HRT-mediated hypersensitive response and resistance to tcv does not require TIP
半导体测序技术
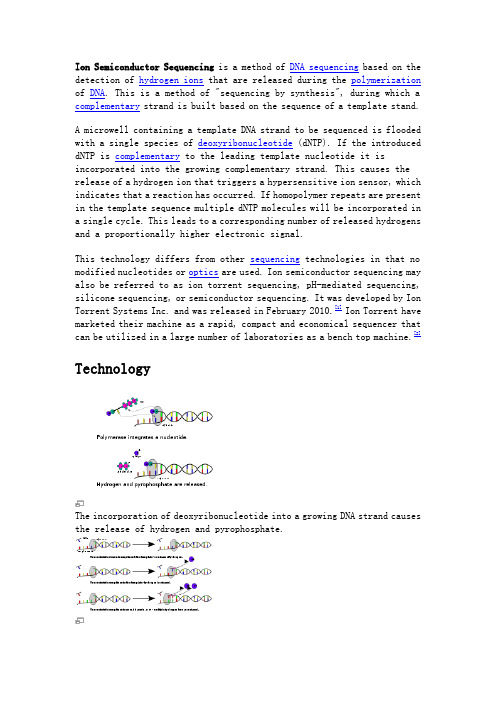
Ion Semiconductor Sequencing is a method of DNA sequencing based on the detection of hydrogen ions that are released during the polymerization of DNA. This is a method of "sequencing by synthesis", during which a complementary strand is built based on the sequence of a template stand.A microwell containing a template DNA strand to be sequenced is flooded with a single species of deoxyribonucleotide (dNTP). If the introduced dNTP is complementary to the leading template nucleotide it is incorporated into the growing complementary strand. This causes the release of a hydrogen ion that triggers a hypersensitive ion sensor, which indicates that a reaction has occurred. If homopolymer repeats are present in the template sequence multiple dNTP molecules will be incorporated in a single cycle. This leads to a corresponding number of released hydrogens and a proportionally higher electronic signal.This technology differs from other sequencing technologies in that no modified nucleotides or optics are used. Ion semiconductor sequencing may also be referred to as ion torrent sequencing, pH-mediated sequencing, silicone sequencing, or semiconductor sequencing. It was developed by Ion Torrent Systems Inc. and was released in February 2010.[1]Ion Torrent have marketed their machine as a rapid, compact and economical sequencer that can be utilized in a large number of laboratories as a bench top machine.[2]TechnologyThe incorporation of deoxyribonucleotide into a growing DNA strand causes the release of hydrogen and pyrophosphate.The release of hydrogen ions indicate if zero, one or more nucleotides were incorporated.Released hydrogens ions are detected by an ion sensor. Multiple incorporations lead to a corresponding number of released hydrogens and intensity of signal.Sequencing ChemistryIn nature, the incorporation of a deoxyribonucleotide (dNTP) into a growing DNA strand involves the formation of a covalent bond and the release of pyrophosphate and a positively charged hydrogen ion.[1] A dNTP will only be incorporated if it is complementary to the leading unpaired template nucleotide. Ion semiconductor sequencing exploits these facts by determining if a hydrogen ion is released upon providing a single species of dNTP to the reaction.Microwells on a semiconductor chip that each contain one single-stranded template DNA molecule to be sequenced and one DNA polymerase are sequentially flooded with unmodified A, C, G or T dNTP.[1][3][4] If an introduced dNTP is complementary to the next unpaired nucleotide on the template strand it is incorporated into the growing complementary strand by the DNA polymerase.[5]If the introduced dNTP is not complementary there is no incorporation and no biochemical reaction. The hydrogen ion that is released in the reaction changes the pH of the solution, which is detected by a hypersensitive ion sensor.[1][3]The unattached dNTP molecules are washed out before the next cycle when a different dNTP species is introduced.[3]Signal DetectionBeneath the layer of microwells is an ion sensitive layer, below which is a hypersensitive ISFET ion sensor.[2] All layers are contained within a CMOS semiconductor chip, similar to that used in the electronics industry.[2][6]Each released hydrogen ion triggers the ISFET ion sensor. The series of electrical pulses transmitted from the chip to a computer is translated into a DNA sequence, with no intermediate signal conversion required.[3][7] Each chip contains an array of microwells with corresponding ISFET detectors.[3] Because nucleotide incorporation events are measured directly by electronics, the use of labeled nucleotides and optical measurements are avoided.[2][6]Sequencing CharacteristicsThe per base accuracy achieved in house by Ion Torrent on the Ion Torrent ion semiconductor sequencer as of February 2011 was 99.6% based on 50 base reads, with 100 Mb per run.[8]The read-length as of February 2011 was 100 base pairs.[8]The accuracy for homopolymer repeats of 5 repeats in length was 98%.[8]. It should be noted that these figures have not yet been independently verified outside of the company.StrengthsThe major benefits of ion semiconductor sequencing are rapid sequencing speed and low upfront and operating costs.[4][7] This has been enabled by the avoidance of modified nucleotides and optical measurements.Because the system records natural polymerase-mediated nucleotide incorporation events, sequencing can occur in real-time. In reality, the sequencing rate is limited by the cycling of substrate nucleotides through the system.[9] Ion Torrent Systems Inc., the developer of the technology, claims that each incorporation measurement takes 4 seconds and each run takes about one hour, during which 100-200 nucleotides are sequenced.[7][10] If the semiconductor chips are improved (as predicted by Moore’s law), the number of reads per chip (and therefore per run) should increase.[7]The cost of acquiring a pH-mediated sequencer from Ion Torrent Systems Inc. at time of launch was priced at around $50,000 USD, excluding sample preparation equipment and a server for data analysis.[4][7][10] The cost per run is also thought to be significantly lower than that of alternative automated sequencing methods.[4][8]LimitationsIf homopolymer repeats of the same nucleotide (e.g. GGGGG) are present on the template strand (strand to be sequenced) multiple introducednucleotides are incorporated and more hydrogen ions are released in a single cycle. This results in a greater pH change and a proportionally greater electronic signal.[7] This is a major limitation of the system in that it is difficult to enumerate long repeats. This is shared by other techniques that detect single nucleotide additions such as pyrosequencing.[11] Signals generated from a high repeat number are difficult to differentiate from repeats of a similar but different number (e.g. 7 from 8 homorepeats).Another limitation of this system is a short read length compared to other sequencing methods such as sanger sequencing or pyrosequencing. Longer read lengths are beneficial for de novo genome assembly. The read length achieved by Ion Torrent Systems Inc. is currently 100 base pairs per run.[1][4] The throughput is currently lower than that of otherhigh-throughput sequencing technologies, although the developers hope to change this by increasing the density of the chip.[1]Comparison to other sequencing methodsIon Torrent[10][12]454Sequencing[13]Illumina [14]SOLiD [15]Sequencing Chemistry IonsemiconductorsequencingPyrosequencingPolymerase-basedsequence-by-synthesisLigation-based sequencingAmplificati on approach Emulsion PCR Emulsion PCRBridgeamplificationEmulsion PCRMb per run 100 Mb 100 Mb 600 Gb 3000 MbTime per run 2 hours 7 hours 9 days 5 daysRead length 100 bp 400 bp 2x100 bp 35-50 bp Cost per run $ 500 USD $ 8,438 USD $ 20,000 USD $ 17,447 USD Cost per Mb $ 5.00 USD $ 84.39 USD $ 0.03 USD $ 5.81 USD Cost perinstrument$ 50,000 USD $ 500,000 USD $ 600,000 USD $ 591,000 USDTable 1. Comparing metrics and performance of next-generation DNA sequencers. [16]ApplicationThe developers of ion semiconductor sequencing have marketed it as a rapid, compact and economical sequencer that can be utilized in a large number of laboratories as a bench top machine.[1][2] The company hopes that their system will take sequencing outside of specialized centers and into the reach of hospitals and smaller laboratories.[17] A January 2011 New York Times article, "Taking DNA Sequencing to the Masses", underlines these ambitions.[17]Due to the ability of alternative sequencing methods to achieve a greater read length (and therefore being more suited to whole genome analysis) this technology may be best suited to small scale applications such as microbial genome sequencing, microbial transcriptome sequencing, targeted sequencing, amplicon sequencing, or for quality testing of sequencing libraries.[1][4]References1.^ a b c d e f g h Rusk, N. (2011). "Torrents of sequence". Nat Meth8(1): 44-44.2.^ a b c d e Ion Torrent Official Webpage.3.^ a b c d e Pennisi, E. (2010). "Semiconductors inspire new sequencingtechnologies". Science 327(5970): 1190.4.^ a b c d e f Perkel, J., "Making contact with sequencing's fourthgeneration". Biotechniques, 2011.5.^ Alberts B, Molecular Biology of the Cell. 5th Edition ed. 2008,New York: Garland Science.6.^ a b Karow, J. (2009) Ion Torrent Patent App Suggests SequencingTech Using Chemical-Sensitive Field-Effect Transistors. In Sequence.7.^ a b c d e f Bio-IT World, Davies, K. It’s “Watson Meets Moore”as Ion Torrent Introduces Semiconductor Sequencing. Bio-IT World 2010.8.^ a b c d Karow, J. (2009) At AGBT, Ion Torrent Customers ProvideFirst Feedback; Life Tech Outlines Platform's Growth. In Sequence.9.^Eid, J., et al., "Real-time DNA sequencing from single polymerasemolecules". Science, 2009. 323(5910): p. 133-8.10.^ a b c Karow, J. (2010) Ion Torrent Systems Presents $50,000Electronic Sequencer at AGBT. In Sequence.11.^Metzker, M.L., "Emerging technologies in DNA sequencing". GenomeRes, 2005. 15(12): p. 1767-76.12.^/products-ion-pgm/13.^/applications/transcriptome-sequencing.asp14.^/pages.ilmn?ID=20415.^/16.^ Shendure, J. and H. Ji, "Next-generation DNA sequencing." NatBiotech, 2008. 26(10): p. 1135-1145.17.^ a b Pollack, A., Taking DNA Sequencing to the Masses, in New YorkTimes. 2011: New York.。
Hypersensitive Reactions
The nasohprynx Rhinorrhea
二、Several Components of Type I Reactions
ALLERGENS REAGINIC ANTIBODY (IGE) MAST CELLS AND BASOPHILS IgE-BINDING Fc RECEPTORS
C48/80 elicits release of mast cell renin, triggering ANG II-induced contraction of rat bronchial rings.
compound 48/80 (C48/80) is mast cell degranulating agent. cromolyn: the mast cell stabilizer BILA2157: the renin inhibitor EXP3174: an AT1R antagonist AT1R: ANG II type 1 receptor renin
When some subpopulations of activated TH cells encounter certain types of antigens, they secrete cytokines that induce a localized inflammatory reaction called delayed-type hypersensitivity (DTH). 一、常见临床病例: 1. 传染性迟发型超敏反应 2. 接触性皮炎 3. 移植排斥反应 4. 脏器特异性自身免疫病
植物-病原菌互作的分子机制

M. grisea
Plant disease
C. fulvum
B. cinerea
P. infestans
I 植物病原菌的侵染机理
侵染途径 特征 寄主范围
植物病原菌寄生方式
腐生 (necrotroph)
活体寄生 (biotroph)
半活体寄生 (semibiotroph)
分泌胞壁降解酶、毒 菌体进入寄主细胞内 先活体寄生,
有菌系均有抗性。是植物防御潜在病原菌的主要机制,也是 植物最基础最普遍的抗病类型
• 抗病(resistance):植株能限制病原菌在侵染点附近、病斑不 扩展或只产生小斑点: 非亲和性反应(incompatibility))
• 感病(susceptibility)病斑扩大形成典型病斑: 亲和性反应 (compatibility)
素
后腐生
寄主组织死亡、病原 寄主细胞一般保持成 侵染早期寄主
菌定殖、大面积组织 活状态
组织仍成活,
软化
而后死亡
广
窄,一般侵染个别植 两者之间
物
病原真菌在植物表皮细胞内形成吸器(Haustorium)从寄主内吸收营养
病原细菌定殖于寄主细胞间隙
植物病毒可在寄主细胞内大量增殖并通过 胞间连丝进行“cell to cell”扩散
抗病反应。
I. PAMP-Triggered Immunity (PTI) PAMP的作用:病原菌的适应性与生存
已知的主要PAMP
Bacterial flagellin (flg22) Bacterial PAMPs EF-Tu Xoo Ax21 Fungal xylanase Fungal chitin Oomycete glucans
超敏反应《医学免疫学》人卫版

超敏反应
Hypersensitivity
超敏反应 (Hypersensitivity):
(变态反应Allergy、 过敏反应Anaphylaxis )
指机体接受某些抗原刺激时,出现生理功能紊乱或组 织细胞损伤的异常特异性免疫应答。
消除Ag
Ag
免疫应答
组织损伤、生理功能改变
超敏反应的分类
I 型超敏反应 Ⅱ型超敏反应 Ⅲ型超敏反应
其机理为:多巴改变了红细胞膜上的Rh系统的e抗原 产生抗RBC抗体 Ⅱ型超敏反应发生机制导致自身 免疫性溶血性贫血。
•药物(青霉素)过敏性血细胞减少症
青霉素 (半抗原)
+
蛋白 (血细胞 膜或血浆)
完全抗原
抗体
免疫复合物+补体
溶血性贫血、粒细胞减少症、血小板减少性紫癜
新生儿溶血症
Rh– 初孕
Rh+
➢ IgE为亲细胞抗体,未结合Ag时即可通过其Fc段与肥大 细胞和嗜碱性粒细胞表面IgE Fc受体结合,使机体处于致 敏状态。
➢ IgE 的产生依赖于细胞因子IL-4
Th2分泌的IL-4能增强IgE的合成
(1)IL-4是一个Ig类转换因子,为B细胞提 供活化信 号,从而 使B细胞由产生IgM转换为产生IgE。
刺激支气管、 子宫和膀胱 等处的平滑 肌收缩
促进粘 膜、腺 体分泌
增多
2.新合成的介质及其作用
新合成介质:主要是细胞膜磷脂代谢产物、细胞因子 及嗜酸性粒细胞产生的酶类物质和脂类介质。
白三烯(leukotrienes,LTs) 前列腺素D2(PGD2) 血小板活化因子(platelet activating factor,PAF) 多种细胞因子:TNF、IL-1、 IL-4、IL-5、IL-10和CSF
药物英文说明书的写法-不良反应

第七节不良反应不良反应这一项中包括药物的副作用、症状及体征、毒性作用及耐受力、过敏反应、停药等。
现分述如下:1.“不良反应”的常用的英语表示法有:Adverse Reaction(s)不良反应Unwanted(Untoward)Reaction(s)不良反应2“副作用”的常用的英语表示法有:Side-effect(s) 副作用Unwanted(Undesirable)Effects 副作用Side Reaction(s) 副作用By-effects 副作用3.常见的霉副反应的症状及体征的词语很多,但记住下列词语很有必要:allergic (hypersensitive,anaphylactic)reaction(s) 过敏反应allergy(hypersensitivity) 过敏dizzziness 眩晕gastrointestinal tract 胃肠道fever 发热local reaction(s) 局部反应flush 潮红skin reaction(s)皮肤反应headache 头痛symptom(s)症状nausea 恶心systematic 全身的pruritus 瘙痒anorexia 厌食rash 皮疹blood count 白细胞计数spasm 痉挛blood pressure 血压thirst 口渴coma 昏迷tiredness 疲倦diarrhea 腹泻vomiting 呕吐4.“霉性”、“耐受性”的英语表示法:toxicity 毒性tolerance(tolerability) 耐受力,耐药性tolerate(toleration)耐受5.“停药”的英语表示法举例:abandon 停药discontinue(discontinuance,discontinuation) 停药,中断(治疗)cease(cessation)(stop)停药(停止治疗)don't use 勿使用(停药)suspend 停药terminate 停止,结束withdraw(be withdrawn,withdrawal)停药6.其他一些常见的基本词汇及短语:acute 急性的control(be controlled)控制chronic 慢性的diminish(reduce,reduction)减少common 常见的disappear 消失irreversible 不可逆的encounter 遇到,见到mild 轻微的give rise to 产生normal 正常的include 包括rare(ly)罕见的(地)lead to 导致reverside 可逆的manifest 表明,显示severe 严重的observe(be observed)(被)观察到temporary 暂时的produce 产生transient 一过性的,短暂的react to 对……反应appear(develop,happen,occur)出现(产生)result from 由……引起be reported (被)报道result in 导致cause (be caused by) (由……)引起treat(treatment)治疗special care(caution)should be exercised 特别小心(注意)例 1 Ciprofloxactin is generally well tolerated. The most frequently reported adverse reactions are: nausea, diarrhoea, vomiting, dyspepsia, abdomina pain, headache, restlessness, rash, dizziness and pruritus.环丙沙星一般耐受良好。
JAs、SAs介导的植物防御反应及在药用植物中的应用

0引言茉莉酸(JA )类物质是一类具有共同的环戊烷酮结构的新型天然植物激素,由十八烷途径合成,在植物体内具有广泛的生理功能。
水杨酸(SA )类物质是一类酚类化合物,在植物等生长发育过程中也起着重要的调节功能。
这两类物质对诱导植物防御反应均有很大作用。
植物防御反应是指植物抵御外部侵染、机械损伤等的细胞生理反应,包括活性氧的产生、病程相关蛋白和其他防御相关蛋白的合成、过敏反应、植保素的合成、防御屏障的形成等。
近几年来,关于茉莉酸类和水杨酸类物质调节植物抗病虫害的研究已经有了很大进展,但寄生植物引起的防御反应仍知之甚少[1]。
文章主要综述JA 途径、SA 途径和JA/SA 交互作用介导的抗病虫害和寄生物基金项目:云南省重大产业项目(云发改高技[2007]1718号);云南省财政厅科技项目;云南省中药现代化科技专项(2002ZY-24)。
第一作者简介:刘艳艳,女,1985年出生,山东人,在读硕士生,主要研究方向:药用植物资源评价与利用,通信地址:650201云南农业大学农学院73#信箱,E-mail :liuyuxiu07@ 。
通讯作者:萧凤回,男,1960年出生,教授,博士生导师,E-mail :fenghuixiao@ 。
收稿日期:2010-03-08,修回日期:2010-04-15。
JAs 、SAs 介导的植物防御反应及在药用植物中的应用刘艳艳1,萧凤回1,2(1云南农业大学中药材研究所,云南省中药材规范化种植技术指导中心,昆明650201;2浙江林学院林业与生物技术学院,杭州311300)摘要:茉莉酸(JA )和水杨酸(SA )介导的信号网络能调节植物防御反应。
一般JA 信号通路涉及抗虫反应,而SA 通路则与抗病有关,JA 和SA 通路之间的交互作用在防御反应的微调中起重要作用。
研究表明,JA 和SA 也能有效调节抗寄生植物的防御反应。
文章综述了一些防御信号分子,尤其是JA 和SA 在植物防御中的作用,包括JA 、SA 介导的途径和JA/SA 交互作用在抗病虫害和抗植物寄生中的作用;介绍茉莉酸、水杨酸类物质在药用植物研究中的初步应用。
FDA工业指南 非青霉素β内酰胺类药品防止交叉污染的指导原则2013.4

工业指南非青霉素β-内酰胺类药品防止交叉污染的指导原则目录I.简介II.背景III.建议You can use an alternative approach if the approach satisfies the requirements of the applicable statutes and regulations. If you want to discuss an alternative approach, contact the FDA staff responsible for implementing this guidance. If you cannot identify the appropriate FDA staff, call the appropriate number listed on the title page of this guidance.本指南代表当前FDA对本议题的看法。
本指南并不授予任何人任何特权,也不对FDA或公众起任何约束作用。
如果有替代的方法能满足法律法规的要求,则可以使用该替代方法。
如果想要讨论替代方法,请与FDA负责实施该指南的工作人员联系。
如果不能与指定的合适FDA工作人员联系,请拨打该指南标题页列出的电话号码。
I. INTRODUCTIONI. 简介This guidance describes the importance of implementing manufacturing controls to prevent cross-contamination of finished pharmaceuticals and active pharmaceutical ingredients (APIs) with non-penicillin beta-lactam drugs. This guidance also provides information regarding the relative health risk of, and the potential for, cross-reactivity in the classes of sensitizing beta-lactams (including both penicillins and non-penicillin beta-lactams). Finally, this guidance clarifies that manufacturers generally should utilize separate facilities for the manufacture of non-penicillin beta-lactams because those compounds pose health risks associated with cross-reactivity.本指南描述了对非青霉素β-内酰胺类成品和原料药的生产过程进行控制,以防止交叉污染的重要性。
E3泛素连接酶调控植物抗病分子机理研究进展

E3泛素连接酶调控植物抗病分子机理研究进展作者:杨玖霞张浩王志龙王旭丽王国梁来源:《植物保护》2015年第04期摘要生物胁迫是影响我国农作物生产的重要因素,也是当前植物界研究方向涉及最为广泛的领域之一。
由泛素介导的降解途径是生物体内最为精细的调控体系,涉及对生物体的生长发育以及生物体对周围环境适应的调控等过程。
E3泛素连接酶因对底物有特异性识别作用,被认为是泛素化过程中最重要的组成部分。
依据其结构和功能的不同可以将E3泛素连接酶分为4个家族。
越来越多的研究表明这些不同的E3家族成员可以参与植物抗病免疫反应的各个过程。
本文在简要概括E3泛素连接酶分类的基础上综述了目前E3泛素连接酶参与调控植物抗病害方面研究进展,并对今后研究方向进行了展望,以期对抗病机理解析及抗病品种研发提供新思路。
关键词UPS;泛素化;E3泛素连接酶;PTI;ETI中图分类号:S 432.1文献标识码:ADOI:10.3969/j.issn.05291542.2015.04.001Recent progresses in the regulation mechanism ofE3 ligases in plant disease resistanceYang Jiuxia1,2,Zhang Hao1,2,Wang Zhilong1,Wang Xuli2,Wang Guoliang1,2(1. College of Agronomy, Hunan Agriculture University, Changsha410128, China; 2. State KeyLaboratory for Biology of Plant Diseases and Insect Pests, Institute of Plant Protection,Chinese Academy of Agricultural Sciences, Beijing100193, China)AbstractBiotic stress is an important factor that affects crop production in China. It is also one of the most widely studied areas in plant sciences. Degradation mediated by the ubiquitin proteasome system (UPS) is one of the most sophisticated regulation systems in eukaryotes, which is involved in plant growth and development and in response to abiotic and biotic stresses. E3 ligase is considered as a key enzyme in the UPS due to its specific interactions with its substrates. Based on the differences in structure and function, E3 ligases can be divided into four main classes. In this paper, we review the recent progresses in the regulation mechanism of E3 ligases in plant disease resistance and propose new research directions.Key wordsUPS;ubiquitination;E3 ligase;PTI;ETI植物与病原物在长期相互作用过程中协同进化。
- 1、下载文档前请自行甄别文档内容的完整性,平台不提供额外的编辑、内容补充、找答案等附加服务。
- 2、"仅部分预览"的文档,不可在线预览部分如存在完整性等问题,可反馈申请退款(可完整预览的文档不适用该条件!)。
- 3、如文档侵犯您的权益,请联系客服反馈,我们会尽快为您处理(人工客服工作时间:9:00-18:30)。
MPMI Vol. 21, No. 10, 2008, pp. 1316–1324. doi:10.1094/MPMI-21-10-1316. © 2008 The American Phytopathological SocietyHRT-Mediated Hypersensitive Response and Resistance to Turnip crinkle virus in Arabidopsis Does Not Require the Function of TIP, the Presumed Guardee ProteinRae-Dong Jeong,1 A. C. Chandra-Shekara,1 Aardra Kachroo,1 Daniel F. Klessig,2 and Pradeep Kachroo1 1Department of Plant Pathology, University of Kentucky, Lexington, KY 40514, U.S.A.; 2Boyce Thompson Institutefor Plant Research, Tower Road, Ithaca, NY 40053, U.S.A.Submitted 29 April 2008. Accepted 6 June 2008.The Arabidopsis resistanc e protein HRT rec ognizes the Turnip crinkle virus (TCV) c oat protein (CP) to induc e a hypersensitive response (HR) in the resistant ec otype Di-17. The CP also interac ts with a nuc lear-targeted NAC family of host transcription factors, designated TIP (TCV-interacting protein). Because binding of CP to TIP prevents nuclear localization of TIP, it has been proposed that TIP serves as a guardee for HRT. Here, we have tested the requirement for TIP in HRT-mediated HR and resistanc e by analyzing plants carrying knockout mutation in the TIP gene. Our results show that loss of TIP does not alter HR or resistanc e to TCV. Furthermore, the mutation in TIP neither impaired the salic ylic ac id–mediated induc tion of HRT expression nor the enhanced resistance conferred by overexpression of HRT. Strikingly, the mutation in TIP resulted in inc reased replic ation of TCV and Cucumber mosaic virus, suggesting that TIP may play a role in basal resistance but is not required for HRT-mediated signaling. Additional keywords: defense.Plants have evolved some highly specific mechanisms to re-sist pathogens. The most studied of these involve the deploy-ment of resistance (R) proteins, which generally impart protec-tion against specific races of pathogens carrying corresponding avirulence (Avr) genes (gene-for-gene interactions) (Flor 1971). R proteins are believed to function as direct or indirect receptors for the appropriate Avr proteins. The tomato Pto and the rice Pi-ta proteins were shown to interact with their cognate Avr pro-teins, AvrPto and Avr-Pita, respectively, in a yeast two-hybrid screen (Jia et al. 2000; Scofield et al. 1996; Tang et al. 1996), and the Arabidopsis RPS2 protein formed an in vivo complex with AvrRpt2 (Leister and Katagiri 2000). However, other R-Avr protein pairs have not yielded a detectable interaction (Nimchuk et al. 2000). Thus, it was suggested that R proteins “guard” other plant proteins that are targets of Avr proteins (V an der Biezen and Jones 1998; Innes 2004). Supporting this guard model, RIN4 was shown to physically interact with both the nucleotide binding–leucine rich repeat (NB-LRR) protein RPM1 and its avirulence factors AvrB and AvrRPM1 (Mackey et al. 2002). RIN4 also interacts with RPS2 and its avirulence factor AvrRpt2 (Axtell and Staskawicz 2003; Mackey et al. 2003). Thus, it was proposed that RIN4 is guarded by at least two different R pro-teins; modifications of RIN4 brought about by the various Avr proteins would lead to activation of the respective R proteins. Similarly, cleavage of protein kinase PBS1 by AvrPphB acti-vates the R protein RPS5 (Shao et al. 2003). In addition, Avr2 inhibits the tomato Rcr3 protease required for Cf-2–mediated resistance (Rooney et al. 2005). These data strongly support an indirect mechanism of pathogen recognition by a majority of NB-LRR proteins.Downstream of the recognition event, the signals activated by various Arabidopsis R proteins appear to converge into a small number of pathways (Kachroo et al. 2006; Parker et al. 1996). The pathway activated by toll interleukin 1 receptor-NB-LRR proteins generally requires the EDS1 gene (Parker et al. 1996), while that activated by most coiled coil (CC)-NB-LRR proteins requires the NDR1 gene (Century et al. 1995). However, several CC-NB-LRR R genes, including RPP8, RPP13-Nd, and HRT, as well as RPP7, signal resistance via one or more pathways that are independent of NDR1 (Bittner-Eddy and Beynon et al. 2001; Chandra-Shekara et al. 2004; McDowell et al. 2000). Strikingly, the CC-NB-LRR gene HRT, which confers hypersensitive response (HR) and resistance to Turnip crinkle virus (TCV), is dependent on EDS1 (Chandra-Shekara et al. 2004).Resistance to TCV is also dependent on EDS5, PAD4, and SID2 genes of the salicylic acid (SA) pathway but does not require NPR1, RAR1, SGT1, or the defense hormones jasmonic acid or ethylene (Chandra-Shekara et al. 2004; Kachroo et al. 2000). Exogenous application of SA or the SA analog benzo (1,2,3) thiadiazole-7-carbothioic acid (BTH) restores resis-tance to TCV in SA-deficient Di-17 plants containing the eds1, eds5, or sid2 mutations. However, exogenous application of SA or BTH does not confer enhanced resistance in plants lack-ing HRT or PAD4 (Chandra-Shekara et al. 2004). In contrast to resistance, TCV-induced HR and PR-1 gene expression are not affected by mutations in the SA pathway, suggesting that these phenotypes are independent of SA.In addition to HRT, resistance to TCV requires a recessive allele at a second locus, designated rrt (regulates resistance to TCV) (Chandra-Shekara et al. 2004, 2006; Kachroo et al. 2000). Exogenous application of SA or high endogenous levels of SA can overcome a requirement for rrt and confer resis-tance to TCV in an HRT-dependent manner (Chandra-Shekara et al. 2004, 2006). Transgenic overexpression of HRT can also overcome a requirement for rrt but is not associated with anCorresponding author: P. Kachroo; E-mail: pk62@; Telephone:+1.859.257.7445 ext. 80729; Fax: +1.859.323.1961.Current address for A. C. Chandra-Shekara: DOW Agrosciences, Indian-apolis, IN, U.S.A.*The e-X tra logo stands for “electronic extra” and indicates that a supple-mental figure is available online.e-X tra*1316 / Molecular Plant-Microbe InteractionsVol. 21, No. 10, 2008 / 1317increase in endogenous SA (Chandra-Shekara et al. 2004; Cooley et al. 2000).HR to TCV is initiated upon direct or indirect interaction between HRT and viral coat protein (CP) (Cooley et al. 2000). TCV CP, which acts as an avirulence factor (Zhao et al. 2000), also interacts with a protein belonging to the NAC family of transcription activators, which is designated TIP (TCV-inter-acting protein) (Ren et al. 2000, 2005). Furthermore, transient coexpression of TIP and CP in tobacco cells has shown that TCV CP prevents nuclear localization of TIP (Ren et al. 2005). These observations have prompted the suggestion that HRT guards TIP and is activated when CP retains TIP in the cytoso-lic compartment (Ren et al. 2005; Soosaar et al. 2005).In this report, we have analyzed the requirement of TIP for basal resistance to TCV and Cucumber mosaic virus (CMV) and for HRT-mediated HR and resistance to TCV . We show that TIP is required for basal resistance to CMV but not for HRT-mediated HR and resistance to TCV . Furthermore, absence of TIP does not affect the SA-mediated induction of HRT or alter any of the phenotypes associated with overexpression of HRT . Our results suggest that the interaction between CP and TIP does not govern downstream signaling leading to HR for-mation and resistance. RESULTSA mutation in TIP does not abolish HR or resistance to TCV . To study the role of TIP in HRT-mediated signaling, we examined the SALK insertional database for lines that carry T-DNA insertion within TIP . One line was obtained in which the T-DNA was inserted 15 bases upstream of the translational start site (Fig. 1A). Plants homozygous for the T-DNA inser-tion were obtained and analyzed for TIP expression. At least five different homozygous lines were analyzed, and none of these showed any detectable expression of TIP (Fig. 1B). The TIP knock-out (KO) plants exhibited a normal morphology (Fig. 1C), which was consistent with basal levels of expression of PR-1 and absence of microscopic cell death in tip plants (Fig. 1D and data not shown).Since retention of TIP in the cytosol has been proposed to induce HRT-mediated signaling (Ren et al. 2005), we analyzed the role of TIP in an HRT background. TCV resistant Di-17 (Dijon ecotype) was crossed with wild-type (wt) Columbia ecotype (Col-0) or tip KO (Col-0) plants. The F2 progeny de-rived from these crosses were genotyped for the presence of HRT and tip . The HRT tip plants were morphologically similar to wt plants (Fig. 2A), did not show any visible or microscopic cell death (Fig. 2B), and showed basal expression of various defense genes (Fig. 2C). Upon TCV-inoculation, HRT tip plants developed HR within 3 days postinoculation (dpi), and the size and distribution of these lesions were similar to those in Di-17 or HRT TIP plants (Fig. 2A and B). The HR-forming leaves from HRT tip plants also expressed increased levels of PR-1, PR-2, PR-5, and GST1 genes, similar to those in Di-17 or HRT TIP leaves (Fig. 2C).The role of TIP in HRT -dependent HR formation was fur-ther evaluated in planta by analyzing F1 progeny derived out of a cross between Di-17 or HRT tip with plants overexpress-ing the TCV CP (35S-CP ) (Cooley et al. 2000). Previously, we have shown that TCV CP acts as the avirulence factor, and plants expressing both CP and HRT show massive necrosis resulting in premature death at the seedling stage (Cooley et al. 2000). F1 seeds derived from Di-17 × 35S-CP or HRT tip × 35S-CP crosses (16 each) were sown in soil, and all the F1 seeds showed massive necrosis resulting in eventual death of the F1 seeds (Fig. 2D). Together, these data suggest that a KO mutation in TIP does not impede HR formation or defense-gene induction in response to TCV .To determine if TIP was required for resistance to TCV , we studied the segregation of resistant plants in a TCV-inoculated F2 population derived from Di-17 × Col-0 and Di-17 × tip crosses (Table 1). Unlike HR, which cosegregates with HRT , resistance to TCV is dependent upon the presence of least one copy of HRT and a recessive locus, rrt (Chandra-Shekara et al. 2004; Kachroo et al. 2000). Only HRT -containing plants showed resistance to TCV , and approximately 25% of F2 plants from both crosses developed resistance (Table 1). Fur-thermore, approximately 25% of the HRT tip plants were resis-tant to TCV . This was further confirmed by RNA gel-blot analysis; HRT tip plants scored as susceptible showed presence of viral transcript in the systemic tissues (Fig. 2E). Further, the susceptible HRT tip accumulated a similar amount of viral transcript in the systemic tissues as the Col-0 plants and showed typical stunted and crinkling phenotype (Fig. 2F). By comparison, the HRT tip scored as resistant did not show anyviral transcript in the systemic tissues and developed normalFig. 1. Isolation of a knock-out (KO) mutation in TIP . A, Line diagram showing structure of the TIP gene and the site of T-DNA insertion. Black boxes rep-resent exons and intervening lines represent introns. B, Reverse transcriptase-polymerase chain reaction analysis showing transcript levels of TIP in Di-17, Col-0, and the tip KO plants. The level of β-tubulin was used as an internal control to normalize the amount of cDNA template. C, Morphological phenotype of four-week-old Col-0 and tip plants. D, Microscopy of trypan blue–stained leaves from Col-0 and tip plants showing absence of any microscopic death in tip plants.1318 / Molecular Plant-Microbe Interactionsbolts (Figs. 2E and F). Together, these data suggest that the null mutation in tip does not impair resistance to TCV . TIP is not required for SA-mediated induction of HRT. Exogenous application of SA or BTH upregulates HRT ex-pression and this increase in expression suppresses HR and enhances resistance to TCV in plants containing the RRT allele (Chandra-Shekara et al. 2004). We evaluated HRT tip plants for their abilities to induce SA-mediated HRT expression, sup-press HR due to overexpression of HRT , or induce HRT -derived resistance in response to exogenous application of BTH. As expected, HR to TCV was suppressed in Di-17 plants pre-treated with BTH 2 days prior to inoculation (Fig. 3A). This, in turn, correlated with increased levels of HRTtranscript in theFig. 2. Morphological and molecular phenotypes in HRT tip plants. A, Visible hypersensitive response formation in Turnip crinkle virus (TCV)-inoculated plants at 3 days postinoculation (dpi). All mock-inoculated plants showed absence of any visible lesions, similar to HRT tip . B, Microscopy of trypan blue–stained leaves of mock- or TCV-inoculated plants. The Di-17 and HRT tip plants showed similar extent of cell death. C, Defense-gene expression in indi-cated genotypes at 3 days after mock- or TCV-inoculation. All mock-inoculated plants showed basal level expression of defense genes, similar to HRT tip . Ethidium bromide staining of rRNA was used as a loading control. D, Morphological phenotypes of two-week-old F1 plants derived from the indicated crosses. Arrow indicates F1 plants that remained stunted and eventually aborted. E, Systemic spread of TCV to uninoculated tissue in TCV-inoculated F2 plants derived from a Di-17 × tip cross. RNA was extracted from the uninoculated tissues at 18 dpi and was analyzed for the presence of the viral coat pro-tein (CP ) transcripts. R and S indicate resistance and susceptible phenotypes, respectively. F, Typical morphological phenotypes of TCV-inoculated Di-17, Col-0, and a resistant and a susceptible HRT tip F2 plant derived from a Di-17 × tip cross. The susceptible plants showed crinkling, stunted bolt development,and drooping of bolts. Plants were photographed at 18 dpi.Table 1. Segregation of resistance in Di-17 × Col-0 and Di-17 × tip plants Cross a Number of plants analyzedGenotype b Number of plants obtained HR c R d S e χ2 P fDi-17 × Col-0 148 HRT/–97 + 22 75 0.15 0.69 Di-17 × tip 394 HRT/– tip/ tip 47 + 9 38 0.85 0.35HRT/– TIP/–183 + 40 143 0.96 0.32a The pollen-accepting plant is indicated first and the pollen donor second.bThe genotype at HRT and various mutant loci was determined by cleaved amplified polymorphic sequence analysis. cHR = hypersensitive response. dR = resistant, no disease symptoms. eS = susceptible, disease symptoms include crinkling of leaves and drooping of the bolt. fOne degree of freedom.Vol. 21, No. 10, 2008 / 1319BTH-treated Di-17 plants (Fig. 4B). Similar to the Di-17 plants, exogenous application of BTH on HRT tip plants also sup-pressed HR and increased HRT expression (Fig. 3A and B). Next, we evaluated whether exogenous application of BTH altered susceptibility to TCV in HRT RRT tip plants. As ex-pected, water-treated HRT RRT tip plants were susceptible and accumulated viral transcripts in their systemic tissues (Fig. 3C and D). In comparison, pretreatment with BTH induced resis-tance in approximately 65% of HRT RRT tip plants (Fig. 3C), and this was comparable to the BTH-induced resistance in HRT RRT or HRT RRT sid2 plants (Chandra-Shekara et al. 2004). Exogenous application of BTH on TIP or tip plants did not have any effect in the absence of HRT . Together, these results suggest that SA triggered increase in HRT expression, suppres-sion of HR, and enhanced resistance are independent of TIP. The above results were further confirmed by mobilizing the tip mutation into the HRT ssi2 background, which contains the RRT allele and high endogenous SA (Kachroo et al. 2003, 2004). Previously, we have shown that the increased SA in the mutant ssi2 background lead to increased HRT expression and conferred resistance to TCV in an RRT-independent manner (Chandra-Shekara et al. 2004). The HRT ssi2 tip plants showed stunted morphology (data not shown) and constitutively ex-pressed the PR-1 gene, similar to the HRT ssi2 plants (Fig. 4A). Unlike Di-17 and HRT tip , both HRT ssi2 and HRT ssi2 tip plants expressed high levels of HRT (Fig. 4B), which also correlated with enhanced resistance in these plants; approxi-mately 95 to 98% of HRT ssi2 and HRT ssi2 tip plants were resistant to TCV (Figs. 4C and D). Absence of ssi2 or HRT led to pronounced susceptibility. Analysis of viral transcripts de-tected high levels of TCV in the systemic tissues of susceptible genotypes (HRT tip, ssi2, ssi2 tip , and Col-0) but not in the resistant plants (Di-17, HRT ssi2, or HRT ssi2 tip ) (Fig. 4E). Together, these results suggested that TIP was neither required for the ssi2-induced increase in HRT expression nor ssi2-con-ferred resistance in the HRT RRTbackground.Fig. 3. Hypersensitive response (HR) formation, HRT transcript levels, Turnip crinkle virus (TCV) resistance, and systemic spread of TCV in HRT tip plants. A, Visible HR formation in water- and benzo (1,2,3) thiadiazole-7-carbothioic acid (BTH)-treated plants at 3 days postinoculation (dpi). B, Reverse transcriptase-polymerase chain reaction analyses showing expression of the HRT gene in water- and BTH-treated plants. The level of β-tubulin was used as an internal control to normalize the amount of cDNA template. C, Percentage of TCV-resistant plants obtained after exogenous application of water or BTH. Resistance was analyzed 3 weeks postinoculation. The number of plants tested are indicated above each bar. The HRT tip plants are F3 progeny of a susceptible HRT tip F2 plant that was derived from a Di-17 × tip cross. HRT tip progeny from three different F3 lines were analyzed, and all showed similar results. Asterisk indicates 100% susceptibility. D, Systemic spread of TCV to uninoculated tissue in TCV-inoculated plants. RNA was extracted from the uninoculated tissues at 14 dpi and was analyzed for the presence of the viral coat protein (CP ) transcripts.1320 / Molecular Plant-Microbe InteractionsA mutation in TIP promotes replication of TCV and CMV . Earlier reports showing interaction between CP and TIP (Ren et al. 2000, 2005) and our result that a null mutation in TIP does not impair HRT-mediated HR or resistance to TCV , suggested the possibility that TIP might serve as a host factor that regulates replication or movement, or both, of TCV in sus-ceptible ecotypes like Col-0 and thus affect basal resistance. Therefore, TCV replication, as determined by CP levels, was assessed at 1, 2, and 3 dpi in inoculated leaves of TIP versus tip plants. There were no significant differences in accumula-tion of CP at 1 or 2 dpi (Fig. 5A). However, by 3 dpi tip plants consistently accumulated slightly increased levels of CP protein (Fig. 5A) and its corresponding transcript (data not shown). To assess if TIP plays a role in the systemic movement of TCV or appearance of disease symptoms, transcript levels of CP (Fig. 5B) and symptoms (Fig. 5C) were analyzed in systemic tissues at 7 and 14 dpi. No differences were seen between wt and mu-tant plants. Therefore, TIP does not appear to play a significant role in symptom development to TCV .To determine whether TIP functions in basal resistance to an unrelated viral pathogen, TIP and tip plants were inoculatedwith CMV and were assayed for CMV-CP levels in the inocu-lated leaves and disease symptoms at 1, 2, and 3 dpi. Strik-ingly, significantly higher levels of CMV CP were detected in tip versus TIP leaves throughout the timecourse (Fig. 5D). This correlated with enhanced disease symptoms; in comparison to TIP , tip plants showed severe stunting and drooping of the bolts (Fig. 5E). These data suggest that TIP is required for basal resistance to CMV .To determine if TIP functions in basal- or R gene–mediated resistance to a bacterial pathogen, TIP and tip plants were inoculated with virulent or avirulent (containing AvrRPT2) Pseudomonas syringae pv. tomato. No difference was detected between TIP versus tip in their resistance status to either viru-lent or to avirulent P . syringae pv. tomato (Figs. 5F and G). To-gether, these data suggest that TIP is not required for basal- or RPS2-mediated resistance against P . syringae . DISCUSSIONIn this study, we have evaluated whether TIP plays a role in HRT-mediated HR and resistance to TCV. Earlier work carriedFig. 4. Molecular analysis of HRT ssi2 tip plants. A, PR-1 gene expression in indicated genotypes at 3 days after mock- or Turnip crinkle virus (TCV)-inoculation. Ethidium bromide staining of rRNA was used as a loading control. B, Reverse transcriptase-polymerase chain reaction analyses showing expres-sion of HRT gene in TIP , HRT tip , HRT ssi2, and HRT ssi2 TIP plants. The level of β-tubulin was used as an internal control to normalize the amount of cDNA template. C, Percentage of TCV-resistant plants. Resistance was analyzed 3 weeks postinoculation. The numbers of plants tested are indicated above each bar. Asterisk indicates 100% susceptibility. D, Typical morphological phenotypes of TCV-inoculated plants at 14 days postinoculation (dpi). E, Sys-temic spread of TCV to uninoculated tissue of mock- or TCV-inoculated plants. RNA was extracted at 14 dpi and was analyzed for the presence of the viral coat protein (CP ) transcripts.Vol. 21, No. 10, 2008 / 1321out by Ren and associates (2005) showed that green fluores-cent protein (GFP)-TIP was retained in the cytosol when coin-filtrated with TCV CP. This, together with the finding that TCV CP binds specifically to TIP, led to the assumption that the uneven distribution of TIP in the presence of CP signals the activation of HRT-mediated defense signaling (Ren et al. 2005). An alternate possibility is that HRT-mediated signaling was activated upon loss or degradation of TIP. Precedence for this is provided by the result that P . syringae avirulence protein AvrRPT2 induces degradation of the host-protein RIN4, and loss of RIN4 appears to activate RPS2-mediated resistance against P . syringae (Axtell and Staskawicz 2003; Mackey et al. 2003). Moreover, deletion of RIN4 is lethal in plants contain-ing functional RPS2but has no apparent phenotype in plantsFig. 5. Basal resistance in tip plants. A, Enzyme-linked immunosorbent assay (ELISA) showing levels of Turnip crinkle virus (TCV) coat protein (CP)in the inoculated leaves at 1 to 3 days postinoculation (dpi). B, Systemic spread of TCV to uninoculated tissue in TCV-inoculated plants. RNA was extracted from the uninoculated tissues at 7 and 14 dpi and was analyzed for the presence of the viral CP transcripts. C, Typical morphological phenotypes of TCV-inoculated plants at 14 dpi. D, ELISA showing levels of Cucumber mosaic virus (CMV) CP in the inoculated leaves at 1 to 3 dpi. E, Typical morphological phenotypes of CMV-inoculated plants at 10 dpi. F, Growth of a virulent isolate of Pseudomonas syringae on Col-0 and tip leaves. G, Growth of an avirulent isolate of P . syringae containing AvrRPT2 on Col-0 and tip leaves.lacking RPS2 (Axtell and Staskawicz 2003; Mackey et al. 2003). A similar possibility for the activation of HRT-mediated signaling can be ruled out, since HRT tip plants are wt-like and show basal expression of defense genes. Absence of detectable TIP transcript in tip mutant plants suggests that the KO muta-tion leads to a null phenotype. The normal HR and resistance in HRT tip plants suggest that these defense responses do not require a functional TIP. Therefore, we conclude that neither the binding between TIP and CP nor the cellular distribution of TIP contribute to HRT-mediated HR or resistance to TCV.The possible role of TIP suggested by Ren and associates (2000, 2005) also relied upon the observation that mutations in CP that prevented their interaction with TIP abolished HR to TCV. A likely explanation for breakdown of resistance and HR seen upon inoculation of CP variants, which cannot bind TIP, is that HRT is unable to perceive the altered forms of CP, which results in failure to activate HR and escape of the virus to the systemic tissues.TIP is a member of the NAC family of proteins, which are involved in diverse physiological responses, ranging from de-velopment of plant embryos and flowers (Aida et al. 1997, 1999), lateral root development (Xie et al. 2000), cell division (Kim et al. 2006), defense (Collinge and Boller 2001; Xie et al. 1999), and abiotic stress responses (Fujita et al. 2004; Tran et al. 2004). The Arabidopsis genome contains 105 NAC-like transcription factors (Olsen et al. 2005; Ooka et al. 2003), which show varying levels of homology to TIP. It is, therefore, conceivable that TIP may fulfill redundant functions, and the loss of TIP could be compensated for by other TIP-like NAC proteins. Amino acid comparison and phylogenetic analysis of TIP to other members of the NAC family showed highest simi-larity to AtNTL6 (NAC62, Supplementary Fig. 1A). Further-more, only AtNTL6 showed homology (approximately 60% identity) to the C-terminal domain of TIP that was shown to interact with CP (Ren et al. 2005). However, two-hybrid analy-sis did not detect any interaction between the full-length CP and full-length or C-terminal 150 amino acids from AtNTL6 (data not shown), suggesting that AtNTL6 is unlikely to com-pensate for the loss of TIP.A subclass of the NAC family of transcription factors contain a transmembrane domain at their C-termini and are thereby membrane associated (Kim et al. 2007). The membrane-associ-ated NAC proteins are usually expressed as dormant precursors, which upon controlled proteolytic cleavage are released from the membranes and enter into the nucleus to activate transcrip-tion of target genes. In a recent study conducted by Kim and associates (2007), the authors showed that overexpression of AtNTL6 (NAC62), which shows the highest similarity to TIP, produced a phenotype only if expressed as a C-terminal trun-cated protein. The C-terminal truncation removed the transmem-brane domain of AtNTL6, generating the active form of the tran-scription factor, which was now able to localize to the nucleus and initiate transcription. These results were further substanti-ated by immunolocalization studies, which showed that AtNTL6 localizes to the membranes and could be processed to a smaller molecular weight protein (Kim et al. 2007).Structural analysis of TIP using the ARAMENNON mem-brane protein database predicts a strong ∝-helical transmem-brane domain at the C-terminal region. A comparison of amino acid sequence at the transmembrane domains of TIP and AtNTL6 showed approximately 80% identity, suggesting that, similar to AtNTL6, TIP may localize to membranes. However, the database prediction does not concur with the result that a G FP-TIP fusion protein localizes to the nucleus of tobacco cells (Ren et al. 2005). The altered localization of TIP in these experiments could either be the result of overexpression or misfolding due to presence of GFP.A possible transmembranous nature of TIP and the fact thatpositive-sense viral replicase complexes are assembled on intra-cellular membranes (Ahlquist 2006) suggests the possibilitythat TIP may play a role in replication of CMV, TCV, or both.Increased levels of TCV and CMV CP in the inoculated leavesfurther support this possibility. Furthermore, the NAC familyof transcription factors is known to modulate replication ofWheat dwarf g eminivirus (Xie et al. 1999) and Tomato leaf curl virus (Selth et al. 2005). However, since viruses use dif-ferent intercellular membranes for replication, the relationshipbetween localization of TIP and viral replication remains un-clear. Further examination of the requirement for TIP in CMVresistance could help elucidate the precise role of TIP in plantviral defense.MATERIALS AND METHODSPlant growth conditions, pathogen infections,and genetic analysis.Plants were grown in the MTPS 144 Conviron walk-inchambers (Winnipeg, Canada) at 22°C with 65% relative hu-midity and a 14-h photoperiod. Transcripts synthesized in vitrofrom a cloned cDNA of TCV using T7 RNA polymerase wereused for viral infections (Dempsey et al. 1993; Oh et al. 1995).For inoculations, the viral transcript was suspended in inocula-tion buffer at a concentration of 0.05 μg/μl, and the inocula-tion was performed as described earlier (Dempsey et al. 1993).After viral inoculations, the plants were transferred to a Convi-ron MTR30 reach-in chamber maintained at 22°C with 65%relative humidity and a 14-h photoperiod. Resistance and sus-ceptibility were scored at 7 and 14 to 21 dpi and was confirmedby Northern gel-blot analysis. Susceptible plants showed stuntedgrowth, crinkling of leaves, and drooping of the bolt.Inoculum for CMV strain Fny was prepared as describedpreviously (Hu et al. 1998). Briefly, CMV was maintained ontobacco plants, and extract prepared from infected tobaccoleaves was used as inoculum for Arabidopsis. Susceptible plantsshowed chlorosis on the inoculated leaves, stunted phenotypes,and drooping of the bolt.Bacterial inoculations were carried out by injecting bacterialsolution into the adaxial surface of leaves and two to threeleaves were inoculated each plant. Four leaf disks were har-vested from pathogen- or mock-inoculated leaves at 3 dpi andground in 10 mM MgCl2, and the bacterial numbers were deter-mined by titration. The bacterial numbers ± standard deviation(n = 4) were presented as colony-forming units (CFU) per unitof leaf area (25 mm2). The experiment was independentlyperformed twice with similar results.Crosses were performed by pollinating flowers of Di-17 andHRT ssi2 plants with pollen from tip plants. The genotypes of the F2 plants at the TIP locus were determined by conducting polymerase chain reaction (PCR) analysis using left border and gene-specific primers (Fwd: G G TCCAAAG G ACAAAA GA AGAG; Rev: CGAATTCTCAAAGTCTCACGC). Chemical treatment of plants.Three-week-old plants were sprayed or subirrigated with asolution of 500 μM SA or 100 μM BTH. Control plants weretreated with water, and two days after treatment, three leavesper plant were inoculated with TCV RNA.RNA extraction andreverse transcriptase (RT)-PCR analysis.Small-scale RNA extractions was performed with TRIzolreagent (Invitrogen, Rockville, MD, U.S.A.), according to themanufacturer’s instructions. RNA gel-blot analysis and synthe-sis of random primed probes were performed as described ear-1322 / Molecular Plant-Microbe Interactions。
