WIN_55,212-2_Mesylate_HNMR_08993_MedChemExpress
洋甘菊提取液MSDS英文版

1. IDENTIFICATION OF THE SUBSTANCE/TREPARATION AND THE COMPANY/UNDERTAKING3.HAZARDS IDENTIFICATION4. FIRST AID MEASURESMATERIAL SAFETY DATA SHEETProduct name:Supplier:Tel:EMERGENCY OVERVIEW: May cause skin irritation and/or dermatitisPrinciple routes of exposure: Inhalation: Ingestion: Skin contact: Eye contact:SkinMay cause irritation of respiratory tract May be harmful if swallowed May cause allergic skin reaction Avoid contact with eyesStatements of hazard MAY CAUSE ALLERGIC SKIN REACTION.Statements of Spill of Leak Label Eliminate all ignition sources. Absorb and/or contain spill with inert materials (e.g., sand, vermiculite). Then place in appropriate container. For large spills, use water spray to disperse vapors, flush spill area. Prevent runoff from entering waterways or sewers.General advice:POSITION/INFORMATION ON INGREDIENTSInhalation:Skin contact:Ingestion:Eye contact:Protection of first – aiders:Medical conditions aggravated by exposure: In the case of accident or if you fell unwell, seek medical advice immediately (show the label where possible).Move to fresh air, call a physician immediately.Rinse immediately with plenty of water and seek medical adviceDo not induce vomiting without medical advice.In the case of contact with eyes, rinse immediately with plenty of water and seek medical advice.No information availableNone knownSuitable extinguishing media:Specific hazards:Special protective equipment for firefighters:Flash point:Autoignition temperature:NFPA rating Use dry chemical, CO2, water spray or “alcohol” foam Burning produces irritant fumes.As in any fire, wear self-contained breathing apparatus pressure-demand, MSHA/NIOSH (approved or equivalent) and full protective gearNot determinedNot determinedNFPA Health: 1 NFPA Flammability: 1 NFPA Reactivity: 0Personal precautions: Environmental precautions: Methods for cleaning up: Use personal protective equipment.Prevent product from entering drains.Sweep up and shovel into suitable containers for disposalStorage:7. HANDLING AND STORAGE5.FIRE-FIGHTING MEASURES6. ACCIDENTAL RELEASE MEASURESRoom temperature Handling:Safe handling advice: Incompatible products:Use only in area provided with appropriate exhaust ventilation.Wear personal protective equipment.Oxidising and spontaneously flammable productsEngineering measures: Respiratory protection: Skin and body protection:Eye protection: Hand protection: Hygiene measures:Ensure adequate ventilation.Breathing apparatus only if aerosol or dust is formed. Usual safety precautions while handling the product will provide adequate protection against this potential effect. Safety glasses with side-shieldsPVC or other plastic material glovesHandle in accordance with good industrial hygiene and safety practice.Melting point/range: Boiling point/range: Density: Vapor pressure: Evaporation rate: Vapor density: Solubility (in water): Flash point:Autoignition temperature:No Data available at this time. No Data available at this time. No data available No data available No data available No data available No data available Not determined Not determinedStability: Stable under recommended storage conditions. Polymerization: None under normal processing.Hazardous decomposition products: Thermal decomposition can lead to release of irritating gases and vapours such as carbon oxides.Materials to avoid: Strong oxidising agents.10. STABILITY AND REACTIVITY9. PHYSICAL AND CHEMICAL PROPERTIES8. EXPOSURE CONTROLS/PERSONAL PROTECTION11. TOXICOLOGICAL INFORMATIONConditions to avoid: Exposure to air or moisture over prolonged periods.Product information Acute toxicityChronic toxicity:Local effects: Chronic exposure may cause nausea and vomiting, higher exposure causes unconsciousness.Symptoms of overexposure may be headache, dizziness, tiredness, nausea and vomiting.Specific effects:May include moderate to severe erythema (redness) and moderate edema (raised skin), nausea, vomiting,headache.Primary irritation: Carcingenic effects: Mutagenic effects: Reproductive toxicity:No data is available on the product itself. No data is available on the product itself. No data is available on the product itself. No data is available on the product itself.Mobility:Bioaccumulation: Ecotoxicity effects: Aquatic toxicity:No data available No data available No data availableMay cause long-term adverse effects in the aquatic environment.12. ECOLOGICAL INFORMATION13. DISPOSAL CONSIDERATIONSWaste from residues/unused products:Contaminated packaging:Waste disposal must be in accordance with appropriate Federal, State and local regulations. This product, if unaltered by use, may be disposed of treatment at a permitted facility or as advised by your local hazardous waste regulatory authority. Residue from fires extinguished with this material may be hazardous.Do not re-use empty containers.UN/Id No:Not regulated14. TRANSPORT INFFORMATIONDOTProper shipping name: Not regulatedTGD(Canada)WHMIS hazard class: Non - controlledIMDG/IMOIMDG – Hazard Classifications Not ApplicableIMO – labels:15. REGULATORY INFOTMATION International Inventories16. OTHER INFORMATIONPrepared by: Health & SafetyDisclaimer: The information and recommendations contained herein are based upon tests believed to be reliable.However, XABC does not guarantee the accuracy or completeness NOR SHALL ANY OF THIS INFORMATION CONSTITUTE A WARRANTY, WHETHER EXPRESSED OR IMPLIED, AS TO THE SAFETY OF THE GOOD, THE MERCHANTABILITY OF THE GOODS, OR THE FITNESS OF THE FITNESS OF THE GOODS FOR A PARTICULAR PURPOSE. Adjustment to conform to actual conditions of usage maybe required. XABC assumes no responsibility for results obtained or for incidental or consequential damages, including lost profits arising from the use of these data. No warranty against infringement of any patent, copyright or trademark is made or implied.End of safety data sheet。
酵母表达载体pPICZ手册

pPICZ A, B, and CPichia expression vectors for selection onZeocin™ and purification of recombinant proteins Catalog no. V190-20Rev. Date: 7 July 2010Manual part no. 25-0148MAN00000034User ManualiiTable of ContentsKit Contents and Storage (iv)Accessory Products (v)Introduction (1)Overview (1)Methods (3)Cloning into pPICZ A, B, and C (3)Pichia Transformation (9)Expression in Pichia (13)Purification (15)Appendix (17)Recipes (17)Zeocin™ (19)Map and Features of pPICZ A, B, and C (21)Lithium Chloride Transformation Method (23)Construction of In Vitro Multimers (24)Technical Support (32)Purchaser Notification (33)References (34)iiiKit Contents and StorageContents The following components are included with Catalog no. V190–20. Note that thepPICZ expression vectors are supplied in suspension.Component QuantityCompositionpPICZ A Expression Vector 20 μg 40 μl of 0.5 μg/μl vector in10 mM Tris–HCl, 1 mM EDTA,pH 8.0pPICZ B Expression Vector 20 μg 40 μl of 0.5 μg/μl vector in10 mM Tris–HCl, 1 mM EDTA,pH 8.0pPICZ C Expression Vector 20 μg 40 μl of 0.5 μg/μl vector in10 mM Tris–HCl, 1 mM EDTA,pH 8.0GS115/pPICZ/lacZ Positive1 stab --Control strainShipping/Storage The components included with Catalog no. V190–20 are shipped on wet ice.Upon receipt, store as directed below.For long-term storage of your positive control stab strain, we recommendpreparing a glycerol stock immediately upon receipt and storing at –80°C.Component ShippingStorage pPICZ A Expression Vector Wet ice Store at –20°CpPICZ B Expression Vector Wet ice Store at –20°CpPICZ C Expression Vector Wet ice Store at –20°CGS115/pPICZ/lacZ positive control strain Wet ice Store at 4°CivAccessory ProductsAdditional ProductsThe products listed in this section are intended for use with the pPICZ vectors.For more information, visit our web site at or contactTechnical Support (page 32).Product QuantityCatalogno. X-33 Pichia strain 1 stab C180-00GS115 Pichia strain 1 stab C181-00KM71H Pichia strain 1 stab C182-00SMD1168H Pichia strain 1 stab C184-00pPICZα A, B, and C 20 μg each V195-20pPIC6α A,B, and C 20 μg each V215-20pPIC6 A, B, and C 20 μg each V210-20pPIC6 Starter Kit 1 kit K210-01Original Pichia Expression Kit 1 kit K1710-01EasySelect™Pichia Expression Kit 1 kit K1740-01Pichia EasyComp™ Transformation Kit 1 kit K1730-01Pichia Protocols 1 book G100-01PureLink™ Gel Extraction Kit 50 preps250 prepsK2100–12K2100–25S.N.A.P ™ Gel Purification Kit 25 preps K1999–25PureLink™ Quick Plasmid Miniprep Kit 50 preps250 prepsK2100–10K2100–11PureLink™ HiPure Plasmid Midiprep Kit 25 preps50 prepsK2100–04K2100–13One Shot® TOP10 (chemically competent E. coli) 10 reactions20 reactionsC4040–10C4040–03One Shot® TOP10 Electrocompetent E. Coli 10 reactions20 reactionsC4040-50C4040-52TOP10 Electrocomp™ Kits 20 reactions C664–55Positope™ Control Protein 5 μg R900-50CIAP (Calf Intestinal Alkaline Phosphatase) 1,000 units 18009–019T4 DNA Ligase 100 units500 units15224–01715224–025Zeocin™ 1g5 gR250-01R250-05β-Gal Assay Kit 1 kit K1455-01β-Gal Staining Kit 1 kit K1465-01E-Gel® Agarose Gels E-Gel® Agarose Gels are bufferless, pre-cast agarose gels designed for fast, convenient electrophoresis of DNA samples. E-Gel® agarose gels are available in different agarose percentage and well format for your convenience.For more details on these products, visit our web site at or contact Technical Support (page 32).Continued on next pagevAccessory Products, ContinuedZeocin™Zeocin™ may be obtained from Invitrogen (see above). For your convenience, the drug is prepared in autoclaved, deionized water and available in 1.25 ml aliquotsat a concentration of 100 mg/ml. The stability of Zeocin™ is guaranteed for sixmonths if stored at –20°C.Detection of Fusion Protein A number of antibodies are available from Invitrogen to detect expression ofyour fusion protein from the pPICZ vector. Horseradish peroxidase (HRP)-conjugated antibodies allow one-step detection in Western blots usingcolorimetric or chemiluminescent detection methods. The amount of antibodysupplied is sufficient for 25 Western Blots.Antibody Epitope Catalogno.Anti-myc R950–25 Anti-myc-HRPDetects the 10 amino acid epitopederived from c-myc (Evans et al., 1985):EQKLISEEDLR951–25Anti-His(C-term) R930–25Anti-His(C-term)-HRPDetects the C-terminal polyhistidine(6xHis) tag (requires the free carboxylgroup for detection) (Lindner et al., 1997):HHHHHH-COOHR931–25Purification of Fusion Protein The polyhistidine (6xHis) tag allows purification of the recombinant fusionprotein using metal-chelating resins such as ProBond™. Ordering information forProBond™ resin is provided below.Product QuantityCatalogno. ProBond™ Purification System 6 purifications K850–01ProBond ™ Purification System with Anti-myc-HRP Antibody1 Kit K852–01ProBond ™ Purification System with Anti-His(C-term)-HRP Antibody1 Kit K853–01ProBond™ Nickel-Chelating Resin 50 ml150 mlR801–01R801–15Purification Columns 50 each R640–50viIntroductionOverviewIntroduction pPICZ A, B, and C are 3.3 kb expression vectors used to express recombinantproteins in Pichia pastoris. Recombinant proteins are expressed as fusions to aC-terminal peptide containing the c-myc epitope and a polyhistidine (6xHis) tag.The vector allows high-level, methanol inducible expression of the gene ofinterest in Pichia, and can be used in any Pichia strain including X33, GS115,SMD1168H, and KM71H. pPICZ contains the following elements:•5′ fragment containing the AOX1 promoter for tightly regulated, methanol-induced expression of the gene of interest (Ellis et al., 1985; Koutz et al., 1989;Tschopp et al., 1987a)•Zeocin™ resistance gene for selection in both E. coli and Pichia (Baron et al.,1992; Drocourt et al., 1990)•C-terminal peptide containing the c-myc epitope and a polyhistidine (6xHis)tag for detection and purification of a recombinant fusion protein (if desired)•Three reading frames to facilitate in-frame cloning with the C-terminalpeptideReference Sources The pPICZ A, B, and C expression vectors may be used with the Original Pichia Expression Kit, and are included in the EasySelect™Pichia Expression Kit (see page v for ordering information). Additional general information about recombinant protein expression in Pichia pastoris is provided in the manuals for the Original Pichia Expression Kit and the EasySelect™Pichia Expression Kit. For more information about the Original Pichia Expression Kit, the EasySelect™Pichia Expression Kit, or their manuals, visit our web site at or contact Technical Support (page 32).More detailed information and protocols dealing with Pichia pastoris may also be found in the following general reference:Higgins, D. R., and Cregg, J. M. (1998) Pichia Protocols. In Methods in Molecular Biology, Vol. 103. (J. M. Walker, ed. Humana Press, Totowa, NJ) (see page v for ordering information).Recommended Pichia Host Strain We recommend using the X-33 Pichia strain as the host for expression of recombinant proteins from pPICZ. Other Pichia strains including GS115, KM71H, and SMD1168H are suitable. The X-33 Pichia strain and other strains are available from Invitrogen (see page v for ordering information). The X-33 Pichia strain has the following genotype and phenotype:Genotype: Wild-typePhenotype: Mut+1Overview, ContinuedExperimental Overview The following table describes the basic steps needed to clone and express your gene of interest in pPICZ.Step Action1 Propagate pPICZ A, B, and C by transformation into a rec A, end A1E. coli strain such as TOP10, DH5 , or JM109.2 Develop a cloning strategy and ligate your gene into one of the pPICZvectors in frame with the C-terminal tag.3 TransformintoE. coli and select transformants on Low Salt LB platescontaining 25 μg/ml Zeocin™.4 Analyze 10–20 transformants by restriction mapping or sequencing toconfirm in-frame fusion of your gene with the C-terminal tag.5 Purify and linearize the recombinant plasmid for transformation intoPichia pastoris.6 TransformyourPichia strain and plate onto YPDS plates containing the appropriate concentration of Zeocin™.7 Select for Zeocin™-resistant transformants.8 Optimize expression of your gene.9 Purify your fusion protein on metal-chelating resin (i.e. ProBond™).Continued on next page2MethodsCloning into pPICZ A, B, and CIntroduction The pPICZ vector is supplied with the multiple cloning site in three readingframes (A, B, and C) to facilitate cloning your gene of interest in frame with theC-terminal peptide containing the c-myc epitope and a polyhistidine (6xHis) tag.Use the diagrams provided on pages 5–7 to help you design a strategy to cloneyour gene of interest in frame with the C-terminal peptide. Generalconsiderations for cloning and transformation are discussed in this section.General Molecular Biology Techniques For assistance with E. coli transformations, restriction enzyme analysis, DNA biochemistry, and plasmid preparation, refer to Molecular Cloning: A Laboratory Manual (Sambrook et al., 1989) or Current Protocols in Molecular Biology (Ausubel et al., 1994).E. coli Strain Many E. coli strains are suitable for the propagation of the pPICZ vectorsincluding TOP10, JM109, and DH5 . We recommend that you propagate thepPICZ vectors in E. coli strains that are recombination deficient (rec A) andendonuclease A deficient (end A).For your convenience, TOP10 E. coli are available as chemically competent orelectrocompetent cells from Invitrogen (page v).Transformation Method You may use any method of choice for transformation. Chemical transformation is the most convenient for many researchers. Electroporation is the most efficient and the method of choice for large plasmids.Maintenance of Plasmids The pPICZ vectors contain the Zeocin™ resistance (Sh ble) gene to allow selection of the plasmid using Zeocin™. To propagate and maintain the pPICZ plasmids, we recommend using the following procedure:e 10 ng of your vector to transform a rec A, end A E. coli strain like TOP10,DH5 , JM109, or equivalent (see above).2.Select transformants on Low Salt LB plates containing 25 μg/ml Zeocin™ (seepage 17 for a recipe).3.Prepare a glycerol stock from each transformant containing plasmid forlong-term storage (see page 8).Continued on next page3Cloning into pPICZ A, B, and C, ContinuedGeneral Considerations The following are some general points to consider when using pPICZ to express your gene of interest in Pichia:•The codon usage in Pichia is believed to be similar to Saccharomyces cerevisiae.•Many Saccharomyces genes have proven to be functional in Pichia.•The premature termination of transcripts because of "AT rich regions" has been observed in Pichia and other eukaryotic systems (Henikoff & Cohen, 1984; Irniger et al., 1991; Scorer et al., 1993; Zaret & Sherman, 1984). If you have problems expressing your gene, check for premature termination by northern analysis and check your sequence for AT rich regions. It may be necessary to change the sequence in order to express your gene (Scorer et al., 1993).•The native 5´ end of the AOX1 mRNA is noted in the diagram for each multiple cloning site. This information is needed to calculate the size of the expressed mRNA of the gene of interest if you need to analyze mRNA for any reason.Cloning Considerations For proper initiation of translation, your insert should contain an initiation ATG codon as part of a yeast consensus sequence (Romanos et al., 1992). An example of a yeast consensus sequence is provided below. The ATG initiation codon is shown underlined.(G/A)NNATG GTo express your gene as a recombinant fusion protein, you must clone your gene in frame with the C-terminal peptide containing the c-myc epitope and the polyhistidine tag. The vector is supplied in three reading frames to facilitate cloning. Refer to the diagrams on pages 5–7 to develop a cloning strategy.If you wish to express your protein without the C-terminal peptide, be sure to include a stop codon.Construction of Multimeric Plasmids pPICZ A, B, and C contain unique Bgl II and Bam H I sites to allow construction of plasmids containing multiple copies of your gene. For information on how to construct multimers, refer to pages 24–31.Continued on next page4Multiple CloningSite of pPICZ A Below is the multiple cloning site for pPICZ A. Restriction sites are labeled to indicate the cleavage site. The boxed nucleotides indicate the variable region.The multiple cloning site has been confirmed by sequencing and functionaltesting.You can download the complete sequence of pPICZ A from our web site at or by contacting Technical Support (see page 32).For a map and a description of the features of pPICZ, refer to the Appendix(pages 21–22). AAT AGC GCC GTC GAC CAT CAT CAT CAT CAT CAT TGTTCCTCAG TTCAAGTTGG GCACTTACGA GAAGACCGGT CTTGCTAGAT TCTAATCAAG AGGATGTCAG AATGCCATTT GCCTGAGAGA TGCAGGCTTC ATTTTTGATA CTTTTTTATTTGTAACCTAT ATAGTATAGG ATTTTTTTTG TCATTTTGTT 1218Asn Ser Ala Val Asp His His His His His His ***3´ AOX1 priming site TGA GTTTTAGCCT TAGACATGAC AACCTTTTTT TTTATCATCA TTATTAGCTT ACTTTCATAA TTGCGACTGG TTCCAATTGA CAAGCTTTTG ATTTTAACGA CTTTTAACGA CAACTTGAGA AGATCAAAAA ACAACTAATT ATTCGAAACG AGGAATTCAC GTGGCCCAGC CGGCCGTCTC GGATCGGTAC CTCGAGCCGC GGCGGCCGCC AGCTT GGGCCC GAA CAA AAA CTC ATC TCA GAA GAG GAT CTG 811Glu Gln Lys Leu Ile Ser Glu Glu Asp Leu 5´ AOX1 priming sitemyc epitope3´polyadenylation sitePolyhistidine tag5´ end of AOX1 mRNA Sfu I Eco R I Pml I Sfi I Bsm B I Asp 718 I Kpn I Xho ISac II Not I Apa I 104210981158871931991Continued on next pageMultiple CloningSite of pPICZ B Below is the multiple cloning site for pPICZ B. Restriction sites are labeled to indicate the cleavage site. The boxed nucleotides indicate the variable region.The multiple cloning site has been confirmed by sequencing and functionaltesting.You can download the complete sequence of pPICZ B from our web site at or by contacting Technical Support (see page 32).For a map and a description of the features of pPICZ, refer to the Appendix(pages 21–22). AAT AGC GCC GTC GAC CAT CAT CAT CAT CAT CAT TGA GTTTGTAGCC TTAGACATGA CTGTTCCTCA GTTCAAGTTG GGCACTTACG AGAAGACCGG TCTTGCTAGA TTCTAATCAA GAGGATGTCA GAATGCCATT TGCCTGAGAG ATGCAGGCTT CATTTTTGAT ACTTTTTTAT TTGTAACCTA TATAGTATAG GATTTTTTTT GTCATTTTGT TTC 1216Asn Ser Ala Val Asp His His His His His His ***3´ AOX1 priming site TGA GTTTGTAGCC TTAGACATGA AACCTTTTTT TTTATCATCA TTATTAGCTT ACTTTCATAA TTGCGACTGG TTCCAATTGA CAAGCTTTTG ATTTTAACGA CTTTTAACGA CAACTTGAGA AGATCAAAAA ACAACTAATTATTCGAAACG AGGAATTCAC GTGGCCCAGC CGGCCGTCTC GGATCGGTAC CTCGAGCCGC GGCGGCCGCC AGCTT TCTA GAA CAA AAA CTC ATC TCA GAA GAG GAT CTG 811Glu Gln Lys Leu Ile Ser Glu Glu Asp Leu 5´ AOX1 priming sitemyc epitope3´ polyadenylation site Polyhistidine tag5´ end of AOX1 mRNA Sfu I Eco R I Pml I Sfi I Bsm B I Asp 718 I Kpn I Xho ISac II Not I Xba I 104010961156871931991Continued on next pageMultiple CloningSite of pPICZ C Below is the multiple cloning site for pPICZ C. Restriction sites are labeled to indicate the cleavage site. The boxed nucleotides indicate the variable region.The multiple cloning site has been confirmed by sequencing and functionaltesting.You can download the complete sequence of pPICZ C from our web site at or by contacting Technical Support (see page 32).For a map and a description of the features of pPICZ, refer to the Appendix(pages 21–22). AAT AGC GCC GTC GAC CAT CAT CAT CAT CAT CAT TGA GTTTGTAGCC TTAGACATGA CTGTTCCTCA GTTCAAGTTG GGCACTTACG AGAAGACCGG TCTTGCTAGA TTCTAATCAA GAGGATGTCA GAATGCCATT TGCCTGAGAG ATGCAGGCTT CATTTTTGAT ACTTTTTTAT TTGTAACCTA TATAGTATAG GATTTTTTTT GTCATTTTGT TTC 1217Asn Ser Ala Val Asp His His His His His His ***3´ AOX1 priming siteTGA GTTTGTAGCC TTAGACATGA AACCTTTTTT TTTATCATCA TTATTAGCTT ACTTTCATAA TTGCGACTGG TTCCAATTGA CAAGCTTTTG ATTTTAACGA CTTTTAACGA CAACTTGAGA AGATCAAAAA ACAACTAATT ATTCGAAACG AGGAATTCAC GTGGCCCAGC CGGCCGTCTC GGATCGGTAC CTCGAGCCGC GGCGGCCGCC AGCTT ACGTA GAA CAA AAA CTC ATC TCA GAA GAG GAT CTG 811Glu Gln Lys Leu Ile Ser Glu Glu Asp Leu 5´ AOX1 priming sitemyc epitope3´ polyadenylation site Polyhistidine tag5´ end of AOX1 mRNA Sfu I Eco R I Pml I Sfi I Bsm B I Asp 718 I Kpn I Xho I Sac II Not I Sna B I 104110971157871931991Continued on next pageE. coli Transformation Transform your ligation mixtures into a competent rec A, end A E. coli strain(e.g. TOP10, DH5, JM109) and select on Low Salt LB agar plates containing25 μg/ml Zeocin™ (see below). Note that there is no blue/white screening for the presence of insert with pPICZ A, B, or C. Once you have obtained Zeocin™-resistant colonies, pick 10 transformants and screen for the presence and orientation of your insert.Important To facilitate selection of Zeocin™-resistant E. coli, the salt concentration of the medium must remain low (<90 mM) and the pH must be 7.5. Prepare Low Salt LB broth and plates using the recipe in the Appendix, page 17.Failure to lower the salt content of your LB medium will result in non-selection due to inhibition of the drug.C We recommend that you sequence your construct to confirm that your gene is in the correct orientation for expression and cloned in frame with the C-terminal peptide (if desired). Refer to the diagrams on pages 5–7 for the sequences and location of the priming sites.Preparing a Glycerol Stock Once you have identified the correct clone, be sure to purify the colony and make a glycerol stock for long-term storage. It is also a good idea to keep a DNA stock of your plasmid at –20°C.1.Streak the original colony out on an Low Salt LB plate containing 25 μg/mlZeocin™. Incubate the plate at 37°C overnight.2.Isolate a single colony and inoculate into 1–2 ml of Low Salt LB containing25 μg/ml Zeocin™.3.Grow the culture to mid-log phase (OD600 = 0.5–0.7).4.Mix 0.85 ml of culture with 0.15 ml of sterile glycerol and transfer to acryovial.5.Store at –80°C.Plasmid Preparation Once you have cloned and sequenced your insert, generate enough plasmid DNA to transform Pichia (5–10 μg of each plasmid per transformation). We recommend isolating plasmid DNA using the PureLink™ Quick Plasmid Miniprep Kit or the PureLink™ HiPure Plasmid Midiprep Kit (page v), or CsCl gradient centrifugation.Once you have purified plasmid DNA, proceed to Pichia Transformation, next page.Pichia TransformationIntroduction You should now have your gene cloned into one of the pPICZ vectors. Yourconstruct should be correctly fused to the C-terminal peptide (if desired). Thissection provides general guidelines to prepare plasmid DNA, transform yourPichia strain, and select for Zeocin™-resistant clones.Zeocin™ Selection We generally use 100 μg/ml Zeocin™ to select for transformants when using the X-33 Pichia strain. If you are transforming your pPICZ construct into anotherPichia strain, note that selection conditions may vary. We recommendperforming a dose response curve to determine the appropriate concentration ofZeocin™ to use for selection of transformants in your strain.Method of Transformation We do not recommend spheroplasting for transformation of Pichia with plasmids containing the Zeocin™ resistance marker. Spheroplasting involves removal of the cell wall to allow DNA to enter the cell. Cells must first regenerate the cell wall before they are able to express the Zeocin™ resistance gene. For this reason, plating spheroplasts directly onto selective medium containing Zeocin™ does not yield any transformants.We recommend electroporation for transformation of Pichia with pPICZ A, B, or C. Electroporation yields 103 to 104 transformants per μg of linearized DNA and does not destroy the cell wall of Pichia. If you do not have access to an electroporation device, use the LiCl protocol on page 23 or the Pichia EasyComp™Transformation Kit available from Invitrogen (see below).PichiaEasyComp™Transformation Kit If you wish to perform chemical transformation of your Pichia strain with pPICZ A, B, or C, the Pichia EasyComp™ Transformation Kit is available from Invitrogen (see page v for ordering information). The Pichia EasyComp™ Transformation Kit provides reagents to prepare 6 preparations of competent cells. Each preparation will yield enough competent cells for 20 transformations. Competent cells may be used immediately or frozen and stored for future use. For more information, visit our web site at or contact Technical Support (page 32).Important Since pPICZ does not contain the HIS4 gene, integration can only occur at the AOX1 locus. Vector linearized within the 5´ AOX1 region will integrate by gene insertion into the host 5´ AOX1 region. Therefore, the Pichia host that you use will determine whether the recombinant strain is able to metabolize methanol (Mut+) or not (Mut S). To generate a Mut+ recombinant strain, you must use a Pichia host that contains the native AOX1 gene (e.g. X-33, GS115, SMD1168H). If you wish to generate a Mut S recombinant strain, then use a Pichia host that has a disrupted AOX1 gene (i.e. KM71H).Continued on next pageHis4 Host Strains Host strains containing the his4 allele (e.g. GS115) and transformed with thepPICZ vectors require histidine when grown in minimal media. Add histidine toa final concentration of 0.004% to ensure growth of your transformants.The pPICZ vectors do not contain a yeast origin of replication. Transformantscan only be isolated if recombination occurs between the plasmid and the Pichiagenome.Materials Needed You will need the following items:Note: Inclusion of sorbitol in YPD plates stabilizes electroporated cells as they appear tobe somewhat osmotically sensitive.•5–10 μg pure pPICZ containing your insert•YPD Medium•50 ml conical polypropylene tubes• 1 liter cold (4°C) sterile water (place on ice the day of the experiment)•25 ml cold (4°C) sterile 1 M sorbitol (place on ice the day of the experiment)•30°C incubator•Electroporation device and 0.2 cm cuvettes•YPDS plates containing the appropriate concentration of Zeocin™ (seepage 18 for recipe)Linearizing YourpPICZ ConstructTo promote integration, we recommend that you linearize your pPICZ constructwithin the 5′ AOX1 region. The table below lists unique sites that may be used tolinearize pPICZ prior to transformation. Other restriction sites are possible.Note that for the enzymes listed below, the cleavage site is the same for versionsA, B, and C of pPICZ. Be sure that your insert does not contain the restriction siteyou wish to use to linearize your vector.Enzyme Restriction Site (bp) SupplierSac I 209 ManyPme I 414 New England BiolabsBst X I 707 ManyRestriction Digest 1.Digest ~5–10 μg of plasmid DNA with one of the enzymes listed above.2.Check a small aliquot of your digest by agarose gel electrophoresis forcomplete linearization.3.If the vector is completely linearized, heat inactivate or add EDTA to stopthe reaction, phenol/chloroform extract once, and ethanol precipitate using1/10 volume 3 M sodium acetate and 2.5 volumes of 100% ethanol.4.Centrifuge the solution to pellet the DNA, wash the pellet with 80% ethanol,air-dry, and resuspend in 10 μl sterile, deionized water. Use immediately orstore at –20°C.Continued on next pagePreparation of Pichia for Electroporation Follow the procedure below to prepare your Pichia pastoris strain for electroporation.1. Grow 5 ml of your Pichia pastoris strain in YPD in a 50 ml conical tube at30°C overnight.2. Inoculate 500 ml of fresh medium in a 2 liter flask with 0.1–0.5 ml of theovernight culture. Grow overnight again to an OD600 = 1.3–1.5.3. Centrifuge the cells at 1500 × g for 5 minutes at 4°C. Resuspend the pelletwith 500 ml of ice-cold (0–4°C), sterile water.4. Centrifuge the cells as in Step 3, then resuspend the pellet with 250 ml ofice-cold (0–4°C), sterile water.5. Centrifuge the cells as in Step 3, then resuspend the pellet in 20 ml of ice-cold (0–4°C) 1 M sorbitol.6. Centrifuge the cells as in Step 3, then resuspend the pellet in 1 ml of ice-cold(0–4°C) 1 M sorbitol for a final volume of approximately 1.5 ml. Keep the cells on ice and use that day. Do not store cells.Transformation by Electroporation 1.Mix 80 μl of the cells from Step 6 (above) with 5–10 μg of linearized pPICZDNA (in 5–10 μl sterile water) and transfer them to an ice-cold (0–4°C)0.2 cm electroporation cuvette.2.Incubate the cuvette with the cells on ice for 5 minutes.3.Pulse the cells according to the parameters for yeast (Saccharomycescerevisiae) as suggested by the manufacturer of the specific electroporation device being used.4.Immediately add 1 ml of ice-cold 1 M sorbitol to the cuvette. Transfer thecuvette contents to a sterile 15 ml tube.5.Let the tube incubate at 30°C without shaking for 1 to 2 hours.6.Spread 50-200 μl each on separate, labeled YPDS plates containing theappropriate concentration of Zeocin™.7.Incubate plates for 2–3 days at 30°C until colonies form.8.Pick 10–20 colonies and purify (streak for single colonies) on fresh YPD orYPDS plates containing the appropriate concentration of Zeocin™.Continued on next pageGenerally, several hundred Zeocin™-resistant colonies are generated using theprotocol on the previous page. If more colonies are needed, the protocol may bemodified as described below. Note that you will need ~20, 150 mm plates withYPDS agar containing the appropriate concentration of Zeocin™.1. Set up two transformations per construct and follow Steps 1 through 5 ofthe Transformation by Electroporation protocol, page 11.2. After 1 hour in 1 M sorbitol at 30°C (Step 5, previous page), add 1 ml YPDmedium to each tube.3. Shake (~200 rpm) the cultures at 30°C.4. After 1 hour, take one of the tubes and plate out all of the cells by spreading200 μl on 150 mm plates containing the appropriate concentration ofZeocin™.5. Optional: Continue incubating the other culture for three more hours (for atotal of four hours) and then plate out all of the cells by spreading 200 μl on150 mm plates containing the appropriate concentration of Zeocin™.6. Incubate plates for 2–4 days at 30°C until colonies form.Mut Phenotype If you used a Pichia strain containing a native AOX1 gene (e.g. X-33, GS115,SMD1168H) as the host for your pPICZ construct, your Zeocin™-resistanttransformants will be Mut+. If you used a strain containing a deletion in theAOX1 gene (e.g. KM71H), your transformants will be Mut S.If you wish to verify the Mut phenotype of your Zeocin™-resistant transformants,you may refer to the general guidelines provided in the EasySelect™PichiaExpression Kit manual or the Original Pichia Expression Kit manual or topublished reference sources (Higgins & Cregg, 1998).You are now ready to test your transformants for expression of your gene ofinterest. See Expression in Pichia, next page.。
碧云天生物技术聚甲基丙烯酸2-羟乙酯产品说明书
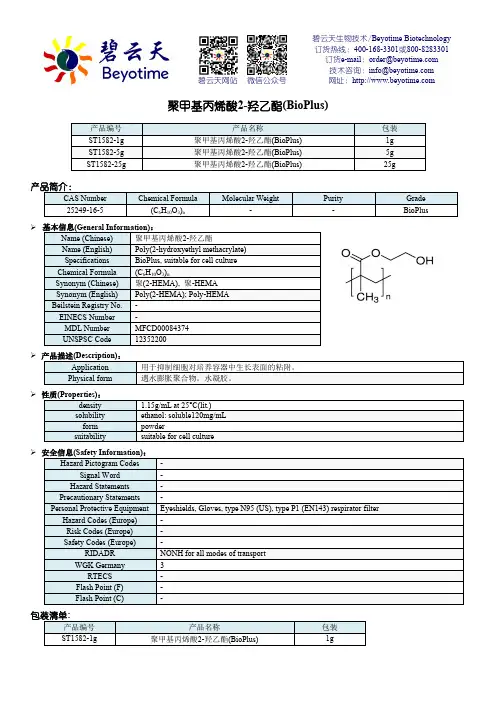
碧云天生物技术/Beyotime Biotechnology 订货热线:400-168-3301或800-8283301 订货e-mail :******************技术咨询:*****************网址:碧云天网站 微信公众号聚甲基丙烯酸2-羟乙酯(BioPlus)产品编号 产品名称包装 ST1582-1g 聚甲基丙烯酸2-羟乙酯(BioPlus) 1g ST1582-5g 聚甲基丙烯酸2-羟乙酯(BioPlus) 5g ST1582-25g聚甲基丙烯酸2-羟乙酯(BioPlus)25g产品简介:CAS Number Chemical FormulaMolecular WeightPurity Grade 25249-16-5(C 6H 10O 3)n--BioPlus基本信息(General Information):Name (Chinese) 聚甲基丙烯酸2-羟乙酯 Name (English) Poly(2-hydroxyethyl methacrylate) Specifications BioPlus, suitable for cell culture Chemical Formula (C 6H 10O 3)n Synonym (Chinese) 聚(2-HEMA), 聚-HEMA Synonym (English) Poly(2-HEMA); Poly-HEMA Beilstein Registry No. - EINECS Number - MDL Number MFCD00084374 UNSPSC Code 12352200 产品描述(Description): Application 用于抑制细胞对培养容器中生长表面的粘附。
Physical form 遇水膨胀聚合物。
水凝胶。
性质(Properties):density 1.15g/mL at 25°C(lit.) solubility ethanol: soluble120mg/mL form powdersuitabilitysuitable for cell culture安全信息(Safety Information):Hazard Pictogram Codes- Signal Word - Hazard Statements - Precautionary Statements-Personal Protective Equipment Eyeshields, Gloves, type N95 (US), type P1 (EN143) respirator filter Hazard Codes (Europe) - Risk Codes (Europe) - Safety Codes (Europe)-RIDADR NONH for all modes of transport WGK Germany3 RTECS - Flash Point (F) - Flash Point (C)-包装清单:产品编号 产品名称包装 ST1582-1g聚甲基丙烯酸2-羟乙酯(BioPlus)1gST1582-5g 聚甲基丙烯酸2-羟乙酯(BioPlus) 5gST1582-25g 聚甲基丙烯酸2-羟乙酯(BioPlus) 25g-说明书1份保存条件:室温保存。
伯乐生化质控靶值表310

封闭添加试剂(按字母顺序排列)

询价
4、BMPA
改变巯基基团为羧基基团,用于制备肽段-蛋白质交联物。
产品特点:
•反应基团:马来酰亚胺和羧基
•反应指向:巯基基团和胺(当与EDC结合使用时)
参考文献
Hermanson, G.T. (2008). Bioconjugate Techniques, 2nd ed., Elsevier Inc., pp.111-113. (Product # 20036)
25g
473
相关产品:
产品货号
产品名称
价格¥
24510
Sulfo-NHS
1848
22980
EDC
1005
20320
DCC
488
14、STAT,SATP和巯基添加试剂盒
添加保护性巯基防止二肽形成。
产品特点:
•与伯胺反应添加保护性巯基
•简单的脱保护步骤即可产生游离的巯基
•Thermo Scientific巯基添加试剂盒包含硫醇化作用,脱保护,纯化和定量所有需要的组分
订购信息:
产品货号
产品名称
包装
价格¥
23031
二盐酸乙二胺
10g
399
8、碘乙酰胺(Iodoacetamide),一次性使用
质谱前可靠的烷基化试剂。产品ຫໍສະໝຸດ 点:•与巯基基团形成共价键
•质谱级别(高纯度)
•方便,一次性使用形式(3个微型管,每管含有9.4mg)
•当用132μl碳酸氢铵(ammonium bicarbonate)溶解时,每管产生375mM的溶液
500mg
620
10、MMTS
可与巯基反应的可逆试剂。
产品特点:
•将-SH基团转化为-S-S-CH3
美索巴莫注射液标准

美索巴莫注射液标准美索巴莫注射液是一种中枢性肌肉松弛剂,主要用于治疗急性骨骼肌疼痛或不适症状。
以下是关于美索巴莫注射液的一些标准信息:1. 中文通用名:美索巴莫注射液2. 英文通用名:Methocarbamol Injection3. 标准号:ws-10001-(hd-0269)-20024. 药品名称:美索巴莫注射液5. 药品英文名:Methocarbamol Injection6. 主要成分:本品为美索巴莫的灭菌聚乙二醇,400水溶液。
含美索巴莫(c11h15no5)应为标示量的95.0%~105.%。
7. 处方:具体处方应根据医生建议和患者病情来确定。
8. 性状:本品为无色或几乎无色略带黏稠的澄明液体。
9. 鉴别:通过化学分析和物理测试等方法来鉴别美索巴莫注射液的真伪和质量。
10. 作用类别:中枢性肌肉松弛剂11. 药理毒理:本品对中枢神经系统有选择作用,特别对脊椎中神经元作用明显。
抑制与骨骼肌痉挛有关的神经突触反射,有抗士的宁和电刺激所致惊厥的作用,并有解痉、镇痛和抗炎作用。
其作用机制主要是阻断脊髓内中枢神经元从而使骨骼肌松弛。
12. 药代动力学:美索巴莫注射液在体内的吸收、分布、代谢和排泄过程。
13. 适应症:主要用于急性骨骼肌疼痛或不适症状的辅助治疗。
14. 用法用量:美索巴莫注射液的用法主要是采用静脉滴注或者是静脉推注的方式给药。
如果是用于静脉推注时,患者在静卧的条件下缓慢推注,给药的速度每分钟不可以超过3ml,注射之后应该至少休息10~15分钟。
如果是用于静脉滴注时,将药品配在9%的氯化钠注射液或者是5%的葡萄糖注射液当中,滴注的速度不宜过快。
使用剂量和次数根据病情和治疗效果来决定。
成人一次使用剂量为1.0g,一日最大剂量为3.0g,连续使用不得超过3天。
轻度病例静注后应改为口服给药以维持治疗。
15. 不良反应:使用美索巴莫注射液可能出现的不良反应,如头痛、恶心、呕吐、皮疹等。
16. 禁忌症:对美索巴莫过敏者、肝肾功能不全者、哺乳期妇女禁用。
阿司咪唑
合成方法
合成方法
化合物(I)和碘甲烷在乙醇中回流8h,环合得到化合物(Ⅱ)。再水解脱去酯基,得到化合物(Ⅲ)。用对甲氧 基苯乙基溴进行N-烷基化,得化合物(Ⅳ)。再用对氟苄基溴烷基化,得阿司咪唑。
1. 1-[(4-氟苯基)甲基]-苯并咪唑-2-(3H)-酮的制备
在反应瓶中加入2-羟基苯并咪唑5.0g(37.3mmol)和NaH 1.6g(53mmol)(NaH含量大约为80%,浸入矿物油中) 的DMF 100ml的悬浮液.加毕.在60ºC.(最好有N2保护)搅拌反应1h.再加入4-氟苄基氯(FBC)5.4g(37mmol),加热 ( 6 0 ºC ) 搅 拌 反 应 5 . 5 h . 冷 却 至 室 温 后 加 入 冰 水 7 0 0 m l , 用 二 氯 甲 烷 ( 5 0 0 m l × 2 ) 提 取 . 有 机 层 用 食 盐 水 洗 . 无 水 N a 2 S O 4 干燥.过滤.滤液减压浓缩.剩余物用石油醚析晶.得1-[(4-氟苯基)甲基]-苯并咪唑-2-(3H)-酮固体8.0g,为无色 结 晶 m p 1 7 8 ~ 1 7 9 ºC , 收 率 8 8 % .
治疗措施
阿司咪唑中毒的治疗要点为: 1.大量摄入者予洗胃,后灌服活性炭和导泻。 2.对心肌抑制和Q-T间期延长者予5%碳酸氢钠250ml静注可能有效。 3.对症、支持治疗。
专家点评
专家点评
阿司咪阿司咪唑自1983年上市以来,在许多国家得到了广泛应用。国外研究显示阿司咪唑治疗荨麻疹的总有 效率为74%。国内的一项多中心双盲安慰剂对照试验表明阿司咪唑对急性荨麻疹的总有效率为82.9%,对慢性荨麻 疹的总有效率为86.0%,均显著高于安慰剂,主要不良反应为嗜睡、倦怠、口干等,连续用药3个月的患者中,半 数有食欲及体重增加。阿司咪唑的心脏毒性虽然发生率较低,但由于后果严重,已限制了它的应用。阿司咪唑为 强效和长效的H1受体拮抗剂,无中枢镇静和抗毒蕈碱样作用。代谢产物去甲阿司咪唑仍有抗胆胺作用。长期服用 可增进食欲和增加体重,服用过量可引起心脏Q-T间期延长和室性心律失常。适用于各种原因引起过敏性疾病。
安捷伦产品目录
15
Real-Time PCR
16
Mx3000P QPCR System
17
Brilliant III Ultra-Fast SYBR Green QPCR and QRT-PCR Reagents
18
Brilliant III Ultra-Fast QPCR and QRT-PCR Reagents
Agilent / STRATAGENE
Agilent website: /genomics
Welgene | Agilent Stratagene
威健股份有限公司 | Stratagene 總代理
Table of Content
Table of Contents
/ XL1-Red Competent Cells SoloPack Gold Supercompetent Cells
/ TK Competent Cells Specialty Cells
/ Classic Cells / Fine Chemicals For Competent Cells
適用於 UNG 去汙染或 bisulphite
sequencing
適用於 TA Cloning
最高敏感性
取代傳統 Taq 的好選擇
-
2
威健股份有限公司 | Stratagene 總代理
PCR Enzyme & Instrument
Agilent SureCycler 8800
市場上領先的 cycling 速度和 sample 體積 10 ~ 100 μL 簡易快速可以選擇 96 well 和 384 well 操作盤 優秀的溫控設備讓各個 well 都能保持溫度的穩定 七吋的高解析度觸控螢幕讓操作上更為簡便 可以透過網路遠端操控儀器及監控儀器 Agilent 專業的技術支援可以幫助您應對各種 PCR 的問題
2-甲基丙烯酸羟乙酯密度
2-甲基丙烯酸羟乙酯密度
2-甲基丙烯酸羟乙酯(简称Methacrylic Acid Hydroxyethyl Ester,缩写为HEMA)是一种无色透明的液体。
它的密度通常在
1.05克/毫升到1.11克/毫升之间,具体数值取决于温度和压力。
在常温下(约20摄氏度),它的密度大约为1.07克/毫升。
需要注意的是,密度值可能会因不同的生产商或供应商而略有不同。
密度的测量对于工业生产和实验室研究中的配方和计量非常重要,因为它直接影响着该化合物在各种应用中的使用量和性能。
密度的准确测量有助于确保产品质量和生产过程的稳定性。
重组贻贝粘蛋白的表征及功效评价
生物技术进展 2023 年 第 13 卷 第 4 期 596 ~ 603Current Biotechnology ISSN 2095‑2341研究论文Articles重组贻贝粘蛋白的表征及功效评价李敏 , 魏文培 , 乔莎 , 郝东 , 周浩 , 赵硕文 , 张立峰 , 侯增淼 *西安德诺海思医疗科技有限公司,西安 710000摘要:为了推进重组贻贝粘蛋白在医疗、化妆品领域的应用,对大肠杆菌规模化发酵及纯化生产获得的重组贻贝粘蛋白进行了表征及功效评价。
经Edman 降解法、基质辅助激光解吸电离飞行时间质谱、PITC 法、非还原型SDS -聚丙烯酰胺凝胶电泳法、凝胶法、改良的Arnow 法对重组贻贝粘蛋白进行氨基酸N 端测序、相对分子量分析、氨基酸组成分析、蛋白纯度分析、内毒素含量测定、多巴含量测定;通过细胞迁移、斑马鱼尾鳍修复效果对重组贻贝粘蛋白进行功效评价。
结果显示,获得的重组贻贝粘蛋白与理论的一级结构一致,蛋白纯度达95%以上,内毒素<10 EU ·mg -1,多巴含量大于5%;重组贻贝粘蛋白浓度为60 μg ·mL -1时能够显著促进细胞增殖的活性(P <0.01);斑马鱼尾鳍面积样品组与模型对照组相比极显著增加(P <0.001)。
研究结果表明,重组贻贝粘蛋白具有显著的促细胞迁移和修复愈合的功效,具备作为生物医学材料的潜质。
关键词:贻贝粘蛋白;基因重组;生物材料;表征;功效评价DOI :10.19586/j.20952341.2023.0021 中图分类号:S985.3+1 文献标志码:ACharacterization and Efficacy Evaluation of Recombinant Mussel Adhesive ProteinLI Min , WEI Wenpei , QIAO Sha , HAO Dong , ZHOU Hao , ZHAO Shuowen , ZHANG Lifeng ,HOU Zengmiao *Xi'an DeNovo Hith Medical Technology Co., Ltd , Xi'an 710000, ChinaAbstract :In order to promote the application of recombinant mussel adhesive protein in the medical and cosmetics field , the recombi⁃nant mussel adhesive protein obtained from scale fermentation and purification of Escherichia coli was characterized and its efficacy was evaluated. Amino acid N -terminal sequencing , relative molecular weight analysis , amino acid composition analysis , protein purityanalysis , endotoxin content , dihydroxyphenylalanine (DOPA ) content of recombinant mussel adhesive protein were determined by the following methods : Edman degradation , matrix -assisted laser desorption ionization time -of -flight mass spectrometry (MALDI -TOF -MS ), phenyl -isothiocyanate (PITC ), nonreductive SDS -polyacrylamide gel electrophoresis (SDS -PAGE ), gel method , modified Ar⁃now. The efficacy of recombinant mussel adhesive protein was evaluated by cell migration and repairing effect of zebrafish tail fin. Re⁃sults showed that the obtained recombinant mussel adhesive protein was confirmed to be consistent with the theoretical primary structure , protein purity of more than 95%, endotoxin <10 EU ·mg -1, DOPA content above 5%. When the recombinant mussel adhesive protein concentration was 60 μg ·mL -1, the effect of promoting cell proliferation was the most obvious , and it had very significant activity (P <0.01). The caudal fin area of zebrafish in sample group was significantly increased compared with model control group (P <0.001). The results indicated that recombinant mussel adhesive protein can promote cell migration and repair healing and has the potential to be used as biomedical materials.Key words :mussel adhesive protein ; gene recombination ; biological materials ; representation ; efficacy evaluation贻贝粘蛋白(mussel adhesive protein , MAP )也称作贻贝足丝蛋白(mussel foot protein ,Mfps ),收稿日期:2023⁃02⁃24; 接受日期:2023⁃03⁃31联系方式:李敏 E -mail:*******************;*通信作者 侯增淼 E -mail:***********************.cn李敏,等:重组贻贝粘蛋白的表征及功效评价是海洋贝类——紫贻贝(Mytilus galloprovincalis)、厚壳贻贝(Mytilus coruscus)、翡翠贻贝(Perna viri⁃dis)等分泌的一种特殊的蛋白质,贻贝中含有多种贻贝粘蛋白,包括贻贝粘蛋白(Mfp 1~6)、前胶原蛋白(precollagens)和基质蛋白(matrix proteins)等[1]。
