;3)化放疗呵护;4)糖尿病:并发症,神经病变;5)缺血缺氧性脑病;各种低氧血症.【阿拓莫兰用法用量】成人经常使用量为每次口服400mg(4片),每日三次.疗程12周.【阿拓莫兰不良反应】1、过敏症:偶有皮疹等过敏症状,应停药.2、偶有食欲不振,恶心、呕吐、上腹痛等症状.【阿拓莫兰禁忌】对阿拓莫兰成分过敏者应禁用.【阿拓莫兰留意事项】放在儿童不容易触及的地方.【阿拓莫兰妊妇及哺乳期妇女用药】小鼠及家兔于孕期经静脉给予谷胱甘肽,未见生殖毒性反应.阿拓莫兰对妊妇及哺乳期妇女的影响尚不明确.【阿拓莫兰儿童用药】尚不明确.【阿拓莫兰老年用药】尚不明确.【阿拓莫兰药物彼此感化】阿拓莫兰不得与维生素B12、维生素K3、甲萘醌、泛酸钙、乳清酸、抗组胺制剂、磺胺药及四环素等混合使用.【阿拓莫兰药物过量】尚不明确.【阿拓莫兰储藏】密封.【阿拓莫兰无效期】24个月【阿拓莫兰批准文号】国药准字H0667【阿拓莫兰生产企业】企业名称:重庆药友制药无限义务公司【康德乐大药房网上药店友谊提示】商品图片信息展现仅供参考,终极包装以商品实物为准.欢迎纠错!说明书内容仅供查阅参考,终极以商品包装内说明书为准.欢迎纠错!使用商品时,请细心浏览说明书,并按说明书使用;如药品,请在医师指点下服用.。
戈那瑞林

化合物简介
基本信息
物化性质
基本信息
中文名称:戈那瑞林 中文别名:黄体激素释放激素 英文名称:gonadorelin 英文别名:Glp-His-Trp-Ser-Tyr-Gly-Leu-Arg-Pro-Gly-NH2;(pyroglutamic acid)-His-Trp-SerTyr-Gly-Leu-Arg-Pro-Gly-NH2;Gonadorelin;GONADERELIN;luteinizing hormone-releasing hormone isoform I; CAS号:-09-2 分子式:C55H75N17O13 分子量:1182. 结构式: 精确质量:1181. PSA:472.
适应症
1.用于诊断下丘脑-垂体-生殖腺功能障碍。 2.治疗闭经与促性腺激素分泌不足和多滤泡性卵巢引起的不孕症。 3.戈那瑞林或其同类物布舍瑞林、戈舍瑞林、亮丙瑞林、那法瑞林和曲普瑞林还可用于避孕、隐睾症、恶性 肿瘤(尤其前列腺癌)、延迟的和提前的青春期。 4.还可用于子宫内膜异位。 5.用于促排卵以治疗下丘脑性闭经所致不孕、原发性卵巢功能不足,特别是对氯米芬无效的患者。 6.还用于小儿隐睾症及雄激素过多、垂体肿瘤等。 。
氨基酸比值取本品约2mg,置10ml量瓶中,加6mol/L盐酸溶液使溶解并稀释至刻度,摇匀,量取1ml置2ml硬 质安瓶中,在-5℃减压封口,置110℃加热24小时,冷却,启封,将内容物移至蒸馏瓶中,在减压下蒸干,残留物 加0.02mol/L盐酸溶液1ml溶解,摇匀,作为供试品溶液;另取与供试品溶液浓度相应的各个氨基酸对照品混合溶 液,作为对照品溶液。精密量取上述两种溶液各20μl,分别注入氨基酸分析仪,记录色谱图,按外标法以峰面 积计算,即得。以L一组氨酸、L一谷氨酸、L一亮氨酸、L一脯氨酸、甘氨酸和L一精氨酸摩尔数之和的七分之一 值为1,供试品溶液中各个氨基酸相应的摩尔比应符合以下规定:L一丝氨酸为0.7—1.05,L一谷氨酸为0.9—l.
- 1、下载文档前请自行甄别文档内容的完整性,平台不提供额外的编辑、内容补充、找答案等附加服务。
- 2、"仅部分预览"的文档,不可在线预览部分如存在完整性等问题,可反馈申请退款(可完整预览的文档不适用该条件!)。
- 3、如文档侵犯您的权益,请联系客服反馈,我们会尽快为您处理(人工客服工作时间:9:00-18:30)。
Inhibitors, Agonists, Screening Libraries
Data Sheet
BIOLOGICAL ACTIVITY:
Galanthamine hydrobromide is a long–acting, centrally active acetylcholinesterase(AChE) inhibitor (IC50 = 410 nM) and allosteric potentiator at neuronal nicotinic ACh receptors.
IC50 Value: 410 nM
Target: AChE
Galanthamine hydrobromide prevents β–amyloid–induced apoptosis in SH–SY5Y and bovine chromaffin cells. Long–term
administration reduces amyloid precursor protein deposition and neurodegeneration in a mouse model of Alzheimer's disease.References:
[1]. Kita Y, Ago Y, Takano E, Fukada A, Takuma K, Matsuda T.Galantamine increases hippocampal insulin–like growth factor 2 expression via α7 nicotinic acetylcholine receptors in mice.Psychopharmacology (Berl). 2012 Aug 30.
[2]. Berkov S, Viladomat F, Codina C, Suárez S, Ravelo A, Bastida J.GC–MS of amaryllidaceous galanthamine–type alkaloids.J Mass Spectrom. 2012 Aug;47(8):1065–73.
[3]. Park CW, Son DD, Kim JY, Oh TO, Ha JM, Rhee YS, Park ES.Investigation of formulation factors affecting in vitro and in vivo characteristics of a galantamine transdermal system.Int J Pharm. 2012 Oct 15;436(1–2):32–40. Epub 2012 Jul 5.
[4]. Park YS, Kim SH, Kim SY, Kim YH, Lee MH, Yang SC, Shaw LM, Kang JS.Quantification of Galantamine in Human Plasma by Validated Liquid
Chromatography–Tandem Mass Spectrometry using Glimepride as an Internal Standard: Application to Bioavailability Studies in 32 Healthy Korean Subjects.J Chromatogr Sci. 2012 Jun 28.
[5]. Arias et al Galantamine prevents apoptosis induced by b–amyloid and thapsigargin: involvement of nicotinic acetycholine receptors. Neuropharmacology (2004) 46 103.
Product Name:
Galanthamine (hydrobromide)Cat. No.:
HY-A0009CAS No.:
1953-04-4Molecular Formula:
C 17H 22BrNO 3Molecular Weight:
368.27Target:
AChE Pathway:
Neuronal Signaling Solubility:
DMSO: ≥ 3.5 mg/mL; DMSO < 7.8 mg/mL
Caution: Product has not been fully validated for medical applications. For research use only.
Tel: 609-228-6898 Fax: 609-228-5909 E-mail: tech@ Address: 1 Deer Park Dr, Suite Q, Monmouth Junction, NJ 08852, USA。
