剪切

Double Nicking by RNA-Guided CRISPR Cas9for Enhanced
Genome Editing Speci?city
F.Ann Ran,1,2,3,4,5,11Patrick D.Hsu,1,2,3,4,5,11Chie-Yu Lin,1,2,3,4,6Jonathan S.Gootenberg,1,2,3,4
Silvana Konermann,1,2,3,4Alexandro E.Trevino,1David A.Scott,1,2,3,4Azusa Inoue,7,8,9,10Shogo Matoba,7,8,9,10 Yi Zhang,7,8,9,10and Feng Zhang1,2,3,4,*
1Broad Institute of MIT and Harvard,7Cambridge Center,Cambridge,MA02142,USA
2McGovern Institute for Brain Research
3Department of Brain and Cognitive Sciences
4Department of Biological Engineering
Massachusetts Institute of Technology,Cambridge,MA02139,USA
5Department of Molecular and Cellular Biology,Harvard University,Cambridge,MA02138,USA
6Harvard/MIT Division of Health Sciences and Technology
7Howard Hughes Medical Institute
8Program in Cellular and Molecular Medicine
9Department of Genetics
10Harvard Stem Cell Institute
Harvard Medical School,Boston,MA02115,USA
11These authors contributed equally to this work
*Correspondence:zhang@https://www.360docs.net/doc/7d12645802.html,
https://www.360docs.net/doc/7d12645802.html,/10.1016/j.cell.2013.08.021
SUMMARY
Targeted genome editing technologies have enabled a broad range of research and medical applications. The Cas9nuclease from the microbial CRISPR-Cas system is targeted to speci?c genomic loci by a20 nt guide sequence,which can tolerate certain mis-matches to the DNA target and thereby promote undesired off-target mutagenesis.Here,we describe an approach that combines a Cas9nickase mutant with paired guide RNAs to introduce targeted dou-ble-strand breaks.Because individual nicks in the genome are repaired with high?delity,simultaneous nicking via appropriately offset guide RNAs is required for double-stranded breaks and extends the number of speci?cally recognized bases for target cleavage.We demonstrate that using paired nicking can reduce off-target activity by50-to 1,500-fold in cell lines and to facilitate gene knockout in mouse zygotes without sacri?cing on-target cleav-age ef?ciency.This versatile strategy enables a wide variety of genome editing applications that require high speci?city.
INTRODUCTION
The ability to perturb the genome in a precise and targeted fashion is crucial for understanding genetic contributions to biology and disease.Genome engineering of cell lines or animal models has traditionally been accomplished through random mutagenesis or low-ef?ciency gene targeting.To facilitate genome editing,programmable sequence-speci?c DNA nuclease technologies have enabled targeted modi?cation of endogenous genomic sequences with high ef?ciency,particu-larly in species that have proven traditionally genetically intrac-table(Carlson et al.,2012;Geurts et al.,2009;Takasu et al., 2010;Watanabe et al.,2012).The RNA-guided Cas9nucleases from the microbial CRISPR(clustered regularly interspaced short palindromic repeat)-Cas systems are robust and versatile tools for stimulating targeted double-stranded DNA breaks(DSBs)in eukaryotic cells(Chang et al.,2013;Cho et al.,2013;Cong et al.,2013;Deltcheva et al.,2011;Deveau et al.,2010;Friedland et al.,2013;Gratz et al.,2013;Horvath and Barrangou,2010; Jinek et al.,2013;Mali et al.,2013b;Wang et al.,2013),where the resulting cellular repair mechanisms—nonhomologous end-joining(NHEJ)or homology-directed repair(HDR)pathways—can be exploited to induce error-prone or de?ned alterations (Hsu and Zhang,2012;Perez et al.,2008;Urnov et al.,2010). The Cas9nuclease from Streptococcus pyogenes can be directed by a chimeric single-guide RNA(sgRNA)(Jinek et al., 2012)to any genomic locus followed by a50-NGG protospacer-adjacent motif(PAM).A20nt guide sequence within the sgRNA directs Cas9to the genomic target via Watson-Crick base pairing and can be easily programmed to target a desired genomic locus (Deltcheva et al.,2011;Deveau et al.,2010;Gasiunas et al.,2012; Jinek et al.,2012).Recent studies of Cas9speci?city have demon-strated that,although each base within the20nt guide sequence contributes to overall speci?city,multiple mismatches between the guide RNA and its complementary target DNA sequence can be tolerated depending on the quantity,position,and base identity of mismatches(Cong et al.,2013;Fu et al.,2013;Hsu et al.,2013;Jiang et al.,2013),leading to potential off-target Cell154,1–10,September12,2013a2013Elsevier Inc.1
DSBs and indel formation.These unwanted mutations can poten-tially limit the utility of Cas9for genome editing applications that require high levels of precision,such as generation of isogenic cell lines for testing causal genetic variations (Soldner et al.,2011)or in vivo and ex vivo genome-editing-based therapies.To improve the speci?city of Cas9-mediated genome editing,we developed a strategy that combines the D10A mutant nickase version of Cas9(Cas9n)(Cong et al.,2013;Gasiunas et al.,2012;Jinek et al.,2012)with a pair of offset sgRNAs complementary to opposite strands of the target site.Whereas nicking of both DNA strands by a pair of Cas9nickases leads to site-speci?c DSBs and NHEJ,individual nicks are predominantly repaired by the high-?delity base excision repair pathway (BER)(Dianov and Hu
¨bscher,2013).A paired nickase strategy was described while this manuscript was under review,which suggests the possibility for engineering a system to ameliorate off-target activity (Mali et al.,2013a ).In a manner analogous to dimeric zinc ?nger nucle-ases (ZFNs)(Miller et al.,2007;Porteus and Baltimore,2003;Sander et al.,2011;Wood et al.,2011)and transcription-acti-vator-like effector nucleases (TALENs)(Boch et al.,2009;Chris-tian et al.,2010;Miller et al.,2011;Moscou and Bogdanove,2009;Reyon et al.,2012;Sanjana et al.,2012;Wood et al.,2011;Zhang et al.,2011),wherein DNA cleavage requires syner-gistic interaction of two independent speci?city-encoding DNA-binding modules directing FokI nuclease monomers,this double-nicking strategy minimizes off-target mutagenesis
by
C
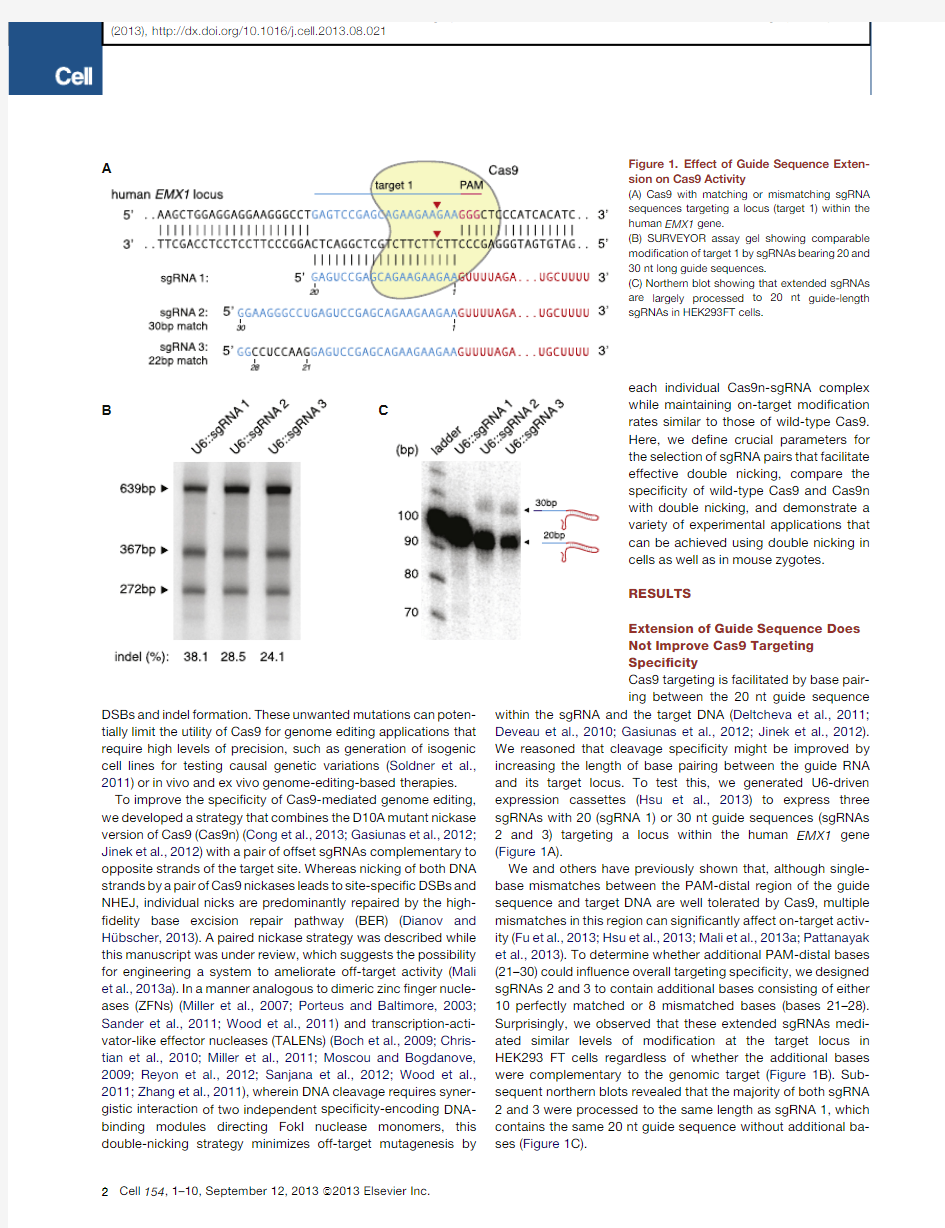
B Figure 1.Effect of Guide Sequence Exten-sion on Cas9Activity
(A)Cas9with matching or mismatching sgRNA sequences targeting a locus (target 1)within the human EMX1gene.
(B)SURVEYOR assay gel showing comparable modi?cation of target 1by sgRNAs bearing 20and 30nt long guide sequences.
(C)Northern blot showing that extended sgRNAs are largely processed to 20nt guide-length sgRNAs in HEK293FT cells.
each individual Cas9n-sgRNA complex while maintaining on-target modi?cation rates similar to those of wild-type Cas9.Here,we de?ne crucial parameters for the selection of sgRNA pairs that facilitate effective double nicking,compare the speci?city of wild-type Cas9and Cas9n with double nicking,and demonstrate a variety of experimental applications that can be achieved using double nicking in cells as well as in mouse zygotes.RESULTS
Extension of Guide Sequence Does Not Improve Cas9Targeting Speci?city
Cas9targeting is facilitated by base pair-ing between the 20nt guide sequence
within the sgRNA and the target DNA (Deltcheva et al.,2011;Deveau et al.,2010;Gasiunas et al.,2012;Jinek et al.,2012).We reasoned that cleavage speci?city might be improved by increasing the length of base pairing between the guide RNA and its target locus.To test this,we generated U6-driven expression cassettes (Hsu et al.,2013)to express three sgRNAs with 20(sgRNA 1)or 30nt guide sequences (sgRNAs 2and 3)targeting a locus within the human EMX1gene (Figure 1A).
We and others have previously shown that,although single-base mismatches between the PAM-distal region of the guide sequence and target DNA are well tolerated by Cas9,multiple mismatches in this region can signi?cantly affect on-target activ-ity (Fu et al.,2013;Hsu et al.,2013;Mali et al.,2013a;Pattanayak et al.,2013).To determine whether additional PAM-distal bases (21–30)could in?uence overall targeting speci?city,we designed sgRNAs 2and 3to contain additional bases consisting of either 10perfectly matched or 8mismatched bases (bases 21–28).Surprisingly,we observed that these extended sgRNAs medi-ated similar levels of modi?cation at the target locus in HEK293FT cells regardless of whether the additional bases were complementary to the genomic target (Figure 1B).Sub-sequent northern blots revealed that the majority of both sgRNA 2and 3were processed to the same length as sgRNA 1,which contains the same 20nt guide sequence without additional ba-ses (Figure 1C).
2Cell 154,1–10,September 12,2013a2013Elsevier
Inc.
Cas9Nickase Generates Ef?cient NHEJ with Paired,Offset Guide RNAs
Given that extension of the guide sequence failed to improve Cas9targeting speci?city,we sought an alternative strategy for increasing the overall base-pairing length between the guide sequence and its DNA target.Cas9enzymes contain two conserved nuclease domains,HNH and RuvC,which cleave the DNA strand complementary and noncomplementary to the guide RNA,respectively.Mutations of the catalytic residues (D10A in RuvC and H840A in HNH)convert Cas9into DNA nickases (Cong et al.,2013;Gasiunas et al.,2012;Jinek et al.,2012).As single-strand nicks are preferentially repaired by the
high-?delity BER pathway (Dianov and Hu
¨bscher,2013),we reasoned that two Cas9-nicking enzymes directed by a pair of sgRNAs targeting opposite strands of a target locus could mediate DSBs while minimizing off-target activity (Figure 2A).A number of factors may affect cooperative nicking leading to indel formation,including steric hindrance between two adjacent Cas9molecules or Cas9-sgRNA complexes,overhang type,
and
C
B
A
Figure 2.Double Nicking Facilitates Ef?-cient Genome Editing in Human Cells
(A)Schematic illustrating DNA double-stranded breaks using a pair of sgRNAs guiding Cas9D10A nickases (Cas9n).The D10A mutation renders Cas9able to cleave only the strand complemen-tary to the sgRNA;a pair of sgRNA-Cas9n com-plexes can nick both strands simultaneously.sgRNA offset is de?ned as the distance between the PAM-distal (50)ends of the guide sequence of a given sgRNA pair;positive offset requires the sgRNA complementary to the top strand (sgRNA a)to be 50of the sgRNA complementary to the bottom strand (sgRNA b),which always creates a 50overhang.
(B)Ef?ciency of double-nicking-induced NHEJ as a function of the offset distance between two sgRNAs.Sequences for all sgRNAs used can be found in Table S1.n =3;error bars show mean ±SEM.
(C)Representative sequences of the human EMX1locus targeted by Cas9n.sgRNA target sites and PAMs are indicated by blue and magenta bars,respectively.(Bottom)Selected sequences showing representative indels.See also Tables S1and S2.
sequence context;some of these may be characterized by testing multiple sgRNA pairs with distinct target sequences and offsets (the distance between the PAM-distal [50]ends of the guide sequence of a given sgRNA pair).To systematically assess how sgRNA offsets might affect subsequent repair and generation of in-dels,we ?rst designed sets of sgRNA pairs targeted against the human EMX1genomic locus separated by a range of offset distances from approximately à200to 200bp to create both 50and 30
overhang products (Figure 2A and Table S1available online).We then assessed the ability of each sgRNA pair with the D10A Cas9mutant (referred to as Cas9n;H840A Cas9mutant is referred to as Cas9H840A)to generate indels in human HEK 293FT cells.Robust NHEJ (up to 40%)was observed for sgRNA pairs with offsets from à4to 20bp,with modest indels forming in pairs offset by up to 100bp (Figure 2B,left).We subsequently recapitulated these ?ndings by testing similarly offset sgRNA pairs at two other genomic loci,DYRK1A and GRIN2B (Figure 2B,right).Of note,across all three loci examined,only sgRNA pairs creating 50overhangs with less than 8bp overlap between the guide sequences (offset greater than à8bp)were able to mediate detectable indel formation (Figure 2C).
Importantly,each guide used in these assays is able to ef?-ciently induce indels when paired with wild-type Cas9(Table S1),indicating that the relative positions of the guide pairs are the most important parameters in predicting double-nicking activity.Because Cas9n and Cas9H840A nick opposite strands of DNA,substitution of Cas9n with Cas9H840A with a given
Cell 154,1–10,September 12,2013a2013Elsevier Inc.
3
A
B
C D
E F
(legend on next page) 4Cell154,1–10,September12,2013a2013Elsevier Inc.
sgRNA pair should result in the inversion of the overhang type.For example,a pair of sgRNAs that will generate a 50overhang with Cas9n should,in principle,generate the corresponding 30overhang instead.Therefore,sgRNA pairs that lead to the gener-ation of a 30overhang with Cas9n might be used with Cas9H840A to generate a 50overhang.Further work will be needed to identify the necessary design rules for sgRNA pairing to allow double nicking by Cas9H840A.
Double Nicking Mediates Ef?cient Genome Editing with Improved Speci?city
Having established that double nicking (DN)mediates high-ef?-ciency NHEJ at levels comparable to those induced by wild-type Cas9(Table S1),we next studied whether DN has improved speci?city over wild-type Cas9by measuring their off-target ac-tivities.We co-delivered Cas9n with sgRNAs 1and 9,spaced by a +23bp offset,to target the human EMX1locus in HEK 293FT cells (Figure 3A).This DN con?guration generated on-target indel levels similar to those generated by the wild-type Cas9paired with each sgRNA alone (Figure 3B,left).Strikingly,unlike with wild-type Cas9,DN did not generate detectable modi?cation at a previously validated sgRNA 1off-target site,OT-4,by SUR-VEYOR assay (Hsu et al.,2013;Figure 3B,right),suggesting that DN can potentially reduce the likelihood of off-target modi?cations.
Using deep sequencing to assess modi?cation at ?ve different sgRNA 1off-target loci (Figure 3A),we observed signi?cant mutagenesis at all sites with wild-type Cas9+sgRNA 1(Fig-ure 3C).In contrast,cleavage by Cas9n at 5off-target sites tested was barely detectable above background sequencing https://www.360docs.net/doc/7d12645802.html,ing the ratio of on-to off-target modi?cation levels as a metric of speci?city,we found that Cas9n with a pair of sgRNAs was able to achieve >100-fold greater speci?city relative to wild-type Cas9with one of the sgRNAs (Figure 3D).We conducted additional off-target analysis by deep sequencing for two sgRNA pairs (offsets of +16and +20bp)targeting the VEGFA locus,with similar results (Figure 3E).DN at these off-target loci (Table S5)was able to achieve 200-to >1,500-fold greater speci?city than the wild-type Cas9(Figure 3F and Table S1).Taken together,these results demonstrate that Cas9-mediated double nicking minimizes off-target mutagenesis and is suitable for genome editing with increased speci?city.
Double Nicking Facilitates High-Ef?ciency Homology-Directed Repair,NHEJ-Mediated DNA Insertion,and Genomic Microdeletions
DSBs can stimulate homology-directed repair (HDR)to enable highly precise editing of genomic target sites.To evaluate DN-induced HDR,we targeted the human EMX1locus with pairs of sgRNAs offset by à3and +18bp (generating 31and 52bp 50overhangs),respectively,and introduced a single-stranded oligodeoxynucleotide (ssODN)bearing a Hin dIII restriction site as the HDR repair template (Figure 4A).Each DN sgRNA pair successfully induced HDR at frequencies higher than those of single-guide Cas9n nickases and comparable to those of wild-type Cas9(Figure 4B).Furthermore,genome editing in embry-onic stem cells or patient-derived induced pluripotent stem cells represents a key opportunity for generating and studying new disease paradigms as well as developing new therapeutics.Because single-nick approaches to inducing HDR in human embryonic stem cells (hESCs)have met with limited success (Hsu et al.,2013),we attempted DN in the HUES62hES cell line and observed successful HDR (Figure 4C).
To further characterize how offset sgRNA spacing affects the ef?ciency of HDR,we next tested in HEK 293FT cells a set of sgRNA pairs in which the cleavage site of at least one sgRNA is situated near the site of recombination (overlapping with the HDR ssODN donor template arm).We observed that sgRNA pairs generating 50overhangs and having at least one nick occur-ring within 22bp of the homology arm are able to induce HDR at levels comparable to those of wild-type Cas9-mediated HDR and signi?cantly greater than those of single Cas9n-sgRNA nick-ing.In contrast,we did not observe HDR with sgRNA pairs that generated 30overhangs or double nicking of the same DNA strand (Figure 4D).
The ability to create de?ned overhangs could enable precise insertion of donor repair templates containing compatible over-hangs via NHEJ-mediated ligation (Maresca et al.,2013).To explore this alternative strategy for transgene insertion,we tar-geted the EMX1locus with Cas9n and an sgRNA pair designed to generate a 43bp 50overhang near the stop codon and sup-plied a double-stranded oligonucleotide (dsODN)duplex with matching overhangs (Figure 5A).The annealed dsODN insert,containing multiple epitope tags and a restriction site,was suc-cessfully integrated into the target (1out of 37screened by Sanger sequencing of cloned amplicons).This ligation-based strategy thus illustrates an effective approach for inserting dsODNs encoding short modi?cations such as protein tags or recombination sites into an endogenous locus.
Additionally,we targeted combinations of sgRNA pairs (four sgRNAs per combination)to the DYRK1A locus in HEK 293FT cells to facilitate genomic microdeletions.We generated a set of sgRNAs to mediate 0.5kb,1kb,2kb,and 6kb deletions (Fig-ure 5B and Table S2;sgRNAs 32,33,and 54–61)and veri?ed successful multiplex nicking-mediated deletion over these ranges via PCR screen of predicted deletion sizes.
Figure 3.Double Nicking Facilitates Ef?cient Genome Editing in Human Cells
(A)Schematic illustrating Cas9n double nicking (red arrows)the human EMX1locus.Five off-target loci with sequence homology to EMX1target 1were selected to screen for Cas9n speci?city.
(B)On-target modi?cation rate by Cas9n and a pair of sgRNAs is comparable to those mediated by wild-type Cas9and single sgRNAs (left).Cas9-sgRNA 1complexes generate signi?cant off-target mutagenesis,whereas no off-target locus modi?cation is detected with Cas9n (right).
(C)Five off-target loci of sgRNA 1are examined for indel modi?cations by deep sequencing of transfected HEK 293FT cells.n =3;error bars show mean ±SEM.(D)Speci?city comparison of Cas9n with double nicking and wild-type Cas9with sgRNA alone at the off-target sites.Speci?city ratio is calculated as on-target/off-target modi?cation rates.n =3;error bars show mean ±SEM.
(E and F)Double nicking minimizes off-target modi?cation at two additional human VEGFA loci while maintaining high speci?city (on/off-target modi?cation ratio).n =3;error bars show mean ±SEM.
Cell 154,1–10,September 12,2013a2013Elsevier Inc.
5
A
B C
D
Figure4.Double Nicking Allows Insertion into the Genome via HDR in Human Cells
(A)Schematic illustrating HDR mediated via a single-stranded oligodeoxynucleotide(ssODN)template at a DSB created by a pair of Cas9n enzymes.A12nt sequence(red),including a Hin dIII restriction site,is inserted into the EMX1locus at the position marked by the gray dashed lines;distances of Cas9n-mediated nicks from the HDR insertion site are indicated on top in italics.
(B)Restriction digest assay gel showing successful insertion of Hin dIII cleavage sites by double-nicking-mediated HDR in HEK293FT cells.Top bands are unmodi?ed template;bottom bands are HindIII cleavage product.
(C)Double nicking promotes HDR in the HUES62human embryonic stem cell line.HDR frequencies are determined by deep sequencing.n=3;error bars show mean±SEM.
(legend continued on next page) 6Cell154,1–10,September12,2013a2013Elsevier Inc.
Double Nicking Enables Ef?cient Genome Modi?cation in Mouse Zygotes
Recent work demonstrated that co-delivery of wild-type Cas9mRNA along with multiple sgRNAs can mediate single-step gen-eration of transgenic mice carrying multiple allelic modi?cations (Wang et al.,2013).Given the ability to achieve genome modi?-cation in vivo using several sgRNAs at once,we sought to assess the ef?ciency of multiple nicking by Cas9n in mouse zygotes.Cytoplasmic coinjection of wild-type Cas9or Cas9n mRNA and sgRNAs into single-cell mouse zygotes allowed successful targeting of the Mecp2locus (Figure 6A).To identify the optimal concentration of Cas9n mRNA and sgRNA for ef?cient gene tar-geting,we titrated Cas9mRNA from 100to 3ng/ul while main-taining the sgRNA levels at a 1:20Cas9:sgRNA molar ratio.All concentrations tested for Cas9double-nicking-mediated modi-?cations in at least 80%of embryos screened,similar to levels achieved by wild-type Cas9(Figure 6B).Taken together,these results suggest a number of applications for double-nicking-based genome editing.
(D)HDR ef?ciency depends on the con?guration of Cas9or Cas9n-mediated nicks.HDR is facilitated when a nick occurs near the center of the ssODN homology arm (HDR insertion site),leading to a 50resulting overhang.Nicking con?gurations are annotated with position and strand (red arrows)and length of overhang (black lines)(left).The distance (bp)of each nick from the HDR insertion site is indicated at the end of the black lines in italics,and the positions of the sgRNAs are illustrated in bold on the schematic of the EMX1locus.HDR ef?ciency mediated by double nicking with paired sgRNAs (top)or single sgRNAs with either Cas9or Cas9n are shown (bottom and Table S2).n =3;error bars show mean ±
SEM.
A
B
Figure 5.Multiplexed Nicking Facilitates Non-HR-Mediated Gene Integration and Genomic Deletions
(A)Schematic showing insertion of a double-stranded oligodeoxynucleotide (dsODN)donor fragment bearing overhangs complementary to 50overhangs created by Cas9double nicking.The dsODN was designed to remove the native EMX1stop codon and contains a HA tag,33FLAG tag,Hin dIII restriction site,Myc epitope tag,and a stop codon in frame,totaling 148bp.Successful insertion was veri?ed by Sanger sequencing as shown (1out of 37clones screened).Amino acid translation of the modi?ed lo-cus is shown below the DNA sequence.
(B)Co-delivery of four sgRNAs with Cas9n generates long-range genomic deletions in the DYRK1A locus (from 0.5to 6kb).Deletion was detected using primers (Table S6)spanning the target region.
DISCUSSION
Given the permanent nature of genomic modi?cations,speci?city is of paramount importance to sensitive applications such as studies aimed at linking speci?c genetic variants with biological processes or disease phenotypes and gene therapy.Here,we have explored strategies to improve the tar-geting speci?city of Cas9.Although simply extending the guide sequence length of sgRNA failed to improve targeting speci-?city,combining two appropriately offset sgRNAs with Cas9n effectively generated in-dels while minimizing unwanted cleavage because individual off-target single-stranded nicks are repaired with high ?delity via base excision repair.Given that signi?cant off-target mutagen-esis has been previously reported for Cas9nucleases in human cells (Fu et al.,2013;Hsu et al.,2013),the DN approach could provide a generalizable solution for rapid and accurate genome editing.The characterization of spacing parameters governing successful Cas9double-nickase-mediated gene targeting re-veals an effective offset window >100bp long,allowing for a high degree of ?exibility in the selection of sgRNA pairs.Previous computational analyses have revealed an average targeting range of every 12bp for the Streptococcus pyogenes Cas9in the human genome based on the 50NGG PAM (Cong et al.,2013),suggesting that appropriate sgRNA pairs should be readily identi?able for most loci within the genome.We have additionally demonstrated DN-mediated indel frequencies com-parable to wild-type Cas9modi?cation at multiple genes and loci in both human and mouse cells,con?rming the reproducibility of this strategy for high-precision genome engineering (Table S1).
Cell 154,1–10,September 12,2013a2013Elsevier Inc.
7
The Cas9double nicking approach is,in principle,similar to ZFN-and TALEN-based genome editing systems,in which cooperation between two heminuclease domains is required to achieve double-stranded break at the target site.Systematic studies of ZFN and TALEN systems have revealed that the tar-geting speci?city of a given ZFN and TALEN pair can be highly dependent on the nuclease architecture (homo-or heterodimeric nucleases)or target sequence,and in some cases TALENs can be highly speci?c (Ding et al.,2013).Although the wild-type Cas9system has been shown to exhibit high levels of off-target mutagenesis,the DN system is a promising solution and brings RNA-guided genome editing to similar speci?city levels as ZFNs and TALENs.
Additionally,the ease and ef?ciency with which Cas9can be targeted renders the DN system especially attractive.However,DNA targeting using DN will likely face similar off-target challenges as ZFNs and TALENs,in which cooperative nicking at off-target sites might still occur,albeit at a signi?cantly reduced likelihood.Given the extensive characterization of Cas9speci-?city and sgRNA mutation analysis (Fu et al.,2013;Hsu et al.,2013),as well as the NHEJ-mediating sgRNA offset range identi-?ed in this study,computational approaches may be used to evaluate the likely off-target sites for a given pair of sgRNAs.To facilitate sgRNA pair selection,we developed an online web tool that identi?es sgRNA combinations with optimal spacing for dou-ble nicking applications (https://www.360docs.net/doc/7d12645802.html,/).Although Cas9n has been previously shown to facilitate HDR at on-target sites (Cong et al.,2013),its ef?ciency is substantially lower than that of wild-type Cas9.The double nicking strategy,by comparison,maintains high on-target ef?ciencies while reducing off-target modi?cations to background levels.
Never-A
B
Figure 6.Cas9Double Nicking Mediates Ef?cient Indel Formation in Mouse Embryos
(A)Schematic illustrating Cas9n double nicking the mouse Mecp2locus.Representative indels are shown for mouse blastocysts coinjected with in-vitro-transcribed Cas9n-encoding mRNA and sgRNA pairs matching targets 92and 93.
(B)Ef?cient blastocyst modi?cation is achieved at multiple concentrations of sgRNAs (1.5to 50ng/ul)and wild-type Cas9or Cas9n (3to 100ng/ul).
theless,further characterizations of DN off-target activity,particularly via whole-genome sequencing and targeted deep sequencing of cells or whole organisms generated using the DN approach,are urgently needed to evaluate the utility of Cas9n DN in biotechnological or clinical applications that require ultrahigh-preci-sion genome editing.Additionally,Cas9n has been shown to induce low levels of in-dels at on-target sites for certain sgRNAs (Mali et al.,2013b ),which may result from residual double-strand break activ-ities and may be circumvented by further structure-function studies of Cas9cata-lytic activity.Overall,Cas9n-mediated multiplex nicking serves as a customizable platform for highly precise and ef?cient tar-geted genome engineering and promises to broaden the range of applications in biotechnology,basic science,and medicine.
EXPERIMENTAL PROCEDURES
Cell Culture and Transfection
Human embryonic kidney (HEK)cell line 293FT (Life Technologies)cell line was maintained in Dulbecco’s modi?ed Eagle’s medium (DMEM)supple-mented with 10%fetal bovine serum (HyClone),2mM GlutaMAX (Life Technol-ogies),100U/ml penicillin,and 100m g/ml streptomycin at 37 C with 5%CO 2incubation.
Cells were seeded onto 24-well plates (Corning)at a density of 120,000cells/well,24hr prior to transfection.Cells were transfected using Lipofect-amine 2000(Life Technologies)at 80%–90%con?uency following the manu-facturer’s recommended protocol.A total of 500ng Cas9plasmid and 100ng of U6-sgRNA PCR product was transfected.
Human embryonic stem cell line HUES62(Harvard Stem Cell Institute core)was maintained in feeder-free conditions on GelTrex (Life Technologies)in mTesR medium (StemCell Technologies)supplemented with 100ug/ml Nor-mocin (InvivoGen).HUES62cells were transfected with Amaxa P3Primary Cell 4-D Nucleofector Kit (Lonza)following the manufacturer’s protocol.SURVEYOR Nuclease Assay for Genome Modi?cation
HEK 293FT and HUES62cells were transfected with DNA as described above.Cells were incubated at 37 C for 72hr posttransfection prior to genomic DNA extraction.Genomic DNA was extracted using the QuickExtract DNA Extrac-tion Solution (Epicenter)following the manufacturer’s protocol.In brief,pel-leted cells were resuspended in QuickExtract solution and were incubated at 65 C for 15min,68 C for 15min,and 98 C for 10min.
The genomic region ?anking the CRISPR target site for each gene was PCR ampli?ed (Table S3),and products were puri?ed using QiaQuick Spin Column (QIAGEN)following the manufacturer’s protocol.400ng total of the puri?ed PCR products were mixed with 2m l 103Taq DNA Polymerase PCR buffer
8Cell 154,1–10,September 12,2013a2013Elsevier
Inc.
(Enzymatics)and ultrapure water to a?nal volume of20m l and were subjected to a reannealing process to enable heteroduplex formation:95 C for10min;
95 C to85 C ramping at–2 C/s;85 C to25 C at–0.25 C/s;and25 C hold for1min.After reannealing,products were treated with SURVEYOR nuclease and SURVEYOR enhancer S(Transgenomics)following the manufacturer’s recommended protocol were and analyzed on4%–20%Novex TBE polyacryl-amide gels(Life Technologies).Gels were stained with SYBR Gold DNA stain (Life Technologies)for30min and were imaged with a Gel Doc gel imaging system(Bio-Rad).Quanti?cation was based on relative band intensities.Indel percentage was determined by the formula1003(1–(1–(b+c)/(a+b+c))1/2), wherein a is the integrated intensity of the undigested PCR product and b and c are the integrated intensities of each cleavage product.
Northern Blot Analysis of TracrRNA Expression in Human Cells Northern blots were performed as previously described(Cong et al.,2013).In brief,RNAs were extracted using the mirPremier microRNA Isolation Kit (Sigma)and were heated to95 C for5min before loading on8%denaturing polyacrylamide gels(SequaGel,National Diagnostics).Afterward,RNA was transferred to a prehybridized Hybond N+membrane(GE Healthcare)and was crosslinked with Stratagene UV Crosslinker(Stratagene).Probes were labeled with[g-32P]ATP(Perkin Elmer)with T4polynucleotide kinase(New England Biolabs).After washing,membrane was exposed to phosphor screen for1hr and scanned with phosphorimager(Typhoon).
Deep Sequencing to Assess Targeting Speci?city
HEK293FT cells were plated and transfected as described above72hr prior to genomic DNA extraction.The genomic region?anking the CRISPR target site for each gene was ampli?ed(see Table S4for primer sequences)by a fusion PCR method to attach the Illumina P5adapters as well as unique sample-speci?c barcodes to the target.PCR products were puri?ed using EconoSpin 96-well Filter Plates(Epoch Life Sciences)following the manufacturer’s recom-mended protocol.
Barcoded and puri?ed DNA samples were quanti?ed by Qubit2.0Fluorom-eter(Life Technologies)and were pooled in an equimolar ratio.Sequencing libraries were then sequenced with the Illumina MiSeq Personal Sequencer (Life Technologies).
Sequencing Data Analysis,Indel Detection,and Homologous Recombination Detection
MiSeq reads were?ltered by requiring an average Phred quality(Q score)of at least30,as well as perfect sequence matches to barcodes and amplicon for-ward primers.Reads from on-and off-target loci were analyzed by performing Ratcliff-Obershelp string comparison,as implemented in the Python dif?ib module,against loci sequences that included30nt upstream and downstream of the target site(a total of80bp).The resulting edit operations were parsed, and reads were counted as indels if insertion or deletion operations were found.Analyzed target regions were discarded if part of their alignment fell outside of the MiSeq read itself or if more than?ve bases were uncalled. Negative controls for each sample provided a gauge for the inclusion or exclusion of indels as putative cutting events.For quanti?cation of homo-logous recombination,reads were?rst processed as in the indel detection work?ow and were then checked for presence of homologous recombination template CCAGGCTTGG.
Microinjection into Mouse Zygotes
Cas9mRNA and sgRNA templates were ampli?ed with T7promoter sequence-conjugated primers.After gel puri?cation,Cas9and Cas9n were transcribed with mMESSAGE mMACHINE T7Ultra Kit(Life Technologies). sgRNAs were transcribed with MEGAshortscript T7Kit(Life Technologies). RNAs were puri?ed by MEGAclear Kit(Life Technologies)and frozen at–80 C. MII-stage oocytes were collected from8-week-old superovulated BDF1fe-males by injecting7.5I.U.of PMSG(Harbor,UCLA)and hCG(Millipore).They were transferred into HTF medium supplemented with10mg/ml bovine serum albumin(BSA;Sigma-Aldrich)and were inseminated with capacitated sperm obtained from the caudal epididymides of adult C57BL/6male mice.Six hours after fertilization,zygotes were injected with mRNAs and sgRNAs in M2media (Millipore)using a Piezo impact-driven micromanipulator(Prime Tech Ltd.,Ibaraki,Japan).The concentrations of Cas9and Cas9n mRNAs and sgRNAs are described in the text and Figure6B.After microinjection,zygotes were cultured in KSOM(Millipore)in a humidi?ed atmosphere of5%CO2and 95%air at37 C.
Genome Extraction from Blastocyst Embryos
Following in vitro culture of embryos for6days,the expanded blastocysts were washed with0.01%BSA in PBS and were individually collected into 0.2ml tubes.Five microliters of genome extraction solution(50mM Tris-HCl [pH8.0],0.5%Triton X-100,and1mg/ml Proteinase K)were added and the samples were incubated in65 C for3hr followed by95 C for10min.Samples were then ampli?ed for targeted deep sequencing as described above.
SUPPLEMENTAL INFORMATION
Supplemental Information includes Extended Experimental Procedures and six tables and can be found with this article online at https://www.360docs.net/doc/7d12645802.html,/10. 1016/j.cell.2013.08.021.
ACKNOWLEDGMENTS
We thank Joshua Weinstein and Yinqing Li for statistical consultation,Xuebing Wu and Phillip Sharp for assistance with northern blotting experiments,Su Vora for help with the manuscript,and the entire Zhang lab for their support and advice.P.D.H.is a James Mills Pierce Fellow.C.-Y.L.is supported by T32GM007753from the National Institute of General Medical Sciences.
D.A.S.is an NSF predoctoral fellow.A.I.and S.M.are research fellows for Research Abroad of the Japan Society for the Promotion of Science.Y.Z.is supported by NIH grants GM68804and U01DK089565and is an Investigator of the Howard Hughes Medical Institute.F.Z.is supported by an NIH Director’s Pioneer Award(1DP1-MH100706),a NIH Transformative R01grant(1R01-DK097768),the Keck,McKnight,Damon Runyon,Searle Scholars,Klingen-stein,Vallee,and Simons Foundations,Bob Metcalfe,and Jane Pauley.
Received:July25,2013
Revised:August13,2013
Accepted:August14,2013
Published:August29,2013
REFERENCES
Boch,J.,Scholze,H.,Schornack,S.,Landgraf,A.,Hahn,S.,Kay,S.,Lahaye, T.,Nickstadt,A.,and Bonas,U.(2009).Breaking the code of DNA binding speci?city of TAL-type III effectors.Science326,1509–1512.
Carlson,D.F.,Tan,W.F.,Lillico,S.G.,Stverakova,D.,Proudfoot,C.,Christian, M.,Voytas,D.F.,Long,C.R.,Whitelaw,C.B.A.,and Fahrenkrug,S.C.(2012). Ef?cient TALEN-mediated gene knockout in livestock.Proc.Natl.Acad.Sci. USA109,17382–17387.
Chang,N.,Sun,C.,Gao,L.,Zhu,D.,Xu,X.,Zhu,X.,Xiong,J.W.,and Xi,J.J. (2013).Genome editing with RNA-guided Cas9nuclease in zebra?sh embryos. Cell Res.23,465–472.
Cho,S.W.,Kim,S.,Kim,J.M.,and Kim,J.S.(2013).Targeted genome engi-neering in human cells with the Cas9RNA-guided endonuclease.Nat. Biotechnol.31,230–232.
Christian,M.,Cermak,T.,Doyle,E.L.,Schmidt,C.,Zhang,F.,Hummel,A., Bogdanove, A.J.,and Voytas, D.F.(2010).Targeting DNA double-strand breaks with TAL effector nucleases.Genetics186,757–761.
Cong,L.,Ran,F.A.,Cox,D.,Lin,S.,Barretto,R.,Habib,N.,Hsu,P.D.,Wu,X., Jiang,W.,Marraf?ni,L.A.,and Zhang,F.(2013).Multiplex genome engineering using CRISPR/Cas systems.Science339,819–823.
Deltcheva,E.,Chylinski,K.,Sharma,C.M.,Gonzales,K.,Chao,Y.,Pirzada, Z.A.,Eckert,M.R.,Vogel,J.,and Charpentier,E.(2011).CRISPR RNA matura-tion by trans-encoded small RNA and host factor RNase III.Nature471, 602–607.
Cell154,1–10,September12,2013a2013Elsevier Inc.
9
Deveau,H.,Garneau,J.E.,and Moineau,S.(2010).CRISPR/Cas system and its role in phage-bacteria interactions.Annu.Rev.Microbiol.64,475–493.Dianov,G.L.,and Hu
¨bscher,U.(2013).Mammalian base excision repair:the forgotten archangel.Nucleic Acids Res.41,3483–3490.
Ding,Q.,Lee,Y.K.,Schaefer,E.A.,Peters,D.T.,Veres,A.,Kim,K.,Kuper-wasser,N.,Motola,D.L.,Meissner,T.B.,Hendriks,W.T.,et al.(2013).A TALEN genome-editing system for generating human stem cell-based disease models.Cell Stem Cell 12,238–251.
Friedland,A.E.,Tzur,Y.B.,Esvelt,K.M.,Colaia
′covo,M.P.,Church,G.M.,and Calarco,J.A.(2013).Heritable genome editing in C.elegans via a CRISPR-Cas9system.Nat.Methods 10,741–743.
Fu,Y.,Foden,J.A.,Khayter,C.,Maeder,M.L.,Reyon,D.,Joung,J.K.,and Sander,J.D.(2013).High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells.Nat.Biotechnol.Published online June 23,2013.https://www.360docs.net/doc/7d12645802.html,/10.1038/nbt.2623.
Gasiunas,G.,Barrangou,R.,Horvath,P.,and Siksnys,V.(2012).Cas9-crRNA ribonucleoprotein complex mediates speci?c DNA cleavage for adaptive immunity in https://www.360docs.net/doc/7d12645802.html,A 109,E2579–E2586.Geurts,A.M.,Cost,G.J.,Freyvert,Y.,Zeitler,B.,Miller,J.C.,Choi,V.M.,Jen-kins,S.S.,Wood,A.,Cui,X.X.,Meng,X.D.,et al.(2009).Knockout rats via embryo microinjection of zinc-?nger nucleases.Science 325,433–433.Gratz,S.J.,Cummings,A.M.,Nguyen,J.N.,Hamm,D.C.,Donohue,L.K.,Har-rison,M.M.,Wildonger,J.,and O’Connor-Giles,K.M.(2013).Genome engineering of Drosophila with the CRISPR RNA-guided Cas9nuclease.Genetics 194,1029–1035.
Horvath,P.,and Barrangou,R.(2010).CRISPR/Cas,the immune system of bacteria and archaea.Science 327,167–170.
Hsu,P.D.,and Zhang,F.(2012).Dissecting neural function using targeted genome engineering technologies.ACS Chem.Neurosci.3,603–610.Hsu,P.D.,Scott,D.A.,Weinstein,J.A.,Ran,F.A.,Konermann,S.,Agarwala,V.,Li,Y.,Fine,E.J.,Wu,X.,Shalem,O.,et al.(2013).DNA targeting speci?city of RNA-guided Cas9nucleases.Nat.Biotechnol.Published online July 21,2013.https://www.360docs.net/doc/7d12645802.html,/10.1038/nbt.2647.
Jiang,W.,Bikard,D.,Cox,D.,Zhang,F.,and Marraf?ni,L.A.(2013).RNA-guided editing of bacterial genomes using CRISPR-Cas systems.Nat.Biotechnol.31,233–239.
Jinek,M.,Chylinski,K.,Fonfara,I.,Hauer,M.,Doudna,J.A.,and Charpentier,E.(2012).A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity.Science 337,816–821.
Jinek,M.,East,A.,Cheng,A.,Lin,S.,Ma,E.,and Doudna,J.(2013).RNA-pro-grammed genome editing in human cells.eLife 2,e00471.
Mali,P.,Aach,J.,Stranges,P.B.,Esvelt,K.M.,Moosburner,M.,Kosuri,S.,Yang,L.,and Church,G.M.(2013a).CAS9transcriptional activators for target speci?city screening and paired nickases for cooperative genome engineer-ing.Nat.Biotechnol.Published online August 1,2013.https://www.360docs.net/doc/7d12645802.html,/10.1038/nbt.2675.
Mali,P.,Yang,L.,Esvelt,K.M.,Aach,J.,Guell,M.,DiCarlo,J.E.,Norville,J.E.,and Church,G.M.(2013b).RNA-guided human genome engineering via Cas9.Science 339,823–826.
Maresca,M.,Lin,V.G.,Guo,N.,and Yang,Y.(2013).Obligate ligation-gated recombination (ObLiGaRe):custom-designed nuclease-mediated targeted integration through nonhomologous end joining.Genome Res.23,539–https://www.360docs.net/doc/7d12645802.html,ler,J.C.,Holmes,M.C.,Wang,J.,Guschin,D.Y.,Lee,Y.L.,Rupniewski,I.,Beausejour, C.M.,Waite, A.J.,Wang,N.S.,Kim,K.A.,et al.(2007).An
improved zinc-?nger nuclease architecture for highly speci?c genome editing.Nat.Biotechnol.25,778–785.
Miller,J.C.,Tan,S.,Qiao,G.,Barlow,K.A.,Wang,J.,Xia,D.F.,Meng,X.,Paschon,D.E.,Leung,E.,Hinkley,S.J.,et al.(2011).A TALE nuclease architec-ture for ef?cient genome editing.Nat.Biotechnol.29,143–148.
Moscou,M.J.,and Bogdanove,A.J.(2009).A simple cipher governs DNA recognition by TAL effectors.Science 326,1501.
Pattanayak,V.,Lin,S.,Guilinger,J.P.,Ma,E.,Doudna,J.A.,and Liu,D.R.(2013).High-throughput pro?ling of off-target DNA cleavage reveals RNA-pro-grammed Cas9nuclease speci?city.Nat.Biotechnol.Published online August 11,2013.https://www.360docs.net/doc/7d12645802.html,/10.1038/nbt.2673.
Perez,E.E.,Wang,J.B.,Miller,J.C.,Jouvenot,Y.,Kim,K.A.,Liu,O.,Wang,N.,Lee,G.,Bartsevich,V.V.,Lee,Y.L.,et al.(2008).Establishment of HIV-1resis-tance in CD4+T cells by genome editing using zinc-?nger nucleases.Nat.Biotechnol.26,808–816.
Porteus,M.H.,and Baltimore,D.(2003).Chimeric nucleases stimulate gene targeting in human cells.Science 300,763.
Reyon,D.,Tsai,S.Q.,Khayter,C.,Foden,J.A.,Sander,J.D.,and Joung,J.K.(2012).FLASH assembly of TALENs for high-throughput genome editing.Nat.Biotechnol.30,460–465.
Sander,J.D.,Dahlborg,E.J.,Goodwin,M.J.,Cade,L.,Zhang,F.,Cifuentes,D.,Curtin,S.J.,Blackburn,J.S.,Thibodeau-Beganny,S.,Qi,Y.,et al.(2011).Selection-free zinc-?nger-nuclease engineering by context-dependent assembly (CoDA).Nat.Methods 8,67–69.
Sanjana,N.E.,Cong,L.,Zhou,Y.,Cunniff,M.M.,Feng,G.,and Zhang,F.(2012).A transcription activator-like effector toolbox for genome engineering.Nat.Protoc.7,171–192.
Soldner,F.,Laganiere,J.,Cheng,A.W.,Hockemeyer,D.,Gao,Q.,Alagappan,R.,Khurana,V.,Golbe,L.I.,Myers,R.H.,Lindquist,S.,et al.(2011).Generation of isogenic pluripotent stem cells differing exclusively at two early onset Par-kinson point mutations.Cell 146,318–331.
Takasu,Y.,Kobayashi,I.,Beumer,K.,Uchino,K.,Sezutsu,H.,Sajwan,S.,Carroll,D.,Tamura,T.,and Zurovec,M.(2010).Targeted mutagenesis in the silkworm Bombyx mori using zinc ?nger nuclease mRNA injection.Insect Biochem.Mol.Biol.40,759–765.
Urnov,F.D.,Rebar,E.J.,Holmes,M.C.,Zhang,H.S.,and Gregory,P.D.(2010).Genome editing with engineered zinc ?nger nucleases.Nat.Rev.Genet.11,636–646.
Wang,H.,Yang,H.,Shivalila,C.S.,Dawlaty,M.M.,Cheng,A.W.,Zhang,F.,and Jaenisch,R.(2013).One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering.Cell 153,910–918.
Watanabe,T.,Ochiai,H.,Sakuma,T.,Horch,H.W.,Hamaguchi,N.,Nakamura,T.,Bando,T.,Ohuchi,H.,Yamamoto,T.,Noji,S.,et al.(2012).Non-transgenic genome modi?cations in a hemimetabolous insect using zinc-?nger and TAL effector https://www.360docs.net/doc/7d12645802.html,mun.3.Published online August 21,2012.https://www.360docs.net/doc/7d12645802.html,/10.1038/ncomms2020.
Wood,A.J.,Lo,T.W.,Zeitler,B.,Pickle,C.S.,Ralston,E.J.,Lee,A.H.,Amora,R.,Miller,J.C.,Leung,E.,Meng,X.,et al.(2011).Targeted genome editing across species using ZFNs and TALENs.Science 333,307.
Zhang,F.,Cong,L.,Lodato,S.,Kosuri,S.,Church,G.M.,and Arlotta,P.(2011).Ef?cient construction of sequence-speci?c TAL effectors for modulating mammalian transcription.Nat.Biotechnol.29,149–153.
10Cell 154,1–10,September 12,2013a2013Elsevier
Inc.
剪切力的计算方法
第3章 剪切和挤压的实用计算 3.1 剪切的概念 在工程实际中,经常遇到剪切问题。剪切变形的主要受力特点是构件受到与其轴线相垂直的大小相等、方向相反、作用线相距很近的一对外力的作用(图3-1a),构件的变形主要表现为沿着与外力作用线平行的剪切面(n m -面)发生相对错动(图3-1b)。 图3-1 工程中的一些联接件,如键、销钉、螺栓及铆钉等,都是主要承受剪切作用的构件。构件剪切面上的力可用截面法求得。将构件沿剪切面n m -假想地截开,保留一部分考虑其平衡。例如,由左部分的平衡,可知剪切面上必有与外力平行且与横截面相切的力Q F (图3-1c)的作用。Q F 称为剪力,根据平衡方程∑=0Y ,可求得F F Q =。 剪切破坏时,构件将沿剪切面(如图3-la 所示的n m -面)被剪断。只有一个剪切面的情况,称为单剪切。图3-1a 所示情况即为单剪切。 受剪构件除了承受剪切外,往往同时伴随着挤压、弯曲和拉伸等作用。在图3-1中没有完全给出构件所受的外力和剪切面上的全部力,而只是给出了主要的受力和力。实际受力和变形比较复杂,因而对这类构件的工作应力进行理论上的精确分析是困难的。工程中对这类构件的强度计算,一般采用在试验和经验基础上建立起来的比较简便的计算方法,称为剪切的实用计算或工程计算。 3.2 剪切和挤压的强度计算 3.2.1 剪切强度计算 剪切试验试件的受力情况应模拟零件的实际工作情况进行。图3-2a 为一种剪切试验装置的简图,试件的受力情况如图3-2b 所示,这是模拟某种销钉联接的工作情形。当载荷F 增大至破坏载荷b F 时,试件在剪切面m m -及n n -处被剪断。这种具有两个剪切面的情况,称为双剪切。由图3-2c 可求得剪切面上的剪力为 2 F F Q =
选择性剪接中的剪接模式
选择性剪接中得剪接模式 有几种不同得剪接模式(见图1—1)[1,6].最常见得模式就是在成熟得mRNA中跳过外显子,使其包括或者剔除盒式外显子(也称为跳过得外显子)。跳过外显子得一个著名得例子就是果蝇性别致死得基因(SXL),这就是一个由性别决定得转变。跳过SXL基因得第3外显子,可以保持雌性得分化。SXL得第3外显子包含一个早提前得终止密码子,这个外显子得存在合成出截短得、也有可能就是非功能得蛋白质[7,8]。另一个剪接模式就是外显子互斥,这使得两个相邻外显子中,仅有一个出现在最终产物中.人类成纤维细胞生长因子受体二号(FGFR—2)基因含有外显子IIIB与IIIC,这两者就是互斥得。从外显子IIIB得到得得基因产物,具有比纤维细胞生长因子低得多得聚合吸引力[9]。 不仅可以作用于整个外显子,不同得剪接方式也可以只剪接外显子得某一部分。5’或3'选择性剪接位点得选择,通过加上、或者不加上与外显子侧面相连得支链而生成,从而造成多样性。果蝇无子(FRU)与双性别(DSX)基因包含了雌性特有得选择性剪接位点,前者在5‘端,而后者在3'端.由于选择性剪接位点得不同,造成支链得细小差异[10,11]。 选择性剪接可发生在转录体得任意一端。选择性终止外显子 不仅改变最后一个外显子得包含性,而且还影响聚腺苷酸化位点得选择. 在许多情况下,它可以在最后得外显子中生成提前得终止密码子,并且生成 功能性截短得多肽或者产生无意义介导衰变(NMD,
即,由于终止密码子位于最后外显子与外显子得结点上游超过50-55碱基对处,从而造成得mRNA得降解) [2,12,13]。钙调节激素(降钙素)基因包括6个外显子。 成熟得得降钙素转录体包括前四个外显子,并使用位于第4外显子上得多聚腺苷酸化位点,从 而生成甲状腺C细胞中超过98%得基因产物。同时,在大脑与周围神经系统中,通过将前三个、第五与第六个外显子编码成降钙素相关肽得前体,并利用下游得腺苷酸化位点(CGRP),从而产生差异[14,15]。同样,选择性启动子得使用使得可以选择不同得转录启动子,这样通常会影响到第一个外显子。尽管人们普遍将其当作转录调控,选择性启动子得使用与选择性剪接有很 大得关联。人们已经观察到,有选择性启动子得基因更容易进行选择性剪接,同时,选择性启动子得数量与不同选择性得剪接方式得数量正相关[16]。鼠标单羧酸转运蛋白二号 (MCT2)基因有几种选择性得启动子,由此形成五种独特得首外显子(1A- 1E).外显子1C用于各种组织中,而其她得外显子则就是有组织特异性得[17]。 除此以外,内含子同样可以参与选择性剪接.在内含子保留性中,整个内含子可以被包括或排除。对于人类来说,这种模式就是罕见得[1]。然而,最近得研究表明,在已知得人类基因中,这个频率比想象中要高得多(约15%)[18]。内含子得保留性在植物中比在其她真核生物中常 见[19]。例如,对于拟南芥,50%以上得案例都就是关于内含子保留性得[20]。人类FosB(FBJ小鼠骨肉瘤病毒致癌基因对等质B)基因得最后一个外显子包含一个140碱基对得序列,它可以剪接出来,产生一个截断得产物——ΔFosB.通过观察动物长期得药物依赖性,检测ΔFosB得表达[21]。 基本剪接机制 无论就是选择性还就是组成型剪接,用得都就是同一种基本剪接机制,称为剪接体。剪接体识别与选择得剪接位点(外显子与内含子得结点),同时,催化打破并重组RNA链。剪接体主要由五个小核核糖核蛋白(snRNP)-—U1,U2,U4,U5与U6组成。其中包括尿苷丰富得小核RNA与多种蛋白质。它们能识别前体mRNA上得剪接信号,并与其她辅助剪接因子交互[22-24]。 在剪接中,三个保留序列元素就是必须得。这些保留序列元素可以就是经典或异常得剪接节点,多嘧啶束与分支点(见图1—2)[22,23]。剪接位点含有包括外显子与内含子结点得短序列。GU与AG二核苷酸在外显子得5'与3'端通常来说就是分别不变得。在剪接结合中,这种类型得GU-AG对被称为经典剪接位点。它存在于超过98%得哺乳动物基因组中[25]。异常得剪接位点含有如GC-AG、AT—AC之类得二核苷酸对,AT—AC等。这种情况比较罕见.多嘧啶束就是指位于内含子得3’端,且紧邻3‘剪接位点得UC丰富得片段。这就是几种剪接因子得结合位点,如U2snRNP辅助因子(U2AF)与多嘧啶束结合蛋白(PTB)[26].分支点(也称为分支位点)位于多嘧啶束得上游。对于人与老鼠来说,分支点与3'剪接结点间得平均距离大约就是30至40个碱基对[27]。虽然分支点序列在哺乳动物中就是可变得,分支点得突变可以促使由组成型剪接到选择性剪接得转变[27]。
高剪切均质机总体设计
1 绪论 剪切式均质技术作为一种新型微米技术,已广泛应用于食品、医药、轻工、微生物等诸多行业,并得到迅速发展,已成为这些行业对有关流体、半流体产品品质所必不可少的工艺过程。 国外早在30年前就产生并使用均质机,且应用于生产。目前,已有美国、日本、德国等10多个国家生产均质机。剪切式均质机作为均质机械中的佼佼者,也被广泛的认识和研究。自从1948年德国FLUKO公司首次发明了应用高剪切原理制成分散乳化设备,高剪切分散乳化设备已经出现了多种系列产品,在世界均质机械行业处于领先地位。近40年来,国外,特别是欧洲一些国家在高剪切分散均质机行业得到迅速发展,并在很多领域发挥着重大作用,如化装品、制药、食品、涂料、黏合剂等。国外所研究制造的剪切式均质设备基本上上是采用定一转子型(stator-rotor)结构作为均质头,在电机的高速驱动下(300-10000r/min),物料在转子与定子之间的间隙内高速运动,形成强烈的液力剪切和湍流,使物料在同时产生的离心、挤压、碰撞等综合作用力的协调作用力下,得到充分的分散、乳化、破碎,达到要求的的效果。美国和德国在剪切式均质机的研究和开发方面都取得了显著进展。如美国IKA-WERKE GMHB CO.KG生产的多系列分散均制设备;美国ROSS 公司研制的高剪切混合乳化机;德国IKA-MASCHINENBAU公司研制的ULTRA分散机;德国YSTRAL公司生产的X40型分散搅拌机;德国公司研制的系列高剪切分散乳化剂、管线式高剪切分散乳化剂、管式分散乳化剂、间歇式高剪切与间歇式无轴承分散乳化剂、高效强力分散乳化剂等世界领先高科技产品。 我国的均质机研究产品是从50年代个别厂家开始的,最早是上海烟草机械厂仿制美国产品,直到80年代才开始逐渐的生产均质机,而且大多是传统的高压均质设备。随着国外剪切式均质机的迅速发展,近年来,国内许多科研人员,制造和使用厂家也开始重视对剪切式均质机的研究工作。目前,已建立了与国外厂商联营、合资研制生产剪切式均质机的公司。如上海菲鲁克(FLUKO)机电设备有限公司;中美合资南通罗斯(ROSS)混合设备有限公司等。现在国内有许多厂家开始生产高剪切均质机,如东市长江机电有限公司、上海环保设备总厂、上海威宇机电有限公司、上海市化工装备研究所生产的集混合、分散、乳化、溶解、粉碎等功能为一体的系列剪切式均质机。 1.1 高剪切均质机的均质原理 剪切均质机基于超剪切原理,实现固相的微化和液相的乳化。目前采用剪切式均质机主要工作部件为一级或多极的相互啮合的定转子又有数层齿圈。其均质乳化有以下方面: 1 液力剪切作用 液力剪切是指高速流动的流体本身会对流体内粒子产生强大的剪切作用,而且由于高速流动产生剧烈的微湍流,在湍流边缘出现很高的局部速度梯度,处于这种局部速度梯度下的粒子会受剪切而微粒化,液力剪切分层流剪切和湍流剪切。在层流区域,流体在定转子槽道内流动时,流体内的最大流速及所受到的最大剪切力与流体流动方向上的压力梯度成正比。当施以周期性高频脉动压力梯度时,最大速度在槽道壁面与机理道中心之间,偏离中心,且频率增大,最大速度增大,且向壁面趋近,剪切力增大。流体在同轴圆筒之间成为旋转流,由于两圆筒速度不同,间隙内流体层之间存在速度梯度,产生剪切力。如圆
剪切机械安全管理制度
剪切机械安全规程GB 6077-85 UDC 621.965:65.08 SAFETY REGULATIONS FOR SHEARING MACHINERY 国家标准局1985-06-06发布,1986-05-01实施 1 总则 1.1 为保护工人在剪切劳动生产过程中的安全和健康,保证剪切设备安全生产,特制订本标准 1.2 本标准适用于机械、液压剪切机械中剪切钢板、型材、钢坯和类似纸质材料等普通及专门化的剪切机械(以下简称剪切机)。 1.3 本标准是剪切机设计、制造、使用、维修和管理等部门的安全基本法规。 当地安全监察部门对本标准的贯彻执行情况负责监督检查。 1.4 剪切机的设计制造,应按本标准和现行有关技术条件等规定进行,必须保证安全、可靠和操作维修方便。 1.5 新设计制造的剪切机,进行技术鉴定时,必须对其安全、可靠性作出结论,鉴定合格后方可正式生产。 鉴定会须有国家劳动安全部门授权的剪切机安全技术管理单位和地方安全监察部门参加。 1.6 剪切机设计制造单位,必须向使用单位提供安全技术说明(包括搬运、安装、使用及维修时应采取的安全与卫生措施和易损件明细表等)。 1.7 劈切机在设计制造、使用改造中,如遇安全技术措施和经济利益发生矛盾时,必须优先考虑安全技术的要求。 2 主要结构、部件的安全要求 2.1 一般要求 2.1.1 在不影响功能的情况下,机架及其他零、部件外露的表面,不准有锯齿状及锐利的棱角或突起等危险部分。 2.1.2 操作者站立平面至工作台面的高度应便于操作,一般应在750 ̄900MM之间。 2.1.3 主要受力构件,如机架、刀架、压料装置等用焊接连接时,必须保证结构强度要求,焊缝表面不得有裂纹、气孔、夹渣、弧坑等缺陷,焊缝的内在质量应检查。
基本操作,剪切复制粘贴关闭窗口等等,快捷键(热键)
基本操作,剪切复制粘贴关闭窗口等等,快捷键(热键) 一、常见用法: F1 显示当前程序或者windows的帮助内容。 F2 当你选中一个文件的话,这意味着“重命名” F3 当你在桌面上的时候是打开“查找:所有文件” 对话框 F10或ALT 激活当前程序的菜单栏 windows键或CTRL+ESC 打开开始菜单 CTRL+ALT+DELETE 在win9x中打开关闭程序对话框 DELETE 删除被选择的选择项目,如果是文件,将被放入回收站 SHIFT+DELETE 删除被选择的选择项目,如果是文件,将被直接删除而不是放入回收站 CTRL+N 新建一个新的文件 CTRL+O 打开“打开文件”对话框 CTRL+P 打开“打印”对话框 CTRL+S 保存当前操作的文件 CTRL+X 剪切被选择的项目到剪贴板 CTRL+INSERT 或CTRL+C 复制被选择的项目到剪贴板 SHIFT+INSERT 或CTRL+V 粘贴剪贴板中的内容到当前位置 ALT+BACKSPACE 或CTRL+Z 撤销上一步的操作 ALT+SHIFT+BACKSPACE 重做上一步怀废 牟僮?br> Windows键+M 最小化所有被打开的窗口。 Windows键+CTRL+M 重新将恢复上一项操作前窗口的大小和位置 Windows键+E 打开资源管理器 Windows键+F 打开“查找:所有文件”对话框 Windows键+R 打开“运行”对话框 Windows键+BREAK 打开“系统属性”对话框 Windows键+CTRL+F 打开“查找:计算机”对话框 SHIFT+F10或鼠标右击打开当前活动项目的快捷菜单 SHIFT 在放入CD的时候按下不放,可以跳过自动播放CD。在打开word的时候按下不放,可以跳过自启动的宏 ALT+F4 关闭当前应用程序 ALT+SPACEBAR 打开程序最左上角的菜单 ALT+TAB 切换当前程序 ALT+ESC 切换当前程序 ALT+ENTER 将windows下运行的MSDOS窗口在窗口和全屏幕状态间切换PRINT SCREEN 将当前屏幕以图象方式拷贝到剪贴板 ALT+PRINT SCREEN 将当前活动程序窗口以图象方式拷贝到剪贴板
连续函数图象的分解与一类剪切集
连续函数图象的分解与一类剪切集 本文主要研究连续函数图象的分解与分形维数(豪斯多夫维数,填充维数)的关系以及一类剪切集的分形测度.在第一章介绍本文的背景,第二章给出预备知识的基础上,用了三章的篇幅分别对上述三方面的问题展开了详细的论述.在第三章,我们考虑区间[0,1]上的连续函数的图象的分解与豪斯多夫维数之间的关系,我们回答了Bayart和Heurtaeux提出的一个问题.具体的,证明了:任意f∈C([0,1]),β∈[1,2],存在连续函数h,g∈C([0,1])使得.f=h+g并且 dimHG9([0,1])=dimH Gh([0,1])=β,其中Gg([0,1]),Gh([0,1])表示函数g,h的图像:Gg([0,1])={(x,g(x)):x∈[0,1]},Gh([0,1])=.{(x,h(x)):x∈[0,1]}.在第四章,我们分两部分内容:第一部分,我们利用填充维数与上盒维数的关系,把Humke和Petruska的结果推广到高维空间中,即如果X是Rn中的不可数紧子集,那么是C(X)中的拓扑普适集;第二部分讨论连续函数图象的分解与填充维数的关系.首先,我们得到:对任意f,g∈C(X),如果dimp(Gg)≠dimp(Gf),那么把该结果应用到函数分解上,我们有:假设β∈[1,2],f∈C([0,1]),那么存在连续函数g,h∈C([0,1])满足当且仅当dimp(Gf([0,1]))≤β.最后,还证明了是1-普适集(1-prevalent).在第五章,我们给出一类剪切集的h-填充测度与h-豪斯多夫测度的上下界估计,其中h是加倍的维数函数.最后,我们在第六章总结了本文的主要结果,并提出了一些可以进一步研究的问题.
可变剪接
可变剪接:有些基因的一个mRNA前体通过不同的剪接方式(选择不同的剪接位点)产生不同的mRNA剪接异构体,这一过程称为可变剪接(或选择性剪接,alternative splicing) 。可变剪接是调节基因表达和产生蛋白质组多样性的重要机制,是导致人类基因和蛋白质数量较大差异的重要原因。 基本内容 大多数真核基因转录产生的mRNA前体是按一种方式剪接产生出一种成熟mRNA分子,因而只翻译成一种蛋白质。但有些基因的一个mRNA前体通过不同的剪接方式(选择不同的剪接位点)产生不同的mRNA剪接异构体,这一过程称为可变剪接(或选择性剪接, alternative splicing)。由于RNA的可变剪接不牵涉到遗传信息的永久性改变.所以是真核基因表达调控中一种比较灵活的方式。可变剪接是调节基因表达和产生蛋白质组多样性的重要机制, 是导致人类基因和蛋白质数量较大差异的重要原因。 可变剪接形式的识别 真核细胞核内前体mRNA加工通过5’加帽、剪接(移除内含子)、3’末端切割加尾.从而形成成熟的mRNA.成熟的mRNA和hnRNP及其他蛋白质形成复合体输出核外再经过选择性降解参与翻译。这些步骤并不是简单的线性顺序.而是在转录物延伸期和转录同时发生的。从而形成一个大型的“生产链。 一般认为,可变剪接有5种基本形式:①内含子保留;②可变的5’端;③可变的3’端; ④外显子盒;⑤互斥外显子(一组外显子中只选其一)。也有分为7种形式的,加上可变的起始或末端外显子,而这两种形式更有可能是可变启动子、可变polyA位点造成的。可进行专门分析。 可变剪接的意义和作用 可变剪接被认为是导致蛋白质功能多样性的重要原因之一,它使一个基因可编码多个不同转录产物和蛋白产物。 可变剪接也是产生基因组规模与生物复杂性之间的矛盾根源之一。 已有实验研究表明,可变剪接在产生受体多样性、控制调节生长发育等方面起决定性作用。尤其表现在神经系统和免疫系统,这与该类系统的功能多样性和反应敏感性是密切相关的。许多遗传疾病都与剪接繁盛异常紧密相关据估计。导致疾病的变异中约15%会影响pre—mRNA的剪接。
剪切闸板使用规范
冀东油田剪切闸板防喷器使用规范 1. 剪切闸板防喷器的使用条件 由于钻具内防喷工具失效或井口处钻具弯曲变形等原因造成井喷失控而无法关井,且采取其它措施也无法控制井口时,使用剪切闸板剪断井内钻具,控制井口。 2. 使用剪切闸板防喷器实施剪切关井的指挥权限 钻井队队长在同甲方钻井监督协商一致后,请示钻井公司井控第一责任人(井控第一责任人不在时,请示井控负责人)同意,立即组织实施剪断钻具关井;若情况紧急,来不及请示,钻井队队长经与甲方钻井监督协商一致后,可以决定并组织实施剪断钻具关井。 3.剪切闸板防喷器剪断钻具关井的操作程序 (1)在确保井内管柱接头和接头过渡带不在剪切闸板剪断位置后,锁定钻机绞车刹车系统。 (2)打开主放喷管线泄压,并确认剪切闸板防喷器以上的半封闸板防喷器和环形防喷器已经关闭。 (3)在钻杆上适当位置安装相应的钻杆死卡,用钢丝绳分4个方向对角与钻机连接并固定牢靠。 (4)打开防喷器远程控制台储能器旁通阀,关剪切闸板防喷器,直至剪断井内钻具关井;若21兆帕未能剪断井内钻具,应由气动泵直接打超高压,直至剪断井内钻具。 (5)关闭液控系统旁通阀,将远控台管汇压力调整到规定值。 (6)打开上部剪切闸板防喷器以上的半封闸板防喷器和环形防喷器,打开钻杆死卡起出被剪断钻具离开全封闸板位置。 (7)关闭全封闸板防喷器,控制井口。
4.使用剪切闸板防喷器的安全注意事项 (1)钻井队应加强对防喷器远程控制台的管理,避免因误操作而导致钻具损坏或更严重的事故。 (2)作业现场应建立完善、有效地剪切闸板防喷器管理制度,至少包括:现场检查、保养管理制度。 (3)剪切闸板防喷器操作指定专人负责,操作剪切闸板时,除远程控制台操作人员外,其余人员全部撤至安全位置,同时按应急预案布置警戒、人员疏散、防喷点火及之后的应急处理工作。 (4)恢复正常工作后,剪切闸板应及时更换。 (5)试验过的剪切刀片不再安装使用。 (6)剪切闸板防喷器原则上不应作全封闸板使用,在全封闸板防喷器失效情况下,剪切闸板防喷器可在空井状态下关井。 (7)井场防喷演习和日常防喷器操作中不允许进行剪切试验。
剪切力的计算方法
第3章剪切和挤压的实用计算 3.1剪切的概念 在工程实际中,经常遇到剪切问题。剪切变形的主要受力特点是构件受到与其轴 线相垂直的大小相等、方向相反、作用线相距很近的一对外力的作用(图3-1a),构件 的变形主要表现为沿着与外力作用线平行的剪切面(m - n面)发生相对错动(图3- 1b)。 图3-1 工程中的一些联接件,如键、销钉、螺栓及铆钉等,都是主要承受剪切作用的构 件。构件剪切面上的内力可用截面法求得。将构件沿剪切面m-n假想地截开,保留一 部分考虑其平衡。例如,由左部分的平衡,可知剪切面上必有与外力平行且与横截面相切的内力F Q (图3-1C)的作用。F Q称为剪力,根据平衡方程',=0,可求得F Q二F。剪切破坏时,构件将沿剪切面(如图3-la所示的m-n面)被剪断。只有一个剪切面的情况,称为单剪切。图3-1a所示情况即为单剪切。 受剪构件除了承受剪切外,往往同时伴随着挤压、弯曲和拉伸等作用。在图3-1中没有完全给出构件所受的外力和剪切面上的全部内力,而只是给出了主要的受力和内力。实际受力和变形比较复杂,因而对这类构件的工作应力进行理论上的精确分析是困难的。工程中对这类构件的强度计算,一般采用在试验和经验基础上建立起来的比较简便的计算方法,称为剪切的实用计算或工程计算。
3.2剪切和挤压的强度计算3.2.1剪切强度计算
剪切试验试件的受力情况应模拟零件的实际工作情况进行。图 试验装置的简图,试件的受力情况如图 3-2b 所示,这是模拟某种销钉联接的工作情 形。当载荷F 增大至破坏载荷 F b 时,试件在剪切面 m - m 及n - n 处被剪断。这种具 有两个剪切面的情况,称为双剪切。由图 3-2c 可求得剪切面上的剪力为 F Q 图3-2 由于受剪构件的变形及受力比较复杂,剪切面上的应力分布规律很难用理论方法 确定,因而工程上一般采用实用计算方法来计算受剪构件的应力。 在这种计算方法中, 假设应力在剪切面内是均匀分布的。若以 A 表示销钉横截面面积,则应力为 F Q A ?与剪切面相切故为切应力。以上计算是以假设“切应力在剪切面上均匀分布”为基础 的,实际上它只是剪切面内的一个“平均切应力”,所以也称为名义切应力。 当F 达到F b 时的切应力称剪切极限应力, 记为-b 。对于上述剪切试验, 剪切极限 应力为 _ Fb ■b - 2A 3-2a 为一种剪切 (3-1) bj
4、选择性剪切
分子机制研究套路(四) 选择性剪切 课题:激酶A通过RNA结合蛋白B影响C的选择性剪切 1.概念介绍: 真核生物结构基因的DNA序列由编码序列和非编码序列两部分组成,编码序列是不连续的,被非编码序列分割开来,成为断裂基因(Split gene)。在结构基因中,编码序列称为外显子(Exon),是表达多肤链序列,非编码序列称为内含子(Intron),是不表达多肤链序列,又称插入序列。 真核生物DNA转录为前mKNA(Pre-mRNA)后经过mRNA的剪切,切去内含子,将有编码意义的外显子连接起来,转变为成熟mRNA。真核基因转录产生的mRNA前体,在细胞分化、发育阶段和生理状态下,可按不同的方式剪切产生出两种或者更多种mRNA,进而翻译出两种或多种蛋白质,此过程为选择性剪切或称可变剪切(Altemative Splicing)。选择性剪接的形式多样,最常见的主要是以下几种:1)外显子跳过,从而导致外显子保留或者不保留在成熟的mRNA中;2)外显子具有多个5’或者3’剪接位点,以此可能产生多种选择性剪切异构体;3)单个或者多个选择性剪接外显子可以位于组成型外显子(constitutive exon)中,以便选择性外显子可以有选择的保留或者不保留于成熟的mRNA中;4)内含子不剪切,内含子可以选择性保留在成熟mRNA中以便被翻译出来。mRNA这种选择性剪切是少量基因产生大量mRNA和蛋白质的重要机制,也使得机体仅少量基因就能对千变万化的复杂的生物性状进行调控成为可能。mRNA这种选择性剪切对扩充生物细胞遗传信息和增强生物细胞功能有着重要作用,并且大量研究证实,选择性剪切对基因表达的调节作用,在干细胞分化过程以及肿瘤的发生、发展过程中均发挥重要作用。 2.示意图:
剪切控制取向注射成型
剪切控制取向注射成型 剪切在制取向注射成型实质是通过浇口将动态的压力施加给熔体,使模腔内的聚合物熔体产生振动剪切流动,在其作用下不同熔体层中的分子链或纤维产生取向并冻结在制件中,从而控制制品的内部结构和微观形态,达到控制制品力学性能和外观质量的目的。将振动引入模腔的方法有螺杆和辅助装置加振两种。 1)螺杆加振 螺杆加振的工作原理是给注射油缸提供脉动油压,使注射螺杆产生往复移动而实现振动,注射螺杆产生的振动作用于熔体,并通过聚合物馆体把振动传入模腔,从而使模腔中的熔体产生振动,这种振动作用可持续到模具绕口封闭。此种装置比较简单,可以利用注塑机的控制系统,或对注塑机的液压和电气控制系统加以改造来实现。 2)辅助装置加振,辅助装置加振是将加振装置安装在模具与注塑机喷嘴之间,注射阶段与普遍注塑一样,通常熔体仅通过一个浇口,此浇口活塞后退以保持流道通畅,另一活塞则切断另一流道;模腔充满后,两个保压活塞在独立的液压系统驱动下开始以同样的频率振动,但其相位差180O。通过两个活塞的往复运动,把振动传入模腔,使模腔中的熔体一边冷却,一边产生振动剪切流动。实验证明这种工艺有助于消除制品的常见缺陷(如缩孔、裂纹、表面沉陷等),提高熔接线强度;利用剪切控制取向成型技术、通过合理设置浇口位置和数量,可以控制分子或纤维的取向,获得比普通注射成型制品强度更高的制品。 剪切控制取向注射成型过程中聚合物熔体被注入模腔后,模腔内开始出现固化层。由于固化层附近速度梯度最大,此处的熔体受到强烈的剪切作用,取向程度最大。中心层附近速度梯度小,剪切作用小,因而取向程度也小。在保压过程中引入振动,使模腔中的聚合物熔体一边冷却,一边受振动的剪切作用,振动剪切产生的取向因模具的冷却作用而形成一定厚度的取向层。同没有振动作用相比,振动剪切流动所产生的取向层厚度远远大于普通注射所具有的取向层厚度,这就是模腔内引入振动剪切流动能使制品的力学性能得到提高的原因。此外,由于振动产生的周期性的压缩增压和释压膨胀作用,可在薄壁部分产生较大的剪切内热,延缓这些部分的冷却,从而使厚壁部分的收缩能从浇口得到足够的补充,有效防止缩孔、凹陷等缺陷。
剪切计算及常用材料强度
2.剪切强度计算 (1) 剪切强度条件 剪切强度条件就是使构件的实际剪应力不超过材料的许用剪应力。 [] s F A ττ =≤ (5-6) 这里[τ]为许用剪应力,单价为Pa或MPa。 由于剪应力并非均匀分布,式(5-2)、(5-6)算出的只是剪切面上的平均剪应力,所以在使用实验的方式建立强度条件时,应使试件受力尽可能地接近实际联接件的情况,以确定试样失效时的极限载荷τ0,再除以安全系数n,得许用剪应力[τ]。 [] n τ τ= (5-7) 各种材料的剪切许用应力应尽量从相关规范中查取。 一般来说,材料的剪切许用应力[τ]与材料的许用拉应力[σ]之间,存在如下关系: 对塑性材料: []0.60.8[] τσ = 对脆性材料: []0.8 1.0[] τσ = (2) 剪切实用计算 剪切计算相应地也可分为强度校核、截面设计、确定许可载荷等三类问题,这里就不展开论述了。但在剪切计算中要正确判断剪切面积,在铆钉联接中还要正确判断单剪切和双剪切。下面通过几个简单的例题来说明。 例5-1 图5-12(a)所示电瓶车挂钩中的销钉材料为20号钢,[τ]=30MPa,直径d=20mm。挂钩及被连接板件的厚度分别为t=8mm和t1=12mm。牵引力F=15kN。试校核销钉的剪切强度。 图5-12 电瓶车挂钩及其销钉受力分析示意图 解:销钉受力如图5-12(b)所示。根据受力情况,销钉中段相对于上、下两段沿m-m和n-n两个面向左错动。所以有两个剪切面,是一个双剪切问题。由平衡方程容易求出: 2 s F F= 销钉横截面上的剪应力为: 3 32 1510 23.9MPa<[] 2(2010) 4 s F A ττ π - ? === ?? 故销钉满足剪切强度要求。 例5-2如图5-13所示冲床,F max=400KN,冲头[σ]=400MPa,冲剪钢板的极限剪应力τb=360 MPa。试设计冲头的最小直径及钢板最大厚度。
(完整版)1.6MN上切式剪切机机构设计(1).docx
机械系统设计 活动连杆剪切机项目汇报 学院:机械工程学院 班级:机设 2 班 小组成员:郝岩李逸然李俊杰 王岩贾庆超李博 陈冲商周一凡 指导教师 : 翟富刚 2016年 10 月 21号
目录 绪论 (3) 一、工作原理 (4) 1.1 活动连杆上切式平行刀片剪切机工作原理 (4) 二、剪切机结构参数 (5) 2.1 刀片行程 (5) 2.2、刀片尺寸 (6) 2.3 剪切机的理论空行程次数 (6) 三、力能参数计算 (7) 3.1 剪切过程分析 (7) 3.2 单位剪切阻力曲线与剪切力、剪切功 (8) 3.3 静力矩 (10) 3.4 电动机功率的预选 (10) 四、实例分析 (11) 五、参考资料 (12)
绪论 剪切机是机床的一种,它采用液压驱动,安全性能可靠,操作方便。剪切机适 用于金属回收加工厂、报废汽车拆解场、冶炼铸造行业,对各种形状的型钢及各种金属材料进行冷态剪断、压制翻边,以及粉末状制品、塑料、玻璃钢、绝缘材料、橡胶的压制成型。 在轧制生产过程中,大断面钢锭和钢坯经过轧制后,其断面变小,长度增加。为了满足后续工序和产品尺寸规格的要求,各种钢材生产工艺过程中必须有剪切工序,剪切机的用途就是用来剪切定尺切头、切尾、切边、切试样及切除轧件的 局部缺陷等。 剪切机特点具有: 1、采用液压驱动,安全性能可靠,操作方便。 2、电机按需求采用可编程控制器。 3、液压系统采用先进的插装阀或滑阀系统控制,实行按钮集中操作的液压机。 4、其压力、速度和行程可根据工艺需要进行调节,并能完成压制成型和定型两种工艺方式。 5、安装不须底脚螺丝。 6、无电源的地方可用柴油机作动力 根据剪切机刀片形状、配置以及剪切方式。剪切机可分为平行刀片剪切机、 斜刀片剪切机、圆盘式剪切机和飞剪机。根据剪切轧件时刀片的运动特点,平行刀片剪切机可分为上切式和下切式两大类。其中活动连杆上切式剪切机具有操作 速度快,实际剪切次数多,活连杆取代传统离合器,适应不同的剪切工况要求(如弯头轧件)等优点。
GB6077-85剪切机械安全规程
剪切机械安全规程GB6077-85 1总则 1.1为保护工人在剪切劳动生产过程中的安全和健康,保证剪切设备安全生产,特制订本标准1.2本标准适用于机械、液压剪切机械中剪切钢板、型材、钢坯和类似纸质材料等普通及专门化的剪切机械(以下简称剪切机)。 1.3本标准是剪切机设计、制造、使用、维修和管理等部门的安全基本法规。当地安全监察部门对本标准的贯彻执行情况负责监督检查。 1.4剪切机的设计制造,应按本标准和现行有关技术条件等规定进行,必须保证安全、可靠和操作维修方便。 1.5新设计制造的剪切机,进行技术鉴定时,必须对其安全、可靠性作出结论,鉴定合格后方可正式生产。 鉴定会须有国家劳动安全部门授权的剪切机安全技术管理单位和地方安全监察部门参加。 1.6剪切机设计制造单位,必须向使用单位提供安全技术说明(包括搬运、安装、使用及维修时应采取的安全与卫生措施和易损件明细表等)。 1.7劈切机在设计制造、使用改造中,如遇安全技术措施和经济利益发生矛盾时,必须优先考虑安全技术的要求。 2主要结构、部件的安全要求 2.1一般要求 2.1.1在不影响功能的情况下,机架及其他零、部件外露的表面,不准有锯齿状及锐利的棱角或突起等危险部分。 2.1.2操作者站立平面至工作台面的高度应便于操作,一般应在750 ̄900mm之间。 2.1.3主要受力构件,如机架、刀架、压料装置等用焊接连接时,必须保证结构强度要求,焊缝表面不得有裂纹、气孔、夹渣、弧坑等缺陷,焊缝的内在质量应检查。 2.1.4主要受力构件的焊接,必须在0℃以上的环境温度中进行,并不得在非焊接区引弧。焊后应进行退火处理。 2.1.5剪切机的重要部件上所使用的螺栓、螺母、销钉等紧固件,必须采取严格的防松措施。 2.2机架与刀架 2.2.1机架结构必须有足够的强度、刚度和稳定性。 2.2.2机架与刀架的导轨间隙,应调整方便、锁紧可靠。 2.2.3刀架(指运动刀架或滑块,以下简称刀架)应满足设计所规定的刚度、强度和刀片承压面的抗压强度。 2.2.4刀架操纵机构动作应相互协调,所有工作规范内的动作应平稳、灵活、可靠。当离合器脱开后刀架应可靠地停留在设计所规定的位置。在设计上,必须排除刀架自行滑车的危险性。 2.2.5刀架和压料装置的危险部位,必须至少设置一种可靠的安全装置 2.3压料装置 2.3.1剪切过程必须有先压紧而后剪切的顺序动作。应有足够的压料力和足够数量的压料脚,其底面应平直、完整。 2.3.2对于剪切厚度小于6.3mm的剪板机,压料防护装置在高度上必须是可以调整的,底面与工作台之间的距离不能超过被剪钢板厚度+8mm。 2.4挡料装置 2.4.1挡料装置(包括前、后挡料装置)应具有挡料准确可靠,送料方便和安全的结构 2.4.2挡料装置应便于剪料,应设有当被剪材料卡死时,不直接用手搬动就能顺利排除的后挡料退让机构。 2.4.3必要时前挡料装置应设有角度挡料器,以便剪切带有角度的工件。
剪切机械 噪声限值(标准状态:现行)
I C S25.120.10 J62 中华人民共和国国家标准 G B24389 2009 剪切机械噪声限值 N o i s e l i m i t s f o r s h e a r i n g m a c h i n e r y 自2017年3月23日起,本标准转为推荐性 标准,编号改为G B/T24389 2009三 2009-09-30发布2010-07-01实施中华人民共和国国家质量监督检验检疫总局
前言 本标准的3.1.1二3.2.1二3.3.1二3.4.1二3.5.1二第4章为推荐性的,其他为强制性的三 本标准由中国机械工业联合会提出三 本标准由全国锻压机械标准化技术委员会归口三 本标准负责起草单位:佛山市南海力丰机床有限公司二沈阳锻压机械有限公司二天水锻压机床有限公司三 本标准主要起草人:周建军二曾立泉二陈文进二岳汉生二蔡礼泉三 本标准为首次发布三
剪切机械噪声限值 1范围 本标准规定了剪切机械的声功率级和声压级的噪声限值三 本标准适用于剪切机械,包括各种类型的棒料剪断机二鳄鱼式剪断机二剪板机二冲型剪切机二联合冲剪机三 2规范性引用文件 下列文件中的条款通过本标准的引用而成为本标准的条款三凡是注日期的引用文件,其随后所有的修改单(不包括勘误的内容)或修订版均不适用于本标准,然而,鼓励根据本标准达成协议的各方研究是否可使用这些文件的最新版本三凡是不注日期的引用文件,其最新版本适用于本标准三 G B/T23281 2009锻压机械噪声声压级测量方法 G B/T23282 2009锻压机械噪声声功率级测量方法 3噪声限值 剪切机械的噪声分为声功率级二声压级要求,并根据剪切机械的类型分别给出了噪声限值要求三3.1棒料剪断机噪声限值 3.1.1棒料剪断机声功率级限值 棒料剪断机在连续空运转时的噪声A计权声功率级L W A不应超过表1的规定三 表1 棒料剪断机的公称力/k N声功率级限值L W A/d B(A) <5000110 ?5000114 3.1.2棒料剪断机声压级限值 3.1.2.1棒料剪断机在连续空运转时在规定位置的噪声A计权声压级L p A不应超过95d B(A)三3.1.2.2棒料剪断机在空运转单次行程时在规定位置的脉冲噪声A计权声压级L p AⅠ不应超过106d B(A)三 3.2鳄鱼式剪断机噪声限值 3.2.1鳄鱼式剪断机声功率级限值 鳄鱼式剪断机在连续空运转时的噪声A计权声功率级L W A不应超过105d B(A)三 3.2.2鳄鱼式剪断机声压级限值 鳄鱼式剪断机在连续空运转时在规定位置的噪声A计权声压级L p A不应超过90d B(A)三 3.3剪板机噪声限值 3.3.1剪板机声功率级限值 机械传动剪板机在连续空运转时的噪声A计权声功率级L W A不应超过表2的规定,液压传动剪板机在连续空运转时的噪声A计权声功率级L W A不应超过表3的规定三
切割方法
科技名称定义 科技名称:钢结构切割,又名钢板切割。科技定义:钢结构切割是在工业生产中,根据需要对钢材结构进行切割加工的过程。通常的切割技述有火焰切割、水切割、等离子切割等。目前使用最广泛的是火焰切割。 编辑本段钢结构切割概述 是针对被切割钢材结构而言的,一般是指工业燃气和氧气混合燃烧并达到切割要求的温度,对钢材结构进行熔化、吹渣和分割的过程。目前所用的技述有火焰切割、水切割、等离子切割、数控切割等。最常用的是火焰切割,它具有成本低,操作简便,技述成熟,使用广泛等特点,是目前工业中使用最广泛的切割技述,火焰切割指利用工业然气与氧气混合燃烧火焰将被切割的金属加热到钢材的溶点,再释放出高压氧气流,使金属进一步剧烈氧化并将燃烧产生的熔渣吹掉形成切口的过程。目前使用的切割气主要有乙炔,丙烷,氢氧气,霞浦气,天然气等。乙炔具有污染严重、对工作人员伤割大、易回火、造价高等缺点,现在国家已明令淘汰使用。丙烷、氢氧气、霞浦气成本相对比较高。天然气切割是近几年发展起来的新技述,具有低碳环保、安全稳定、成本低廉、气源丰富等优点,是国家大力推广的技述,具有广扩的前景,普通天然气带氧燃烧的火焰温度达不到乙炔带氧燃烧的火焰温度,必须添加增温助燃添加剂(如神麒天然气增效剂等)与燃气发生络合反应,经崔化、活化、聚集热量之后才能实现天然气切割所要求达到的切割温度。 编辑本段钢结构切割工艺 切割下料标准 1.范围:本标准适用于原材料切割下料的加工过程。适用于以火焰切割及等离子切割作为切割方式的切割下料过程。 2.施工准备:2.1材料要求: 2.1.1用于切割下料的钢板应经质量部门检查验收合格,其各项指标满足国家规范的相应规定。 2.1.2钢板在下料前应检查钢板的牌号、厚度和表面质量,如钢材的表面出现蚀点深度超过国标钢板负偏差的部位不准用于产品。小面积的点蚀在不减薄设计厚度的情况下,可以采用焊补打磨直至合格。 2.1.3在下料时必须核对钢板的牌号、规格和表面质量情况,在确认无疑后才可下料。 2.2施工设备及工具: 2.2.1切割下料设备主要包括数控火焰切割机、数控等离子切割机、直条切割机、半自动切割机等。 2.2.2在气割前,
选择性剪接中的剪接模式
选择性剪接中的剪接模式 有几种不同的剪接模式(见图1-1)[1,6]。最常见的模式是在成熟的mRNA中跳过外显子,使其包括或者剔除盒式外显子(也称为跳过的外显子)。跳过外显子的一个著名的例子是果蝇性别致死的基因(SXL),这是一个由性别决定的转变。跳过SXL基因的第3外显子,可以保持雌性的分化。SXL的第3外显子包含一个早提前的终止密码子,这个外显子的存在合成出截短的、也有可能是非功能的蛋白质[7,8]。另一个剪接模式是外显子互斥,这使得两个相邻外显子中,仅有一个出现在最终产物中。人类成纤维细胞生长因子受体二号(FGFR-2)基因含有外显子IIIB和IIIC,这两者是互斥的。从外显子IIIB得到的的基因产物,具有比纤维细胞生长因子低得多的聚合吸引力[9]。 不仅可以作用于整个外显子,不同的剪接方式也可以只剪接外显子的某一部分。5'或3'选择性剪接位点的选择,通过加上、或者不加上与外显子侧面相连的支链而生成,从而造成多样性。果蝇无子(FRU)和双性别(DSX)基因包含了雌性特有的选择性剪接位点,前者在5‘端,而后者在3'端。由于选择性剪接位点的不同,造成支链的细小差异[10,11]。 选择性剪接可发生在转录体的任意一端。选择性终止外显子 不仅改变最后一个外显子的包含性,而且还影响聚腺苷酸化位点的选择。 在许多情况下,它可以在最后的外显子中生成提前的终止密码子,并且生成 功能性截短的多肽或者产生无意义介导衰变(NMD,
即,由于终止密码子位于最后外显子与外显子的结点上游超过50-55碱基对处,从而造成的mRNA的降解) [2,12,13]。钙调节激素(降钙素)基因包括6个外显子。 成熟的的降钙素转录体包括前四个外显子,并使用位于第4外显子上的多聚腺苷酸化位点,从而生成甲状腺C细胞中超过98%的基因产物。同时,在大脑和周围神经系统中,通过将前三个、第五和第六个外显子编码成降钙素相关肽的前体,并利用下游的腺苷酸化位点(CGRP),从而产生差异[14,15]。同样,选择性启动子的使用使得可以选择不同的转录启动子,这样通常会影响到第一个外显子。尽管人们普遍将其当作转录调控,选择性启动子的使用和选择性剪接有很大的关联。人们已经观察到,有选择性启动子的基因更容易进行选择性剪接,同时,选择性启动子的数量与不同选择性的剪接方式的数量正相关[16]。鼠标单羧酸转运蛋白二号(MCT2)基因有几种选择性的启动子,由此形成五种独特的首外显子(1A - 1E)。外显子1C用于各种组织中,而其他的外显子则是有组织特异性的[17]。 除此以外,内含子同样可以参与选择性剪接。在内含子保留性中,整个内含子可以被包括或排除。对于人类来说,这种模式是罕见的[1]。然而,最近的研究表明,在已知的人类基因中,这个频率比想象中要高得多(约15%)[18]。内含子的保留性在植物中比在其他真核生物中常见[19]。例如,对于拟南芥,50%以上的案例都是关于内含子保留性的[20]。人类FosB(FBJ 小鼠骨肉瘤病毒致癌基因对等质B)基因的最后一个外显子包含一个140碱基对的序列,它可以剪接出来,产生一个截断的产物——ΔFosB。通过观察动物长期的药物依赖性,检测ΔFosB的表达[21]。 基本剪接机制 无论是选择性还是组成型剪接,用的都是同一种基本剪接机制,称为剪接体。剪接体识别和选择的剪接位点(外显子和内含子的结点),同时,催化打破并重组RNA链。剪接体主要由五个小核核糖核蛋白(snRNP)——U1,U2,U4,U5和U6组成。其中包括尿苷丰富的小核RNA 和多种蛋白质。它们能识别前体mRNA上的剪接信号,并与其他辅助剪接因子交互[22-24]。 在剪接中,三个保留序列元素是必须的。这些保留序列元素可以是经典或异常的剪接节点,多嘧啶束和分支点(见图1-2)[22,23]。剪接位点含有包括外显子和内含子结点的短序列。GU 和AG二核苷酸在外显子的5'和3'端通常来说是分别不变的。在剪接结合中,这种类型的GU-AG对被称为经典剪接位点。它存在于超过98%的哺乳动物基因组中[25]。异常的剪接位点含有如GC-AG、AT-AC之类的二核苷酸对,AT-AC等。这种情况比较罕见。多嘧啶束是指位于内含子的3’端,且紧邻3‘剪接位点的UC丰富的片段。这是几种剪接因子的结合位点,如U2 snRNP 辅助因子(U2AF)和多嘧啶束结合蛋白(PTB)[26]。分支点(也称为分支位点)位于多嘧啶束的上游。对于人和老鼠来说,分支点和3'剪接结点间的平均距离大约是30至40个碱基对[27]。虽然分支点序列在哺乳动物中是可变的,分支点的突变可以促使由组成型剪接到选择性剪接的转变[27]。
PLC在木板自动剪切机中的应用_毕业设计
石家庄铁道大学四方学院毕业设计 PLC在木板自动剪切机中的应用 The Application of PLC in Automatic Shearing Machine for Wood 2011届电气工程系 专业自动化 完成日期 2011年5月25日
毕业设计成绩单 学生姓名刘举红学号20076749班级0753-1专业自动化毕业设计题目PLC在木板自动剪切机中的应用 指导教师姓名冯涛李文娟 指导教师职称讲师助教 评定成绩 指导教师得 分 评阅人得分 答辩小组组长得分 成绩: 院长(主任) 签字: 年月日
毕业设计任务书 题目PLC在木板自动剪切机中的应用 学生姓名刘举红学号20076749班级0753-1专业自动化 承担指导任务单位电气工程系导师 姓名 冯涛 导师 职称 讲师 一、主要内容 主要设备有行程开关、光电开光、木板、压块、剪刀、工作台、送料传送带、送料小车电动机等组成。具体工艺流程如下:按下启动按钮→电动机送料,板料右行,至指定位置停止→压块下行→将板料压紧→剪刀下行剪切→剪落板料→压块和剪刀上行复位→等到下次启动。 二、基本要求 1.先检测相关设备的初始状态,如电源是否完好,小车是否到位以及行程开关和光电开关是否有故障,然后根据要求剪切木板,当剪切完一块木板以后木板落下,此时光电开关被遮断一下,木板计数一次,并将木板累计数送去显示,当系统计数器记满9999次后报警器发出报警声并重新复位计数器,如此循环进行生产,当按下停止按钮时,结束工作。 2.上位机采用组态王软件进行监控,下位机采用日本三菱公司的FX2N系列。 三、主要技术指标(或研究方法) 1.设计应贯彻最新国家标准; 2.根据控制选择PLC型号,分配I/O端口; 3.设计I/O电路,选择电器元件; 4.绘制电气控制系统图,梯形图,绘制用户程序短语表并模拟调试; 5.编制元件清单; 6.编写设计、使用说明书。 四、应收集的资料及参考文献 [1]《电气控制原理与设计》方承运宁夏人民出版社 [2]《工厂电器控制设备》赵明机械工业出版社 [3]《电气控制技术》韩顺杰中国林业出版社 [4]《可编程控制器原理与应用》高等教育出版社 [5]《小型可编程序控制器原理与实践》辽宁科技出版社 [6]《可编程控制器应用技术》机械工业出版社 五、进度计划 准备、搜集资料、写开题报告第1- 2周 分析、确定方案第3- 4周 系统软硬件设计、模拟调试第5-10周 整理、撰写、编辑论文(打印)第11-15周 答辩第16周 教研室主任签字时间年月日
