Protein Expression
protein expression and purification

Advantages
Fast growth Cheap medium and equipment for growing High expression level Good knowledge of the host
Disadvantages
different
frequencies with which the different codons appear in genes of these organisms
No
post-translational modifications glycosylation etc)
Recombinant
(SS bonds,
proteins expressed in E. coli easily form inclusion bodies
Protein Expression in Bacteria
Protein Expression and purification
黄义德 2013-09-29
Why do we express proteins
Clinical application
Research application
Can I express proteins that I want in bacteria or ……?
I want to say you all can do it after the lectures
Before you start, Things need to be considered
How to produce?
choose for protein expression system (vector and host)
crt蛋白表达 -回复

crt蛋白表达-回复"CRP Protein Expression: Unraveling the Mechanisms and its Significance"Introduction:CRP (C-reactive protein) is an important biomarker that plays a crucial role in inflammation and the immune response. It is synthesized mainly in the liver and is released into the bloodstream in response to tissue damage, infection, or inflammation. This article aims to unravel the mechanisms behind CRP protein expression and explore its significance in various physiological and pathological conditions.1. Regulation of CRP Gene Expression:1.1 Transcriptional Regulation:The expression of the CRP gene is mainly controlled by transcription factors. Various cytokines, such as interleukin-1 beta (IL-1β), interleukin-6 (IL-6), and tumor necrosis factor-alpha (TNF-α), stimulate the production of these transcription factors. For example, IL-6 induces the activation of signal transducer andactivator of transcription 3 (STAT3), which binds to the CRP gene promoter and initiates its transcription.1.2 Epigenetic Regulation:Epigenetic modifications, including DNA methylation and histone acetylation, also influence CRP gene expression. DNA methylation at specific CpG sites within the CRP gene promoter region can suppress its transcription, leading to decreased CRP protein expression. On the other hand, histone acetylation, which relaxes the chromatin structure, facilitates access of transcription factors to the CRP gene promoter, thus enhancing CRP expression.2. Significance of CRP Protein Expression:2.1 Inflammation Marker:CRP is a sensitive and non-specific marker of inflammation. Elevated levels of CRP in the blood often indicate the presence of an underlying inflammatory condition, such as rheumatoid arthritis, cardiovascular diseases, or infections. Monitoring CRP levels can help diagnose and monitor the progression of these diseases.2.2 Predictor of Cardiovascular Risk:High CRP levels have been associated with an increased risk of cardiovascular events, including heart attack and stroke. CRP reflects the overall burden of chronic low-grade inflammation, which contributes to the development and progression of atherosclerosis. Measurement of CRP levels can provide valuable prognostic information in assessing cardiovascular risk.2.3 Role in Immune Responses:CRP, as a member of the pentraxin family, plays a crucial role in the innate immune response. It promotes complement activation, opsonization, and phagocytosis, which aid in the clearance of pathogens and damaged cells. Additionally, CRP modulates the production of various cytokines, regulating the inflammatory response and immune cell activation.2.4 Therapeutic Target:Given the significance of CRP in inflammation and immune responses, specific targeting of CRP or its downstream signaling pathways may have therapeutic potential. Interfering with CRP expression or function could potentially regulate the inflammatory response and mitigate the severity of various diseases.Conclusion:CRP protein expression is tightly regulated at the transcriptional level by various cytokines and transcription factors. Epigenetic modifications further modulate CRP gene expression. Elevated CRP levels serve as a reliable marker for inflammation and cardiovascular risk, and monitoring CRP levels has clinical implications in various diseases. Understanding the mechanisms behind CRP expression and its functional significance opens avenues for developing therapeutic strategies targeting this important protein. Further research in this field will continue to unveil the complexity of CRP biology and its role in health and disease.。
Protein Expression and purification

Protein Expression and Purification22,159–164(2001)160SCOTT A.LESLEYTABLE1Genomic versus Proteomic TechnologiesGenomic technologies(DNA)Proteomic technologies(protein) Identification Determined experimentally,bioinformatics Predicted from genomic informationFunction1-dimensional information storage3-dimensional organization of chemicalfunctionalitiesBuilding blocks4bases20ϩamino acidsDetection sensitivity PCR amplification techniques Direct detection methodsSynthetic approaches Cheap and efficient oligonucleotide synthesis Limited capacity of peptide synthesismethods combined with PCRSequence determination500–700bases common by automated sequencing Direct sequencing difficult,mass spectrometry Purification Generic methods Generic methods require modification of proteinthrough gene fusionAnalysis methods Typically employ enzymes,hybridization Chemical,biophysical,biochemicaland mutagenesis),interactions between proteins(two-are of primary interest and typically are expressed in hybrid),and global protein changes(2D gels and LC–a bacterial host.Often this approach leads to problems MS).Purified protein is often required in these studies associated with expression levels and proper folding of and defines the outputs of any parallel expression and the protein of interest.Flexibility in expression options purification process.is a key parameter.Pichia or baculovirus expressionsystems can offer effective alternatives to bacterial sys-tems.Each expression scenario requires a specific vec-GENE CLONING FOR EXPRESSIONtor.Recloning cDNAs into each of these specific vectors Determining gene function through genomics typi-is extremely labor-intensive.Recombinatorial cloning cally starts from a query of a database.Sequence infor-methods provide an opportunity to minimize the effort mation for the3.9billion bases of sequence from the required for alternate expression.human genome is now available(1,2).Access and inter-Two systems are commonly used for recombinatorial pretation of this information often require sophisticated cloning and shown in Fig.1.The cre–lox recombination bioinformatics software outside the scope of this dis-system described by Elledge utilizes a single recombina-cussion.Public archives such as the Unigene Data-tion to introduce the gene of interest into a recipient base(/)or TIGR(http://vector(3).This is an in vitro reaction combined with a /)provide bioinformatic access to many in-genetic selection for the recombinant vector.In this way teresting plete genomic sequence infor-the gene of interest is cloned once into a donor vector mation is now available through Celera(http://and can then be moved into any of a number of recipient /index.cfm).Future annotation of fullplasmids for expression in different hosts or to utilize sequence information will greatly expand the access todifferent purification tags.A similar system utilizes full-length cDNA sequences.The first requirement is converting this genomic in-lambda Int/Xis/IHF recombination at att sites(4)to formation into an actual cDNA clone of that gene.Am-achieve transfer of open reading frames(ORFs).This plification of full-length cDNAs via PCR is the typical system has the advantage of a precise ORF transfer to first step.Both reverse transcriptase and amplification the expression vector rather than the cointegrant vector polymerases typically are lacking in proofreading activ-product of the cre–lox system.With either system,the ity.Care must be taken to limit the number of steps of primary limitation is that translational fusion of the amplification and to use proofreading enzymes where recombination sites is typically required to maintain possible to minimize the probability of introducing un-the flexibility and utility of the recombination method wanted mutations.Tissue selection for cDNA librariesfor expression.In those cases,such as crystallography, also is an important consideration for attempting towhere translational fusions are potentially detrimental isolate genes as they must be expressed within thatto the protein,a conventional cloning approach is library source for successful amplification.This infor-more appropriate.mation is often obtained from cDNA or oligonucleotideRegardless of the cloning method,parallel expression expression arrays.and purification requires utilization of purification Amplified gene products are cloned into appropriatetags.Many options exist for this purpose.A comprehen-vectors for expression.Depending on the source of thesive review is beyond the scope of this article.A list of gene,the host,and the end use of the protein,manydifferent vectors may be appropriate.Eukaryotic genes some commonly used tags is shown in Table2.By farHIGH-THROUGHPUT PROTEOMICS161FIG.1.Strategies of recombinatorial cloning.Individual cDNAs are cloned into a donor vector that can then be recombined into any number of recipient vectors through recombinatorial cloning.One option is to form a cointegrant plasmid through Cre-mediated recombination across a lox site.In the second scenario,a cDNA is flanked by phage lambda att sites which direct recombination into an expression vector through the use of the INT/XIS/IHF proteins.In these ways,a single donor clone can easily be transferred into any number of recipient vectors. the most common fusion is the histidine tag for purifica-analysis studies into small-scale for analysis of expres-tion on metal-chelate resins.This tag provides a sub-sion levels and properties and into large-scale for use stantial purification handle while being relatively un-with many of the proteomic applications.obtrusive as a fusion partner.Beyond purification,Small-scale expression is most useful for identifying translational fusions often provide a means to enhance those clones which express recombinant protein to high expression.The larger fusion tags such as thioredoxin levels and for evaluating the folding state of the protein. and GST often are superior in this respect.Crude expression testing is typically done by simpleSDS–PAGE analysis of whole cells.Evaluation of thefolding state is typically done by centrifugal fraction-RECOMBINANT EXPRESSION OF PROTEINation of a lysate,requiring a more gentle lysis proce-dure.Several lysozyme and mild detergent methods are Bacterial expression is most common for recombinantcommercially available for this purpose.proteins because of its ease of use and the high levelsof protein obtained.It is useful to divide expression For large-scale production for many applications,TABLE2Common Purification TagsBasis of purification Elution Reference Small tagsHistidine tag Metal affinity resin Imidizole(5)S-tag Interaction with S-protein Temperature(6) Calmodulin-binding protein Interaction with Calmodulin Calcium(7) Large tagsGlutathione S-transferase Glutathione agarose Reduced glutathione(8) Thioredoxin Phenylarsine oxide resin-Mercaptoethanol(9) Biotinylation domain Monomeric avidin resin Biotin(10) Maltose binding protein Amylose resin Maltose(11) Chitin binding protein/intein Chitin affinity Thiol(12)162SCOTT A.LESLEYtens of milligrams of protein typically are required.PURIFICATION STRATEGYEven with common bacterial expression levels,500–Proteins are highly diverse in their properties mak-1000ml of culture typically is required to provide these ing generic methods of purification difficult.Purifica-amounts.While such methods are commonplace in labo-tion tags such as described in Table2are a typical ratories,systems for parallel processing large numbers solution for purifying proteins in parallel.His-tag fu-of cultures at this level are not commercially available.sions are very common and provide a single-step chro-By developing instrumentation and optimizing media matographic purification that yields protein of suffi-and aeration conditions for high-density cell growth,cient purity for most applications.In addition,the his-our laboratory can parallel process96cultures at this tag sequence requires the addition of only six amino scale.Optical density(OD)values at600nm can reach acids to the recombinant protein,reducing the likeli-a value of40with logarithmic growth through at least hood that such a fusion will adversely affect gene func-30OD units.These cell densities allow us to producetion.A typical purification strategy is outlined in Fig.2. sufficient cell mass with65-ml culture volumes to yieldtens of milligrams of recombinant protein,sufficient for PURIFICATION AUTOMATIONmost applications.Such instrumentation is not com-Parallel processing typically involves instrumenta-monly available,but common shaking incubators cantion for automation.Lysis methods such as sonication or substitute with larger volumes.using a French press are not simple automation tasks. Recombinant expression of proteins is achievedLikewise,centrifugation is not easily integrated into through induction of a strong promoter system.Manyautomation due to problems of locating and indexing options exist in this regard including tac,T7,lambdathe rotor position.Automation of this process typically P L,and ara B promoters.It is important for parallelinvolves protocol modifications.This can easily be processing that growth and induction characteristicsachieved in small-scale methods.are consistent.For this reason,it is important to retainOn small-scale,parallel processing usually involves tight repression of expression and have a simple induc-use of a96-well plate format.Lysis is typically achieved tion procedure for high-level expression.For T7sys-using a combination of lysozyme and freeze–thaw cy-tems,the lac operator and T7lysozyme(pLysS)provide cles.Phage lysozymes are more effective than hen egg an extra level of repression.The arabinose promoter is white lysozyme for this purpose and can be combined tightly repressed in the absence of inducer and is our with nucleases to reduce viscosity and facilitate re-preferred system for parallel growth.With all of the moval of cell debris at the low g forces commonly used promoters listed,recombinant expression levels of10–with microtiter plates.Alternatively,nonionic deter-50%of total cell protein are common.gents can be employed for nondenaturing lysis.FIG.2.Generalized purification strategy of recombinant fusion protein.A common purification strategy is shown here.Proteins are purified from fermentation cultures by affinity purification.Isolated cell pellets are resuspended in an appropriate lysis buffer and disrupted by high-intensity sonication.Cell walls and insoluble debris are pelleted by centrifugation and the soluble supernatant containing the recombinant fusion protein is applied to chromatography resin containing an immobilized metal for affinity purification.Fractions containing the recombinant protein can be used directly or further purified using conventional chromatographic techniques.HIGH-THROUGHPUT PROTEOMICS163TABLE3Robotic Systems with Capabilities Adaptable to ProteinPurificationManu-facturer Instrument WebsiteQiagen BioRobot3000/Tomtec Quadra96/Matrix PlateMate /Hamilton Microlab4200/Beckman Biomek2000/Packard PlateTrak /index2.htmGilson Nebula215/index.htmlRobotic systems for nucleic acid purification are rela-tively commonplace and have recently been adapted forprotein purification.The Qiagen BioRobot3000per-forms multiple functions relevant to protein purifica-tion.It provides aspirate,dispense,pipet,vacuum fil-tration,and plate-shaking functions on a relativelycompact platform.These functions can be adapted to FIG.4.SDS–PAGE analysis of purified protein.Metal affinity chro-perform cell lysis and chromatography steps from1–2matography yields highly purified protein from a single chromato-graphic separation.This gel shows typical yields and purity obtained ml of bacterial culture.Specialized96-well plates clearfrom parallel purification using an automated purification system. cell debris via vacuum filtration and are also used toSuch proteins have been incorporated directly in successful crystalli-retain resin for chromatographic separations.The Wal-zation trials.Ten-microliter samples of12ml protein eluates from Ni-lac Quadra96also has most of these capacities and resin were separated by10%SDS–PAGE.Samples are recombinant can parallel process96or384samples.Both of thesefusions of thioredoxin to human proteins as indicated by accessionnumber.systems have been used with success in our laboratoryfor small-scale protein purification of proteins in micro-titer plates.Table3lists some robotic systems that maybe applied to small-scale protein purification.providing the throughput needed for proteomic studies Despite the difficulties,large-scale protein purifica-involving tens of thousands of proteins.We are cur-tion also can be automated.In our laboratory we simul-rently able to process approximately96–192proteins taneously process96bacterial cultures of65–70ml.per day with this system with average yields of around Instrumentation for processing96parallel cultures at10mg of purified protein.Affinity purification results that scale required development of custom roboticsin recombinant protein that is typically80–90%pure shown in Fig.3.These robotics incorporate liquid aspi-(see Fig.4)which is sufficient for most applications. rate and dispense,centrifugation,and sonication capa-Subsequent purification is sometimes necessary,for ex-bilities required for purification.Automation is key to ample,in protein crystallography,and is achieved usingFIG.3.Protein purification automation.Custom robotics for performing the purification strategy outlined in Fig.2are shown.(a)The instrument has capacity for automated liquid aspiration and dispensing,sonication,centrifugation,and fractionation.Ninety-six cultures are processed in parallel,giving up to10–50mg of purified protein per culture.(b)Expanded view of aspirate/dispense/sonicate head accessing rotor.164SCOTT A.LESLEYREFERENCESstandard ion-exchange and size-exclusion chromatog-raphy.Automation of these methods is relatively1.Venter,J.C.et al.(2001)The sequence of the human genome. straightforward employing standard FPLC and au-Science291,1304–1351.tosampler instrumentation.2.International Human Genome Sequencing Consortium(2001)Initial sequencing and analysis of the human genome.Nature SUMMARY409,860–921.3.Liu,Q.,Li,M.Z.,Leibham,D.,Cortez,D.,and Elledge,S.J. Determining gene function and understanding the(1999)The univector plasmid-fusion system,a method for rapid relationships and interactions of the gene products are construction of recombinant DNA without restriction enzymes.a global effort in biological studies.The approach toCurr.Biol.8,1300–1309.performing this immense task is driven by the availabil- 4.Hatley,J.L.,Temple,G.F.,and Brasch,M.A.(2000)DNA cloningity of genomic information.To utilize this informationusing in vivo site-specific recombination.Genome Res.10,pp.1788–1795.for experimentation,however,significant effort isneeded to actually isolate and express proteins from5.Petty,K.J.(1996)Metal-chelate affinity chromatography,in“Current Protocols in Molecular Biology,”Vol.2,Wiley,New York. the genes of interest for study.The complexity of this6.Kim,J.S.,and Raines,R.T.(1993)Ribonuclease S-peptide as a effort is compounded by the large number of gene prod-carrier in fusion proteins.Protein Sci.2,348–356.ucts comprising the proteome.Parallel processing and7.Stofko-Hahn,R.E.,.Carr,D.W,and Scott,J.D.(1992)A single generic methods are required to achieve a systematicstep purification for recombinant proteins.FEBS Lett.302, and thorough evaluation of gene function.274–278.Experimental uses of proteins for structural and8.Smith,D.B.,and Johnson,K.S.(1988)Single-step purification functional studies typically require milligram amounts of polypeptides expressed in Escherichia coli as fusions within purified form.Unlike genomic technologies that pri-glutathione S-transferase.Gene67,31–40.marily involve the study of nucleic acids,proteomic9.Lu,Z.,DiBlasio-Smith,E.A.,Grant,K.L.,Warne,N.W.,LaVallie,studies focus on proteins.Proteins are by nature much E.R.,Collins-Racie,L.A.,Follettie,M.T.,Williamson,M.J.,more diverse in composition and properties than nucleicand McCoy,J.M.(1996)Histidine patch thioredoxins.Mutantforms of thioredoxin with metal chelating affinity that provide acids.In many ways,this makes them more interestingfor convenient purifications of thioredoxin fusion proteins.J. but also less amenable to generic methods and technolo-Biol.Chem.271,5059–5065.gies for parallel processing.Nonetheless,methods and10.Cronan,J.E.(1990)Biotination of proteins in vivo.A post-trans-instrumentation are currently available to meet this lational modification to label,purify,and study proteins.J.Biol.ing these advances will allow a systematic Chem.265,10327–33.effort at understanding biological pathways at the11.Maina,C.V.,Riggs,P.D.,Grandea,A.G.,Slatko,B.E.,Moran,molecular level.L.S.,Tagliamonte,J.A.,McReynolds,L.A.,and Guan,C.D.(1988)An Escherichia coli vector to express and purify foreign ACKNOWLEDGMENTS proteins by fusion to and separation from maltose-binding pro-tein.Gene74,365–373.The author acknowledges the help of Marc Nasoff,Heath Klock,Dan McMullan,Tanya Shin,Juli Vincent,Mike Hornsby,Mark12.Chong S.,Mersha F.B.,Comb D.G.,Scott M.E.,Landry D.,Vence L.M.,Perler F.B.,Benner J.,Kucera R.B.,Hirvonen C. Knuth,Loren Miraglia,and Jeremiah Gilmore for their contributionsto the high-throughput cloning and expression efforts.He also recog- A.,Pelletier J.J.,Paulus H.,and Xu M.Q.(1997)Single-column nizes Bob Downs,Mark Weselak,Andy Meyer,and Jim Mainquistpurification of free recombinant proteins using a self-cleavable and the rest of the GNF engineering staff for their contributions to affinity tag derived from a protein splicing element.Gene192, the custom robotics that make this effort possible.271–281.。
大肠杆菌表达蛋白基础知识l
program
Advantages
• Fast growth • Cheap medium and equipment for growing • Good knowledge of the host
RBS
RBS 5-9 n START
GAAGGAATTCAGGAGCCCTTCACCATG ... ...
START codons: E. coli uses 77% ATG (AUG), 14% GTG (GUG), 8% TTG (UUG) and a few others STOP codons: TAG (UAG), TGA (UGA), TAA (UAA)
Combinations
Genetic Elements Essential for Expression
Replication Origin
Plasmid pBR322
pUC pACYC pSC101 colE1
Replicon pMB1 pUC p15A pSC101 colE1
Copy Number 15-20
Key features
Plac Low level up to middle (IPTG)
Ptac Ptrc
Moderately high (IPTG)
(trp-lac)
Weak, regulated. Suitable for expression of gene products at very low intracellular level. Comparatively expensive induction.
蛋白融合表达

蛋白融合表达全文共四篇示例,供读者参考第一篇示例:蛋白融合表达(Protein Fusion Expression)是生物技术领域中一种常用的蛋白表达技术,通过将不同蛋白基因序列进行融合,使其能够在目标宿主细胞中表达出含有多个功能区域的融合蛋白。
蛋白融合表达技术是从基因水平上控制蛋白质的结构和功能,为蛋白质的生物学功能研究、药物研发和生物制药等领域提供了有效的手段。
一、蛋白融合表达的原理蛋白融合表达技术是利用基因工程技术将两个或多个蛋白基因的编码序列连接在一起,形成一个新的融合蛋白基因,然后通过转染或转化等手段将其导入目标宿主细胞中,使其表达出融合蛋白。
蛋白融合表达的基本原理是将两个或多个不同功能的蛋白通过融合技术合并在一起,达到协同作用或增强某一功能的效果。
蛋白融合表达可通过多种途径实现,常见的方法包括直接连接两个蛋白的编码序列、利用核酶切割和PCR等技术进行DNA重组,以及通过载体和质粒等载体介导融合蛋白的表达。
不同的蛋白融合表达技术具有各自的特点和适用范围,选择合适的融合表达策略可提高蛋白表达效率和提取纯度。
1. 生物学功能研究:蛋白融合表达技术可用于研究蛋白质的结构和功能,通过融合不同功能区域的蛋白进行功能分析和蛋白相互作用研究,揭示蛋白质的生物学特性和作用机制。
2. 药物研发:蛋白融合表达技术在药物研发中具有广泛的应用,可用于合成重组蛋白、多肽和抗体等生物制剂,提高药物的活性和稳定性,开发新型药物和治疗方法。
3. 生物制药:蛋白融合表达技术是生物制药领域中最常用的生产方法之一,可用于大规模生产融合蛋白、重组蛋白和生物药物,提高生产效率和产品质量,满足临床需求。
4. 技术创新:蛋白融合表达技术在生物技术领域具有重要的技术意义,可以用于开发新型蛋白表达系统、优化蛋白表达和纯化工艺、改良蛋白结构和功能等方面,推动生物技术的发展和进步。
1. 提高蛋白表达效率:蛋白融合表达技术可以利用多个功能区域相互作用增强蛋白的稳定性和可溶性,提高蛋白的表达水平和纯度。
质粒图谱大全

(转载)一. 九种表达载体Pllp-OmpA, pllp-STII, pMBP-P, pMBP-C,pET-GST, pET-Trx, pET-His, pET-CKS, pET-DsbA二. 克隆载体pTZ19RDNApUC57DNAPMD18TPQE30pUC18pUC19pTrcHisApTrxFuspRSET-ApRSET-BpVAX1PBR322pbv220pBluescriptIIKS( )L4440pCAMBIA-1301pMAL-p2XpGD926三.PET 系列表达载体ProteinExpression?ProkaryoticExpression?pETDsbFusionSystems39band40b ProteinExpression?ProkaryoticExpression?pETExpressionSystem33b ProteinExpression?ProkaryoticExpression?pETExpressionSystems ProteinExpression?ProkaryoticExpression?pETExpressionSystemsplusCompetentCells ProteinExpression?ProkaryoticExpression?pETGSTFusionSystems41and42 ProteinExpression?ProkaryoticExpression?pETNusAFusionSystems43.1and44 ProteinExpression?ProkaryoticExpression?pETVectorDNAProteinPurification?PurificationSystems?Strep?TactinResinsandPurificationKits四.PGEX 系列表达载体TEcoR?pGEX-1I/BAPpGEX-2TpGEX-2TKpGEX-3XpGEX-4T-1pGEX-4T-2pGEX-4T-3pGEX-5X-1pGEX-5X-2pGEX-5X-3pGEX-6P-1pGEX-6P-2pGEX-6P-3五.PTYBsystemPTYB1PTYB2PTYB11PTYB12六. 真核表达载体pCDNA3.1(-)pCDNA3.1( )pPICZalphaApGAPZα APYES2.0pBI121pEGFP-N1pEGFP-C1pPIC9KpPIC3.5K如何阅读分析质粒图谱载体主要有病毒和非病毒两大类, 其中质粒DNA是一种新的非病毒转基因载体。
信号肽及其在蛋白质表达中的应用
蛋白质组学-无细胞蛋白表达三折页-笔记精简版
产品
E. coli S30 Extract System for Circular DNA
E. coli S30 Extract System for Linear Templates TNT® T7 Insect Cell Extract Protein Expression System TNT® T8 Insect Cell Extract Protein Expression System
L5540 L5600 L5800 L5900
TNT® T7 Quick for PCR DNA Gold TNT® T7 Express 96 System Gold TNT® SP6 Express 96 System
T7 Sample System
兔网织红细胞表达
系统,适合PCR片
段模板
规格 30 reactions 30 reactions 10 reactions 40 reactions 24 reactions 8 reactions 30 reactions 40 reactions 5 reactions 40 reactions 5 reactions 40 × 50µl reactions 10 × 50µl reactions 40 × 50µl reactions 10 × 50µl reactions 40 reactions 40 reactions 40 reactions 30 × 50µl reactions 12 reactions 5 × 200µl 5 × 200µl 40 reactions 8 reactions 40 reactions 8 reactions 40 reactions 5 × 200µl 40 reactions 40 reactions 40 reactions 40 reactions 40 reactions 30 reactions 30 reactions 175µl 40 reactions 1 × 96 wells 1 × 96 wells 1each
cap生物化学名词解释
cap生物化学名词解释
CAP 是"Cell-free Protein Expression and Purification"的缩写,是一种用于纯化细胞质中的蛋白质的技术。
这项技术利用细胞质中的细胞器 (如线粒体和质体) 被排出或失活的特点,通过离心、沉淀和离子交换色谱等方法,将细胞质中的蛋白质纯化出来。
CAP 技术可以用于生产大规模蛋白质纯品,用于研究蛋白质结构、功能、相互作用等方面。
CAP 技术的主要优点是可以去除细胞器和细胞质中的其他杂质,从而纯化出高纯度的蛋白质。
此外,CAP 技术还可以用于生产大规模蛋白质库,用于筛选蛋白质相互作用和对蛋白质进行结构生物学研究。
这项技术在生物化学和生物物理学研究中得到了广泛应用,并且已经被拓展用于生产各种人类疫苗和药物分子。
大肠杆菌系统蛋白表达纯化流程
大肠杆菌系统蛋白表达纯化流程英文回答:Introduction:The expression and purification of proteins in Escherichia coli (E. coli) is a commonly used method in molecular biology research. E. coli is a well-studied and easily manipulated organism, making it an ideal host for protein expression. In this article, we will discuss the general workflow for the expression and purification of proteins in E. coli.Expression of the target protein:1. Gene cloning: The first step is to clone the gene encoding the target protein into an expression vector. This vector contains a promoter that drives the expression of the gene in E. coli.2. Transformation: The recombinant expression vector is then introduced into E. coli cells through a process called transformation. This results in the production of many E. coli cells carrying the target gene.3. Expression induction: The transformed E. coli cells are grown in a suitable culture medium until they reach a specific growth phase. At this point, expression of the target gene is induced by adding a chemical inducer or by changing the growth conditions.4. Protein expression: The induced E. coli cells produce the target protein, which can either be present in the soluble fraction or form insoluble aggregates called inclusion bodies.Protein purification:1. Cell lysis: The E. coli cells are harvested by centrifugation and then lysed to release the proteins. Various methods can be used for cell lysis, such as sonication, freeze-thaw cycles, or enzymatic digestion.2. Removal of cell debris: The cell lysate is then clarified by centrifugation to remove cell debris and insoluble material. The resulting supernatant contains the target protein along with other cellular components.3. Protein purification: Different purification techniques can be employed to isolate the target protein from the crude lysate. These techniques include affinity chromatography, ion exchange chromatography, size exclusion chromatography, and hydrophobic interaction chromatography. The choice of purification method depends on the properties of the target protein.4. Protein concentration: After purification, thetarget protein is often in a dilute solution. Concentration can be achieved by using techniques such as ultrafiltration or precipitation with ammonium sulfate.5. Protein characterization: The purified protein should be characterized to confirm its identity and purity. Techniques such as SDS-PAGE, western blotting, and massspectrometry can be used for protein analysis.Conclusion:The expression and purification of proteins in E. coli is a well-established and widely used technique in molecular biology research. The workflow involves gene cloning, protein expression, cell lysis, protein purification, concentration, and characterization. By following this general procedure, researchers can obtain purified proteins for further analysis and functional studies.中文回答:简介:大肠杆菌(E. coli)中的蛋白表达和纯化是分子生物学研究中常用的方法。
- 1、下载文档前请自行甄别文档内容的完整性,平台不提供额外的编辑、内容补充、找答案等附加服务。
- 2、"仅部分预览"的文档,不可在线预览部分如存在完整性等问题,可反馈申请退款(可完整预览的文档不适用该条件!)。
- 3、如文档侵犯您的权益,请联系客服反馈,我们会尽快为您处理(人工客服工作时间:9:00-18:30)。
Promoters from phages
T7, T3, SP6, T5, PL - Highly efficient and specific expression
Plac: Regulation
Plac, Ptac, Ptrc: Characteristics
Level of expression (inductor) Plac Low level up to middle (IPTG) Key features
Cloning Using A-overhangs
TA-Cloning with Topoisomerase
Directional Cloning
CACC
Gateway Technology
Expression of Fusion Proteins
We may fuse the target protein with various tags to facilitate purification or protein detection HHHHHH-target, epitope-target highly soluble proteins to improve solubility and to facilitate purification Thioredoxin-target, GST-target signal peptides or other proteins to promote secretion SP-target
pTtrc99
Translational vector
pQE
Translational vector + CDR
pET
pCR&pEXP
pBAD
Expression strains
# Strain 1 BL21 2 BL21* /STAR Key features Deficient in lon and ompT proteases #1 + deficient in RNaseE Improves the stability of mRNA transcripts and increases
‘Short’ Fusion Protein Construction
ATG CAT CAC CAT CAC CAT CAC
‘Long’ Fusion Protein Construction
NcoI HindIII HindIII PstI
pUC18/19
ApaLI (178)
ALPHA P(BLA)
Combinations
Genetic Elements Essential for Expression
Replication Origin
Plasmid pBR322 pUC pACYC pSC101 colE1 Replicon pMB1 pUC p15A pSC101 colE1 Copy Number 15-20 500-700 18-22 5 15-20
Accumulation of lipopolysaccharides (generally referred to as endotoxins) …
Goals
To obtain as much as possible soluble folded protein in a form that is easy to purify /good expression+good cell growth /reduced aggregation /use of secretion and tags
Types of Expression Vectors
1
2
3
Insertion into Transcriptional Vectors
Insertion into Translational Vectors
Cloning Using Restriction Enzymes
NcoI HindIII
Genetic Elements Essential for Expression
Promoters
Host’s promoters
2500 in the entire genome of E. coli K12 strain Most frequently used: Plac / Ptac / Ptrc, PPBAD, rhaPBAD - Regulation of expression
RBS RBS 5-9 n START GAAGGAATTCAGGAGCCCTTCACCATG ... ...
START codons: E. coli uses 77% ATG (AUG), 14% GTG (GUG), 8% TTG (UUG) and a few others STOP codons: TAG (UAG), TGA (UGA), TAA (UAA)
Limitation for expression of eukaryotic proteins due to differences in the distribution of tRNAs and posttranslational modifications …
Rear codons, SS bonds, gilitates cytoplasmic disulfide bond formation
Expression optimization
To optimase: Level of inducer (e.g. arabinose)
Time of induction
Temperature of the induction step (popular - 18oC overnight)
Co-expression from two plasmids
Protein Expression in Bacteria Part2
1. Cloning strategies 2. Overview of the available expression systems and expression strains 3. Design of cloning procedures using the VNTI program
protein expression yield
3 BL21*(DE3)
#1 or 2 + carry T7 polymerase under Plac Enables T7 expression 4 BL21*(DE3)pLysS/E #3 + plasmid pLysS or pLysE expressing T7 lysozyme
HindIII (400) PstI (416) XbaI (424) BamHI (430) AvaI (435)
ApaLI (2367)
APr
pUC18
XmaI (435) SmaI (437) EcoRI (451)
P(LAC) ORI
2686 bp
ApaLI (1121)
Transcriptional vector
Ptac Moderately high (IPTG) Ptrc (trp-lac)
PPBAD: Regulation
PPBAD and RhaPBAD
Level of expression (inductor) PPBAD Variable from low to high level (L-arabinose) Variable from low to high level (L-rhamnose) Key features
Weak, regulated. Suitable for expression of gene products at very low intracellular level. Comparatively expensive induction. High-level, but lower than T7 system. Regulated expression still paratively expensive induction. High basal level.
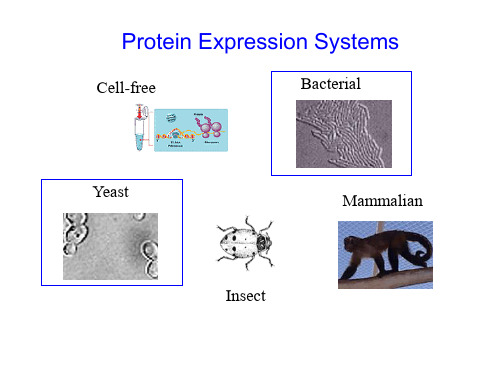
Protein Expression Systems
Cell-free Bacterial
Yeast
Mammalian
Insect
Protein Expression in Bacteria
1. Advantages/disadvantages 2. Genetic elements essential for the expression 3. Cloning strategies 4. Overview of the available expression and expression strains systems
5. Design of cloning procedures using the VNTI program
Advantages
Fast growth Cheap medium and equipment for growing Good knowledge of the host
Disadvantages
rhaPBAD
Phage Promoters
Level of expression (inductor) T7 T5 PL Very high High Moderately high (temperature shift) Key features
Utilizes T7 RNA polymerase. Utilizes E. coli RNA polymerase. Temperature-sensitive host required. Less likelihood of "leaky" un-induced expression. Basal level; high basal level by temperatures below 30°C. No inducer.
