Distinct signaling pathways after higher or n in three closely related human lymphoblast cell lines
癌症肿瘤学:死亡结构沉默子在急性淋巴细胞白血病中的研究进展

死亡结构沉默子在急性淋巴细胞白血病中的研究进展吴星综述;徐之良校审(武汉大学人民医院儿科,湖北武汉430060)【摘要】急性淋巴细胞白血病(ALL)是儿童最常见的恶性肿瘤,其发生与细胞凋亡相关基因(Bcl-2,NFκB)的表达及调控异常有关。
死亡结构域沉默子(silencer of death domains。
SODD)是新近发现的一个凋亡调控基因,是细胞表面死亡受体配体凋亡途径中的主要负调节蛋白。
在一些人类肿瘤细胞株中SODD呈持续高表达状态,抑制细胞的凋亡,导致化疗耐药。
在急性淋巴细胞白血病的发生发展治疗及预后具有重要意义【关键词】 ALL ;SODD; Bcl-2;NFκ B儿童急性淋巴细胞白血病(acute lymphocytic leukemia,ALL)是当今疗效最好、治愈率最高的恶性肿瘤性疾病之一,完全缓解(CR)率可达95%以上,5年以上无病生存(DFS)率可达80%~90%,治愈率可达到70%,但仍有30%患儿对化疗反应不佳,耐药性的产生是这类患儿化疗失败的主要原因[1]。
这类难治性白血病因自身特性难达完全缓解( CR) 和长期无病生存,是目前白血病研究的重点和难点。
在白血病及其他癌细胞中,化疗耐药性增加与细胞凋亡敏感性降低之间的关系已引起注意。
凋亡是一个受调节的过程,凋亡调节基因的分子改变可能导致细胞对凋亡敏感性的改变,进而影响细胞对化疗药物的反应[2]。
因此,诱导凋亡是抗肿瘤治疗的重要手段。
白血病细胞不仅增殖和分化异常,同时存在凋亡受抑,而且凋亡的抑制与恶性肿瘤的发生发展以及耐药有密切关系[3]。
死亡结构域沉默子(silencer of death domains,SODD)是新近发现的一个凋亡调控基因,定位于8q12,属于BAG蛋白家族成员,又名BAG-4,广泛表达于多种细胞的胞浆内[4],SODD特异识别并结合于TNFR1、HSP/HSC70、DR3及Bcl-2胞浆段死亡结构域(death domains, DD),目前认为SODD是TNFR1介导的凋亡信号途径中的关键负调节蛋白,在肿瘤细胞异常增殖及逃避凋亡中发挥了重要作用[5]。
高迁移率族蛋白B1( HMGB1)和肿瘤

高迁移率族蛋白B1( HMGB1)和肿瘤章俊强;梅晓冬【摘要】HMGB1是一种非常古老的核蛋白,在悠久的进化史中具有高度保守的特点,哺乳动物中HMGB1具有99%的同源特质[1].HMGB1作为一种核蛋白可以稳定核小体,调节多种基因的转录,同时作为一种炎症因子,它可被多种炎症细胞释放或分泌,参与多种疾病的发生,发展和转归,如脓毒症,缺血-再灌注损伤,关节炎和肿瘤.作为一种危险警告因子,HMGB1在无菌性损伤性炎症和感染性炎症中作用机制和生物学效应已被国内外学者系统阐述[2],而HMGB1在肿瘤发生发展中的作用为现今研究的热点,其作用机制尚不甚明确.本文就目前HMGB1和肿瘤的相关研究做一简要综述.【期刊名称】《临床肺科杂志》【年(卷),期】2011(016)012【总页数】2页(P1930-1931)【作者】章俊强;梅晓冬【作者单位】230000 安徽合肥,安徽医科大学附属省立医院呼吸科;230000 安徽合肥,安徽医科大学附属省立医院呼吸科【正文语种】中文HMGB1是一种非常古老的核蛋白,在悠久的进化史中具有高度保守的特点,哺乳动物中HMGB1具有99%的同源特质[1]。
HMGB1作为一种核蛋白可以稳定核小体,调节多种基因的转录,同时作为一种炎症因子,它可被多种炎症细胞释放或分泌,参与多种疾病的发生,发展和转归,如脓毒症,缺血-再灌注损伤,关节炎和肿瘤。
作为一种危险警告因子,HMGB1在无菌性损伤性炎症和感染性炎症中作用机制和生物学效应已被国内外学者系统阐述[2],而 HMGB1在肿瘤发生发展中的作用为现今研究的热点,其作用机制尚不甚明确。
本文就目前HMGB1和肿瘤的相关研究做一简要综述。
一、HMGB1的结构HMG超家族成员均含有能和DNA结合的同源性碱性结构域,即HMG盒(HMG boxes),分子量约25-30千道达尔。
人类HMGB1基因由六个外显子组成,定位于人类染色体13q12,编码一条215个氨基酸序列的单链多肽。
Jagged-1抑制小鼠胚胎干细胞分化为造血干祖细胞

January 2021Vol.41 No.12021年 1月 第41卷第1期基础医学与临床Basic & Clinical Medicine文章编号:1001-6325 ( 2021 ) 01-0055-07研究论文Jagged-1抑制小鼠胚胎干细胞分化为造血干/祖细胞陈日玲1,郑伟荣2,谭 霖3,刘东强3,史 惠3,陈启康1*收稿日期:2019-05-16 修回日期:2020-03-30基金项目:湛江市科技竞争性项目(2014A01022)*通信作者(corresponding author ) : chen.qikang@ (1.广东医科大学附属顺德妇女儿童医院儿科,广东顺德528300; 2.泉州市第一医院儿科,福建泉州362000;3.广东医科大学附属医院儿童医学中心,广东湛江524023)摘要:目的探讨Notch 信号通路在小鼠胚胎干细胞(ESC )分化为造血干/祖细胞(HSC/HPC )中的作用。
方法1)体外培养小鼠拟胚体细胞(EBs ),使用Jagged-1蛋白活化Notch 信号通路及DAPT (7-分泌酶抑制剂)抑制Notch信号通路。
实验分为拟胚体细胞复苏组(EB 组)、对照组、Jagged-1组、DAPT 组和Jagged-1-DAPT 组。
2)流式细胞计量术检测ESC 特异性表型和HSC/HPC 特异性表型的表达。
3)实时定量PCR 检测各组Notch 信号通路基因、小 鼠ESC 表型基因和HSC/HPC 表型基因的表达。
结果1) Jagged-1组细胞胚胎干细胞数较对照组和Jagged-1-DAPT组明显增多(P <0. 05) , 2)Jagged-1-DAPT 组分化的 HSC/HPC 数较 control 组及 Jagged-1 组明显增多(P <0. 05);3)Jagged-1 组 Notch1、Notch2、Notch4 mRNA 表达量较对照组明显升高(P <0. 05) ;4) DAPT 组和 Jagged-1-DAPT 组中 Notch1、Notch4 mRNA 表达量较对照组降低(P <0.05)。
MDH1介导苹果酸-天冬氨酸NADH穿梭维持胎儿肝造血干细胞的活性水平。(Sonarsignal)
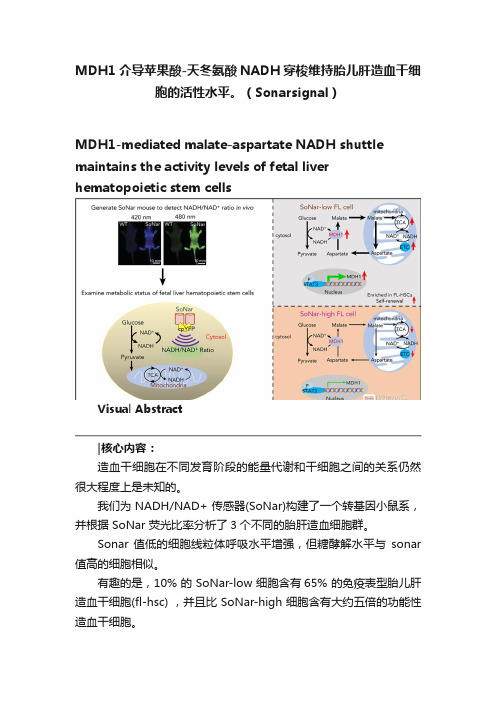
MDH1介导苹果酸-天冬氨酸NADH穿梭维持胎儿肝造血干细胞的活性水平。
(Sonarsignal)MDH1-mediated malate-aspartate NADH shuttle maintains the activity levels of fetal liver hematopoietic stem cellsVisual Abstract造血干细胞在不同发育阶段的能量代谢和干细胞之间的关系仍然很大程度上是未知的。
我们为 NADH/NAD+ 传感器(SoNar)构建了一个转基因小鼠系,并根据 SoNar 荧光比率分析了3个不同的胎肝造血细胞群。
Sonar 值低的细胞线粒体呼吸水平增强,但糖酵解水平与sonar 值高的细胞相似。
有趣的是,10% 的 SoNar-low 细胞含有65% 的免疫表型胎儿肝造血干细胞(fl-hsc) ,并且比 SoNar-high 细胞含有大约五倍的功能性造血干细胞。
Sonar 能够敏感地监测体内外造血干细胞能量代谢的动态变化。
STAT3通过反式激活 MDH1维持苹果酸-天冬氨酸 NADH 的穿梭活性和 HSC 的自我更新和分化。
我们揭示了一个意想不到的fl-hsc 代谢程序,为造血干细胞或其他类型干细胞的代谢研究提供了一个强有力的遗传工具。
原文摘要:The connections between energy metabolism and stemness of hematopoietic stem cells (HSCs) at different developmental stages remain largely unknown.We generated a transgenic mouse line for the genetically encoded NADH/NAD+ sensor (SoNar) and demonstrate that there are 3 distinct fetal liver hematopoietic cell populations according to the ratios of SoNar fluorescence.SoNar-low cells had an enhanced level of mitochondrial respiration but a glycolytic level similar to that of SoNar-high cells. Interestingly, 10% of SoNar-low cells were enriched for 65% of total immunophenotypic fetal liver HSCs (FL-HSCs) and contained approximately fivefold more functional HSCs than their SoNar-high counterparts.SoNar was able to monitor sensitively the dynamic changes of energy metabolism in HSCs both in vitro and in vivo.Mechanistically, STAT3 transactivated MDH1 to sustain the malate-aspartate NADH shuttle activity and HSC self-renewal and differentiation.We reveal an unexpected metabolic program of FL-HSCs and provide a powerful genetic tool for metabolic studies of HSCs or other types of stem cells.Key Points•FL-HSCs mainly use oxidative phosphorylation but with normal glycolysis, as indicated by a highly responsive NADH/NAD+ sensor.•FL-HSC activities are tightly regulated by the STAT3/MDH1-mediated malate-aspartate NADH shuttle.Subjects:Hematopoiesis and Stem CellsTopics:aspartate, fetus, fluorescence, liver, malates, mice, transgenic, mitochondria, nicotinamide adenine dinucleotide (nad), stat3 protein, hematopoietic stem cellsIntroductionHematopoietic stem cells (HSCs) originate from the aorta-gonad-mesonephros region1 and migrate into the fetal liver (FL) and undergo dramatic expansion,2,3 gradually localizing to and residing in the bone marrow niche after birth.4HSCs can self-renew to maintain the stem cell pool and generate all downstream progenitors and terminally differentiate into multiple lineages.5,6Increasing evidence indicates that the metabolic state is tightly connected to HSC activity.7-9Adult HSCs preferentially undergo glycolysis, rather than oxidative phosphorylation, in the hypoxic niche,7,10,11 which is extensively regulated by several signaling pathways, including HIF1A,12 MYC,13 PDK,14 DLK-GTL2,15 and vitamin A–retinoic acid signaling.16We have also shown that both murine and human HSCs adopt a glycolytic metabolic profile under certain conditions andthat this profile is fine-tuned by MEIS1/PBX1/HOXA9/HIF1A signaling pathways.17-19Interestingly, recent studies have suggested that adult HSCs also have high mitochondrial mass and enhanced dye efflux but possess limited respiratory and turnover capacity,20 which indicates that mitochondria are likely required for the function of adult HSCs, as evidenced by the fact that FOXO3 serves as a regulator to couple mitochondrial metabolism with HSC homeostasis.21The metabolic profiles of FL-HSCs and the effects of metabolism on HSC function, however, remain largely unknown.FL-HSCs undergo rapid division/expansion, conceivably through an increased demand on energy sources compared with that needed by adult HSCs, which are usually maintained in a relatively quiescent state.It is also possible that distinct microenvironments in different hematopoietic organs may affect the metabolism of HSCs.Interestingly, a recent report showed that loss of Rieske iron-sulfur protein, a mitochondrial complex III subunit, impairs the quiescent status of adult HSCs and the differentiation capacity of FL-HSCs.22FL-HSCs seem to have increased expression levels of many mitochondrial respiration–related genes, although whether metabolic status determines the cell fate of FL-HSCs remains unknown.23Results from previous studies indicate that mitochondrial activity may play a role in HSCs in the FL stage, although the detailed metabolic profiles and their underlying mechanisms await further investigation.Because of limitations in the availability of HSCs, moststudies related to the nutrient metabolism of HSCs have depended heavily on flow cytometric analysis with MitoTracker dyes, TMRE, and DCFDA to determine mitochondrial mass, membrane potential, and ROS level, respectively.Improved techniques have been used to measure several metabolic features of HSCs, such as oxygen consumption and lactate generation9,24 ; however, these studies may not directly reflect the true extent of glycolysis, oxidative phosphorylation, or other metabolic processes in HSCs.Recent studies have provided interesting evidence showing that it is feasible to perform a metabolomic analysis with fewer than 104 HSCs to explore the metabolic networks of different types of nutrients.25Nevertheless, it remains difficult to detect all of the metabolites sensitively with a limited number of HSCs using conventional metabolomic analysis.Few tools are available for real-time imaging of metabolic states in live HSCs, either in vitro or in vivo. Therefore, alternative approaches, such as metabolite biosensors, are required for the direct, precise, and real-time detection of subtle changes in nutrient metabolism in HSCs.Recently, we developed a highly responsive NADH/NAD+ sensor, called SoNar,26 which was designed by inserting cpYFP into the NAD(H)-binding domain of T-Rex.SoNar shows distinct fluorescence responses to NADH and NAD+.Inside the cell under physiological conditions, the total intracellular pool of NAD+ and NADH in the range of hundreds of micromolars27-30 far exceeds the dissociation constants of SoNar for NAD+ (K d, 5.0 μM) and NADH (K d, 0.2 μM);thus, the sensor would be occupied by either NAD+ or NADH molecules, and its steady-state fluorescence would report the NAD+/NADH ratio rather than the absolute concentrations of either of the 2 nucleotides according to equilibrium thermodynamics.26In addition, SoNar fluorescence is intrinsically ratiometric (比率计), with 2 excitation wavelengths, and its fluorescence excited at 420 (or 405) and 485 nm shows opposing responses to ligand binding.26,31This ratiometric property of a sensor is highly desired for quantitative imaging in live cells and in vivo,32,33 because it eliminates the differences in instrumental efficiency, environmental effects, and probe concentration, enabling it to be widely used in different biological samples.The SoNar sensor has a 15-fold (or 1500%) dynamic range, enabling us to measure the cytosolic NAD+/NADH ratio from 0.8 to 2000.26,31,34Interestingly, SoNar has many desirable properties that make it an ideal sensor; it has a rapid response, high sensitivity, intense fluorescence, and large dynamic range, and it is capable of reporting subtle perturbations in many pathways affecting energy metabolism, including glycolysis and mitochondrial respiration.We generated SoNar transgenic mice and examined the metabolic profiles of FL-HSCs and their contributions/connections to cell fate determinations, as well as the underlying mechanisms governing FL-HSC function.参考文献:/10.1182/blood.2019003940HEMATOPOIESIS AND STEM CELLS| JULY 30, 2020----------------------------------------------------------------------------------------------------------------------------MethodsMiceSoNar DNA consists of the sequence of cpYFP, truncated T-Rex (78-211), and the linkers between them.26 Its coding gene (1.2 kbp) is much smaller than those of the first-generation NADH sensors Peredox (2.8 kbp) and Frex35 (1.8 kbp). For transgenic studies, smaller is better for the expression of the sensor in cells and in vivo.31 To generate SoNar transgenic mice, SoNar DNA was cloned into the pCAG vector with chicken β-actinpromoter. The targeting construct was linearized, purified, and microinjected into FVB blastocysts. SoNar DNA was randomly incorporated into the genome and determined by polymerase chain reaction (PCR) assay. Messenger RNA (mRNA) and protein expressions of SoNar in different tissues of SoNar mice were further evaluated by both reverse transcription PCR (RT-PCR) and fluorescence microscopy. The resulting chimeric mice were bred with FVB mice to obtain germ line transmission. These mice were next backcrossed with a C57BL/6 CD45.2 background, and germ line transmission was checked by PCR and flow cytometry. Heterozygote transgenic SoNar mice were used for most of the experiments in the current study. C57BL/6 CD45.2 mice were purchased from the Shanghai SLAC Laboratory Animal Co., Ltd. CD45.1 mice were provided by Dr Jiang Zhu at Shanghai Jiao Tong University School of Medicine. All animal experiments were conducted according to the Guidelines for Animal Care at Shanghai Jiao Tong University School of Medicine. All these materials, including SoNar sensor, are available upon request.In vivo imaging of SoNar transgenic miceGenotyping, mRNA expression, and histology of SoNar transgenic miceCompetitive reconstitution assayMetabolic imaging and quantification of cytosolicNAD+/NADH ratio in living cellsReal-time metabolic imaging in the BM nicheFlow cytometryUltrahigh-performance LC–qTOF–MS analysisMicroarray and quantitative RT-PCRMetabolic analysisOxygen consumption rate (OCR) and extracellularacidification rate were determined in CD45.2+ SoNar-high and -low FL hematopoietic cells with the XF Cell Mito Stress Test Kit (Seahorse #103015-100) and XF Glycolysis Stress Test Kit (Seahorse #103020-100) according to the manufacturer’s instructions using a Seahorse XF96 analyzer. In brief, for the OCR analysis, 3 × 105 SoNar-high and -low FL hematopoietic cells were incubated in the 37°C carbon dioxide–free incubator in 175 μL of assay medium (XF Base Medium with 2 mM of glutamine, 1 mM of pyruvate, and 10 mM of glucose [pH, 7.4]; 37°C); 1.5 μM of oligomycin, 2 μM of FCCP, and 0.5 μM of rotenone/antimycin A were loaded in injection ports A, B, and C, respectively. For the detection of extracellular acidification rate, 3 × 105 CD45.2+ SoNar-high and -low FL hematopoietic cells were incubated in the 37°C carbon dioxide–free incuba tor in 175 μL of assay medium (XF Base Medium with 1 mM of glutamine [pH, 7.4]; 37°C); 10 mM of glucose, 1.5 μM of oligomycin, and 100 mM of 2-DG were loaded into injection ports A, B, and C, respectively. ATP level was analyzed using the ATP Bioluminescence Assay Kit HS II (Roche) according to the manufacturer’s protocol, and data were normalized to cell count. T o analyze the mitochondrial DNA (mtDNA) copy numbers, total genomic DNA was extracted from the indicated cells for comparing the copies of the mitochondrial-specific mt-ND4 gene with those of the nuclear B2m gene. The primer sequences used are shown in supplemental Table 1.For intracellular and extracellular pyruvate and lactate assays, the extracts were prepared from 5 × 106 CD45.2+ FL hematopoiet ic cells using 300 μL of ice-cold 0.5-M perchloric acid for each sample. Extracts were centrifuged at 10 000 g for 5 minutes at 4°C, and the supernatant was neutralized with 5 M ofKOH and centrifuged at 10 000 g for 5 minutes at 4°C. The supernatant was removed for assay. For the pyruvate assay, 180 μL of assay buffer (100 mM of potassium chloride [pH, 6.7], 1 mM of EDTA, 0.1% bovine serum albumin, 10 μM of flavin adenine dinucleotide, 0.2 mM of thiamine pyrophosphate, 0.5 U of pyruvate oxidase, 0.2 U of h orseradish peroxidase, and 50 μM of AmplexRed) was added to a 96-well plate containing 20 μL of the cell extract or medium containing extracellular pyruvate. Changes in fluorescence were measured every 30 seconds for 15 minutes at 37°C by a Synergy 2 Multi-Mode Microplate Reader with an excitation filter of 530 BP 40 nm and emission filter of 590 BP 35 nm at 37°C. Calibration experiments were performed with 20 μL of pyruvate standards (0, 10, 20, 40, 60, 100, and 200 μM per well). For the lactate assay, 180μL of assay buffer (PBS [pH, 7.4], 0.1% bovine serum albumin, 500 μM of NAD+, 0.5 U of lactate dehydrogenase (LDH), 0.2 U of diaphorase, and 10 μM of resazurin) was added to a 96-well plate containing 20 μL of the cell extract or medium containing extracellular lactate. Changes in fluorescence were measured every 30 seconds for 15 minutes at 37°C by a Synergy 2 Multi-Mode Microplate Reader with an excitation filter of 540 BP 25 nm and emission filter of 590 BP 35 nm at 37°C. Calibration experiments were performed with 20 μL of lactate standards (0, 10, 20, 40, 60, 100, and 200 μM per well). All samples were diluted to fit within the range of the standard curve and run in triplicate. For the evaluation of NADH/NAD+ ratios by a biochemical assay, 1 million SoNar-high and -low FL hematopoietic cells were sorted from E14.5 FLs and subjected to measurement of NADH/NAD+ level using a commercially available NADH/NAD+ assay kit (Sigma #MAK037) according to the manufacturer’s instructions.Immunoblot analysisSingle cell colony formingThe single CD45.2+ SoNar-high or -low FL hematopoietic cell was freshly isolated and plated onto a 35-mm poly-D-lysine hydrobromide-coated glass-bottom dish (Cellvis) and cultured in Stemspan serum-free medium (Stemcell Technologies) containing 10 ng/mL of murine SCF and 10 ng/mL of murine TPO (Peprotech) for 96 hours. The ratios of daughter cells derived from a single parent cell were recorded with excitation at 405 and 488 nm using Nikon A1 confocal microscopy and analyzed with Image J software.Luciferase reporter assaysThe luciferase reporter vector pGL4.27 containing the Mdh1 promoter was constructed to identify transcriptional activation of Mdh1 by STAT3. Indicated doses of pLVX-Stat3 (or negative control vector) plasmid along with pGL4.27-mdh1 promoter vector were cotransfected into 293T cells. Luciferase activities were measured according to the manufacturer’s instructions (Promega #E1910) by using a luciferase reporter system (GloMax Multi Instrument) 24 hours after transfection. ChIP assaysChromatin immunoprecipitation (ChIP) assays were performed using the ChIP Assay Kit (Beyotime #P2078). Briefly, 293T cells were overexpressed with pGL4.27-mdh1 promoter vector and STAT3 (with Strep II tag) crosslinked with 1% formaldehyde (S igma) at 37°C for 10 minutes, and precleared DNA was then used for immunoprecipitation with 4 mL of anti–Strep II antibody (Genescript) or rabbit control immunoglobulin G (CST) at 4°C overnight. For the sample input, 1% of the sonicated pre-cleared DNA was purified at the same time withthe precipitated immune complex. The ChIP samples were purified by the Gel and PCR Clean Up Kit (Necleospin). The STAT3-binding sequence was amplified by semiquantitative PCR using primers specific for the Mdh1 promoter region as listed in supplemental Table 1.Methylation-specific PCR assayGenomic DNA was extracted from 500 000 CD45.2+ SoNar-high and -low FL hematopoietic cells using a DNA extraction kit (Generay Biotech #GK0122). The promoter methylation status of MDH1 was determined by sodium bisulfate to convert unmethylated (but not methylated) cytosine to uracil, followed by analysis by methylation-specific PCR to amplify specifically either methylated or unmethylated DNA using the Zymoresearch kit (EZ DNA Methylation-Direct Kit D5020) according to the manufacturer’s instruction. The methylation-specific PCR primers are listed in supplemental Table 1.Statistical analysisStatistical analysis was performed using GraphPad and SPSS software (version 19.0). Data are represe nted as mean ± standard error of the mean. n represents the number of independent experiments or the number of cells or mice per group from independent experiments. All experiments were performed independently 3 to 5 times. Data were analyzed with a Student t test (2 tailed), 1-way analysis of variance with Tukey’s multiple comparison test, or 2-way analysis of variance with Sidak’s multiple comparison test according to the experimental design, and statistical significance was set at P < .05.Establishment of pan-tissue SoNar transgenic mice.Figure 2.SoNar indicates metabolically distinct populations of FL hematopoietic cells.Figure 3.SoNar-low FL hematopoietic cells exhibit similar glycolytic but enhanced mitochondrial activity compared with SoNar-high cells.Figure 4.Functional HSCs are enriched in SoNar-low FLhematopoietic cells.Figure 5.FL-HSCs respond differently to AOA stimulation in the BM niche compared with adult HSCs.Figure 6.MDH1 enhances the malate-aspartate NADH shuttle and decreases NADH/NAD+ level in SoNar-low FL-HSCs.Figure 7.STAT3 transactivates Mdh1 expression to maintain FL-HSC activities.。
泛素化组学英文

泛素化组学英文全文共四篇示例,供读者参考第一篇示例:The development of high-throughput mass spectrometry techniques has greatly facilitated ubiquitinomics research. Mass spectrometry allows for the rapid identification and quantification of thousands of proteins in a single experiment. By combining mass spectrometry with specific ubiquitin-affinity purification methods, researchers can isolate ubiquitinated proteins from a cell lysate and analyze their ubiquitin modification sites.第二篇示例:One of the key techniques used in ubiquitinomics is mass spectrometry, a powerful analytical tool that allows for the identification and quantification of proteins in complex samples. By coupling mass spectrometry with advanced proteomics methodologies, researchers can uncover the intricacies of ubiquitin signaling pathways and elucidate the functional consequences of protein ubiquitination.第三篇示例:In cancer research, ubiquitin proteomics has been used to identify novel biomarkers for early detection and prognosis of cancer. By profiling the ubiquitinome of cancer cells, researchers have been able to identify specific ubiquitinated proteins that are dysregulated in cancer and may serve as potential targets for therapy. In neurodegenerative diseases, ubiquitin proteomics has shed light on the role of aberrant protein aggregation and clearance mechanisms in disease progression, offering new insights into potential therapeutic strategies.第四篇示例:The study of ubiquitinomics is challenging due to the dynamic and reversible nature of ubiquitination. Ubiquitin is rapidly added and removed from target proteins in response to various stimuli, making it difficult to capture the full landscape of ubiquitinated proteins in a cell. Additionally, ubiquitination can occur on multiple lysine residues within a protein, leading to a complex pattern of ubiquitin modifications that can be difficult to analyze.。
药物化学进展全解

任何学科的形成和发展,都与当时的科学技术水平。
经济建设要求以及相关学科的促进分不开的。
早期的药物化学以化学学科为主导,包括天然和合成药物的性质、制备方法和质量检测等内容。
随着科技发展,天然药物化学、合成药物化学和药物分析等学科相继建立。
现代药物化学是化学和生物学科相互渗透的综合性学科。
主要任务是创制新药、发现具有进一步研究开发前景的先导物。
研究内容主要有:基于生命科学研究揭示的药物作用靶点(受体、酶、离子通道、核酸等),参考天然配体或底物的结构特征。
设计药物新分子,以期发现选择性地作用于耙点的新药;通过各种途径和技术寻找先导物,如内源性活性物质的发掘,天然有效成分或现有药物的结构改造和优化,活性代谢物的发现等,其次计算机在药物研究中的应用日益广泛,计算机辅助药物设计(CADD)和构效关系也是药物化学的研究内容。
如今信息科学迅猛发展,利用各种数据库和信息技术,比如Reaxys,可广泛收集药物化学的文献资料,有利于扩展思路,开拓视野,丰富药物化学的内容。
药物化学既要研究化学药物的结构、性质和变化规律,又要了解药物用于人体的生理生化效应和毒副反应以及构效关系才能完成它的任务。
有人比喻,如果现代药物化学是一只鼎,那么支撑这只鼎的分别是化学、生物学科和计算机技术。
创制新药是涉及多学科。
多环节的探索性系统工程。
是集体研究的成果,基于药物化学首先要发现先导物,为后续学科研究提供物质基础,在研究过程中起着十分重要的作用,因此药物化学在药学科学领域处于带头学科的地位。
Burger的名著《药物化学》现已改为(药物化学与药物发现)(Medicinal Chemistry and Drug Discovery),以突出药物化学的任务是创制新药和发现先导物,从而达到促进医药工业发展,保护人类健康的目的。
80年代初诺氟沙星用于临床后医学教|育网搜集整理,迅速掀起喹诺酮类抗菌药的研究热潮,相继合成了一系列抗菌药物,这类抗菌药和一些新抗生素的问世,认为是合成抗菌药发展史上的重要里程碑。
免疫细胞学英语

IntroductionImmunocytology, a specialized branch of cellular biology, delves into the intricate world of immune cells, their structure, function, and interactions within the complex network of the immune system. These cells, often referred to as leukocytes or white blood cells, play a pivotal role in defending our bodies against a myriad of pathogens, foreign substances, and even aberrant cells that arise from within. This essay provides a comprehensive, high-quality analysis of immunocytology, examining various aspects of immune cells, including their classification, development, activation mechanisms, effector functions, and the emerging therapeutic applications that harness their power.Classification and Development of Immune CellsThe immune system is composed of a diverse array of cell types, each with distinct roles and characteristics. Broadly, immune cells can be classified into two main categories: innate immune cells and adaptive immune cells. Innate immune cells, such as neutrophils, monocytes/macrophages, dendritic cells (DCs), natural killer (NK) cells, and mast cells, provide the first line of defense against invading pathogens. They recognize conserved pathogen-associated molecular patterns (PAMPs) via pattern recognition receptors (PRRs) and respond rapidly but non-specifically.In contrast, adaptive immune cells, comprising B cells and T cells, offer a highly specific, long-lasting defense. B cells produce antibodies, while T cells execute cytotoxic or helper functions depending on their subsets (CD4+ T helper cells, CD8+ cytotoxic T cells, regulatory T cells, etc.). The development of these immune cells occurs primarily in the bone marrow (for B cells and myeloid cells) and the thymus (for T cells). A tightly regulated process involving hematopoietic stem cell (HSC) differentiation, gene rearrangements, positive and negative selection, and maturation ensures the generation of a diverse and self-tolerant immune repertoire.Activation Mechanisms and Signal TransductionThe activation of immune cells is a finely orchestrated process triggeredby the recognition of antigens or danger signals. For innate immune cells, PRR engagement initiates signaling cascades involving adaptor proteins like MyD88 and TRIF, leading to the activation of transcription factors such as NF-κB and IRF3/7, which drive the expression of pro-inflammatory cytokines, chemokines, and antimicrobial peptides.Adaptive immune cells, particularly T and B cells, require antigen recognition through their unique antigen receptors (TCR for T cells, BCR for B cells). This interaction, when accompanied by appropriate co-stimulatory signals, activates intracellular signaling pathways involving kinases such as Lck, Zap70, and PI3K, ultimately leading to the activation of transcription factors like NF-κB, AP-1, and NFAT. These transcription factors orchestrate the expression of genes involved in cell proliferation, differentiation, and effector function.Effector Functions of Immune CellsInnate immune cells execute various effector functions to combat infections. Neutrophils phagocytose and kill pathogens through the release of reactive oxygen species (ROS) and granule contents. Monocytes/macrophages display similar phagocytic abilities and also present antigens to T cells, produce inflammatory cytokines, and participate in tissue repair. DCs are professional antigen-presenting cells (APCs) that capture, process, and present antigens to naïve T cells, initiating adaptive immune responses. NK cells directly eliminate virus-infected or transformed cells without prior sensitization, relying on the balance of activating and inhibitory receptors interacting with cell surface ligands.Adaptive immune cells contribute to immunity through antibody production and cell-mediated responses. B cells differentiate into plasma cells that secrete antibodies, which neutralize pathogens, opsonize them for enhanced phagocytosis, or activate complement. T cells, upon activation, differentiate into effector subsets: CD4+ T helper cells (Th1, Th2, Th17, Tfh, etc.) that provide help to other immune cells, and CD8+ cytotoxic T cells that directlykill infected or transformed cells. Regulatory T cells (Tregs) maintain immune homeostasis by suppressing excessive immune responses and preventing autoimmunity.Emerging Therapeutic ApplicationsRecent advances in immunocytology have paved the way for innovative therapeutic strategies targeting immune cells. Cancer immunotherapy, for instance, has revolutionized cancer treatment, with approaches such as immune checkpoint inhibitors (e.g., anti-PD-1, anti-CTLA-4 antibodies) that unleash the cytotoxic potential of T cells suppressed by tumor microenvironment. Chimeric antigen receptor (CAR)-T cell therapy involves engineering patient's T cells to express CARs, enabling targeted recognition and destruction of tumor cells. Additionally, adoptive transfer of ex vivo expanded or genetically modified NK cells is being explored for cancer therapy due to their inherent ability to recognize and kill malignant cells.In autoimmune diseases and transplant rejection, therapies targeting immune cells aim to suppress pathogenic immune responses. These include the use of monoclonal antibodies against pro-inflammatory cytokines or their receptors, T cell-depleting agents, and Treg-based therapies. Moreover, modulation of innate immune cells, particularly DCs, through targeted delivery of antigens or immunomodulatory molecules, holds promise for the induction of tolerance in autoimmune and allergic disorders.ConclusionImmunocytology offers a rich tapestry of knowledge, elucidating the complexities of immune cells and their integral role in maintaining host defense. From the classification and development of these cells to the intricate mechanisms governing their activation and effector functions, understanding immunocytology is crucial for both fundamental biological insights and translational applications. The ongoing advancements in this field continue to fuel the development of novel therapeutic strategies that harness the power of immune cells, transforming the landscape of modern medicine in the fight againstinfectious diseases, cancer, and autoimmune disorders.。
JBC-细胞核蛋白提取参考文献

Nerve Growth Factor Stimulates Proliferation and Survival of Human Breast Cancer Cells through Two Distinct Signaling Pathways*Received for publication,November 20,2000,and in revised form,February 14,2001Published,JBC Papers in Press,February 28,2001,DOI 10.1074/jbc.M010499200Simon Descamps‡§,Robert-Alain Toillon‡,Eric Adriaenssens§¶,Vale ´rie Pawlowski§ʈ,Simon M.Cool**,Victor Nurcombe**,Xuefen Le Bourhis‡,Be ´noni Boilly‡,Jean-Philippe Peyrat ʈ,and Hubert Hondermarck‡‡‡From the ‡Equipe Facteurs de Croissance,UPRES EA-1033Biologie du De ´veloppement,Universite ´des Sciences et Technologies de Lille,59655Villeneuve d’ASCQ France,the ¶Immunopathologie Cellulaire des Maladies Infectieuses,CNRS,UMR 8527,Institut de Biologie de Lille,59000France,the **Department of Anatomical Sciences,University of Queensland,St.Lucia,Queensland 4072,Australia,and the ʈLaboratoire d’Oncologie Mole ´culaire Humaine,Centre Oscar Lambret,59020Lille,FranceWe show here that the neurotrophin nerve growth factor (NGF),which has been shown to be a mitogen for breast cancer cells,also stimulates cell survival through a distinct signaling pathway.Breast cancer cell lines (MCF-7,T47-D,BT-20,and MDA-MB-231)were found to express both types of NGF receptors:p140trkA and p75NTR .The two other tyrosine kinase receptors for neu-rotrophins,TrkB and TrkC,were not expressed.The mitogenic effect of NGF on breast cancer cells required the tyrosine kinase activity of p140trkA as well as the mitogen-activated protein kinase (MAPK)cascade,but was independent of p75NTR .In contrast,the anti-apo-ptotic effect of NGF (studied using the ceramide ana-logue C2)required p75NTR as well as the activation of the transcription factor NF-kB,but neither p140trkA nor MAPK was necessary.Other neurotrophins (BDNF,NT-3,NT-4/5)also induced cell survival,although not proliferation,emphasizing the importance of p75NTR in NGF-mediated survival.Both the pharmacological NF-B inhibitor SN50,and cell transfection with IkBm,resulted in a diminution of NGF anti-apoptotic effect.These data show that two distinct signaling pathways are required for NGF activity and confirm the roles played by p75NTR and NF-B in the activation of the survival pathway in breast cancer cells.Nerve growth factor (NGF)1is the archetypal member of theneurotrophin superfamily,which also includes brain-derived neurotrophic factor (BDNF),neurotrophin-3(NT-3),NT-4/5,and NT-6(1).NGF interacts with two classes of membrane receptor:the TrkA proto-oncogene product p140trkA ,which pos-sesses intrinsic tyrosine kinase activity,and a secondary re-ceptor,p75NTR ,that belongs to the tumor necrosis factor (TNF)receptor family (2).The stimulation of cell survival and cell differentiation by NGF and other neurotrophins have been described primarily in neuronal cell systems (3).Although the neurotrophic effect through p140trkA is known to involve the MAPK cascade,the role of p75NTR is still controversial;there is evidence that it can both positively and negatively regulate neuronal cell death and differentiation,depending on the cell type examined (4).In some cases,p75NTR is an inducer of apoptosis,even without NGF stimulation (5),whereas in other cases the activation of p75NTR by NGF results in a protection from cell death (6).In addition to its neurotrophic function,other activities of NGF have been described.For example,NGF can modulate gene expression in monocytes (7),it is chemotac-tic for melanocytes (8),and its inhibition on p75NTR can block the migration of Schwann cells (9).NGF also stimulates the proliferation of chromaffin cells (10),lymphocytes (11),and keratinocytes (12).We have previously shown that NGF is mitogenic for cancerous but not normal human breast cells (13),and these data,as well as others showing a role for NGF in the stimulation of prostatic cancer cells (14–17),implicate NGF in non-neuronal carcinogenesis.Both cellular proliferation as well as tumor cell survival are crucial for malignant progression.The effect of NGF on the survival of cancer cells through the p75NTR receptor has been shown for neuroblastoma (18)and schwannoma (6).In prostate cancer,p75NTR has been shown to be a mediator of NGF’s effects during critical phases of developmental cell death and carcinogenic progression (19).To date only the mitogenic effect of NGF for breast cancer cells has been described (13),with its roles in the control of breast cancer cell survival unknown.In this study,we have shown that,in addition to its mito-genic effect,NGF is also an anti-apoptotic factor for breast cancer cells.These cells express mRNA for both p140trkA and p75NTR receptors.Our results indicate that the mitogenic effect of NGF requires p140trkA and the MAPK cascade,but not the p75NTR receptor,whereas the promotion of cell survival strictly requires p75NTR as well as NF-B,but not p140trkA and MAPK.Thus the mitogenic and anti-apoptotic effects of NGF on breast*This work was supported in part by a grant from the Ligue Na-tionale Contre le Cancer (Comite ´du Nord)and by the French Ministry of Research and Education.The costs of publication of this article were defrayed in part by the payment of page charges.This article must therefore be hereby marked “advertisement ”in accordance with 18U.S.C.Section 1734solely to indicate this fact.§Recipients of an Association pour la Recherche sur la Cancer fellowship.‡‡To whom correspondence should be addressed:EA-1033,batiment SN3,Universite ´des Sciences et Technologies de Lille,59655Villeneuve d’Ascq cedex,France.Tel.:33-3-20-43-40-97;Fax:33-3-20-43-40-38;E-mail:hubert.hondermarck@univ-lille1.fr.1The abbreviations used are:NGF,nerve growth factor;PAGE,poly-acrylamide gel electrophoresis;NF-B,nuclear factor-B;BDNF,brain-derived neurotrophic factor;NT,neurotrophin;PARP,polyADP-ribose polymerase;TNF,tumor necrosis factor;TBP,TATA box binding pro-tein;RT-PCR,reverse transcriptase-polymerase chain reaction;FCS,fetal calf serum;DTT,dithiothreitol;PBS,phosphate-buffered saline;ERK,extracellular signal-regulated kinase;GFP,green fluorescence protein;I Bm,dominant-negative I B ␣mutant;bp,base pair(s);PD98059,Park Davis 98059.T HE J OURNAL OF B IOLOGICAL C HEMISTRYVol.276,No.21,Issue of May 25,pp.17864–17870,2001©2001by The American Society for Biochemistry and Molecular Biology,Inc.Printed in U.S.A.This paper is available on line at 17864cancer cells are mediated through two different signaling pathways.EXPERIMENTAL PROCEDURESMaterials—Cell culture reagents were purchased from BioWhittaker (France)except insulin,which was obtained from Organon(France). Recombinant human nerve growth factor,brain derived growth factor (BDNF),and neurotrophins3(NT-3)and4(NT-4)were from R&D Systems(UK).K-252a(inhibitor of trk-tyrosine kinase activity)and PD98059(inhibitor of MAPK cascade)were from Calbiochem(France). The mouse monoclonal anti-NGF receptor(p75NTR)antibody was from Euromedex(France)and was previously described for its ability to block the interaction between p75NTR and NGF(20).The anti-lamin B(C-20), goat polyclonal IgG,and the polyclonal anti-p140trkA(trk763)were from Santa Cruz Biotechnology.C2ceramide analogue(N-acetyl-D-sphingo-sine),Hoechst33258,and electrophoresis reagents were from Sigma Chemical Co.(France).The SN50NF-B inhibitor peptide,the rabbit polyclonal anti-NF-B p65antibody,was obtained from TEBU(France). Anti-PARP antibody was from Oncogene Research Products(UK). Primers and probes for TrkA and p75NTR,probe for TATA box binding protein(TBP)were from Eurogentec(Belgium).RT-PCR reagents were from Applied Biosystems(France).Lipofectin reagent and Opti-MEM were provided by Life Technologies,Inc.(France).The green fluores-cence protein plasmid(EGFPC1)was purchased from CLONTECH,and the dominant-negative IB␣mutant(IBm)expression vectors(in PCDNA3)containing a Ser to Ala substitution at residues32and36 were obtained from Dr.Jean Feuillard(UPRES EA1625,Bobigny, France).p65(rel-A)and c-rel cDNA were cloned at Eco RI site in PSVK3 expression plasmid.All vectors were obtained from Dr.Pascale Cre´pieux(McGill University,Montreal).The SY5Y subclone of SK-N-SH neuroblastoma cell line was a kind gift of Dr.Luc Bue´e(INSERM, U422,Lille,France).NT-2(Ntera/D1)human neural precursor cells (Stratagene)are derived from a clone of the NT-2teratocarcinoma.Cell Culture—Breast cancer cell lines(MCF-7,T47-D,BT-20,and MDA-MB-231)were obtained from the American Type Culture Collec-tion and routinely grown as monolayer cultures.Cells were maintained in minimal essential medium(Earle’s salts)supplemented with20m M Hepes,2g/liter sodium bicarbonate,2m M L-glutamine,10%fetal calf serum(FCS),100units/ml penicillin-streptomycin,50g/ml gentami-cin,1%of non-essential amino acids,and5g/ml insulin.Detection of Neurotrophin Receptors mRNA Expression—The reverse transcription reaction mixture contained2g of purified total RNA (extracted from breast cancer cell lines,NT-2cells,or SY5Y cells),1ϫreverse transcription reaction buffer,10m M DTT,400m M dNTP each, 2.5M oligo(dT)18primer,40units of RNasin,and200units of Moloney murine leukemia virus reverse transcriptase were added to25l of total reaction volume.All the reaction mixtures were incubated at37°C for1h and then inactivated at95°C for5min.Polymerase chain reaction was performed on cDNAs after RT or corresponding total RNA samples without the RT step for negative controls.The primers used for trkA and p75RT-PCR detection in breast cancer cell lines were as follows:trkA sense primer,5Ј(291)-CATCGTGAAGAGTGTCTCCG-3Ј(311)and antisense primer,5Ј(392)-GAGAGAGACTCCAGAGCGTT-GAA-3Ј(370)or p75sense primer,5Ј(442)-CCTACGGCTACTACCAG-GATGAG-3Ј(462)and antisense primer,5Ј(588)-TGGCCTCGTCG-GAATACG-3Ј(571).The primers used for RT-PCR comparative detection of trks in MCF-7cells were as follows:trkA sense primer,5Ј(118)-AGGCGGTCTGGTGACTTCGTTG-3Ј(139)and antisense primer, 5Ј(1162)-GGCAGCCAGCAGGGTGTAGTTC-3Ј(1141)or trkB sense primer,5Ј(134)-CGAGGTTGGAACCTAACAGCATTG-3Ј(157)and an-tisense primer,5Ј(1182)-GTCAGTTGGCGTGGTCCAGTCTTC-3Ј(1159)or trkC sense primer,5Ј(219)-CACGGACATCTCAAGGAAGA-GCA-3Ј(241)and antisense primer,5Ј(1078)-CTGAGAACTTCACCC-TCCTGGTAG-3Ј(1056).Each pair of primers was used in RT-PCR reaction to amplify trks or p75.To PCR tubes were added5l of PCRbuffer(200m M Tris-HCl,pH8.4,500m M KCl),10l of15m M MgCl2,1l of10m M dNTP mix,1l of cDNA or total mRNA(for negative control),1l of50m M respective primers,1l of2.5units/l Taq DNA polymerase,and water to a total volume of50l.The PCR conditions were as follows:after95°C for3min for denaturing cDNA,30cycles were run at94°C for1min,57°C for2min,and72°C for3min.The PCR tubes were incubated for a further10min at72°C for the exten-sion of cDNA fragments after the final cycle,and the PCR products were electrophoresed in an agarose gel.Cell Growth Assay—Experiments were performed as previously de-scribed(13).35-mm diameter dishes were inoculated with2ϫ104 cells/dish in2ml of medium containing10%FCS.After24h,cells were washed twice with serum-free medium.Next day,the medium wasreplaced with2ml of serum free medium containing100ng/ml NGF orvarious concentrations of other neurotrophins(BDNF,NT-3,NT-4/5).To study the effect of pharmacological inhibitors or blocking antibodies,various concentrations were added simultaneously with NGF(100ng/ml).After2days of NGF exposure,cells were harvested by trypsiniza-tion and counted using an hemocytometer.Determination of the Percentage of Apoptotic Cell Nuclei—Apoptosisof breast cancer cells was induced by the ceramide analogue C2,whichhas been described as a pro-apoptotic agent for human breast cancercells(21,22).Apoptosis was obtained by treatment with2M C2for 24h.To evaluate the anti-apoptotic activity of NGF,various concen-trations of this factor were tested;we found that the maximal effect wasobtained for100ng/ml.Consequently,this concentration was used in allexperiments with pharmacological inhibitors or blocking antibody.Fordetermination of apoptotic cell percentage,cells were fixed with coldmethanol(Ϫ20°C)for10min and washed twice with phosphate-buff-ered saline(PBS)before staining with1g/ml Hoechst33258for10min at room temperature in the dark.Cells were then washed with PBS andmounted with coverslips using Glycergel(Dako).The apoptotic cellsexhibiting condensed and fragmented nuclei were counted under anOlympus-BH2fluorescence microscope in randomly selected fields.Aminimum of500–1000cells was examined for each condition,andresults were expressed as a ratio of the total number of cells counted.Statistical Analysis and Software—The statistical analysis of thedata gathered from cell and apoptotic nuclei counting was performedusing SPSS version9.0.1(SPSS inc.,Chicago,IL).Analyses of variancewere followed by the Tukey’s test to determine the significance.NGF Receptors and PARP Immunoblotting—Subconfluent cell cul-tures were harvested by scraping in serum-free medium.After centrif-ugation(1000ϫg,5min),the pellet was treated with lysis buffer(0.3%SDS,200m M dithiothreitol)and boiled5min.In the case of PARP,thepellet was lysed with urea-rich buffer(62.5m M Tris-HCl,pH6.8,6Murea,10%glycerol,2%SDS),sonicated and incubated at65°C for15min.The lysates were subjected to SDS-PAGE,transferred onto anitrocellulose membrane(Immobilon-P,Millipore)by electroblotting(100V,75min),and probed with anti-trkA,anti-p75NTR or anti-PARPantibodies at4°C overnight.The membranes were then incubated atroom temperature for3h with biotin-conjugated anti-rabbit(TrkA)oranti-mouse(p75NTR and PARP)immunoglobulin G.After1h of incu-bation with extravidin,the reaction was revealed using the chemilumi-nescence kit ECL(Amersham Pharmacia Biotech)with Kodak X-OmatAR film.Detection of p140trkA and MAPK Activation—Proteins were extractedin lysis buffer(150m M NaCl,50m M Tris,pH7.5,0.1%SDS,1%NonidetP-40,100M sodium orthovanadate)prior to immunoprecipitation. Preclearing was done with protein A-agarose(10l/250l,60min, 4°C).After centrifugation(10,000ϫg,2min),the supernatant wasincubated with monoclonal anti-MAPK(anti-ERK2)antibody(10l/250l,60min,4°C).Protein A-agarose(10l)was added for60min(4°C) and then pelleted by centrifugation(10,000ϫg,2min).The pellet wasthen rinsed three times with lysis buffer and boiled for5min inLaemmli buffer.After SDS-PAGE and electroblotting,nitrocellulosemembranes were blocked with3%bovine serum albumin.Membraneswere then incubated with PY20anti-phosphotyrosine antibody over-night at4°C,rinsed,and incubated with a horseradish peroxidase-conjugated anti-mouse IgG for3h at room temperature.Membraneswere rinsed overnight at4°C before visualization with ECL.Cell Fractionation and NF-B Detection—Cell nuclear extracts were prepared as described by Herrmann et al.(23).Cells were trypsinized and then pelleted in minimal essential medium containing10%FCS. After washing with ice-cold PBS,cells were repelleted and resuspended in400l of ice-cold hypotonic buffer(10m M Hepes,pH7.8,10m M KCl,2m M MgCl2,0.1m M EDTA,10g/ml aprotinin,0.5g/ml leupeptin,3 m M phenylmethylsulfonyl fluoride,and3m M DTT).After10min on ice, 25l of10%Nonidet P-40was added and crude nuclei were collected by centrifugation for5min.The nuclear pellet was resuspended in high salt buffer(50m M Hepes,pH7.4,50m M KCl,300m M NaCl,0.1m M EDTA,10%(v/v)glycerol,3m M DTT,and3m M phenylmethylsulfonyl fluoride).After30min on ice with frequent agitation,the insoluble nuclear material was pelleted in a microcentrifuge for10min.Crude nuclear protein was collected from the supernatant and snap-frozen in a dry ice/ethanol bath.After thawing and boiling for5min in Laemmli buffer,the nuclear extracts were subjected to SDS-PAGE and probed with an anti-NF-B p65antibody.A control was established with anti-lamin B antibody.Transfection of I,c-rel,and rel-A—Cotransfection experiments were carried out using Lipofectin reagent,as described by the manu-Intracellular Signaling Pathways of NGF in Breast Cancer Cells17865facturer.Briefly,MCF-7cells were incubated for 5h in 1ml of Opti-MEM transfection medium containing 8l of Lipofectin reagent,0.8g of green fluorescence protein (GFP)-carrying vector and 0.2g of empty vector PCDNA3or 0.2g of I Bm.In the case of c-rel or rel-A,cells were cotransfected with 0.8g of GFP-carrying GFP and 0.6g of PSVK 3(empty plasmid),c-rel,or rel-A.Cells were then grown for 24h with 10%FCS minimal essential medium and rinsed for 2h in serum-free me-dium before incubation in serum-free medium in the presence or ab-sence of 100ng/ml NGF and/or 2M C2for another 24h.Cells were then fixed with paraformaldehyde 4%(4°C)for 30min,and the per-centage of apoptotic cell nuclei in GFP-stained cells was determined as described above.RESULTSNGF Mitogenic and Anti-apoptotic Activity for Breast Cancer Cells—The effects of 100ng/ml NGF on cell proliferation and C2-induced apoptosis were evaluated by cell counting and Hoechst staining,respectively.The results show that NGF induces an increase in cell number for all breast cancer cell lines tested (Fig.1A ).We have previously demonstrated that NGF has a direct mitogenic effect on breast cancer cells by recruiting cells in G 0phase and by shortening the G 1length (Descamps et al.,1998).In addition,NGF rescued breast cancer cells undergoing C2-induced apoptosis;the maximum survival was observed at 200ng/ml (Fig.1B ).The morphology of cells undergoing this NGF-induced anti-apoptotic rescue was quite distinct (Fig.2A ).The induction of apoptosis by C2was found to involve cleavage of poly(A)DP-ribose polymerase (PARP);this cleavage was reversed by NGF (Fig.2B ).TrkA and p75NTR Expression—RT-PCR was used to show the expression of mRNA for both high and low affinity NGF recep-tors in MCF-7,T47-D,BT-20,and MDA-MB-231cells (Fig.3A );the 102-bp band for the TrkA transcript and a 147-bp band for the p75NTR transcript were readily detectable on 1%agarose gels.Moreover,Western blotting demonstrated that both p140trkA and p75NTR were present in all the breast cancer cell lines (Fig.3B ).Real-time quantitative RT-PCR indicated that there was no significant change in the levels of TrkA and p75NTR mRNAs in the presence of FCS,NGF,or C2(data not shown)and that the levels of mRNA for TrkA and p75NTR in breast cancer cells was between 5and 10times lower than the level observed in SY5Y neuroblastoma cells (data not shown).This indicates that NGF receptor expression in breast cancer cells is relatively limited.It should be noticed that,although mRNA levels of NGF receptors differ between breast cancer cells and SY5Y,the protein levels apparently do not.However,F IG .1.Effect of NGF on the growth and survival of breast cancer cells.A ,breast cancer cells were serum-deprived in minimum essential medium,and after 24h the NGF (100ng/ml)was added.After 48h,cells were harvested and counted.B ,cells were serum-deprived in minimum essential medium and treated with 2M C2with or without 100ng/ml NGF.After 24h,cells were fixed and the proportion of apoptotic nuclei were determined after Hoechst staining under an Olympus-BH2fluorescence microscope.For measurement of both cell number and apoptosis,results are expressed as the means ϮS.D.of five separate experiments.Significance was determined using the Tukey’s test (*,p Ͻ0.01).F IG .2.Anti-apoptotic effect of NGF.A ,Hoechst staining of apop-totic cell nuclei in control,C2and C2ϩNGF-treated MCF-7cells.Cells were serum-deprived in minimum essential medium and treated with C2.NGF was added at 100ng/ml.After 24h,cells were fixed and apoptotic nuclei were observed after Hoechst staining.B ,immunoblot detection of PARP cleavage.C2-induced PARP cleavage was reversed by p75NTR activation mediated by NGF.MCF-7cells were serum-de-prived in minimum essential medium for 24h and were then treated with 100ng/ml NGF in the presence or absence of 2M C2,10n M K-252a,10M PD98059,or 10g/ml anti-p75NTR -blocking antibody (Euromedex)for another 24-h period.Proteins were detected after SDS-PAGE of cell preparations from MCF-7breast cancer cells,electro-blotting onto nitrocellulose,and immunodetection with anti-PARP antibodies.Intracellular Signaling Pathways of NGF in Breast Cancer Cells17866it has been shown before that the level of a given cellular protein cannot be simply deduced from mRNA transcript level (24).One could hypothesize that the stability of mRNA and/or protein for NGF receptors,differs between breast cancer cells and neuroblastoma cells,leading to the observed disproportion-ality between mRNA and protein levels.Involvement of p140trkA and p75NTR in Mitogenic and Sur-vival Activities of NGF—We used a combination of specific antibodies and pharmacological inhibitors to study the puta-tive functions of p140trkA and p75NTR in the stimulation of proliferation and cell survival induced by NGF.The Trk tyro-sine kinase inhibitor K-252a,and the MEK inhibitor PD98059,both strongly inhibited the growth-stimulatory effect of NGF on MCF-7cells,but had no effect on its anti-apoptotic effects (Fig.4).Conversely,neither the anti-p75NTR blocking antibody nor the NF-B inhibitor SN50affected NGF-stimulated prolif-eration,although both strongly reduced the anti-apoptotic ef-fects (Fig.4).The tyrosine kinase activity of p140trkA was inhibited by K-252a but not by the anti-p75NTR or PD98059(Fig.5).On the other hand,the activity of the MAPKs was inhibited by K-252a and PD98059but not by the anti-p75NTR (Fig.5).It should be noted that the SN50peptidic inhibitor of NF-B,similarly to the anti-p75NTR ,inhibited the anti-apo-ptotic effect of NGF but neither its proliferative effect nor its activation of p140trkA and MAPKs.The effect of other neuro-trophins on MCF-7cell growth and survival was also evaluated (Fig.6A ).In contrast to NGF,no proliferative effect was pro-vided by BDNF,NT-3,or NT-4/5.However,all neurotrophins tested exhibited a rescue effect on C2-treated cells that was not altered in the presence of the trk inhibitor K-252a (Fig.6B ).These data suggest that trk receptors are not involved in NGF survival activity.Moreover,the participation of trkB and trkCin these events can be ruled out,because they are not expressed in these breast cancer cells (Fig.6C ).NF-B Involvement in the Anti-apoptotic Effect of NGF—The inhibitory effect of SN50on the NGF anti-apoptotic activity indicated the potential involvement of NF-B in the signaling leading to the protective activity of this growth factor.To fur-ther investigate this phenomenon,we studied the effect of NGF on the nuclear translocation of NF-B,as well as the conse-quence of transfection by IkBm (an inhibitor of NF-B)or by c-rel and rel-A (constitutively active subunits of NF-B)on the NGF-mediated anti-apoptotic activity in MCF-7cells.Western blotting revealed no change in the nuclear levels of NF-B (p65)during apoptosis induced by C2(Fig.7).In contrast,the addi-tion of NGF on C2-treated cells induced a translocation of NF-B from cytoplasm to puterized quantifica-tion revealed a doubling p65band intensity normalized to the total intensity of the lane (data not shown).Moreover,this NF-B nuclear translocation was inhibited by the presence of p75NTR -blocking antibody or SN50,but was not affected by K-252a and PD98059.Interestingly,in the absence of C2-in-duced apoptosis NGF was not able to induce the nuclear trans-location of NF-B,confirming previous observations that p75NTR -mediated NF-B activation requires cell stress (25).Transfection of MCF-7cells with IkBm,an inhibitor of NF-B,reversed the anti-apoptotic effect of NGF (Fig.8A ).As a control,we transfected MCF-7cells with an empty vector;no effect was observed.In addition,transfection with activators of the NF-B pathway,c-rel or rel-A (Fig.8B ),resulted in an inhibition of C2-induced apoptosis of MCF-7cells,even in absence ofNGF,F IG .3.TrkA and p75NTR expression in breast cancer cells.A ,agarose gel electrophoresis of RT-PCR products evidenced a 102-bp band and a 147-bp band,which are characteristic of TrkA and p75NTR ,respectively.Both NGF receptors were found in all cell types tested.B ,p140trkA and p75NTR were immunodetected after SDS-PAGE of breast cancer cell lines.The neuroblastoma cells SY5Y were used as positive control for the expression of NGFreceptors.F IG .4.Pharmacological modulation of the proliferative and anti-apoptotic effect of NGF.MCF-7cells were starved in minimum essential medium,and after 24h,100ng/ml NGF was added with or without inhibitors or antibody.A ,after 48h,cells were harvested and counted.B ,after 24h,cells were fixed and the proportion of apoptotic nuclei determined after Hoechst staining.The following concentrations were used:2M C2,10n M K-252a,10g/ml anti-p75NTR -blocking antibody (Euromedex),10M PD98059,18M SN50.For A and B ,results are expressed as the means ϩS.D.of five separate experiments.Significance was determined using the Tukey’s test (*,p Ͻ0.01).Intracellular Signaling Pathways of NGF in Breast Cancer Cells 17867confirming the involvement of NF-B family members in hu-man breast cancer cell survival.DISCUSSIONThis study shows that,in addition to its mitogenic activity,NGF is anti-apoptotic for breast cancer cells,and that these two biological effects are differentially mediated by the p140trkA and p75NTR receptors,respectively.The growth of breast cancer results from a balance between cell proliferation and apoptosis,both of which can be modulated by various regulatory peptides.For example,epidermal growth factor,fibroblast growth factors,and insulin-like growth factor-1can all stimulate the proliferation and survival of breast cancer cells (26).On the other hand,agents such as transforming growth factor-or tumor necrosis factor-␣can inhibit growth and induce apoptosis in these cells (27).Recently we have shown that NGF,which was primarily described for its neuro-trophic properties,is a strong mitogen for cancerous but not for normal human breast epithelial cells,suggesting a crucial func-tion for this factor in the initiation and progression of human breast tumors (13).In the present study,we have shown that the breast cancer cells express transcripts for both TrkA and p75NTR receptors.In contrast,no expression of TrkB and TrkC was found in any of the breast cancer cells tested,in accordance with the fact that BDNF,NT-3,or NT-4/5have no mitogenic effect for these cells.The presence of NGF receptors has been detected previously in breast cancer cells (28),and low levels of NGF receptor expression have recently been reported in other breast cancer cell lines (29),leading to the hypothesis of a recruitment and cooperation between p140trkA and p185Her-2for the induction of mitogenesis by NGF.Our results indicate a stimulation of p140trkA tyrosine kinase activity and of the MAPK cascade by NGF,and the use of the pharmacological inhibitors K-252a and PD98059demonstrate the requirementfor these signals in NGF-induced MCF-7cell proliferation.The induction of MAPK activity required p140trkA activation,but p75NTR did not appear to be involved,because p75NTR -blocking antibodies did not have any effect on NGF-inducedMAPKF IG .5.p140trkA and MAPK activation.MCF-7cells were treated with 100ng/ml NGF in the presence or absence of 10n M K-252a,10g/ml anti-p75NTR -blocking antibody,or 10M PD98059.p140trkA (A )and MAPK activation (B )were determined after immunoprecipitation using polyclonal anti-TrkA and monoclonal anti-ERK2antibodies,re-spectively.After SDS-PAGE and electroblotting,nitrocellulose mem-branes were counterprobed with the PY20anti-phosphotyrosine anti-body.For detection of TrkA (A )and MAPK (B )activation,the lower panel shows reprobing of the blots with the immunoprecipitatingantibody.F IG .6.Effect of different neurotrophins on MCF-7cells growth and survival.MCF-7cells were serum-deprived in minimum essential medium,and after 24h the neurotrophins (100ng/ml NGF,50ng/ml BDNF,50ng/ml NT-3,100ng/ml NT-4/5)were added.A ,after 48h,cells were harvested and counted.In contrast with NGF,neither BDNF,NT-3,nor NT-4/5displayed significant bioactivity (for concen-trations up to 400ng/ml).B ,MCF-7cells were serum-deprived in minimum essential medium and treated with 2M C2,with or without neurotrophins (100ng/ml NGF,50ng/ml BDNF,50ng/ml NT-3,100ng/ml NT-4/5).After 24h,cells were fixed and apoptotic nuclei percent-age was determined after Hoechst staining under an Olympus-BH2fluorescence microscope.For measurement of both cell number and apoptosis,results are expressed as the means ϮS.D.of five separate experiments.Significance was determined using the Tukey’s test (*,p Ͻ0.01).C ,TrkB and TrkC mRNA expression in MCF-7cells.Agarose gel electrophoresis of RT-PCR products reveals TrkA expression,but no TrkB or TrkC expression in MCF-7breast cancer cells.Human NT2cells were used as positive control for the expression of TrkB and ne 1,NT2-negative control without RT step;lane 2,NT2-positive control;lane 3,MCF-7cells-negative control without RT step;lane 4,MCF-7cells.Intracellular Signaling Pathways of NGF in Breast Cancer Cells17868。
- 1、下载文档前请自行甄别文档内容的完整性,平台不提供额外的编辑、内容补充、找答案等附加服务。
- 2、"仅部分预览"的文档,不可在线预览部分如存在完整性等问题,可反馈申请退款(可完整预览的文档不适用该条件!)。
- 3、如文档侵犯您的权益,请联系客服反馈,我们会尽快为您处理(人工客服工作时间:9:00-18:30)。
BIOLOGY CONTRIBUTIONDISTINCT SIGNALING PATHWAYS AFTER HIGHER OR LOWER DOSES OF RADIATION IN THREE CLOSELY RELATED HUMAN LYMPHOBLAST CELL LINES T ZU-P IN L U,B.S.,*1L IANG-C HUAN L AI,P H.D.,zy1B E-I L IN,B.S.,z L I-H AN C HEN,M.S.,z T ZU-H UNG H SIAO,M.S.,zx H OWARD L.L IBER,P H.D.,jj J OHN A.C OOK,P H.D.,{J AMES B.M ITCHELL,P H.D.,{M ONG-H SUN T SAI,P H.D.,z**AND E RIC Y.C HUANG,S C.D.*yxyyzz From the*Graduate Institute of Biomedical Electronics and Bioinformatics,z Bioinformatics and Biostatistics Core,Research Center for Medical Excellence,y Graduate Institute of Physiology,x Department of Electrical Engineering,**Institute of Biotechnology, yy Department of Life Science,and zz Graduate Institute of Epidemiology,National Taiwan University,Taipei,Taiwan;jj Department of Environmental and Radiological Health Sciences,Colorado State University,Fort Collins,CO;{Radiation Biology Branch,Center for Cancer Research,National Cancer Institute,National Institutes of Health,Bethesda,MDPurpose:The tumor suppressor p53plays an essential role in cellular responses to DNA damage caused by ionizingradiation;therefore,this study aims to further explore the role that p53plays at different doses of radiation.Materials and Methods:The global cellular responses to higher-dose(10Gy)and lower dose(iso-survival dose,i.e.,the respective D0levels)radiation were analyzed using microarrays in three human lymphoblast cell lines withdifferent p53status:TK6(wild-type p53),NH32(p53-null),and WTK1(mutant p53).Total RNAs wereextracted from cells harvested at0,1,3,6,9,and24h after higher and lower dose radiation exposures.Template-based clustering,hierarchical clustering,and principle component analysis were applied to examinethe transcriptional profiles.Results:Differential expression profiles between10Gy and iso-survival radiation in cells with different p53statuswere observed.Moreover,distinct gene expression patterns were exhibited among these three cells after10Gy ra-diation treatment,but similar transcriptional responses were observed in TK6and NH32cells treated with iso-sur-vival radiation.Conclusions:After10Gy radiation exposure,the p53signaling pathway played an important role in TK6,whereasthe NFkB signaling pathway appeared to replace the role of p53in WTK1.In contrast,after iso-survival radiationtreatment,E2F4seemed to play a dominant role independent of p53status.This study dissected the impacts of p53,NFkB and E2F4in response to higher or lower doses of g-irradiation.Ó2010Elsevier Inc.Microarray,Human lymphoblast cells,Radiation,Signaling pathway,p53.INTRODUCTIONIonizing radiation exposure of mammalian cells causes DNA damages associated with a series of cellular responses including cell-cycle arrest,DNA repair,and cell death(1–3). Several studies have demonstrated that the biological effects of lower dose and higher dose ionizing radiation are not lin-early related(1–3)—that is,the transcripts or proteins in-duced at higher doses of radiation cannot be extrapolated to lower or physiological doses(4).Genes involved in dif-ferent functions and pathways were activated in response to different doses of radiation.Lower dose radiation gener-ally induced genes involved in cell–cell signaling,signal transduction,cell/organism defense,and homeostasis(2, 5);in contrast,higher dose radiation induced genes involved in apoptosis and cell proliferation in many cell lines,such as malignant gliomas(7),myeloid(5),and human skinfibro-blasts(2).The tumor suppressor protein p53is an important tran-scription factor in the cellular response to DNA damage and can activate cell-cycle checkpoints and apoptotic path-ways.Problems with checkpoint control can easily lead to in-stability,because replication or mitosis can proceed before DNA repair is completed.Alteration of p53status has been proved to increase spontaneous and X-ray–induced mutation frequencies(6).However,the activation and functions of p53 in response to radiation were most pronounced after higher dose radiation(7).The role of p53in response to lower doses of radiation remained to be elucidated.Reprint requests to:Mong-Hsun Tsai;(motiont@.tw)or Eric Y.Chuang(chuangey@.tw)Supported by95R0066-BM01-01and97R0066-08from National Taiwan University,Taiwan,ROC;NSC95-2314-B-002-112-MY3,NSC97-2627-B-002-009and NSC98-2314-b-002-065-MY3from National Science Council,Taiwan,ROC; DOH96-TD-G-111-014from Department of Health,Taiwan,ROC. 1These authors contributed equally to this work.Conflict of interest:none.Received May15,2009,and in revised form Aug6,2009. Accepted for publication Aug7,2009.212Int.J.Radiation Oncology Biol.Phys.,Vol.76,No.1,pp.212–219,2010CopyrightÓ2010Elsevier Inc.Printed in the USA.All rights reserved0360-3016/10/$–see front matterdoi:10.1016/j.ijrobp.2009.08.015To investigate the effect of radiation-induced p53-depen-dent or p53-independent pathways in response to ionizing ra-diation,three human B-lymphoblast cell lines with different p53status were chosen;TK6is wild-type for p53,WTK1 overexpresses a mutant form of p53,and NH32is a double p53knockout cell line derived from TK6.These lines have different spontaneous mutation rates and respond quite dif-ferently to ionizing radiation.For example,WTK1was10-fold hypermutable at the thymidine kinase locus after2Gy irradiation compared with TK6(8).In our previous study,cDNA microarrays were used to identify gene expression patterns at different times after10 Gy g-ray treatments in these three cell lines(9).In this fol-low-up study,cDNA microarrays were used in TK6, NH32,and WTK1to identify gene expression patterns at dif-ferent times after iso-survival(D0)doses of radiation(10),for comparison with those seen after10Gy.The results indicated that10Gy and iso-survival radiation treatments triggered dif-ferent transcriptional responses.The cells exhibited different signal transduction pathways after10Gy radiation:the p53 signaling pathway was activated in TK6but the NFkB signal-ing pathway was induced and appeared to take over p53func-tion in p53-mutated WTK1.In contrast,all three lines showed similar transcriptional responses after iso-survival ra-diation,with the downregulation of E2F4-related genes being perhaps the most significant change.The results indicated that the E2F4-related pathway plays a dominant role in cells with lower dose radiation regardless of p53status.Based on our results,we propose a model to illustrate differential tran-scriptions in response to higher or lower doses of radiation.MATERIALS AND METHODSCell culture and radiation treatmentThe conditions to culture three human lymphoblastoid cell lines (TK6,WTK1,and NH32)were described previously(9).Cells were treated with g-ray radiation at10Gy and doses of37%iso-sur-vival level(D0).The D0doses for TK6,WTK1,and NH32were81 cGy,150cGy,and88cGy,respectively(10).Total RNA was ex-tracted from10Gy-treated and iso-survival-dose–treated cells at 0,1,3,6,9,and24h after irradiation for cDNA microarrays anal-ysis.Cells were irradiated with an Eldorado860Co teletherapy unit(Theratronics International Ltd.,Ontario,Canada)at dose rates between200and250cGy/min.Each experiment was repeated inde-pendently in triplicate.RNA preparation,probe labeling,and microarray hybridizationRNA was extracted using Trizol reagent(Invitrogen,Carlsbad, CA)and purified using Qiagen Rneasy Mini Kit(Valencia,CA)ac-cording to manufacturers’protocols.Methods for probe labeling and microarray hybridization were described previously(11).Briefly,20 m g of sample RNA was labeled with Cy3and40m g of universal hu-man reference RNA(Stratagene,La Jolla,CA)was labeled with Cy5using Superscript II Reverse Transcriptase(Invitrogen,Carls-bad,CA).The labeled cDNAs were purified and hybridized onto mi-croarray slides containing7,680human cDNA clones.The details of this microarray were described previously(11,12).After16–20h of hybridization,slides were washed and scanned.Microarray image and data analysisHybridized slides were scanned by a GenePix4000B scanner (Axon Ins.,Union City,CA).The ratios of the sample intensity were normalized by adjusting the center of ratio distribution to 1.0.The template-based clustering algorithm was used to select sig-nificantly changed genes after radiation treatment(11).The criteria for gene selection were defined as follows:$60%of time points in each profile had a maximum intensity>500and a minimum inten-sity>200;correlation coefficient offitting r k is>0.85;the maxi-mum intensity over the minimum intensity in each profile is >twofold;and total p of three replicates is<0.001(9).Tight cluster-ing(13)was used to further select genes with similar profiles.Inge-nuity Pathway Analysis(Redwood City,CA)was used for functional analysis of selected genes.Quantitative reverse transcription polymerase chain reaction analysisQuantitative reverse transcription polymerase chain reaction(RT-PCR)analysis was used on selected genes for validation(MAD2L1, PTTG1,STMN1,E2F4,RB1,and ACTB for internal control).In ad-dition,the effects of dose-dependent response were examined by treating cells with0.8,1.5,2,4,and10Gy of radiation.At24h after radiation,cells were harvested,and quantitative RT-PCR was con-ducted with ABI7300system according to the manufacturer’s pro-tocol.RESULTSDistinct gene expression profiles between10Gy and iso-survival radiation exposuresTo compare radiation-induced gene expression profiles be-tween10Gy and iso-survival doses among three different p53status of human lymphoblastoid cell lines(TK6, WTK1,and NH32),differentially expressed genes after irra-diation were identified.Among all three cell lines,494genes met the criteria after the10Gy treatment and827genes were detected after iso-survival treatment.Hierarchical clustering of the selected genes for each cell line is shown in Fig.1a. In TK6,619genes had at least twofold changes after10 Gy or iso-survival irradiations,and103genes(17%)were in common to both treatments(Fig.1a,left column).In WTK1,378genes showed at least twofold changes after10 Gy or iso-survival irradiations,but only11of these genes (3%)changed at both radiation exposures.Thus,as shown in Fig.1a,middle column,the expression profiles at10Gy or iso-survival dose treatments exhibited very distinct pat-terns,with the upregulated genes after one treatment showing minor or even opposite changes after the other treatment.In NH32(Fig.1a,right column),341twofold genes were iden-tified after10Gy or iso-survival treatments,and in this case 31genes(9%)were in common.These results indicate that distinct transcriptional profiles were activated after10Gy and iso-survival doses in these cells with different p53status. To examine the global similarity of gene expression pat-terns at10Gy and iso-survival treatment,principle compo-nent analysis was performed.As shown in Fig.1b,10Gy and iso-survival doses showed a distinct distribution indicat-ing the dose-dependent nature in response to irradiation ex-posure.The radiation responses of three different cell linesDistinct Pathways After Radiation Treatments in Human Cells d T.-P.L U et al.213after 10Gy or iso-survival radiation were analyzed further.For the iso-survival treatments (Fig.1c,right),827genes were identified with twofold changes in at least one cell line.Among these genes,108genes were identified in both TK6and NH32,whereas only 38genes for TK6and WTK1and 29genes for WTK1and NH32were identified.Functional analysis of the 108genes common to TK6and NH32revealed that cell death process were significantly en-riched (p <10–7).Furthermore,the iso-survival doses of TK6and NH32were very similar (81cGy for TK6,88cGy for NH32).The similar enriched functional category and the ac-tivated/repressed genes suggested a common gene set was triggered in TK6and NH32to regulate the cell death process at the transcriptional level.In contrast,when cells were treated with 10Gy irradiation (Fig.1c,left),only a small number of genes ($30)responded at any two cell lines,indi-cating that different radiation responses were activated in these three cell lines after 10Gyirradiation.Fig.1.(A)Hierarchical clustering of RNA expression profiles for TK6,NH32,and WTK1cells after 10Gy and iso-sur-vival doses of radiation.For each cell line,expression levels relative to time 0are shown at 0,1,3,6,9,and 24h after both 10Gy and iso-survival radiation exposures.The Venn diagrams for each cell line are shown on the right side.(B)Principal component analysis of the genes with twofold changes for each cell line.Red spots indicate 10Gy treatment;green spots indicate iso-survival treatments.(C)Venn diagrams of genes in response to 10Gy and iso-survival radiation for each cell line.214I.J.Radiation Oncology d Biology d Physics Volume 76,Number 1,2010Inactivation of E2F4-related genes after iso-survival dose of radiation exposureTo dissect the importance of cell death processes after iso-survival radiation,tight clustering analysis (13)was applied on the 827genes with twofold changes in at least one cell line after iso-survival treatments.Fifty-one genes in the top two tightest clusters were selected for further analysis.As shown in Fig.2a,the expression profiles of these genes in the most confident clusters were consistently downregulated throughout all time points among three cell lines for iso-sur-vival treatments,but not in the 10Gy treatment.To understand biological functions of how these differentially expressed genes were involved,functional analysis using ingenuity path-way analysis revealed that 24genes involved in cell-cycle con-trol were significantly (p =2.03Â10–14)enriched.Moreover,network analysis revealed 16genes with E2F4binding motifs,which enabled the construction of an E2F4-centered gene network (Fig.2b).These results suggested that at iso-survivalradiation exposure,E2F4may play a pivotal role in controlling cell cycle regardless of p53status.Activation of p53signaling pathway in TK6afar 10Gy radiation exposureTo investigate which functional pathways were influenced among three cell lines with 10Gy irradiation,all differen-tially expressed genes in TK6responding to 10Gy radiation (n =270)were analyzed using ingenuity pathway analysis.The results of functional analysis showed that the G1/S checkpoint regulation pathway in TK6was significantly in-duced (p =5.37Â10–4)at 10Gy,but not at iso-survival ra-diation treatment.Because p53is a known key player regulating the G1/S arrest and different apoptosis kinetics in TK6was observed (9),selected p53-dependent apoptotic genes,such as ID3,RB1,BTG1,FAS,and CASP1,were fur-ther investigated to elucidate the importance of p53in re-sponse to 10Gy treatment in TK6.As shown in Fig.3a,Fig.2.Downregulation of E2F4-related cell-cycle genes in TK6,WTK1,and NH32after iso-survival treatments.(A)Hi-erarchical clustering of 36E2F4-related cell-cycle genes.(B)Gene network of the E2F4-related cell-cycle genes was re-vealed by using ingenuity pathway analysis.The solid lines indicate direct evidence of an interaction between the two genes according to published literature reports,whereas the dash lines indicate indirect evidence.Green areas denote genes that were downregulated at 24h after iso-survival irradiation in TK6.Dose–response curves of MAD2L1,PTTG1,and STMN1in TK6(C)and NH32(D)using quantitative reverse transcription polymerase chain reaction (RT-PCR).Cells were harvested at 24h after radiation.The expression values were first normalized to ACTB and presented as the relative ratio to control (0Gy).The data represent an average of three independent quantitative RT-PCR reactions.Distinct Pathways After Radiation Treatments in Human Cells d T.-P.L U et al .215most of these genes showed two-to sixfold change 24h after irradiation.These results indicated that the p53-dependent apoptosis pathway is activated in TK6after 10Gy radiation.Activation of NFkB signaling pathway in WTK1after 10Gy radiation exposureBecause WTK1and NH32do not have functional p53,we explored alternative pathways in these cells after 10Gy radi-ation.All differentially expressed genes in WTK1after 10Gy radiation (n =101)were analyzed using ingenuity pathway analysis.The result of network analysis showed that the NFKB1-and NFKB2-related gene networks were the most significant gene networks,and eight genes directly interacted with NFKB1and NFKB2in the gene network.Because upre-gulation of the NFkB signaling pathway provided survival signals (14)and resulted in the resistance of cancer cells to radio-and chemotherapies (15,16),these eight survival-re-lated genes regulated by NFkB (NFkB1,NFkB2,CD80,CCL3,IRF1,TRAF1,MAP3K14,and TNFRSF8)were inves-tigated in cells without functional p53(i.e.,WTK1and NH32)after 10Gy radiation treatment.As shown inFig.3b,left,transcriptional levels of these genes remained unchanged in TK6after 10Gy radiation,whereas eight NFkB-related genes were upregulated at least twofold in WTK1and downregulated in NH32.Functional analysis re-vealed an NFkB-centered gene network (Fig.3b,right).In contrast to the response after 10Gy radiation treatment,these genes only showed minor and insignificant changes in TK6,WTK1,or NH32after iso-survival radiation treatments (data not shown).These results suggested that NFkB did not play an important role in cells with functional p53after 10Gy ra-diation nor in cells after iso-survival radiation.Conversely,when p53is mutated as in WTK1,NFkB,and its downstream genes were activated to replace the function of p53to cope with the damage caused by 10Gy radiation.Validation of microarray results using quantitative RT-PCRTo confirm the microarray results,three E2F4downstream genes (MAD2L1,PTTG1,and STMN1)were selected for quantitative RT-PCR analysis in TK6(Fig.2c)and NH32(Fig.2d).Generally,the results of RT-PCR were ingoodFig.3.Activation of p53-and NFkB-signaling pathways in TK6and WTK1,respectively,after 10Gy radiation treatment.(A)Hierarchical clustering (left)and p53-centered gene network (right)at 24h after irradiation in TK6of the 25cell death–related genes.(B)Hierarchical clustering (left)and NFkB-centered gene network (right)at 9h (maximum response)after irradiation in WTK1of the eight survival-related genes.Gray (white)areas denote nonresponsive genes.Other details of the gene network are the same as in Fig.2b.216I.J.Radiation Oncology d Biology d Physics Volume 76,Number 1,2010agreement with the microarray results.The genes were down-regulated at0.8Gy radiation and upregulated at10Gy radi-ation in both TK6and NH32.Regarding the effects of dose-dependent response,genes were downregulated below2Gy radiation and upregulated above4Gy radiation in a dose-dependent manner in TK6 (Fig.2c).In NH32,genes were downregulated at different ra-diation treatments except10Gy(Fig.2d).Although minor differences in the expression values existed between TK6 and NH32,the downregulation profiles suggested that E2F4mainly functioned in lower radiation doses,especially below2Gy radiation.As radiation doses increased,the effect of E2F4diminished gradually.DISCUSSIONThis study demonstrated that10Gy and the lower iso-sur-vival irradiation doses lead to distinct gene expression pat-terns in three lymphoblastoid cell lines with different p53 status.Moreover,this study dissected the impacts of several transcription factors,such as p53,NFkB,and E2F4,in re-sponse to different doses of radiation exposure.Results indi-cated that10Gy irradiation and iso-survival treatments trigger distinct transcriptional responses;p53plays a domi-nant role in wild-type TK6after exposure to10Gy radiation but not the lower dose(on the other hand,NFkB plays a more significant role in non-functional p53cells);and E2F4re-presses cell cycle–related genes in response to iso-survival treatment that is independent of p53status.As expected,the gene expression profiles after10Gy and iso-survival irradiation exposure show distinct patterns.The numbers of twofold changed genes in TK6,WTK1,and NH32were very different and relatively few genes were in common,indicating that the differential transcriptional mechanism was triggered in response to higher or lower doses of irradiation in cells with different p53status (Fig.1a,right).A significant portion of genes responding to10Gy radiation were involved in cell death processes,re-flecting that the higher dose of radiation results in severe DNA damage.Indeed,a subset of p53-dependent apopto-sis-related genes were upregulated in TK6after10Gy irradi-ation,but not after iso-survival treatments.This result may suggest that p53-dependent apoptosis is activated in TK6af-ter10Gy treatment.Next,we explored the question of how did cells cope with 10Gy radiation when no functional p53was available?Our preliminary results showed that the kinetics of radiation-in-duced apoptosis in TK6were different from those of WTK1and NH32after10Gy radiation(data not shown).Re-cent studies also reported that upregulation of the NFkB sig-naling pathway provided survival signals(14);thus,NFkB keeps cells proliferating and protects cells from apoptosis when cells are exposed to higher doses of irradiation(17, 18).Therefore,selected survival-related genes regulated by NFkB were explored in cells without functional p53(i.e., WTK1and NH32)after10Gy radiation treatment.Our re-sults showed several NFkB-regulated genes were upregu-lated two-to threefold in WTK1(e.g.,MAP3K14),but remained unchanged in TK6after10Gy radiation exposure. In TK6,p53may override the effect of NFkB,so that NFkB-regulated genes were not transcribed.From our expression data,many genes were upregulated only in WTK1,not in TK6containing intact p53protein nor in NH32containing no p53protein.We speculate that in WTK1,activation of NFkB-regulated genes might be due to the activity of mutant p53molecule,which contains a single point mutation (ile237),and is overexpressed.NFkB may be involved in controlling apoptosis at the10Gy radiation treatment level when functional p53is not present.The regulation of apopto-sis may be controlled by IKK-alpha switching the binding component of CBP from p53to NFkB(19).However,we observed that these genes were downregulated in NH32, which may be due to specific cell response or other unidenti-fied reasons.In addition to NFkB,E2F1may be another important tran-scription factor that determines cell fate by controlling cell cycle in cells without functional p53.Previous studies re-ported that knockdown of NBS1was able to increase radia-tion sensitivity and suppress NFkB and XIAP expression regardless of p53status,consistent with its important role in the response to radiation(20,21).NBS1not only stabilizes p53protein to assist in p53-dependent apoptosis,but also ac-tivates E2F1via ATM to induce p53-independent apoptosis (20,22,23).Therefore,in the absence of functional p53, we hypothesize that E2F1may replace p53to regulate the ap-optosis process after10Gy irradiation treatment in WTK1 and NH32.In terms of cells responding to iso-survival treatments,our data suggested that E2F4plays a dominant role in regulating cell cycle–related genes,regardless of p53status.Cell-cycle Fig.4.The proposed model of differential transcriptional responses to higher and lower doses of ionizing radiation.Solid boxes repre-sent cellular responses.The box with a dotted line indicates evidence inferred from the literature.Question marks denote yet-to-be established connections.Distinct Pathways After Radiation Treatments in Human Cells d T.-P.L U et al.217arrest after radiation treatment has been reported in different cell types(24,25).Radiation usually induces G1/S arrest and initiates DNA repair mechanisms;however,no obvious radi-ation-induced G1arrest has been observed in both TK6and WTK1(26).Instead,a slight G2-phase arrest has been seen (27,28).The mechanism of radiation induced G2arrest re-mains unclear.In this study,we demonstrated that E2F4-re-lated genes were downregulated in the iso-survival radiation among the three cell lines.At least two studies have indicated that E2F4and p130cooperatively repress a common set of genes and maintain G2arrest to prevent en-tering mitosis(29,30).Although our microarray data indicate that transcriptional levels of E2F4did not change,many E2F4-downstream genes were repressed greater than twofold in cells despite their p53status.This indicated E2F4may be important for regulation of G2arrest in a p53-independent manner(31).Although E2F4was reported to be activated in cells ex-posed to8–10Gy radiation(31,32),the repression of E2F4-related genes was only observed in our iso-survival radiation treatments.This discrepancy may be attributable to different radiosensitivities of the cells.For example,in prostate cancer cells,10Gy radiation is sufficient to induce E2F4/p130complex formation,but it is not in MCF-7breast cancer cells(33).The other possibility is that the effects of E2F4are overridden by other transcription factors,such as p53and NFkB,in the10Gy radiation treatment.Besides, E2F4is transcriptionally repressed by E2F1(34,35).Re-pression of E2F4by E2F1may result in inactivation of E2F4-related genes while cells were exposed to10Gy radi-ation treatment.In addition,RB1,an important p53-regulated gene in-volved in an apoptotic pathway,was reported to interact with RB2and E2F1to inhibit transcription of E2F1-down-stream genes(36).We also observed different expression pat-terns of RB1in response to10Gy and iso-survival dose radiation(unpublished data).RB1was immediately upregu-lated(2.5Â)at1h after iso-survival treatment and remained elevated(2.0Â)until24h.Yet,after10Gy radiation expo-sure,RB1wasfirst downregulated(0.5Â)at1h,and then up-regulated(2.5Â)at24h.This further suggested that the important role of E2F4in the iso-survival condition may be due to E2F1inhibition by RB1,resulting in the activation of E2F4.The late induction of RB1at24h with10Gy radi-ation might result from activation of p53-dependent apopto-sis process.Based on our results,we propose a model to illustrate the differential transcription in response to higher or lower doses of radiation(Fig.4).First,after iso-survival radiation treat-ments,the E2F4-regulated genes were all downregulated among all three cell lines,but no changes were observed in the microarray and RT-PCR data after10Gy radiation. This may suggest that E2F4regulates the cell cycle and cell death pathways in the antiapoptosis process to minimize radiation damage in lower dose radiation treatment.Second, E2F1was activated by a p53-independent apoptotic pathway after lethal dose irradiation(20,21).Thus,in contrast to the well-known apoptotic pathway regulated by p53,the10Gy radiation dose may induce E2F1to initiate the process of ap-optosis in human lymphoblast cells without functional p53. Last,a subset of NFkB-related survival genes showed signif-icant changes after10Gy irradiation in WTK1and NH32. These NFkB-regulated genes were up-regulated in WTK1, but downregulated in NH32.Further experiments are needed to explore the opposite postirradiation response of NFkB be-tween WTK1and NH32.REFERENCES1.Short SC,Buffa FM,Bourne S,et al.Dose-and time-dependentchanges in gene expression in human glioma cells after low ra-diation doses.Radiat Res2007;168:199–208.2.Ding LH,Shingyoji M,Chen F,et al.Gene expression profilesof normal humanfibroblasts after exposure to ionizing radia-tion:A comparative study of low and high doses.Radiat Res 2005;164:17–26.3.Amundson SA,Bittner M,Meltzer P,et al.Induction of geneexpression as a monitor of exposure to ionizing radiation.Ra-diat Res2001;156:657–661.4.Criswell T,Leskov K,Miyamoto S,et al.Transcription factorsactivated in mammalian cells after clinically relevant doses of ionizing radiation.Oncogene2003;22:5813–5827.5.Amundson SA,Lee RA,Koch-Paiz CA,et al.Differential re-sponses of stress genes to low dose-rate gamma irradiation.Mol Cancer Res2003;1:445–452.6.Xia F,Liber HL.The tumor suppressor p53modifies mutationalprocesses in a human lymphoblastoid cell line.Mutat Res1997;373:87–97.7.Vogelstein B,Lane D,Levine AJ.Surfing the p53network.Na-ture2000;408:307–310.8.Xia F,Wang X,Wang YH,et al.Altered p53status correlateswith differences in sensitivity to radiation-induced mutation and apoptosis in two closely related human lymphoblast lines.Cancer Res1995;55:12–15.9.Tsai MH,Chen X,Chandramouli GV,et al.Transcriptional re-sponses to ionizing radiation reveal that p53R2protects against radiation-induced mutagenesis in human lymphoblastoid cells.Oncogene2006;25:622–632.10.Chuang YY,Chen Q,Brown JP,et al.Radiation-induced muta-tions at the autosomal thymidine kinase locus are not elevated in p53-null cells.Cancer Res1999;59:3073–3076.11.Chuang YY,Chen Y,Gadisetti,et al.Gene expression aftertreatment with hydrogen peroxide,menadione,or t-butyl hydro-peroxide in breast cancer cells.Cancer Res2002;62: 6246–6254.12.Kimura T,Takeda S,Sagiya Y,et al.Impaired function ofp53R2in Rrm2b-null mice causes severe renal failure through attenuation of dNTP pools.Nat Genet2003;34:440–445. 13.Tseng GC,Wong WH.Tight clustering:a resampling-based ap-proach for identifying stable and tight patterns in data.Biomet-rics2005;61:10–16.14.Mittal A,Papa S,Franzoso G,et al.NF-kappaB-dependent reg-ulation of the timing of activation-induced cell death of T lym-phocytes.J Immunol2006;176:2183–2189.15.Djavaheri-Mergny M,Amelotti M,Mathieu J,et al.NF-kappaBactivation represses tumor necrosis factor-alpha-induced au-tophagy.J Biol Chem2006;281:30373–30382.16.Nakanishi C,Toi M.Nuclear factor-kappaB inhibitors as sensi-tizers to anticancer drugs.Nat Rev Cancer2005;5:297–309.218I.J.Radiation Oncology d Biology d Physics Volume76,Number1,2010。
