wolf EIGRP分解实验三
数值计算方法LU分解法实验

数值计算方法LU分解法实验LU分解法是一种常见的数值计算方法,用于解线性方程组或求解矩阵的逆。
该方法的核心思想是将一个矩阵分解为一个下三角矩阵和一个上三角矩阵的乘积。
在本篇文章中,我们将进行关于LU分解法的实验,并探讨其性能和应用。
首先,我们需要明确LU分解法的数学原理。
假设我们有一个n阶方阵A,LU分解法的目标是找到两个矩阵L和U,使得A=LU。
其中L是一个下三角矩阵,U是一个上三角矩阵。
然后,我们可以将原始的线性方程组Ax=b转化为两个新的方程组Ly=b和Ux=y。
通过求解这两个方程组,我们可以得到原始方程组的解。
接下来,我们将通过一个具体的例子来说明LU分解法的步骤和计算过程。
假设我们有以下方程组:2x+y+z=8-3x-y+2z=-11-2x+y+2z=-3首先,我们将系数矩阵A进行LU分解。
在这个例子中,我们可以得到下三角矩阵L和上三角矩阵U:L=100-1.510-1-11U=21101.52.5001然后,我们将方程组转化为Ly=b和Ux=y的形式。
解这两个方程组,可以得到y和x的值。
最终,我们可以得到方程组的解为x=2,y=3,z=-1通过以上的实例,我们可以看到LU分解法的步骤较为繁琐,但是它的结果是准确的。
那么,接下来我们将进行一系列实验,来评估LU分解法的性能和应用。
首先,我们将进行LU分解法的准确性测试。
我们将随机生成一组方程组,并使用LU分解法求解出它们的解。
然后,我们将使用该解验证原方程组,并计算出其误差。
我们重复这个过程多次,并计算平均误差。
通过这次实验,我们可以判断LU分解法的准确性。
其次,我们将评估LU分解法的计算效率。
我们将随机生成不同规模的方程组,并使用LU分解法求解它们。
然后,我们记录下求解所需的时间,并绘制出问题规模和求解时间的关系图。
通过这个实验,我们可以了解LU分解法在不同规模问题上的计算效率,从而评估其可行性和应用范围。
此外,我们还可以将LU分解法与其他数值计算方法进行比较。
高斯列主元消去法和直接三角分解法(LU分解)

实验六高斯列主元消去法和直接三角分解法(LU分解)一、实验名称:分别用高斯列主元消去法和直接三角分解法(LU分解)求方程组的解系数矩阵:10 7 8 7 常向量:107 5 6 5 88 6 10 9 67 5 9 10 7精确解为:(-60,102,-27,16)二、试验目的:分别用高斯列主元消去法和直接三角分解法(LU分解)求方程组的解,比较二者不同的特点。
三、算法描述:2、直接三角分解法(LU分解)四、源程序:1、高斯列主元消去法#include <stdio.h>void main(){float a[4][4]={{10,7,8,7},{7,5,6,5},{8,6,10,9},{7,5,9,10}}, y[4],c[4][4],x[4],d[4],m,b; int i,n,j,f;printf("请输入右端项:\n");for(i=0;i<=3;i++)scanf("%f",&y[i]);for(n=0;n<=2;n++){ m=a[n][n]; f=n;for(i=(n+1);i<=3;i++){if(m<a[i][n]){m=a[i][n]; f=i;}}if(f!=n){for(j=0;j<=3;j++){c[n][j]=a[n][j]; a[n][j]=a[f][j];a[f][j]=c[n][j];}d[n]=y[n]; y[n]=y[f]; y[f]=d[n];}for(i=(n+1);i<=3;i++){b=-a[i][n]/a[n][n];for(j=0;j<=3;j++)a[i][j]=a[n][j]*b+a[i][j];y[i]=y[n]*b+y[i];}}x[3]=y[3]/a[3][3];x[2]=(y[2]-a[2][3]*x[3])/a[2][2];x[1]=(y[1]-a[1][3]*x[3]-a[1][2]*x[2])/a[1][1];x[0]=(y[0]-a[0][3]*x[3]-a[0][2]*x[2]-a[0][1]*x[1])/a[0][0];printf("x1的值为%f\nx2的值为%f\nx3的值为%f\nx4的值为%f\n",x[0],x[1],x[2],x[3]);}2、直接三角分解法(LU分解)#include <stdio.h>void main (){float a[4][4]={{10,7,8,7},{7,5,6,5},{8,6,10,9},{7,5,9,10}},y[4], l[4][4],x[4],u[4][4],b[4]; int i,n,j;printf("请输入右端项:");for(i=0;i<=2;i++)scanf ("%f",&b[i]);for (i=0;i<=3;i++){ u[0][i]=a[0][i];l[i][0]=a[i][0]/u[0][0];}for (n=1;n<=2;n++){for(j=0;j<=(3-n);j++){u[n][n+j]=a[n][n+j];for (i=0;i<n;i++)u[n][n+j]=u[n][n+j]-l[n+j][i]*u[i][n]; }if (n=1){l[2][1]=(a[2][1]-l[2][0]*u[0][1])/u[1][1]; l[3][1]=(a[3][1]-l[3][0]*u[0][1])/u[1][1]; } else if (n=2){l[3][2]=(float)(a[3][2]-l[3][0]*u[0][2]-l[3][1]*u[1][2])/u[2][2]; } }for (n=0;n<=3;n++){y[n]=b[n];for (i=0;i<n;i++)y[n]=y[n]-l[n][i]*y[i]; }for (n=3;n>=0;n--){x[n]=y[n];for (i=3;i>n;i--)x[n]=(x[n]-u[n][i]*x[i]);x[n]=x[n]/u[n][n]; }for(i=0;i<=3;i++)printf("x%d的的值为:%f\n",i+1,x[i]); }五、输出结果1、高斯列主元消去法(1)请输入右端项:10 8 6 7X1的值为-59.999584X1的值为101.999306X1的值为-26.999817X1的值为15.999890(2)请输入右端项:5 6 7 8X1的值为-98.999306X1的值为163.998840X1的值为-40.999695X1的值为24.999817六、对算法的理解和改进改变右端项会对结果产生明显的影响,高斯列主元消去法仅考虑依次按列选取主元素,然后按行使之变到主元位置,再进行消去运算,消元结果冲掉A,计算解X冲掉常数项b,则在计算过程中由于右端项的不同解必然不同。
DNA光致分解
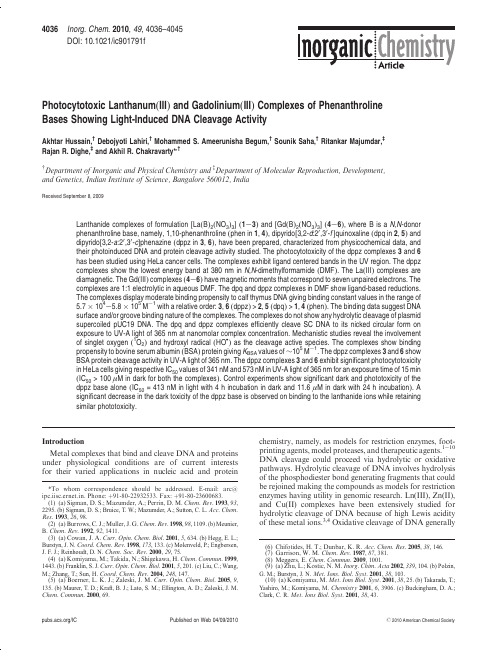
4036Inorg.Chem.2010,49,4036–4045 DOI:10.1021/ic901791fPhotocytotoxic Lanthanum(III)and Gadolinium(III)Complexes of Phenanthroline Bases Showing Light-Induced DNA Cleavage ActivityAkhtar Hussain,†Debojyoti Lahiri,†Mohammed S.Ameerunisha Begum,†Sounik Saha,†Ritankar Majumdar,‡Rajan R.Dighe,‡and Akhil R.Chakravarty*,††Department of Inorganic and Physical Chemistry and‡Department of Molecular Reproduction,Development,and Genetics,Indian Institute of Science,Bangalore560012,IndiaReceived September8,2009Lanthanide complexes of formulation[La(B)2(NO3)3](1-3)and[Gd(B)2(NO3)3](4-6),where B is a N,N-donor phenanthroline base,namely,1,10-phenanthroline(phen in1,4),dipyrido[3,2-d:20,30-f]quinoxaline(dpq in2,5)and dipyrido[3,2-a:20,30-c]phenazine(dppz in3,6),have been prepared,characterized from physicochemical data,and their photoinduced DNA and protein cleavage activity studied.The photocytotoxicity of the dppz complexes3and6 has been studied using HeLa cancer cells.The complexes exhibit ligand centered bands in the UV region.The dppz complexes show the lowest energy band at380nm in N,N-dimethylformamide(DMF).The La(III)complexes are diamagnetic.The Gd(III)complexes(4-6)have magnetic moments that correspond to seven unpaired electrons.The complexes are1:1electrolytic in aqueous DMF.The dpq and dppz complexes in DMF show ligand-based reductions.The complexes display moderate binding propensity to calf thymus DNA giving binding constant values in the range of5.7Â104-5.8Â105M-1with a relative order:3,6(dppz)>2,5(dpq)>1,4(phen).The binding data suggest DNAsurface and/or groove binding nature of the complexes.The complexes do not show any hydrolytic cleavage of plasmid supercoiled pUC19DNA.The dpq and dppz complexes efficiently cleave SC DNA to its nicked circular form on exposure to UV-A light of365nm at nanomolar complex concentration.Mechanistic studies reveal the involvement of singlet oxygen(1O2)and hydroxyl radical(HO•)as the cleavage active species.The complexes show binding propensity to bovine serum albumin(BSA)protein giving K BSA values of∼105M-1.The dppz complexes3and6show BSA protein cleavage activity in UV-A light of365nm.The dppz complexes3and6exhibit significant photocytotoxicity in HeLa cells giving respective IC50values of341nM and573nM in UV-A light of365nm for an exposure time of15min (IC50>100μM in dark for both the complexes).Control experiments show significant dark and phototoxicity of the dppz base alone(IC50=413nM in light with4h incubation in dark and11.6μM in dark with24h incubation).A significant decrease in the dark toxicity of the dppz base is observed on binding to the lanthanide ions while retaining similar phototoxicity.IntroductionMetal complexes that bind and cleave DNA and proteins under physiological conditions are of current interests for their varied applications in nucleic acid and protein chemistry,namely,as models for restriction enzymes,foot-printing agents,model proteases,and therapeutic agents.1-10 DNA cleavage could proceed via hydrolytic or oxidative pathways.Hydrolytic cleavage of DNA involves hydrolysis of the phosphodiester bond generating fragments that could be rejoined making the compounds as models for restriction enzymes having utility in genomic research.Ln(III),Zn(II), and Cu(II)complexes have been extensively studied for hydrolytic cleavage of DNA because of high Lewis acidity of these metal ions.3,4Oxidative cleavage of DNA generally*To whom correspondence should be addressed.E-mail:arc@ ipc.iisc.ernet.in.Phone:þ91-80-22932533.Fax:þ91-80-23600683. (1)(a)Sigman,D.S.;Mazumder,A.;Perrin,D.M.Chem.Rev.1993,93,2295.(b)Sigman,D.S.;Bruice,T.W.;Mazumder,A.;Sutton,C.L.Acc.Chem.Res.1993,26,98.(2)(a)Burrows,C.J.;Muller,J.G.Chem.Rev.1998,98,1109.(b)Meunier,B.Chem.Rev.1992,92,1411.(3)(a)Cowan,J.A.Curr.Opin.Chem.Biol.2001,5,634.(b)Hegg,E.L.;Burstyn,J.N.Coord.Chem.Rev.1998,173,133.(c)Molenveld,P.;Engbersen, J.F.J.;Reinhoudt,D.N.Chem.Soc.Rev.2000,29,75.(4)(a)Komiyama,M.;Takida,N.;Shigekawa,mun.1999, 1443.(b)Franklin,S.J.Curr.Opin.Chem.Biol.2001,5,201.(c)Liu,C.;Wang, M.;Zhang,T.;Sun,H.Coord.Chem.Rev.2004,248,147.(5)(a)Boerner,L.K.J.;Zaleski,J.M.Curr.Opin.Chem.Biol.2005,9, 135.(b)Maurer,T.D.;Kraft,B.J.;Lato,S.M.;Ellington,A.D.;Zaleski,J.M. mun.2000,69.(6)Chifotides,H.T.;Dunbar,K.R.Acc.Chem.Res.2005,38,146.(7)Garrison,W.M.Chem.Rev.1987,87,381.(8)Meggers,mun.2009,1001.(9)(a)Zhu,L.;Kostic,N.M.Inorg.Chim.Acta2002,339,104.(b)Polzin,G.M.;Burstyn,J.N.Met.Ions.Biol.Syst.2001,38,103.(10)(a)Komiyama,M.Met.Ions Biol.Syst.2001,38,25.(b)Takarada,T.; Yashiro,M.;Komiyama,M.Chemistry2001,6,3906.(c)Buckingham,D.A.; Clark,C.R.Met.Ions Biol.Syst.2001,38,43./IC Published on Web04/09/2010r2010American Chemical SocietyArticle Inorganic Chemistry,Vol.49,No.9,20104037causes degradation of the sugar and/or base moiety thus making the process suitable for foot-printing and therapeutic applications.Transition metal complexes with their tunable coordination environments and versatile redox and spectral properties have been used for oxidative cleavage of DNA and as models for the chemotherapeutic agents,namely,iron-bleomycins(FeBLMs).11-13Besides DNA cleaving agents, the chemistry of compounds showing protein cleavage activ-ity has received considerable current interests toward design-ing complexes that could show site-specific protein cleavage or could target cellular kinases that are expressed in the malignant tumor cells causing metastasis.14-17The anti-metastasis agents are required to target the enzymes belong-ing to the protein kinase family.Half-sandwich(η6-arene)-ruthenium(II)complexes and trans-[tetrachlorobis(1H-indazole)ruthenate(III)]are known to show potent antime-tastasis properties.18,19Further,there are reports on com-plexes showing hydrolytic cleavage of proteins.20-22The complexes showing oxidative cleavage of proteins are rela-tively rare.23-25Oxidative cleavage of DNA by metal complexes can be achieved in the presence of external reagents or on photo-activation.Photocleavage of DNA is of importance for its potential phototherapeutic applications as an emerging non-invasive treatment modality of cancer.26-29The photo-drug Photofrin which is an oligomeric mixture of hemato-porphyrin species,undergoes photoactivation at633nm to generate a1(π-π*)state with subsequent conversion to a 3(π-π*)state that activates molecular oxygen to formcytotoxic singlet oxygen.The PDT agents based on metal-loenediynes are known to produce diradical intermediates upon photoactivation.30Non-porphyrinic organic dyes have been used as potent photocytotoxic agents.31-34Use of metal-based agents showing photocytotoxicity is relatively new.Turro and co-workers have reported a ruthenium(II) complex as a cisplatin analogue that binds to DNA in a similar manner as cisplatin upon photoactivation and a rhodium(II)complex that shows significant photocytotoxi-city toward Hs-27human skin cells.35There are few reports on platinum(IV),ruthenium(II),and rhodium(II)complexes showing photocytotoxicity.36-39The reports from our la-boratory have shown that iron(III)and oxovanadium(IV) complexes could be designed as3d metal-based PDT agents showing photocytotoxicity in visible light.40,41The present work stems from our interest to design and study the photoinduced DNA cleavage activity of lanthanide com-plexes.The lanthanide complexes with their varied coordina-tion geometries could be suitably designed to achieve oxidative DNA cleavage activity.The Ln(III)complexes with their redox stability are expected to show poor chemical nuclease activity in the presence of cellular thiols.The hydrolytic DNA cleavage activity of Ln(III)complexes could be controlled with suitable design of the complexes to(11)(a)Burger,R.M.Chem.Rev.1998,98,1153.(b)Wolkenberg,S.E.; Boger,D.L.Chem.Rev.2002,102,2477.(12)(a)Thomas,C.J.;McCormick,M.M.;Vialas,C.;Tao,Z.-F.; Leitheiser,C.J.;Rishel,M.J.;Wu,X.;Hecht,S.M.J.Am.Chem.Soc. 2002,124,3875.(b)Rishel,M.J.;Thomas,C.J.;Tao,Z.-F.;Vialas,C.;Leitheiser, C.J.;Hecht,S.M.J.Am.Chem.Soc.2003,125,10194.(13)(a)Hertzberg,R.P.;Dervan,P.B.J.Am.Chem.Soc.1982,104,313.(b)Mukherjee,A.;Dhar,S.;Nethaji,M.;Chakravarty,A.R.Dalton Trans.2005, 349.(14)Kumar,C.V.;Buranaprapuk,A.;Thota,J.Proc.Indian Acad.Sci.: Chem.Sci.2002,114,579.(15)(a)Suh,J.;Chei,W.S.Curr.Opin.Chem.Biol.2008,12,207.(b)Suh, J.Acc.Chem.Res.1992,25,273.(16)(a)Pacor,S.;Zorzet,S.;Cocchietto,M.;Bacac,M.;Vadori,M.; Turrin,C.;Gava,B.;Castellarin,A.;Sava,G.J.Pharmacol.Exp.Ther.2004, 310,737.(b)Sava,G.;Capozzi,I.;Clerici,K.;Gagliardi,R.;Alessio,E.; Mestroni,G.Clin.Exp.Metastasis1998,16,371.(c)Scolaro,C.;Bergamo, A.;Brescacin,L.;Delfino,R.;Cocchietto,M.;Laurenczy,G.;Geldbach,T.J.; Sava,G.;Dyson,P.J.J.Med.Chem.2005,48,4161.(17)(a)Liu,H.-K.;Berners-Price,S.J.;Wang,F.;Parkinson,J.A.;Xu,J.; Bella,J.;Sadler,P.J.Angew.Chem.,Int.Ed.2006,45,8153.(b)Yan,Y.K.; Melchart,M.;Habtemariam,A.;Sadler,mun.2005,4764.(18)Dale,L.D.;Tocher,J.H.;Dyson,T.M.;Edwards,D.I.;Tocher,D.A.Anti-Cancer Drug Des.1992,7,3.(19)Hoeschele,J.D.;Habtemariam,A.;Muir,J.;Sadler,P.J.Dalton Trans.2007,4974.(20)Rajendiran,V.;Palaniandavar,M.;Swaminathan,P.;Uma,L. Inorg.Chem.2007,46,10446.(21)(a)de Oliveira,M.C.B.;Scarpellini,M.;Neves,A.;Terenzi,H.; Bortoluzzi,A.J.;Szpoganics,B.;Greatti,A.;Mangrich,A.S.;Souza,E.M. De;Fernandez,P.M.;Soares,M.R.Inorg.Chem.2005,44,921.(b)Kumar,C.V.;Buranaprapuk,A.;Cho,A.;Chaudhari,mun.2000,597.(22)(a)Zhu,L.;Kosti c,N.M.J.Am.Chem.Soc.1993,115,4566.(b) Parac,T.N.;Kosti c,N.M.J.Am.Chem.Soc.1996,118,51.(c)Milovi c,N.M.; Dutc a,L.-M.;Kosti c,N.M.Inorg.Chem.2003,42,4036.(d)Milovi c,N.M.; Dutc a,L.-M.;Kostic,N.M.Chem.;Eur.J.2003,9,5097.(23)(a)Kumar,C.V.;Buranaprapuk,A.;Opiteck,G.J.;Moyer,M.B.; Jockusch,S.;Turro,N.J.Proc.Natl.Acad.Sci.U.S.A.1998,95,10361.(b) Buranaprapuk,A.;Leach,S.P.;Kumar,C.V.;Bocarsly,J.R.Biochim.Biophys. Acta1998,1387,309.(c)Kumar,C.V.;Thota,J.Inorg.Chem.2005,44,825.(24)(a)Tanimoto,S.;Matsumura,S.;Toshima,mun.2008, 3678.(b)Suzuki,A.;Hasegawa,M.;Ishii,M.;Matsumura,S.;Toshima,K. Bioorg.Med.Chem.Lett.2005,15,4624.(c)Suzuki,A.;Tsumura,K.;Tsuzuki, T.;Matsumura,S.;Toshima,mun.2007,4260.(25)Roy,M.;Bhowmick,T.;Santhanagopal,R.;Ramakumar,S.; Chakravarty,A.R.Dalton Trans.2009,4671.(26)Bonnett,R.Chemical Aspects of Photodynamic Therapy;Gordon& Breach:London,U.K.,2000.(27)Henderson,B.W.;Busch,T.M.;Vaughan,L.A.;Frawley,N.P.; Babich,D.;Sosa,T.A.;Zollo,J.D.;Dee,A.S.;Cooper,M.T.;Bellnier,D.A.;Greco,W.R.;Oseroff,A.R.Cancer Res.2000,60,525.(28)Detty,M.R.;Gibson,S.L.;Wagner,S.J.J.Med.Chem.2004,47, 3897.(29)(a)Ali,H.;Van Lier,J.E.Chem.Rev.1999,99,2379.(b)Kar,M.; Basak,A.Chem.Rev.2007,107,2861.(c)Henderson,B.W.;Dougherty,T.J. Photochem.Photobiol.1992,55,145.(30)Bhattacharyya,S.;Zaleski,J.M.Curr.Top.Med.Chem.2004,4, 1637.(31)(a)Sessler,J.L.;Hemmi,G.;Mody,T.D.;Murai,T.;Burrell,A.; Young,S.W.Acc.Chem.Res.1994,27,43.(b)Wei,W.-H.;Wang,Z.;Mizuno, T.;Cortez,C.;Fu,L.;Sirisawad,M.;Naumovski,L.;Magda,D.;Sessler,J.L. Dalton Trans.2006,1934.(c)Mao,J.;Zhang,Y.;Zhu,J.;Zhang,C.;Guo,Z. mun.2009,908.(32)(a)Ramaiah,D.;Eckert,I.;Arun,K.T.;Weidenfeller,L.;Epe,B. Photochem.Photobiol.2004,79,99.(b)Ramaiah,D.;Eckert,I.;Arun,K.T.; Weidenfeller,L.;Epe,B.Photochem.Photobiol.2002,76,672.(33)(a)Pandey,R.K.;Sumlin,A.B.;Constantine,S.;Aoudia,M.; Potter,W.R.;Bellnier,D.A.;Henderson,B.W.;Rodgers,M.A.;Smith, K.M.;Dougherty,T.J.Photochem.Photobiol.1996,64,194.(b)Sternberg, E.D.;Dolphin,D.;Br€u ckner,C.Tetrahedron1998,54,4151.(c)Nelson,J.S.; Roberts,W.G.;Berns,J.M.Cancer Res.1987,47,4681.(d)Bonnett,R.;White, R.D.;Winfield,U.-J.;Berenbaum,M.C.Biochem.J.1989,261,277. (34)Atilgan,S.;Ekmeckci,Z.;Dogan,A.L.;Guc,D.;Akkaya,E.U. mun.2006,4398.(35)(a)Singh,T.N.;Turro,C.Inorg.Chem.2004,43,7260.(b)Lutterman,D.A.;Fu,P.K.-L.;Turro,C.J.Am.Chem.Soc.2006,128,738.(36)Mackay,F.S.;Woods,J.A.;Heringov a,P.;Ka s p a rkov a,J.;Pizarro,A.M.;Moggach,S.A.;Parsons,S.;Brabec,V.;Sadler,P.J.Proc.Natl. Acad.Sci.U.S.A.2007,104,20743.(37)Rose,M.J.;Fry,N.L.;Marlow,R.;Hinck,L.;Mascharak,P.K. J.Am.Chem.Soc.2008,130,8834.(38)Angeles-Boza,A.M.;Chifotides,H.T.;Aguirre,J.D.;Chouai,A.; Fu,P.K.-L.;Dunbar,K.R.;Turro,C.J.Med.Chem.2006,49,6841. (39)Brindell,M.;Kuli s,E.;Elmroth,S.K.;Urba n ska,K.;Stochel,G. J.Med.Chem.2005,48,7298.(40)(a)Roy,M.;Saha,S.;Patra,A.K.;Nethaji,M.;Chakravarty,A.R. Inorg.Chem.2007,46,4368.(b)Saha,S.;Majumdar,R.;Roy,M.;Dighe,R.R.; Chakravarty,A.R.Inorg.Chem.2009,48,2652.(41)Sasmal,P.K.;Saha,S.;Majumdar,R.;Dighe,R.R.;Chakravarty,mun.2009,1703.4038Inorganic Chemistry,Vol.49,No.9,2010Hussain et al. minimize cellular dark toxicity.While there are few literaturereports on the lanthanide complexes showing DNA bindingpropensity,the study on photoinduced DNA cleavage activ-ity of lanthanide complexes is relatively unexplored exceptsome lanthanide complexes of macrocyclic organic dyes.Forexample,the Gd(III)and Lu(III)complexes of texaphyrinshave been studied as photodynamic therapy(PDT)agentswith the lutetium(III)texaphyrin(LUTRIN)showing PDTeffect in near-IR light of732nm.42,43The major biomedicalapplications of the lanthanide complexes are directed towardmagnetic resonance imaging(MRI)of cancer.44-47TheGd(III)-based complexes,namely,[Gd(DTPA)(H2O)]2-(Magnevist)and[Gd(DOTA)(H2O)]-(Dotarem)are cur-rently used as clinical MRI agents.47It would be of immenseinterest to develop the chemistry of lanthanide complexesshowing dual functionality as MRI and PDT agents.To explore the photoinduced DNA and protein cleavageactivity of La(III)and Gd(III)complexes,we have usedphotoactive planar phenanthroline bases like dipyridoqui-noxaline(dpq)and dipyridophenazine(dppz).The dpq anddppz ligands are known to generate photoexcited3(n-π*) and/or3(π-π*)state cleaving DNA on photoirradiation with high energy UV light.48Binding of these ligands to metal ions could significantly augment their DNA photo-cleavage activity in low-energy UV-A light.Parker and co-workers have recently shown that lanthanide complexes of a nonadentate ligand having a covalently attached dpq moiety exhibit UV light-induced plasmid DNA cleavage activity.49 A mechanism based on a Ln4þtransient intermediate is proposed for the DNA cleavage reaction.We have syn-thesized and characterized lanthanide complexes[La(B)2-(NO3)3](1-3)and[Gd(B)2(NO3)3](4-6),where B is a N, N-donor phenanthroline base,namely,1,10-phenanthroline (phen in1,4),dipyrido[3,2-d:20,30-f]quinoxaline(dpq in2,5) and dipyrido[3,2-a:20,30-c]phenazine(dppz in3,6),to study their photoinduced DNA cleavage activity and the mecha-nistic pathways involved in the DNA cleavage reactions (Scheme1).We have observed the presence of dual mechan-istic pathways involving type-II and photoredox processes in the DNA photocleavage reactions.Significant results of this report include observation of efficient photoinduced DNA cleavage activity of the dpq and dppz complexes at nanomo-lar concentrations and significant reduction of the cellular dark toxicity of the dppz base on binding to the lanthanide ions.Experimental SectionMaterials and Methods.All the reagents and chemicals were procured from commercial sources(SD Fine Chemicals,India; Aldrich,U.S.A.)and used without any further purification. Solvents used were purified by standard procedures.50Super-coiled(SC)pUC19DNA(cesium chloride purified)was pur-chased from Bangalore Genie(India).Tris-(hydroxymethyl)-aminomethane-HCl(Tris-HCl)buffer solution was prepared using deionized and sonicated triple distilled water using a quartz water distillation setup.Calf thymus(CT)DNA,agarose (molecular biology grade),distamycin-A,methyl green,cata-lase,superoxide dismutase(SOD),2,2,6,6-tetramethyl-4-piper-idone(TEMP),1,4-diazabicyclo[2.2.2]octan(DABCO),ethi-dium bromide(EB),Hoechst33258,bovine serum albumin (BSA)protein,acrylamide,N,N0-methylene-bis-acrylamide, ammonium persulphate,N,N,N0,N0-tetramethylethylenediamine (TEMED),2-mercaptoethanol(MPE),glycerol,sodium dodecyl sulfate(SDS),bromophenol blue,coomassie brilliant blue R-250 were from Sigma(U.S.A.).The N,N-donor heterocyclic bases dipyrido-[3,2-d:20,30-f]-quinoxaline and dipyrido[3,2-a:20,30-c]-phenazine were prepared by literature procedures using1,10-phenanthroline-5,6-dione as a precursor reacted with ethylenedia-mine for dpq and1,2-phenylenediamine for dppz.51,52The elemental analyses were done using a Thermo Finnigan Flash EA1112CHNS analyzer.The infrared spectra were recorded on a Bruker ALPHA FT-IR spectrometer.Electro-nic spectra were recorded on a PerkinElmer Spectrum one55 spectrophotometer.Molar conductivity measurements were performed using a Control Dynamics(India)conductivity meter.Room temperature magnetic susceptibility data for the Gd(III)complexes were obtained from a George Associates Inc. Lewis-coil force magnetometer using Hg[Co(NCS)4]as a stan-dard.Experimental susceptibility data were corrected for dia-magnetic contributions.53Cyclic voltammetric measurements were made at25°C on a EG&G PAR Model253VersaStat potentiostat/galvanostat with electrochemical analysis software 270using a three electrode setup comprising a glassy carbon working,platinum wire auxiliary,and a saturated calomel reference(SCE)electrode.Tetrabutylammonium perchlorate plexes1-6and the Phenanthroline BasesUsed(42)(a)Young,S.W.;Woodburn,K.W.;Wright,M.;Mody,T.D.;Fan, Q.;Sessler,J.L.;Dow,W.C.;Miller,R.A.Photochem.Photobiol.1996,63, 892.(b)Guldi,D.M.;Mody,T.D.;Gerasimchuk,N.N.;Magda,D.;Sessler,J.L. J.Am.Chem.Soc.2000,122,8289.(43)Sessler,J.L.;Miller,R.A.Biochem.Pharmacol.2000,59,733.(44)(a)Werner,E.J.;Datta,A.;Jocher,C.J.;Raymond,K.N.Angew. Chem.,Int.Ed.2008,47,8568.(b)Datta,A.;Raymond,K.N.Acc.Chem.Res. 2009,42,938.(c)Major,J.L.;Meade,T.J.Acc.Chem.Res.2009,42,893.(45)(a)Caravan,P.Acc.Chem.Res.2009,42,851.(b)Caravan,P.Chem. Soc.Rev.2006,35,512.(c)Aime,S.;Botta,M.;Fasano,M.;Terreno,E.Chem. Soc.Rev.1998,27,19.(46)(a)Parker,D.;Dickins,R.S.;Puschmann,H.;Crossland,C.; Howard,J.A.K.Chem.Rev.2002,102,1977.(b)Bottrill,M.;Kwok,L.;Long,N.J.Chem.Soc.Rev.2006,35,557.(c)Ahrens,E.T.;Rothb€a cher,U.;Jacobs,R.E.;Fraser,S.E.Proc.Natl.Acad.Sci.U.S.A.1998,95,8443. (47)(a)Caravan,P.;Ellison,J.J.;McMurry,T.J.;Lauffer,R.B.Chem.Rev.1999,99,2293.(b)Lauffer,R.B.Chem.Rev.1987,87,901.(48)Toshima,K.;Takano,R.;Ozawa,T.;Matsumura,-mun.2002,212.(49)Frias,J.C.;Bobba,G.;Cann,M.J.;Hutchison,C.J.;Parker,D.Org.Biomol.Chem.2003,1,905.(50)Perrin,D.D.;Armarego,W.L.F.;Perrin,D.R.Purification of Laboratory Chemicals;Pergamon Press:Oxford,1980.(51)(a)Dickeson,J.E.;Summers,L.A.Aus.J.Chem.1970,23,1023.(b) Collins,J.G.;Sleeman,A.D.;Aldrich-Wright,J.R.;Greguric,I.;Hambley,T.W. Inorg.Chem.1998,37,3133.(52)Amouyal,E.;Homsi,A.;Chambron,J.-C.;Sauvage,J.-P.J.Chem. Soc.,Dalton Trans.1990,1841.(53)Kahn,O.Molecular Magnetism;VCH:Weinheim,1993.Article Inorganic Chemistry,Vol.49,No.9,20104039(TBAP,0.1M)was used as a supporting electrolyte in DMF.The electrochemical data were uncorrected for junction poten-tials.Electrospray ionization mass spectral measurements were done using Bruker Daltonics make(Esquire300Plus ESIModel).Mass spectral measurements of BSA samples weredone using Bruker Daltonics Ultraflex MALDI-TOF instru-ment.1H NMR spectra of the ligands and the La(III)complexeswere recorded at room temperature on a Bruker400MHz NMRspectrometer.Synthesis of[La(B)2(NO3)3](1-3)and[Gd(B)2(NO3)3](4-6)[B=phen(in1,4);dpq(in2,5),dppz(in3,6)].The complexes 1-6were prepared by following a reported synthetic procedurein modified form in which a hot ethanolic solution of Ln(NO3)33 6H2O(Ln=La(III),0.433g;Gd(III),0.451g;1.0mmol)inboiling ethanol(30mL)was added dropwise to a stirred solution of the respective heterocyclic base B(0.40g phen;0.47g dpq;0.57g dppz;2.0mmol)in boiling ethanol(30mL).54Afterstirring for5min,a crystalline precipitate was obtained.Thesolid was isolated,washed with20mL of hot ethanol followed by20mL of diethyl ether,and finally dried in vacuum over P4O10[Yield:∼70%].The characterization data for the com-plexes are given below.[La(phen)2(NO3)3](1).Anal.Calcd for C24H18N7O9La:C,42.06;H,2.35;N,14.30.Found:C,42.32;H,2.45;N,14.16.ESI-MS in10%aqueous MeOH:m/z623[M-NO3-]þ.IR data/ cm-1:1620w,1591w,1470vs,1408s,1284s,1150w,1102w, 1027s,862w,843s,816m,763w,717vs,637m,417m(vs,very strong;s,strong;m,medium;w,weak).UV-visible in DMF [λmax/nm(ε/M-1cm-1)]:322sh(2420),310sh(4340),266(66 420)(sh,shoulder).1H NMR in DMSO-d6(δ,ppm):9.01(dd, 2H,J=8.0,1.7Hz),8.4(dd,2H,J=4.8,1.6Hz),7.9(s,2H), 7.68(dd,2H,J=8.2,4.4Hz).[La(dpq)2(NO3)3](2).Anal.Calcd for C28H18N11O9La:C,42.60;H,2.04;N,19.52.Found:C,42.86;H,2.26;N,19.31.ESI-MS in1:1MeOH-MeCN mixture:m/z727[M-NO3-]þ.IRdata/cm-1:1634w,1576m,1460vs,1395s,1287vs,1210w, 1120w,1084m,1037m,867w,813s,735vs,696w,634w,434w, 416m.UV-visible in DMF[λmax/nm(ε/M-1cm-1)]:340 (17700),325sh(21700),265(72400).1H NMR in DMSO-d6 (δ,ppm):9.37(dd,2H,J=8.3,1.8Hz),9.15(dd,2H,J=4.4, 1.8Hz),9.09(s,2H),7.88(dd,2H,J=8.3,4.2Hz).[La(dppz)2(NO3)3](3).Anal.Calcd for C36H20N11O9La:C,48.61;H,2.27;N,17.32.Found:C,48.80;H,2.29;N,17.42.ESI-MS in1:1MeOH-MeCN mixture:m/z827[M-NO3-]þ.IRdata/cm-1:1577m,1477vs,1410s,1361m,1287vs,1135w, 1080s,1025m,818s,762s,730vs,703m,637w,615w,575w, 415s.UV-visible in DMF[λmax/nm(ε/M-1cm-1)]:380(19 460),369(15900),362(18150),351(13000),294sh(30000),268 (72400).1H NMR in DMSO-d6(δ,ppm):9.44(dd,2H,J=8.2, 1.8Hz),9.11(dd,2H,J=4.5,2.0Hz),8.29-8.33(m,2H), 7.95-7.99(m,2H),7.85(dd,2H,J=8.4,4.0Hz).[Gd(phen)2(NO3)3](4).Anal.Calcd for C24H18N7O9Gd:C,40.96;H,2.29;N,13.93.Found:C,41.04;H,2.00;N,14.21.ESI-MS in10%aqueous MeOH:m/z642[M-NO3-]þ.IR data/ cm-1:1621w,1590w,1468vs,1410s,1290vs,1143m,1099m, 1027m,855s,812m,765w,717vs,636m,417m.UV-visible in DMF[λmax/nm(ε/M-1cm-1)]:322sh(3230),310sh(4850), 266(68040).μeff=7.94μB at298K.[Gd(dpq)2(NO3)3](5).Anal.Calcd for C28H18N11O9Gd:C,41.63;H,2.00;N,19.07.Found:C,41.71;H,2.15;N,19.20.ESI-MS in1:1MeOH-MeCN mixture:m/z746[M-NO3-]þ.IRdata/cm-1:1580m,1463vs,1400s,1296vs,1215w,1125w,1080 m,1027m,880w,809m,728vs,700m,637w,430m,419m. UV-visible in DMF[λmax/nm(ε/M-1cm-1)]:340(17930), 325sh(22110),265(74100).μeff=7.98μB at298K.[Gd(dppz)2(NO3)3](6).Anal.Calcd for C36H20N11O9Gd:C,47.63;H,2.22;N,16.97.Found:C,47.84;H,2.20;N,16.77.ESI-MS in1:1MeOH-MeCN mixture:m/z846[M-NO3-]þ.IR data/cm-1:1570w,1480vs,1416m,1362m,1295vs,1136w, 1073m,1028s,821s,761s,730vs,703s,641w,613w,572w,415s.UV-visible in DMF[λmax/nm(ε/M-1cm-1)]:380(20270), 369(17260),361(19450),351(14600),293sh(31000),269 (78600).μeff=7.96μB at298K.Solubility and Stability.All the complexes were soluble in DMF and DMSO.The phenanthroline complexes were solublein H2O and MeCN.The complexes showed less solubility in MeOH and EtOH.The complexes were stable in the solid state. The complexes showed decomposition on prolonged storage inthe solution phase.54The solution stability of the dppz complex 3(logβ2=10.4)was measured in10%aqueous DMF following a literature method(vide Supporting Information for details).55DNA Binding Methods.DNA binding experiments were done in Tris-HCl/NaCl buffer(5mM Tris-HCl,5mM NaCl,pH7.2)using DMF solution of the complexes1-6.Calf thymus(CT) DNA(ca.350μM NP)in this buffer medium gave a ratio of UV absorbance at260and280nm of about1.9:1indicating that the DNA is apparently free from protein.The concentration of CTDNA was estimated from its absorption intensity at260nm with a known molar extinction coefficient value(ε)of6600M-1 cm-1.56Absorption titration experiments were made by varying the concentration of the CT DNA while keeping the metalcomplex concentration constant.Due correction was made for the absorbance of CT DNA itself.Each spectrum was recorded after equilibration of the sample for5min.The intrinsicequilibrium binding constant(K b)and the binding site size(s) of the complexes1-6to CT DNA were obtained by the McGhee-von Hippel(MvH)method using the expression of Bard et al.by monitoring the change of the absorption intensityof the spectral bands with increasing concentration of CT DNA by regression analysis using the equation(εa-εf)/(εb-εf)= (b-(b2-2K b2C t[DNA]t/s)1/2)/2K b C t,where b=1þK b C tþK b[DNA]t/2s andεa is the extinction coefficient observed forthe absorption band at a given DNA concentration,εf is the extinction coefficient of the complex free in solution,εb is the extinction coefficient of the complex when fully bound to DNA,K b is the equilibrium binding constant,C t is the total metal complex concentration,[DNA]t is the DNA concentration in nucleotides,and s is the binding site size in base pairs.57,58The non-linear least-squares analyses were done using Origin Lab, version6.1.The apparent DNA binding constant(K app)values of the complexes1-6were obtained from fluorescence spectral measurements using ethidium bromide bound CT DNA solu-tion in Tris-HCl/NaCl buffer(pH,7.2).The fluorescence in-tensities of ethidium bromide at600nm(546nm excitation) were recorded with an increasing amount of the added complex concentration.Ethidium bromide showed no apparent emission in Tris-buffer medium because of fluorescence quenching of the free ethidium bromide by the solvent molecules.59In the presence of CT DNA,ethidium bromide showed significantly enhanced emission intensity.The K app values were obtained from the equa-tion:K appÂ[complex]50=K EBÂ[EB],where K app is the apparent binding constant of the complex studied,[complex]50is the concentration of the complex at50%quenching of DNA-bound ethidium bromide emission intensity,K EB is the binding constant of ethidium bromide(K EB=1.0Â107M-1),and[EB]is the concentration of ethidium bromide(1.3μM).60(54)Hart,F.A.;Laming,F.P.J.Inorg.Nucl.Chem.1965,27,1605.(55)Vallee,B.L.;Coombs,T.L.J.Biol.Chem.1959,234,2615.(56)Reichmann,M.E.;Rice,S.A.;Thomas,C.A.;Doty,P.J.Am. Chem.Soc.1954,76,3047.(57)McGhee,J.D.;von Hippel,P.H.J.Mol.Biol.1974,86,469.(58)Carter,M.T.;Rodriguez,M.;Bard,A.J.J.Am.Chem.Soc.1989, 111,8901.(59)(a)Waring,M.J.J.Mol.Biol.1965,13,269.(b)LePecq,J.-B.;Paoletti,C.J.Mol.Biol.1967,27,87.(60)Lee,M.;Rhodes,A.L.;Wyatt,M.D.;Forrow,S.;Hartley,J.A. Biochemistry1993,32,4237.4040Inorganic Chemistry,Vol.49,No.9,2010Hussain et al.DNA melting experiments were carried out by monitoring the absorbance of CT DNA(200μM)at260nm at various temperatures,both in the absence and presence of the complexes (20μM).Measurements were carried out using a Cary300bio UV-visible spectrometer with a Cary temperature controller at an increase rate of0.5°C per min of the solution.Viscometric titrations were performed with a Schott Gerate AVS310Auto-mated Viscometer that was thermostatted at37°C in a constant temperature bath.The concentration of CT DNA was150μM in NP(nucleotide pair),and the flow times were measured using an automated timer.Each sample was measured3times,and an average flow time was calculated.Data were presented as (η/η0)1/3versus[complex]/[DNA],whereηis the viscosity of DNA in the presence of complex andη0is that of DNA alone. Viscosity values were calculated from the observed flow time of DNA-containing solutions(t)corrected for that of the buffer alone(t0),η=(t-t0)/t0.Due corrections were made for the viscosity of DMF solvent present in the solution.DNA Cleavage Experiments.The cleavage of supercoiled (SC)pUC19DNA(30μM,0.2μg,2686base-pairs)was studied by agarose gel electrophoresis.For photoinduced DNA clea-vage studies,the reactions were carried out under illuminated conditions using UV-A light of365nm(6W,Model LF-206.LS of Bangalore Genei).Eppendorf vials were used for photoclea-vage experiments in a dark room at25°C using SC DNA(1μL, 30μM)in50mM Tris-HCl buffer(pH7.2)containing50mM NaCl and the complex(2μL)with varied concentrations.The concentration of the complexes in DMF or the additives in buffer corresponded to the quantity in2μL stock solution after dilution to the20μL final volume using Tris-HCl buffer.The solution path length in the sample vial was∼5mm.After light exposure,each sample was incubated for1.0h at37°C and analyzed for the photocleaved products using gel electrophore-sis.Mechanistic studies were carried out using different addi-tives(NaN3,0.5mM;TEMP,0.5mM;DABCO,0.5mM; DMSO,4μL;KI,0.5mM;catalase,4units;SOD,4units)prior to the addition of the complex.For the D2O experiment,this solvent was used for dilution of the sample to20μL final volume.The samples after incubation in a dark chamber were added to the loading buffer containing0.25%bromophenol blue,0.25%xylene cyanol,30%glycerol(3μL),and the solution was finally loaded on1%agarose gel containing1.0μg/mL ethidium bromide.Electrophoresis was carried out in a dark room for2.0h at60V in TAE(Tris-acetate EDTA)buffer. Bands were visualized in UV light and photographed.The extent of SC DNA cleavage was measured from the intensities of the bands using the UVITEC Gel Documentation System. Due corrections were made for the low level of nicked circular (NC)form of DNA present in the original SC DNA sample and for the low affinity of ethidium bromide binding to SC compared to NC and linear forms of DNA.61The observed error in measuring the band intensities was in the range3-5%.BSA Interaction and Cleavage Experiments.The protein interaction study was performed by tryptophan fluorescence quenching experiments using BSA(2μM)in phosphate buffer (pH6.8)containing15%DMF.Quenching of the emission intensity of tryptophan residues of BSA at344nm(excitation wavelength at295nm)was monitored using complexes1-6as quenchers with increasing complex concentration.62Stern-Volmer I0/I versus[complex]plots were made using the corrected fluorescence data taking into account the effect of dilution.A linear fit of the data using the equation:I0/I= 1þK BSA[Q],where I0and I are the respective emission intensities of BSA in absence and presence of the quencher of concentration[Q],gave the interaction constant(K BSA)values using Origin6.1.Photoinduced BSA cleavage experiments were carried out following literature procedure.63Freshly prepared solutions of BSA in50mM Tris-HCl buffer(pH7.2)containing0.6%DMF were used for the photochemical protein cleavage studies.The BSA solutions in Tris-HCl buffer medium containing complexes 1-6(10μM and15μM)and BSA(5μM)were photoirradiated at365nm(100W)in Eppendorf vials.The BSA solutions containing the complexes were incubated at37°C for1.0h prior to the photoexposure.The photoirradiated samples (50μL)were dried in a centrifugal vaporizer(EYELA Centri-fugal Vaporizer,Model CVE-200D),and the samples were dissolved in the loading buffer(24μL)containing SDS(7%w/v), glycerol(4%w/v),Tris-HCl buffer(50mM,pH6.8),mercap-toethanol(2%v/v),and bromophenol blue(0.01%w/v).The protein solutions were then denatured by heating on a boiling water bath for3min.The samples were loaded on a3% polyacrylamide stacking gel.The gel electrophoresis was done at an initial applied voltage of60V until the dye passed into the separating gel from the stacking(3%)gel,and then the voltage was increased to110V.The gels were run for1.5h,stained with Coomassie Brilliant Blue R-250solution(acetic acid/ methanol/water=1:2:7v/v)and destained with water/ methanol/acetic acid mixture(5:4:1v/v)for4h.The gels,after destaining,were scanned with a HP Scanjet G3010scanner,and the images were further processed using the Adobe Photoshop 7.0software package.Molecular weight markers were used in each gel to calibrate the molecular weight of the protein.The presence of reactive oxygen species was investigated by carrying out the photoinduced protein cleavage experiments using singlet oxygen quenchers such as NaN3(3mM)and TEMP (3mM)and hydroxyl radical scavengers,namely,DMSO (20μL)and KI(3mM).Cell Cytotoxicity Assay.The photocytotoxicity of the dppz complexes(3,6)and the dppz ligand was studied using3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide(MTT) assay which is based on the ability of mitochondrial dehydro-genases of viable cells to cleave the tetrazolium rings of MTT forming dark purple membrane impermeable crystals of for-mazan that could be quantified at595nm after solubilization in detergent.64Approximately,8000cells of human cervical carci-noma(HeLa)were plated in96wells culture plate in Dulbecco’s Modified Eagle Medium(DMEM)containing10%FBS,and after24h of incubation at37°C in a CO2incubator,various concentrations of the complexes3,6or dppz ligand dissolved in 1%DMSO were added to the cells and incubation was con-tinued for4h in dark.The medium was subsequently replaced with PBS and photoirradiated for15min using UV-A light of 365nm.After photoexposure,PBS was removed and replaced with DMEM-FBS,and incubation was continued for further 24h in dark.At the end of the incubation period,20μL of5mg mL-1MTT was added to each well and incubated for an additional3h.The culture medium was finally discarded,and 100μL of10%SDS/0.01M HCl was added.The plates were then incubated at37°C for6h to dissolve the formazan crystals, and the absorbance at595nm was determined using a BIORAD ELISA plate reader.Cytotoxicity of the dppz ligand and the complexes3and6was measured as the percentage ratio of the absorbance of the treated cells to the untreated controls.The IC50values were determined by non-linear regression analysis using the GraphPad Prism software.To determine the dark cytotoxicity of the complexes3and6and the dppz ligand, various concentrations of the complexes or dppz ligand dis-solved in DMSO(1%)were added to the HeLa cells and incubated for24h in dark,thereafter the media were discarded(61)Bernadou,J.;Pratviel,G.;Bennis,F.;Girardet,M.;Meunier,B.Biochemistry1989,28,7268.(62)Quiming,N.S.;Vergel,R.B.;Nicolas,M.G.;Villanueva,J.A. J.Health Sci.2005,51,8.(63)Kumar,C.V.;Buranaprapuk,A.;Sze,H.C.;Jockusch,S.;Turro, N.J.Proc.Natl.Acad.Sci.U.S.A.2002,99,5810.(64)Mosmann,T.J.Immunol.Methods1983,65,55.。
实验三 高斯消去法和三角分解法1

实验报告实验三 高斯消去法与矩阵的三角分解一、实验目的1、掌握列主元素消去法,并且能够用MATLAB 编写相关程序,实现高斯消去法的求解。
2、能够用矩阵理论理解与研究高斯消去法,通过对矩阵的初等变换实现高斯消去法。
3、学会矩阵的三角分解,并且能够用MATLAB 编写相关程序,实现矩阵的三角分解,解方程组。
二、上机内容⎥⎥⎥⎥⎥⎥⎥⎥⎥⎦⎤⎢⎢⎢⎢⎢⎢⎢⎢⎢⎣⎡=⎥⎥⎥⎥⎥⎥⎥⎥⎥⎦⎤⎢⎢⎢⎢⎢⎢⎢⎢⎢⎣⎡⎥⎥⎥⎥⎥⎥⎥⎥⎥⎦⎤⎢⎢⎢⎢⎢⎢⎢⎢⎢⎣⎡2822171310871234567112345611123451111234111112311111121111111764321x x x x x x1、用列主元素高斯消去法求解方程组。
2、用列主元消去法求解方程组(实现PA=LU) 要求输出: (1)计算解X;(2)L,U;(3)正整型数组IP(i),(i=1,···,n) (记录主行信息)。
三、实验原理1、列主元素消去法用高斯消去法求解方程组时,为了减小误差,在消去的过程中要避免用绝对值较小的主元素。
因此在高斯消去法的每一步应该在系数矩阵货消去后的低阶矩阵中选取绝对值较大的元素作为主元素,保持|m ik |<=1,以减小计算过程中的舍入误差对计算解的影响。
此方法为完全主元素消去法。
完全主元素消去法在选主元素时花费一定的计算机时间,因此实际计算中常用列主元消去法。
列主元消去法在每次选主元时,仅依次按列选取绝对值最大的元素作为主元素,且仅交换两行,再进行消元计算。
装订 线第k步计算如下:对于k=1,2,…,n-1(1)按列选主元:即确定t使(2)如果t≠k,则交换[A,b]第t行与第k行元素。
(3)消元计算(4)回代求解计算流程图回代求解 b=b/a (当a nn ≠0)b ←(b -∑a x )/adet=a nn *det输出计算解及行列式及detk=1,2,…,n-1输入n ,A,b,εdet=1按列主元|a i(k),k |=max|a ik |C 0=a i(k),k换行 a ik a i(k)j(j=k,…n ) b k b j(k), 消元计算 (i=k+1,…,n ) a ik=a ik -a kk *m ik a ij=a ij -a kj *m ik (j=k+1,…,n )|C 0|<εi k =kdet=a kk det否否是是k<=n-1输出det(A)=0停机停机2. 矩阵的三角分解法 (1)定理设 n n R A ⨯∈ 。
运筹学实验报告(F-R共轭梯度法、Wolfe简约梯度法)

f ( X ( k 1) ) min f ( X ( k ) d )
0
(k ) ( k 1) (k ) 0 的解为 如此下去, 得到序列{ X }。不难求得 d , Ad
(k )
X ( k 1) X ( k )
(k )
b AX ( k ) , d ( k ) ( k ) d d ( k 1) , Ad ( k 1)
end
T(j)= new_direction(j) else T(j)=0; end if (T(j)<0) TT=abs([TT,(x_1(j)/T(j))]) end j=j+1;
n=size(TT); for uk=1:n if(tmax>TT(uk)) tmax=TT(uk) n=n+1; end end x=x_1+t*new_direction; xx=[x(1),x(2)]; f_step=subs(f,findsym(f),xx); F=diff(f_step,t); solve(F,t); t0=tmax; x_2=x_1+0.18*new_direction A=[A;x_2]; norm0=norm; search_direction=new_direction x_1=x_2; xx2=[x_2(1),x_2(1)]; Y=subs(f,findsym(f),xx2); HSZ=[HSZ;Y] k=k+1; end k x_2 norm
注意到 d 的选取不唯一,我们可取
d ( k ) f ( X ( k ) ) k 1d ( k 1)
(k ) ( k 1) 0 可得: 由共轭的定义 d , Ad
k 1
06第17讲-过程合成1解析

可能的组合方案数为:
N max
N max
CPN P!/ N!/(P!N!)
N m in
N m in
其中,总费用最小的 方案即为最佳方案
(4)反应器网络的合成
任务:给定动力学和进料条件下寻求适宜的反应器类型、流程 结构和关键设计参数,以使特定的目标函数最优。
组分数N
方法数P
序列数Z N
P
Z
3
1
2
9
1
1430
3
28Leabharlann 95558593750
4
2
40
10
1/2
4862/24893
44
5
1
14
11
1
16792
(3) 换热网络
换热过程是化工生产中的基本过程
化工生产过程中换热网络的作用和意义
CO+3H2
30℃
14
H2
CH4+H2O 高压蒸汽
甲脱烷甲化烷塔塔
B10 B12
第6章 系统综合 (系统合成,Process system synthesis )
0概述
化工系统工程主要研究内容
系统分析 系统优化 系统合成
核心是 优化
化工过程系统工程研究的四个基本问题
操作型问题
设计型问题 操作优化 过程合成
系统分析的任务在于描述或预测系统的 特性;系统优化是为了以最佳的方式实 现系统的目标或功能;系统综合?
分离过程合成中的一些经验规则
举例:
●在所有的分离方法中,优先采用能量分离剂(精馏),避免采用质量 分离剂(如萃取);当关键组分相对挥发度小于1.1时,可采用后者。 ●首先除去腐蚀性、有毒有害的产品; ●最后处理难分离或分离要求高的的产品; ●进料中最多的组分应当首先分离出去;
matlab牛顿拉夫逊法与快速分解法的实现

一、概述MATLAB是一种强大的数学软件工具,它提供了许多优秀的数值计算和数据分析功能。
其中,牛顿拉夫逊法和快速分解法是两种常用的数值计算方法,它们在解决非线性方程组和矩阵分解等问题中具有重要的应用价值。
本文将介绍如何在MATLAB中实现这两种方法,并对它们的优缺点进行详细分析。
二、牛顿拉夫逊法的实现1. 算法原理牛顿拉夫逊法是一种用于求解非线性方程组的迭代算法。
它利用函数的一阶和二阶导数信息来不断逼近方程组的解,直到满足精度要求为止。
算法原理可以用以下公式表示:公式1其中,x表示解向量,F(x)表示方程组的函数向量,J(x)表示方程组的雅可比矩阵,δx表示解的更新量。
通过不断迭代更新x,最终得到方程组的解。
2. MATLAB代码实现在MATLAB中,可以通过编写函数来实现牛顿拉夫逊法。
以下是一个简单的示例代码:在这段代码中,首先定义了方程组的函数向量和雅可比矩阵,然后利用牛顿拉夫逊法进行迭代更新,直到满足精度要求为止。
通过这种方式,就可以在MATLAB中实现牛顿拉夫逊法,并应用于各种实际问题。
三、快速分解法的实现1. 算法原理快速分解法是一种用于矩阵分解的高效算法。
它利用矩阵的特定性质,通过分解为更小的子问题来加速计算过程。
算法原理可以用以下公式表示:公式2其中,A表示要分解的矩阵,L和U分别表示矩阵的下三角和上三角分解。
通过这种分解方式,可以将原始矩阵的计算量大大减小,提高求解效率。
2. MATLAB代码实现在MATLAB中,可以利用内置函数来实现快速分解法。
以下是一个简单的示例代码:在这段代码中,利用MATLAB内置的lu函数进行LU分解,得到矩阵的下三角和上三角分解。
通过这种方式,就可以在MATLAB中实现快速分解法,并应用于各种矩阵计算问题。
四、方法比较与分析1. 算法复杂度牛顿拉夫逊法和快速分解法在计算复杂度上有所不同。
牛顿拉夫逊法的迭代次数取决于所求解问题的非线性程度,通常需要较多的迭代次数。
用矩阵的lu三角分解法编程求解方程组

用矩阵的lu三角分解法编程求解方程组下载提示:该文档是本店铺精心编制而成的,希望大家下载后,能够帮助大家解决实际问题。
文档下载后可定制修改,请根据实际需要进行调整和使用,谢谢!本店铺为大家提供各种类型的实用资料,如教育随笔、日记赏析、句子摘抄、古诗大全、经典美文、话题作文、工作总结、词语解析、文案摘录、其他资料等等,想了解不同资料格式和写法,敬请关注!Download tips: This document is carefully compiled by this editor. I hope that after you download it, it can help you solve practical problems. The document can be customized and modified after downloading, please adjust and use it according to actual needs, thank you! In addition, this shop provides you with various types of practical materials, such as educational essays, diary appreciation, sentence excerpts, ancient poems, classic articles, topic composition, work summary, word parsing, copy excerpts, other materials and so on, want to know different data formats and writing methods, please pay attention!用矩阵的LU三角分解法编程求解方程组在数值计算中,求解线性方程组是一个基础且常见的问题。
- 1、下载文档前请自行甄别文档内容的完整性,平台不提供额外的编辑、内容补充、找答案等附加服务。
- 2、"仅部分预览"的文档,不可在线预览部分如存在完整性等问题,可反馈申请退款(可完整预览的文档不适用该条件!)。
- 3、如文档侵犯您的权益,请联系客服反馈,我们会尽快为您处理(人工客服工作时间:9:00-18:30)。
EIGRP分解实验三
1.实验EIGRP 跨路由器建立邻居关系:
a.R2上no EIGRP进程
b.静态路由先让R1 R3之间互相ping通,R1 R3单播互指邻居,注
意出现的提示信息,分析产生的原因
c.将R1 R3接口地址修改为/16,观察收到路由情况,并分析ping
相互收到的路由能否Ping通
2.如图所示,将所有串口和环回口宣告进EIGRP,在R1 R2 R3上开启
debug eigrp packets query reply ack
然后Shut down R1 LO0口,观察各路由器的query包,reply包的发送和确认情况,理解DUAL的扩散查询行为
3.通过在R2上S1/1口设置ACL,阻止R3过来的所有EIGRP包,通
过设置holddown time到最长,使它们之间邻居关系保持不DOWN,然后shut down R1 LO0 口,观察EIGRP的拓扑表,学习EIGRP的SIA状态,以及cisco设计SIA-QUERY/SIA-REPLY两种报文的意义
4.实验解决SIA的方法一,设定ACTIVE-TIME时间;方法二,汇总路
由
5.实验EIGRP的STUB路由器,理解STUB的几个选项的意义
a.将R3设置成STUB receive-only, R1上观察R3的环回口路由
b.在R3上增加一条静态路由33.3.3.0 指向null0, 通过重分布发
布进EIGRP,观察stub redistributed/static的行为
c.还原,将R2设置成stub,使用ACL/route-map 匹配1.1.1.0/24,
观察stub leek-map选项的行为
6.在R1 R2之间开启EIGRP 的认证
7.在R1上使用distance 255的方法过滤33.3.3.0/24,观察实验现象,
理解没有生效的原因
8.在R3上使用distribute-list 过滤这条33.3.3.0/24,使之在R1上不
可见,但是不能使用基于接口的过滤。
