2012 HKDSE Biology Paper 2
生物科BIOLOGY-九龙塘学校中学部

評核模式
組成部分 試卷一 涵蓋課程必修部分 公開考試 試卷二 涵蓋課程選修部分 校本評核 比重 時間 兩小時三十 60 % 分鐘
20 %
20 %
一小時
試卷一 涵蓋課程必修部分 試卷二 涵蓋課程選修部分
兩小時三十 60 % 分鐘
20 % 一小時
• 試卷一由甲、乙兩部組成, 甲部是多項選擇題,佔本科分數18%; 乙部由短題目、結構式題目和論述題組成, 佔本科分數42%。考生須回答試卷一的全部 試題。
– 細胞與生命分子 – 遺傳與進化 – 生物與環境 – 健康與疾病 – 科學探究
• 選修部分 (50小時)
– 人體生理學:調節與控制 – 應用生態學
細胞與生命分子
• • • • • 生命分子 碳水化合物、脂質、蛋白質、核酸 水和無機離子 細胞組織 細胞的發現、細胞膜 / 亞細胞結構和功能 物質穿越細胞膜的活動 細胞週期和分裂 擴散、滲透和主動轉運 細胞吞噬的現象 細胞能量學 細胞生長、核分裂、胞質分裂
呼吸速率和深度的控制 心輸出量的控制 運動的影響 月經週期激素的相互作用 避孕藥和治療不育
應用生態學
• • • • 人類對環境的影響 人口增長 污染控制 資源的利用 都市化和工業化的影響 保育 4R 的控制策略 污水處理 生物多樣性的重要性 可持續發展
物種的保育 利用可持續發展的概念來管理 生境的保育 資源 漁業、農業和林業
科學探究 (額外20小時)
• 簡單探究活動 • 實驗
評核目標
1 憶述及了解生物學的事實、概念、原理 及「課程架構」內各課題的相互關係; 2 應用生物學知識、概念及原理,解釋現 象和觀察結果,以及解答問題; 3 提出假說、設計並進行實驗以驗證假說; 4 展示有關生物學研習的實驗技巧;
指纹图谱 外文

Journal of Ethnopharmacology 140 (2012) 482–491Contents lists available at SciVerse ScienceDirectJournal ofEthnopharmacologyj o u r n a l h o m e p a g e :w w w.e l s e v i e r.c o m /l o c a t e /j e t h p h a rmThe potential of metabolic fingerprinting as a tool for the modernisation of TCM preparationsHelen Sheridan a ,Liselotte Krenn b ,Renwang Jiang c ,Ian Sutherland d ,Svetlana Ignatova d ,Andreas Marmann e ,Xinmiao Liang f ,Jandirk Sendker g ,∗aTrinity College,Dublin,School of Pharmacy and Pharmaceutical Sciences,East End Development 4/5,Dublin 2,Ireland bUniversity of Vienna,Department of Pharmacognosy,Althanstrasse 14,A-1090Vienna,Austria cGuangdong Province Key Laboratory of Pharmacodynamic Constituents of Traditional Chinese Medicine and New Drugs Research,College of Pharmacy,Jinan University,Guangzhou 510632,People’s Republic of China dBrunel University,Brunel Institute for Bioengineering,Uxbridge,Middlesex UB83PH,United Kingdom eUniversity of Düsseldorf,Institute of Pharmaceutical Biology and Biotechnology,Universitätsstr.1,40225Düsseldorf,Germany fChinese Academy of Sciences,Dalian Institute of Chemical Physics,Bio-technology Department,457Zhongshan Road,Dalian 116023,People’s Republic of China gUniversity of Münster,Institute of Pharmaceutical Biology and Phytochemistry,Hittorfstrasse 56,48149Münster,Germanya r t i c l ei n f oArticle history:Received 31October 2011Received in revised form 30January 2012Accepted 31January 2012Available online 7 February 2012Keywords:Chinese herbal medicines ExtractionCompound analysis MetabolomicsFingerprint analysisa b s t r a c tA vast majority Chinese herbal medicines (CHM)are traditionally administered as individually prepared water decoctions (tang )which are rather complicated in practice and their dry extracts show techno-logical problems that hamper straight production of more convenient application forms.Modernised extraction procedures may overcome these difficulties but there is lack of clinical evidence supporting their therapeutic equivalence to traditional decoctions and their quality can often not solely be attributed to the single marker compounds that are usually used for chemical extract optimisation.As demonstrated by the example of the rather simple traditional TCM formula Danggui Buxue Tang,both the chemical com-position and the biological activity of extracts resulting from traditional water decoction are influenced by details of the extraction procedure and especially involve pharmacokinetic synergism based on co-extraction.Hence,a more detailed knowledge about the traditional extracts’chemical profiles and their impact on biological activity is desirable in order to allow the development of modernised extracts that factually contain the whole range of compounds relevant for the efficacy of the traditional application.We propose that these compounds can be identified by metabolomics based on comprehensive finger-print analysis of different extracts with known biological activity.TCM offers a huge variety of traditional products of the same botanical origin but with distinct therapeutic properties,like differentially pro-cessed drugs and special daodi qualities.Through this variety,TCM gives an ideal field for the application of metabolomic techniques aiming at the identification of active constituents.© 2012 Elsevier Ireland Ltd. All rights reserved.1.IntroductionThere is an increasing interest in the use and application of CHM throughout the Western hemisphere.Notwithstanding the intake of powdered herbal drugs in different delivery forms,the typical preparation of CHM involves some kind of extraction of herbal drug material.Traditionally,the most important delivery form is theAbbreviations:AR,Astragali Radix;ASR,Angelicae sinensis Radix;CHM,Chi-nese herbal medicine;CMM,Chinese Materia Medica;DBT,Danggui Buxue Tang;FP,fingerprint analysis;PCA,principal component analysis;PLS-DA,partial least square discriminant analysis;SCO,supercritical carbon dioxide;SFE,Supercritical Fluid Extraction;TCM,traditional Chinese medicine.∗Corresponding author.Tel.:+492518333379;fax:+492528338341.E-mail address:Jandirk.sendker@uni-muenster.de (J.Sendker).water decoction of a mixture containing typically 2–12different herbal materials (Yi and Chang,2004).It should be explicitly stated that the item which is finally administered to a patient is an extract which is not just represented by the botanical origin of its herbal ingredients but also influenced by any procedure that is applied to the herbal material.Any such procedure may severely impact upon the extract’s chemical composition and hence the products’qual-ity with regard to its therapeutic efficacy.This is especially true for complex TCM formulae where we still lack a comprehensive understanding of the interactions between the thousands of chem-ical compounds that constitute the herbal metabolome(s).Besides showing synergistic effects on a pharmacodynamic level,chem-ical compounds of a TCM formula or even an individual herbal material can also interact with each other (i)on a pharmacoki-netic level,influencing the solubility,stability or resorption of compounds or (ii)on a biochemical level,where residual herbal0378-8741/$–see front matter © 2012 Elsevier Ireland Ltd. All rights reserved.doi:10.1016/j.jep.2012.01.050H.Sheridan et al./Journal of Ethnopharmacology140 (2012) 482–491483enzymes may impact the chemical composition;(iii)a mixture of herbal drugs could even be shown to contain metabolites that none of the individual herbal ingredients displayed(Nüsslein et al.,2000; Spinella,2002;Gu et al.,2004;Nualkaew et al.,2004;Ma et al., 2009;Nahrstedt and Butterweck,2010).Facing this complexity, the recent approaches for modernisation of CHM,aiming at the development of more convenient and better standardised prod-ucts,bear the risk that such features are lost by turning away from the traditional products and their traditional drug processing and extraction procedures.Extract optimisation is frequently guided by the analysis of single compounds which can often be regarded as the active substance(Yan et al.,2010).Yet,lacking the appro-priate knowledge,the activity of many herbal preparations cannot be clearly linked to single chemical compounds(Vlietinck et al., 2009).Even very well established herbal products like extracts of Salicis cortex have shown efficacy beyond that which is explicable by their content of salicin derivatives which have been consid-ered to solely constitute the extract’s active substance for decades (Schmid et al.,2001).With regard to CHM,it seems desirable that the traditional procedures like the unique pàozhìdrug processing and the extraction of specific combinations of herbal drugs are sys-tematically investigated for their impact on the herbal extracts’therapeutic qualities and metabolomes(Yi and Chang,2004;Zhao et al.,2010).In this review we will describe the current situation and problems connected to the extraction and compound anal-ysis of CHM and show possible research approaches to gain the chemical information required for a rational extract modernisation that gives consideration to the complex composition of traditional preparations.2.Traditional extraction of CHMThe majority of CHM for oral use is applied as water decoc-tions(tang).Other oral preparations include macerates in aqueous ethanol,the intake of powdered drugs suspended in water or pre-pared in pills,e.g.with honey,water or rice gruel as excipients(Li et al.,2007;Martin and Stöger,2008).Decoctions,macerates and suspensions are very simple to prepare and allow a highflexibility of the recipes.A factor which fundamentally influences the chem-ical composition of extracts is the pàozhìprocessing of the crude drug prior to extraction.This is of special importance for the detox-ification of toxic materials like Aconitum drugs(Singhuber et al., 2010)but also for the preservation of active constituents,ease of administration,flavour correction or cleansing(Zhu,1998).Many drugs can be processed by various methods like stir-frying,steam-ing or calcining in order to gain a product with altered therapeutic properties and–implicitly–altered chemical composition(Zhao et al.,2010;Zhan et al.,2011).To prepare a typical traditional decoction,a complex TCM for-mula is macerated with water for a period of time before afirst boiling step follows.Afterfiltration,the herbal material is re-extracted a second time(usually with less water)and the combined extracts are ingested and/or kept for further use.The time spans for soaking and cooking depend on the drugs as well as on the indica-tion.As a rule of thumb,decoctions for acute conditions are cooked for a shorter period of time as compared to preparations for chronic diseases.Over thousands of years numerous special instructions for the boiling process have been developed in the use of CHM.These methods seem to be related to the different physical,chemical and pharmacological characteristics of the active compounds(Martin and Stöger,2008;Körfers and Sun,2009).Herbal drugs containing volatile or temperature-sensitive sub-stances are added only a few minutes before the end of thefirst boiling period to avoid losses or decomposition(hòuxià).Expen-sive drugs,e.g.Ginseng Radix or Panacis quinquefolii Radix might be cooked separately to optimise the yield and avoid adsorption of active compounds to other ingredients within the prescription (lìngji¯an,lìngd¯un).In some instances drugs have to be added to the mixture wrapped in a thin cloth(b¯aoji¯an),this occurs with drugs like Typhae Pollen,which might lead to turbid decoctions or Inulae Flos which contains drug particles which could cause intestinal irri-tations.For toxic drugs such as Aconiti radix,which are only used orally after pàozhìprocessing,additional cooking steps or longer boiling times for the preparation are recommended.In prepara-tions containing Acori Rhizoma,an extended decoction process(in summary3h)has been proven to be very efficient for the reduction of genotoxicß-asarone(Chen et al.,2009).3.The example of Danggui Buxue Tang(DBT)In order to evaluate the significance of traditional procedures for the therapeutic value of a CHM,the rather simple two-herb for-mula DBT has been chosen as a model to investigate the influence of numerous extraction parameters on the chemical composition and pharmacological efficacies of a water decoction.The traditional composition of DBT as established over the centuries is reported as five parts Astragali Radix(AR)and one part Angelicae sinensis Radix (ASR).An investigation of several mixtures with different ratios of the two drugs has shown the best pharmacological effects as well as the highest yields of the(active)markers astragaloside IV,caly-cosin,formononetin,ferulic acid,total saponins,totalflavonoids and total polysaccharides for the traditional composition,while the undesired compound ligustilide was found to be least con-centrated with this ratio.The concentrations of these compounds varied over the examined drug ratios(AR:ASR from1:1to10:1) by a factor of∼2,demonstrating a significant impact of the drug ratio on the extraction yield(Dong et al.,2006;Po et al.,2007). Systematic investigations compared different durations of boiling, drug-solvent-ratios and numbers of extractions for DBT and also examined the chemical properties of both herbal ingredients’indi-vidual extracts.It could be shown that the treatment with DBT was up to twice as effective as individual extracts of its herbal ingredients at the same concentration.Interestingly,the extraction corresponding most closely to the traditional prescription(twofold extraction of the drug mixture with the eightfold amount of water and an entire boiling time of2h)resulted in the best extraction of active markers and the best effect in two bio-assays(Song et al., 2004;Gao et al.,2006).A mixture of individually prepared extracts of AR and ASR showed an inferior pharmacological effect when compared to the traditional co-extraction of the drugs as well as a lower concentration of active markers.It is interesting to note that the investigated markers from ASR(ligustilide and ferulic acid) showed an astonishing increase of concentration of more than25 fold when co-extracted with ASR,indicating a massive influence of AR on the extraction of these compounds(Mak et al.,2006).Another study of DBT yielded similar results with respect to biological activ-ity,however,the chromatographicfingerprints did not show such a huge influence of co-extraction on the ASR markers.The authors concluded that other,non-observed compounds influenced by co-extraction would impact the biological activity(Choi et al.,2011). The pàozhìprocessing of ASR with rice wine prior to extraction was also examined and resulted in higher extraction yields of astraga-loside IV,isoflavones and polysaccharides(Dong et al.,2006).As the processing of ASR with rice wine results in a decreased con-centration of ligustilide in the processed product,it was examined if the co-extraction of AR with pure ligustilide would impact the extraction yields of AR markers;in fact,ligustilide lowered the yields of these markers in a dose-dependent manner,indicating that the pàozhìprocessing of ASR influences the activity of DBT by altering its properties with regard to co-extraction with AR(Zheng484H.Sheridan et al./Journal of Ethnopharmacology140 (2012) 482–491et al.,2010).Further,the exchange of ASR with another drug,Radix Chuanxiong,which shares the occurrence of ligustilide and ferulic acid with ASR,was examined,resulting in both an inferior pharma-cological efficacy and lower concentrations of the AR markers when compared to a DBT extract,indicating that other,non-observed compounds of ASR may impact the quality of DBT(Li et al.,2009).It must be stated that the activities in the above mentioned stud-ies were assessed by completely different methods.Nevertheless, the example of DBT impressively demonstrates that the subject of extraction is not a trivial one and that the chemical composition and pharmacological efficacies of the traditional water decoctions are influenced to an astonishing degree by a multitude of factors which apparently have found their optimum in the traditional DBT prescription.4.Modernisation of CHM extractsThe development of modernised extracts and application forms is desired for numerous reasons:(i)individually prepared water decoctions are more likely to entail quality shortcomings caused by improper herbal drugs when compared to herbal medicines produced in industrial scale under best controllable conditions. (ii)Water decoctions are probably the worst possible preparation in terms of stability and may give rise to microbial contamina-tions,decomposition of constituents by hydrolytic or oxidative reactions or precipitations that may impact the product’s qual-ity.(iii)TCM water decoctions are infamous for their unpleasant organoleptic properties.(iv)The rather complicated and time con-suming preparation and storage of water decoctions may cause compliance problems while modernised application forms based on dry extracts(granules,capsules,tablets,etc.)are easily man-ageable for both patients and practitioners.(v)The possibility for standardisation of large-scale extracts is a prerequisite for future evidence-based research.(vi)The blinding of clinical studies employing traditional water extracts is hampered by their strong organoleptic properties(Martin and Stöger,2008;Flower et al., 2011).As evident from the studies on DBT,turning away from traditional procedures bears the risk of altering the product’s therapeutic properties.Therefore some modernisation approaches, directly based on water decoctions,have been developed to over-come the above mentioned problems.One such approach involves the delivery of water decoctions in sealed,separately packaged, daily dosages thus increasing the compliance and stability of the product.Also an unpleasantly tasting herbal placebo composed of nine culinary herbs was designed(Flower et al.,2011).4.1.Pressurised hot waterPressurised hot water extraction at high temperature is some-times applied and can be regarded as a kind of modernisation.This method has been shown to decrease the extractant’s polarity and thus provides the extraction of a wider range of compounds(Deng et al.,2007).Nevertheless,it is reported that the use of pressure cookers for the preparation of decoctions to shorten cooking times leads to less active preparations(Martin and Stöger,2008).4.2.Formation of granulesA major approach to modernise and facilitate the application of CHM was the introduction of granules,which are usually pre-pared from decoctions byfluid-bed granulation or spray-drying. As the technological properties of such extracts are impaired by high amounts of hydrophilic constituents,especially carbohydrates which result in hydroscopic,sticky and hence hardly processable extracts.Thus,excipients are added to the decoction of the herbal material(one step procedure)or mixed with the spray-dried prod-uct(two step procedure)(Martin and Stöger,2008;Ai et al.,2008). Rather high amounts of additives are required(Wang et al.,2011) and these additives further add on to the extract dose which is already quite large due to the presence of polar“bulk material”. Another method to improve the technological properties is the removal of highly polar compounds from the extract by ethanol precipitation prior to drying(Tan et al.,2006).Such preparations can be instantly applied by suspension in hot water or can be used to produce single dosage forms like capsules.An advantage is that these products can be applied quickly avoiding the time consuming preparation of a decoction.e of less polar extractantsOther modernisation approaches employ less polar extractants and hence can yield a dry extract with superior technologi-cal properties when compared to dried water extracts.These approaches comprise the extraction with(hydro)alcoholic or organic extractants performed by different extraction techniques like maceration,Soxhlet extraction,microwave-assisted extrac-tion,ultrasonic extraction or accelerated solvent extraction. Though these different extraction techniques show clear distinct properties(e.g.thermal decomposition during Soxhlet extraction, solvent limitations for microwave-assisted extraction),the dif-ferences can mainly be attributed to extraction speed while the chemical composition of the resulting extracts after establishment of equilibration is mainly influenced by the kind of extractant used and its temperature(Yan et al.,2010).From a general view, the differences in extract composition between these techniques can be regarded as negligible when compared to the differences between any of these techniques employing(hydro)alcoholic or organic solvents and a traditional water decoction.A study com-paring hydroalcoholic extracts of more than30drugs with their respective water decoctions clearly showed that decoction in most cases allows an extraction of phenolics which is similarly efficient as maceration with50%ethanol.The yields after extraction with 80%methanol,a very common method for analytical determina-tion of phenolics and their testing on antioxidant activity,was much lower.In this study,most of the decoctions,among them drugs from Rehmannia glutinosa Libosch,Astragalus membranaceus (Fisch.)Bge.or Atractylodes macrocephala Koidz.,showed better or similar antioxidant activity as compared to the hydroethanolic macerates.The activity of the80%methanolic extracts was worse for almost all tested drugs(Li et al.,2007).In a comparison of aque-ous and ethanolic extracts from different drugs a more pronounced effect on various CYP enzymes was proven for the water extracts (Tang et al.,2006).4.4.Supercritical Fluid Extraction(SFE)A completely different extraction technique that by its techni-cal properties is attractive for large scale extraction,is Supercritical Fluid Extraction(SFE).This is an extraction technique which takes advantage of the enhanced solvating and penetrating capacity of gases or liquids in their critical phases(ASTM,2006).The unique properties of supercriticalfluids were observed more than a cen-tury ago,but only in the last four decades SFE has emerged as an extraction technique(Khosravi-Darani,2010).Advantages of SFE in contrast to conventional extraction are(i)superior extraction efficiency and selectivity for low polar phytochemicals.The extrac-tion efficiency and selectivity can be tuned by optimising pressure and temperature(Wang et al.,2008;Liu et al.,2008a).(ii)Among the many applicable solvents for SFE,the most commonly used for extraction of CHM is supercritical carbon dioxide(SCO)which is inert,easily available,inexpensive,odourless,environmentalH.Sheridan et al./Journal of Ethnopharmacology140 (2012) 482–491485friendly and has mild critical point properties.To increase the sol-ubility of compounds from CHM in SCO,small amounts of polar modifiers,e.g.ethanol,may be added,usually not more than10%of the amount required for conventional extraction techniques(Chen and Ling,2000;Silva et al.,2009;Chen et al.,2011).(iii)SCO is eas-ily removable from the extract by depressurization,and thus no solvent residue is left in the extract(Vagi et al.,2002).(iv)Prefer-able product stability.The extraction is conducted at oxygen and light free operating conditions which prevent oxidation and light dependent changes,for example,a SCO extract of tomatoes could be stored for more than half a year at−20◦C without lycopene loss (Lenucci et al.,2010).Furthermore,the low temperatures minimize thermal degradation of sensitive materials,e.g.volatiles.As a green separation technique,SFE has a promising future in its application in thefields of TCM and natural products(Martinez,2008).However,there are limitations for any of the extraction method-ologies to be considered for the production of more convenient application forms.Because of the“Lipinski rule offive”(Lipinski et al.,2001),it is generally believed that less polar extractants like alcohols,acetone or SCO are capable of extracting pharmacologi-cally relevant analytes while excluding higher amounts of polar, technologically difficult“bulk material”like carbohydrates,pro-teins,amino acids etc.from the extract.However,this is a very reductionistic view which actually applies to a single active com-pound and especially excludes the possibility of pharmacokinetic synergism in herbal extracts,which has been shown to have a major impact on the chemical composition of CHM(e.g.Mak et al., 2006).It cannot be generally ruled out that the extraction of very polar constituents(needing water as extractant)can be essential for producing efficient extracts from specific drugs or for specific appli-cations,polymeric carbohydrates or compatible solutes like ectoine are examples for such highly polar bioactive compounds(Lentzen and Schwarz,2006).When relating any modernised CHM extract to the long experience of TCM it should be considered that its clinical efficacy should at least match the one of the traditional prepara-tion it relates to.It has been shown that the single active markers that are typically used for the quality control of herbal drugs may not be able to fully explain a product’s quality,especially when the drug is used as part of a co-extracted herbal mixture(Schmid et al.,2001;Li et al.,2009;Vlietinck et al.,2009),and frequently not even an active marker is known but analytical markers without known clinical relevance are used as a surrogate.As a consequence, such single markers cannot be generally recommended to guide the optimisation of an extract.Hence,for the rational development of new products it is essential to increase our knowledge about which chemical compounds define the quality of a traditional prepara-tion,so a production chain can be established thatfinally yields an improved product lacking technologically problematic“bulk mate-rial”while conserving any compound that has been shown to be relevant for the traditional product’s quality in terms of clinical efficacy.In summary,the rational modernisation of CHM requires further research to identify chemical compound that can be linked to an herbal preparation’s quality considering pharmacodynami-cally and pharmacokinetically relevant compounds.5.Fingerprint analysis in activity studies of herbal extractsA major challenge in the modernisation of CHM is the incorpo-ration of between one and twelve herbs in a given formula,thus conferring a high degree of complexity and the potential for vari-ation in composition and quality of a preparation.It has also been established that interactions between chemical components may take place during traditional co-extraction of complex herbal mix-tures and thus impact the extract’s chemical composition.Due to the lack of knowledge about the chemical compounds that completely constitute the quality of many CHM,the widespread practice of using single herb monographs and analysing of sin-gle compounds for the characterisation of CHM extracts tested in pharmacological or clinical studies seems to be insufficient.This is of particular issue when dealing with HM that contain multi-ple herbs,and can be illustrated by the fact that sometimes the same,rather unspecific metabolites are used for the quality assess-ment of herbal drugs with distinct clinical properties.Examples are chlorogenic acid,which is used as a marker compound for Caulis Lonicera,Flos Lonicera and Flos Chrysantemi,or berberine,which is used as a marker compound for Rhizoma Coptidis and Cortex Phel-lodendri(Zhou et al.,2008).Consequently,a more comprehensive view on the chemistry of extracts is desirable.It is obvious that the metabolomic techniques currently available forfingerprint analy-sis(FP)of complex biological and herbal samples and those with evolving applications in this area,have the potential to enhance the quality control of CHM and to assist with the correlation of bioactiv-ity with composition.FP can be defined in this context as an analysis aiming at the representation of an extract’s chemical composition to a maximum possible degree;while being largely untargeted,FP can also be used for or as an addition to the quantification of single compounds.Chromatographic FP and the simultaneous determination of multiple compounds is becoming an important trend(Liang et al., 2010).However an analysis of97original papers1assessing bio-logical effects of CHM extracts revealed that only16provided–exclusively chromatographic–FP data while24characterised the extracts by quantifying relevant single compounds and57did not chemically characterise the tested extract at all.We consider the presentation of FP data within activity studies of herbal extracts as fundamental information for chemical characterisation.Consider-ing the possible variability of an extract’s chemical profile that can be caused by differences in the herbal materials(growth conditions, post-harvest treatment,pàozhìprocessing,etc.)or manufacturing (extraction conditions,drying,etc.)it must be stated that the exam-ined item in such studies is literally unknown without at least some basic chemical characterisation.Different methods are used for FP which may be roughly cat-egorised as(i)low resolution techniques like TLC or IR-based methods which are typically applied for assessing the identity or origin of an herbal material by visual or–in case of NIR–computer aided comparison of signal patterns,mostly without(detailed) assignment of signals to chemical compounds(Xie et al.,2006;Sun et al.,2010).(ii)High resolution techniques like GC,HPLC,MS,NMR or hyphenated techniques allow for a detailed assignment of signals to the detected chemical compounds and are commonly accepted as well suited for FP aiming at a comprehensive extract charac-terisation as well as for metabolomic approaches.The advantages and disadvantages of these methods with regard to suitable tar-get metabolites,reproducibility of signal intensities and positions, sensitivity,resolution and sample preparation efforts have been extensively discussed(Verpoorte et al.,2008).It must be stated that no available analytical method is capable of fulfiling the demand for a complete qualitative and quantitative assessment of a biological sample’s whole chemical composition.1The literature research was performed with either Scopus or Pubmed using an OR-conjuncted combination of any botanical identifiers given by the consor-tium’s priority list of species(CP2005Latin binominal species name,taxonomically accepted Latin binominal species name,Latin drug names,Pinyin names,Latin binominal synonyms).The search result was then refined by limiting the hits to the topics“Extraction”and“Chemistry”or adding these terms to the general search term with“AND”conjunctions for Pubmed,respectively.The97papers mentioned here were chosen from the results by the fact that they assessed the biological activity of an herbal extract.。
2012年SCI分区表
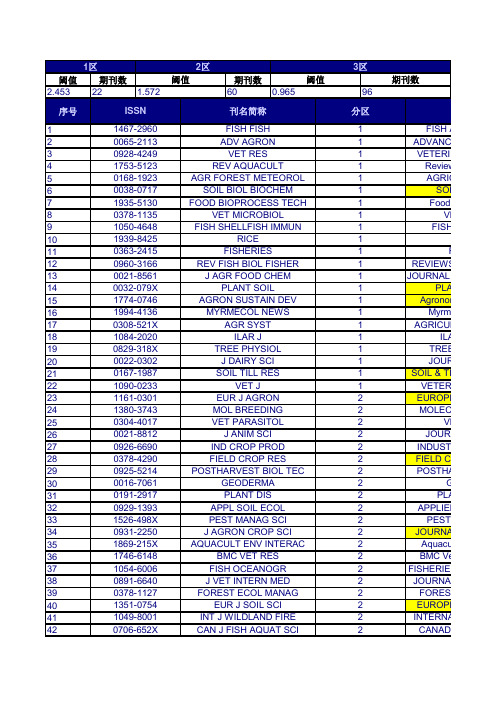
阈值期刊数期刊数2.4532260序号1234567891011121314151617181920212223242526272829303132333435363738394041420706-652XCAN J FISH AQUAT SCI2CANADIAN1351-0754EUR J SOIL SCI 2EUROPEA 0891-6640J VET INTERN MED 2JOURNAL O 1049-8001INT J WILDLAND FIRE 2INTERNATI 1746-6148BMC VET RES 2BMC Vete 0378-1127FOREST ECOL MANAG2FOREST E 0931-2250J AGRON CROP SCI 2JOURNAL 1054-6006FISH OCEANOGR 2FISHERIES O 0929-1393APPL SOIL ECOL 2APPLIED S 1869-215X AQUACULT ENV INTERAC2Aquacultu 0016-7061GEODERMA 2GEO 1526-498X PEST MANAG SCI 2PEST M 0378-4290FIELD CROP RES 2FIELD CRO 0191-2917PLANT DIS 2PLAN 0021-8812J ANIM SCI 2JOURNA 0925-5214POSTHARVEST BIOL TEC2POSTHAR1380-3743MOL BREEDING 2MOLECUL0926-6690IND CROP PROD 2INDUSTRIA 1090-0233VET J1VETERIN 0304-4017VET PARASITOL 2VET 0022-0302J DAIRY SCI 1JOURN 1161-0301EUR J AGRON 2EUROPEA 1084-2020ILAR J1ILAR 0167-1987SOIL TILL RES1SOIL & TILL 1994-4136MYRMECOL NEWS1Myrmec 0829-318X TREE PHYSIOL 1TREE P 0032-079X PLANT SOIL1PLANT 0308-521X AGR SYST 1AGRICULT0960-3166REV FISH BIOL FISHER 1REVIEWS IN1774-0746AGRON SUSTAIN DEV 1Agronomy 1939-8425RICE 10021-8561J AGR FOOD CHEM1JOU 0378-1135VET MICROBIOL 1VET 0363-2415FISHERIES1FIS0038-0717SOIL BIOL BIOCHEM 1SOIL B 1050-4648FISH SHELLFISH IMMUN1FISH &1753-5123REV AQUACULT1Reviews 1935-5130FOOD BIOPROCESS TECH1Food an0065-2113ADV AGRON 1ADVANCES 0168-1923AGR FOREST METEOROL 1AGRICU ISSN 刊名简称分区刊0928-4249VET RES 1VETERINA 阈值阈值期刊数1467-2960FISH FISH 1FISH AN 1区2区3区1.5720.9659643 44 45 46 47 48 49 50 51 52 53 54 55 56 57 58 59 60 61 62 63 64 65 66 67 68 69 70 71 72 73 74 75 76 77 78 79 80 81 82 83 84 85 86 8788 890936-6768REPROD DOMEST ANIM3REPRO 0300-9858VET PATHOL3VETERINAR 0014-2336EUPHYTICA3EUP 0002-9254AM J ENOL VITICULT3AMERICAN 1461-9555AGR FOREST ENTOMOL3AGRICU 0022-0299J DAIRY RES2JOURN 1751-7311ANIMAL3A 0931-2668J ANIM BREED GENET2JOURNA 0043-1745WEED SCI3WEED 0929-1873EUR J PLANT PATHOL2EUROPEA 0931-1890TREES-STRUCT FUNCT2TREES-ST 0002-1962AGRON J2AGRONO 0165-7836FISH RES2FISHERIE 1353-5773AQUACULT NUTR2AQUACULT 0032-5791POULTRY SCI2POULT 0425-1644EQUINE VET J2EQUINE 0378-4320ANIM REPROD SCI2ANIMAL R 0021-8596J AGR SCI2JOU 0005-2086AVIAN DIS2AVIAN 0266-0032SOIL USE MANAGE2SOIL 1385-1314NUTR CYCL AGROECOSYS2NUTRIEN 1936-9751FOOD ANAL METHOD2Food Ana 0959-4493VET DERMATOL2VET 0140-7775J FISH DIS2JOURNAL O 1436-8730J PLANT NUTR SOIL SC2JOURNA 0307-9457AVIAN PATHOL2AVIAN P 0377-8401ANIM FEED SCI TECH2ANIMAL FEE 0011-183X CROP SCI2CROP 0168-1591APPL ANIM BEHAV SCI2APPLI 0342-7188IRRIGATION SCI2IRRIGAT 0003-1488JAVMA-J AM VET MED A2JAVMA-JO 0739-7240DOMEST ANIM ENDOCRIN2DOMES 0177-5103DIS AQUAT ORGAN2DISEASE 1054-3139ICES J MAR SCI2ICES JOUR 0043-1737WEED RES2WEED 0378-3774AGR WATER MANAGE2AGRICULT 1866-7910FOOD ENG REV2Food Engin 0044-8486AQUACULTURE2AQUA 1612-4669EUR J FOREST RES2EUROPEA 1064-1262REV FISH SCI2REVIEWS 0269-283X MED VET ENTOMOL2MEDICAL A 1865-1674TRANSBOUND EMERG DIS2Transbound 0361-5995SOIL SCI SOC AM J2SOIL SCIEN 0178-2762BIOL FERT SOILS2BIOLOGY AN 0093-691X THERIOGENOLOGY2THERIO 0165-2427VET IMMUNOL IMMUNOP2VETERINAR 0167-5877PREV VET MED2PREVENTIV909192939495969798991001011021031041051061071081091101111121131141151161171181191201211221231241251261271281291301311321331341351362042-6496FOOD FUNCT 3Food1437-4781FOREST PATHOL 3FOREST 0167-4366AGROFOREST SYST 3AGROFORE0830-9000CAN J VET RES 3CANADIAN0275-6382VET CLIN PATH3VETERIN 0044-605X ACTA VET SCAND 3ACTA V 1344-7610BREEDING SCI 3BREEDI 1479-3261J VET EMERG CRIT CAR 3JOURNAL O 0022-1201J FOREST3JOURNAL 0037-5330SILVA FENN 3SILVA 1537-5110BIOSYST ENG 3BIOSYSTEM1467-2987VET ANAESTH ANALG3VETERINAR0889-048X AGR HUM VALUES 3AGRICULTU 0015-749X FOREST SCI 3FORES 0304-4238SCI HORTIC-AMSTERDAM 3SCIENTIA H 0022-4561J SOIL WATER CONSERV3JOURNAL 1618-8667URBAN FOR URBAN GREE3URBAN FOR 1936-5802CHEMOSENS PERCEPT 3Chemosen 0090-3558J WILDLIFE DIS3JOURNAL 0004-9573AUST J SOIL RES3AUSTRALIA 1085-3278LAND DEGRAD DEV 3LAND DE 1550-7424RANGELAND ECOL MANAG 3Rangela 0921-4488SMALL RUMINANT RES 3SMALL 0253-1933REV SCI TECH OIE 3REVUE SC 0043-9339WORLD POULTRY SCI J 3WORLDS PO 0179-9541PLANT BREEDING 3PLANT 1871-1413LIVEST SCI 3Livesto 1040-6387J VET DIAGN INVEST 3JOURNAL O1385-2256PRECIS AGRIC 3PRECISION0002-9645AM J VET RES3AMERICAN 0261-2194CROP PROT 3CROP P 0015-752X FORESTRY 3FO 0140-7783J VET PHARMACOL THER 3JOURNAL O 0042-4900VET REC 3VETERIN 0034-5288RES VET SCI 3RESEARCH 1836-0947CROP PASTURE SCI3Crop & Pa 1098-612X J FELINE MED SURG 3JOURNA 0161-3499VET SURG3VETERINA 0920-1742FISH PHYSIOL BIOCHEM 3FISH PHY 0925-9864GENET RESOUR CROP EV3GENETIC R 0002-8487T AM FISH SOC3TRANSAC 1473-5903INT J AGR SUSTAIN 3Internatio 0045-5067CAN J FOREST RES 3CANADIAN 0261-1929ATLA-ALTERN LAB ANIM 3ATLA-ALT 1286-4560ANN FOREST SCI 3ANNALS 1125-7865DENDROCHRONOLOGIA3DENDROC 0906-6691ECOL FRESHW FISH 3ECOLOGY O4190005-9080BERLANDWIRTSCH4BERICHTE UBERLANDWIRTSCHAFT0.1290.1494200032-681XPRAKTTIERARZT4PRAKTISCHETIERARZT0.1280.094区总计期刊数期刊数2684463年平均IF5.5744.2153.8013.7493.2713.2413.1723.1523.0863.0032.7592.7572.7032.6742.6512.6442.6392.612.5242.5082.4692.4532.452.4392.3962.3812.362.3472.3262.3192.3192.2962.2512.2232.1882.1862.1622.1462.1432.1342.1162.11ADIAN JOURNAL OF OPEAN JOURNAL OF 2.213NAL OF VETERINARY 2.34NATIONAL JOURNAL 2.231 Veterinary Research 1.992EST ECOLOGY AND 2.487NAL OF AGRONOMY 2RIES OCEANOGRAPHY 2.044IED SOIL ECOLOGY 2.433aculture Environment 2.188GEODERMA 2.368ST MANAGEMENT 2.251 CROPS RESEARCH 2.318PLANT DISEASE 2.449URNAL OF ANIMAL 2.474THARVEST BIOLOGY 2.411ECULAR BREEDING 2.096STRIAL CROPS AND 2.469ERINARY JOURNAL 2.852VETERINARY 2.579URNAL OF DAIRY 2.239OPEAN JOURNAL OF 2.477ILAR JOURNAL 2.564 TILLAGE RESEARCH 2.425rmecological News 2.333EE PHYSIOLOGY 2.876LANT AND SOIL 2.644CULTURAL SYSTEMS 2.899WS IN FISH BIOLOGY 2.733nomy for Sustainable3.33Rice 2.5JOURNAL OF 2.823VETERINARY 3.105FISHERIES 2.367SOIL BIOLOGY & 3.327SH & SHELLFISH 3.322iews in Aquaculture 3.504od and Bioprocess 3.703NCES IN AGRONOMY4.036RICULTURAL AND 3.389刊名全称5.204RINARY RESEARCH 4.062011年IF H AND FISHERIES 5.82.0792.0772.0722.0372.0272.0162.0082.0031.9661.9321.931.9121.8931.8681.861.8341.821.7991.7881.7771.7681.7671.7591.7581.721.7151.7111.7061.71.6971.6961.6851.6781.6691.6611.641.5911.5781.5771.5721.5711.5551.5541.5481.5451.5231.519EPRODUCTION IN RINARY PATHOLOGY 1.356EUPHYTICA 1.554RICAN JOURNAL OF1.945RICULTURAL AND 1.596URNAL OF DAIRY 1.826Animal1.744URNAL OF ANIMAL 1.566WEED SCIENCE1.733OPEAN JOURNAL OF 1.455S-STRUCTURE AND 1.685RONOMY JOURNAL 1.413HERIES RESEARCH 1.586CULTURE NUTRITION 1.794OULTRY SCIENCE 1.728UINE VETERINARY2.179AL REPRODUCTION 1.75JOURNAL OF1.456VIAN DISEASES1.462SOIL USE AND2.041RIENT CYCLING IN 1.792d Analytical Methods 1.608VETERINARY 1.943AL OF FISH DISEASES 1.943URNAL OF PLANT 1.596IAN PATHOLOGY 2L FEED SCIENCE AND 1.691CROP SCIENCE1.711PPLIED ANIMAL1.918IGATION SCIENCE 1.641A-JOURNAL OF THE 1.791OMESTIC ANIMAL 1.635EASES OF AQUATIC2.201OURNAL OF MARINE 2.056EED RESEARCH 1.924ICULTURAL WATER 2.007Engineering Reviews 1.893AQUACULTURE1.998OPEAN JOURNAL OF 1.982IEWS IN FISHERIES2.041AL AND VETERINARY 1.91oundary and Emerging 1.946CIENCE SOCIETY OF 1.979GY AND FERTILITY OF 1.809ERIOGENOLOGY 1.9632.319RINARY IMMUNOLOGY 2.076ENTIVE VETERINARY 2.0461.5181.5061.5021.5011.4841.461.4561.4541.4511.4431.4411.4251.4211.4181.4171.4111.4111.4051.4041.4031.3981.381.3731.3381.3261.3151.2891.2731.271.2581.2561.2451.2391.2351.2321.2291.2281.221.2071.2071.1961.1941.1921.1881.1871.1821.179Food & Function REST PATHOLOGY 1.179FORESTRY SYSTEMS 1.74ADIAN JOURNAL OF 0.939ERINARY CLINICAL 1.378CTA VETERINARIA 1.367EEDING SCIENCE 1.556NAL OF VETERINARY 2.038RNAL OF FORESTRY 1.248SILVA FENNICA 1.248STEMS ENGINEERING 1.354INARY ANAESTHESIA 0.944ULTURE AND HUMAN 1.354OREST SCIENCE 1.047TIA HORTICULTURAE 1.54RNAL OF SOIL AND 1.265 FORESTRY & URBAN 1.527mosensory Perception 1.643RNAL OF WILDLIFE 1.27RALIAN JOURNAL OF 1.53D DEGRADATION & 1.079ngeland Ecology &1.461MALL RUMINANT 1.402UE SCIENTIFIQUE ET 1.099DS POULTRY SCIENCE 1.295LANT BREEDING1.596ivestock Science 1.104NAL OF VETERINARY 1.214SION AGRICULTURE 1.506RICAN JOURNAL OF 1.269OP PROTECTION 1.549FORESTRY1.337NAL OF VETERINARY 1.402ERINARY RECORD 1.248ARCH IN VETERINARY 1.181p & Pasture Science 1.418URNAL OF FELINE 1.649ERINARY SURGERY 1.265 PHYSIOLOGY AND 1.38TIC RESOURCES AND 1.554NSACTIONS OF THE 1.528rnational Journal of 1.696ADIAN JOURNAL OF 1.592-ALTERNATIVES TO 1.455NALS OF FOREST 1.685DROCHRONOLOGIA 1.5251.788GY OF FRESHWATER 1.5731.1771.1741.1651.1641.1631.1471.131.1191.0921.091.0841.0831.0811.0761.0741.0731.0671.0661.0651.0631.0611.0571.0461.0411.0411.0391.0361.0351.0341.0331.0231.0171.0140.9860.9860.9810.9810.9780.9760.970.970.9650.9620.96RINARY CLINICS OF 1.231ee-Ring Research Science & Technology 0.884ED TECHNOLOGY 1.212ANTHROZOOS1.183RNAL OF AQUATIC 0.833STAN VETERINARY 0.86ADIAN VETERINARY 1.063al Production Science 1.255ADIAN JOURNAL OF 0.821CHIVES OF ANIMAL 0.986and Water Environment 0.986actions of the ASABE 0.987SOIL SCIENCE1.144AWA JOURNAL1.033CAL ANIMAL HEALTH 1.115SCIENCE AND PLANT 1.042H POULTRY SCIENCE 1.005RICAN JOURNAL OF 1.017RALIAN VETERINARY 0.945ORNIA COOPERATIVE 1.234SHERY BULLETIN 1.098URNAL OF ANIMAL 1.074AL OF SMALL ANIMAL 1alian Journal of Crop 0.855RINARY CLINICS OF 1.474 AMERICAN JOURNAL 1.632 and Coastal Fisheries 1.065EALAND VETERINARY 0.943l of Veterinary Science 1.161n Research and Practice 0.887INAVIAN JOURNAL OF 1.197AQUACULTURAL 1.083INARY RADIOLOGY & 1.082RIES MANAGEMENT 1.421NIMAL WELFARE1.135AQUATIC LIVING 1.294 JOURNAL OF ANIMAL 1.079nary and Comparative 1.152VETERINARY0.748ASS AND FORAGE 1.561CULTURE RESEARCH 1.2031.099RINARY CLINICS OF 1.638REST POLICY AND 1.482。
3D脑肿瘤与内部大脑结构的MR图像分割(IJIGSP-V4-N1-5)

I.J. Image, Graphics and Signal Processing, 2012, 1, 35-43Published Online February 2012 in MECS (/)DOI: 10.5815/ijigsp.2012.01.053D Brain Tumors and Internal Brain Structures Segmentation in MR Images1P.NARENDRAN M.Sc., PGDCA, M.Phil., B.Ed,Head & Associate Professor, PG & Research Department of Computer Science,Gobi Arts & Science College (Autonomous),Gobichettipalayam – 638 453, Erode District, Tamil Nadu, India.Email ID:narendranp@2 Mr. V.K. NARENDIRA KUMAR M.C.A., M.Phil.,Assistant Professor, Department of Information Technology,Gobi Arts & Science College (Autonomous),Gobichettipalayam – 638 453, Erode District, Tamil Nadu, India.Email ID: kumarmcagobi@3 Dr. K. SOMASUNDARAMProfessor & Head, Dept. of Computer Science & Applications,Gandhigram rural university,Gandhigram – 624 302, Tamil Nadu, India.Email ID: somasundaramk@Abstract - The main topic of this paper is to segment brain tumors, their components (edema and necrosis) and internal structures of the brain in 3D MR images. For tumor segmentation we propose a framework that is a combination of region-based and boundary-based paradigms. In this framework, segment the brain using a method adapted for pathological cases and extract some global information on the tumor by symmetry based histogram analysis. We propose a new and original method that combines region and boundary information in two phases: initialization and refinement. The method relies on symmetry-based histogram analysis. The initial segmentation of the tumor is refined relying on boundary information of the image. We use a deformable model which is again constrained by the fused spatial relations of the structure. The method was also evaluated on 10 contrast enhanced T1-weighted images to segment the ventricles, caudate nucleus and thalamus.Index Terms—3D, Brain, Tumor, Segmentation, MRI, Image Registration, and Brain Structures.I.INTRODUCTIONTumor is one of the most common brain diseases, so its diagnosis and treatment have a vital importance for more than 400000 persons per year in the world (based on the World Health Organization (WHO) estimates). On the other hand, in recent years, developments in medical imaging techniques allow us to use them in several domains of medicine, for example, computeraided pathologies diagnosis, follow-up of these pathologies, surgical planning, surgical guidance, statistical and time series (longitudinal) analysis. Among all the medical image modalities, Magnetic Resonance Imaging (MRI) is the most frequently used imaging technique in neuroscience and neurosurgery for these applications. MRI creates a 3D image which perfectly visualizes anatomic structures of the brain such as deep structures and tissues of the brain, as well as the pathologies.Segmentation of objects, mainly anatomical structures and pathologies from MR images is a fundamental task, since the results often become the basis for other applications. Methods for performing segmentation vary widely depending on the specific application and image modality. Moreover, the segmentation of medical images is a challenging task, because they usually involve a large amount of data, they have sometimes some artifacts due to patient’s motion or limited acquisition time and soft tissue boundaries are usually not well defined. The accurate segmentation of internal structures of the brain is of great interest for the study and the treatment of tumors. It aims at reducing the mortality and improving the surgical or radio therapeutic management of tumors. In brain oncology it is also desirable to have a descriptive human brain model that can integrate tumor information extracted from MRI data such as its localization, its type, its shape, its anatomo-functional positioning, as well as its influence on other brain structures.Despite numerous efforts and promising results in the medical imaging community, accurate and reproducible segmentation and characterization of abnormalities are still a challenging and difficult task. Existing methods leave significant room for increased automation, applicability and accuracy.II. OBJECTIVESThe first aim of this work is to develop a framework for a robust and accurate segmentation of a large class of brain tumors in MR images. Most existing methods are region-based. They have several advantages, but line and edge information in computer vision systems are also important. The proposed method tries to combine region and edge information, thus taking advantage of both approaches while cancelling their drawbacks. 3D contrast enhanced T1-weighted and FLAIR images are the inputs to perform an automatic segmentation of the solid part of tumor and the potential associated edema and necrosis [3].We first segment the brain to remove non-brain data. However, in pathological cases, standard segmentation methods fail, in particular when the tumor is located very close to the brain surface. Therefore we propose an improved segmentation method, relying on the approximate symmetry plane. Then we developed two new and original methods to detect and initially segment brain tumors. The first one is a fuzzy classification method which combines membership, typicality and neighborhood information [1]. The second one relies on a symmetry-based histogram analysis. The approximate sagittal symmetry plane is first computed, and the tumor is then extracted by comparing the histograms of the two cerebral hemispheres. To refine the initial segmentation, which is not accurate enough, we use edge information.A deformable model constrained by spatial relations is applied for this purpose [8].Segmentation of internal structures of the pathological brain is another aim of this paper. The use of prior knowledge can guide the segmentation task in medical imaging. Due to the existence of different types of tumors and consequently different effects on the brain structures, segmentation using prior knowledge such as an atlas is a difficult task. In this work we use another type of prior knowledge which reserves its properties in pathological cases.III. ANATOMY OF THE BRAINThe nervous system is commonly divided into the Central Nervous System (CNS) and the peripheral nervous system. The CNS is made up of the brain, its cranial nerves and the spinal cord. In this section we briefly study the cell structures and anatomical components of the brain. The brain consists mainly of two tissue types: Gray Matter (GM) and White Matter (WM). Gray matter is made of neuronal and glial cells, also known as neuroglia or glia that control brain activity, while the cortex is a coat of gray matter that covers the brain and the basal nuclei are the gray matter nuclei located deep within the white matter. The basal nuclei include: caudate nucleus, putamen, pallidum and claustrum. White matter fibers are myelinated axons which connect the cerebral cortex with other brain regions. The corpus callosum, a thick band of white matter fibers, connects the left and right hemispheres ofthe brain.Figure 1: Anatomy of the brainAnatomically the brain is composed of the cerebrum, the cerebellum and the brainstem (Figure 1). The cerebrum, which forms the major part of the brain, is divided into two major parts by the longitudinal fissure: the right and left cerebral hemispheres. Each hemisphere is divided into 4 lobes or areas: the frontal lobe in the front of the brain, the parietal lobe behind the frontal lobe, the temporal lobe on each side of the brain and theoccipital lobe at the back of the brain as illustrated in Figure 1.The central structures of the brain, i.e. the diencephalon, include the thalamus, hypothalamus and pituitary gland. The ventricular system that provides the CSF is divided into four cavities called ventricles, which are connected by a series of holes referred to as foramen, and tubes. Two ventricles enclosed in the cerebral hemispheres are called the lateral ventricles (first and second). They communicate with the third ventricle. The third ventricle is in the center of the brain, and its walls are made up of the thalamus and hypothalamus. The third ventricle connects with the fourth ventricle through a long tube.IV. BRAIN TUMORSA brain tumor is an intracranial mass produced by an uncontrolled growth of cells either normally found in the brain such as neurons, lymphatic tissue, glial cells, blood vessels, pituitary and pineal gland, skull, or spread from cancers primarily located in other organs [2].Brain tumors are classified based on the type of tissue involved, the location of the tumor, whether it is benign or malignant, and other considerations. Primary (true) brain tumors are the tumors that originated in the brain and are named for the cell types from which they originated. They can be benign (non cancerous), meaning that they do not spread elsewhere or invade surrounding tissues. They can also be malignant and invasive (spreading to neighboring area). Secondary ormetastasis brain tumors take their origin from tumor cells which spread to the brain from another location in the body. Most often cancers that spread to the brain to cause secondary brain tumors originate in the lumy, breast, and kidney or from melanomas in the skin.(a)(b)(c)(d)Figure 2: MRI of brain. (a) T1-weighted image without contrast enhancement. (b) T1-weighted image with contrast enhancement. (c)T2-weighted image. (d) FLAIR image.Figure 3: One axial slice of a MR image of the brain showing tumorareas.Each primary brain tumor, in addition to thesolid portion of the tumor, may have other associated parts such as edema and necrosis as in Figures 2 and 3. Edema is one of the most important factors leading to mortality associated with brain tumors. By definition, brain edema is an increase in brain volume resultingfrom increased sodium and water content and results from local disruption of the Blood Brain Barrier (BBB). Edema appears around the tumor mainly in white matter regions. Tumor associated edema is visible in MRI, as either hypo intense (darker than brain tissue) or rarely is intense (same intensity as brain tissue) in T1-weighted scans, or hyper intense (brighter than brain tissue) in T2-weighted and FLAIR MRI (Figure 3). Necrosis iscomposed of dead cells in the middle of the brain tumor and is seen hypo intense in T1-weighted images (Figure 3). A brain tumor may also infiltrate the surrounding tissues or deform the surrounding structures [4].V. CLASSIFICATION OF BRAIN TUMORS The classification of primary brain tumors is usually based on the tissue of origin, and occasionally on tumor location. The degree of tumor malignancy is determinedby the tumor’s histopathology features. Because of the substantial variety and unusual biology of brain tumors, it has been extremely difficult to develop a widely accepted histological classification system .The earliest brain tumor classifications were provided by Bailey and Cushing in 1926. Their classification scheme proposed 14 brain tumor types, directed important attention to the process of cell differentiation, and dominated views of gliomas until 1949 when a new system was introduced by Kernohan and Sayre. Kernohan and Sayre made the important realization that different histopathology appearances may not represent separate tumor types but rather different degrees of differentiation of one tumor type.They classified tumors into five subtypes: astrocytoma, oligodendroglioma, ependymoma, gangliocytoma, and medulloblastoma and very importantly added a four-level grading system for astrocytomas [1]. The grading system was based on increasing malignancy and decreasing differentiation with increasing tumor grade. The addition of a grading system was a very important advance in classifying brain tumors, and provided information not only regarding tumors’ bio logic behavior but also information that could be used toguide treatment decisions.VI. BRAIN TUMOR SEGMENTATION Despite numerous efforts and promising results in the medical imaging community, accurate and reproducible segmentation and characterization of abnormalities are still a challenging and difficult task because of the variety of the possible shapes, locations and image intensities of various types of tumors. Some of them may also deform the surrounding structures or may be associated to edema or necrosis that changes the image intensity around the tumor. As we surveyed in the previous chapter, existing methods leave significant room for increased automation, applicability and accuracy. Most of them are usually dedicated to full-enhanced tumors or specific types of tumors, and do not extent easily to more general types [5].The automated brain tumor segmentation method that we have developed consists of two main components: preprocessing and segmentation. The inputs of this system are two different modalities of MR images: CE-T1w and FLAIR that we believe are sufficient for brain tumor segmentation [2]. In the segmentation preprocessing section, operations such as: reduction of intensity inhomogeneity and inter-slice intensity variation of images, spatial registration (alignment) of the input images, segmentation of the brain, computation of the approximate symmetry plane and histogram analysis based on symmetry plane are performed.VII. PREPROCESSINGIn the real MRI data there are some problems that have to be first solved before any segmentation operation. Therefore we first try to reduce the intensity in homogeneity and inter slice intensity variations, two main problems of MRI data, in the input images. Our system uses two different modalities of MRI, usually not spatially aligned and often having different resolutions. Hence it is required to add a registration and interpolation step. The brain is then segmented by a combination of histogram analysis, morphological operations and symmetry analysis. In this step we compute the approximate symmetry plane that will be used in the segmentation and sometimes to correct the brain segmentation result. Finally we analyze the histograms of the right and left hemispheres to detect the pathological hemisphere and the type of tumor.A. Image PreprocessingTwo main problems of MR images are intensity inhomogeneity or bias field and interslice intensity variations which are caused by the limitations of the current MRI equipments (the main factors are RF excitation field inhomogeneity, non-uniform reception coil sensitivity, eddy currents driven by field gradients, RF penetration and standing wave effects). In today MR images, the bias field is not always visible to the human observer, but it causes significant tissue misclassification problems when intensity-based segmentation is used [7]. Therefore, it is required tocorrect intensity inhomogeneity in the image volume.(a) (b) (c)Figure 4: Bias field correction. (a) An axial slice of the original image.(b) Same bias field corrected slice. (c) Applied bias field.An automatic method based on entropy minimization is used (as seen in Figure 4). In addition to a smoothly varying field inhomogeneity, two-dimensional multislice sequence MR images, which are acquired in an interleaved way, are typically also corrupted with a slice by slice constant intensity offset. This is usually due to gradient eddy currents and crosstalk between slices. Hence, it is required to normalize interslice intensity to have a correct 3D segmentation. Here a method based on scale-space analysis of histogram is used [9].B.Image RegistrationImage registration is the operation of aligning images in order to relate corresponding features. For most kinds of image processing on two or more images, it is required that the images are aligned, so that one voxel position represents the same anatomical position in all images [10]. This step allows the use of modalities that are not in perfect alignment. An image registration program has typically four modules: the transformation model, feature extraction, similarity measure, and an optimization method. In our system, the CE-T1w image is used as reference or target image (R) and the FLAIR image as test or source image (T).Several transformation models can be used to transform the test image T, such as rigid, affine, projection and curved transformations. Here, the registration concerns 3D head images from the same person, which makes it reasonable to assume that the head will not be deformed, and thus can be considered a rigid body. Hence, the rigid transformation model (rotation and translation) is therefore sufficient for our purpose. By using a rigid transformation, we are assuming that the two images can be aligned using a parameterization with 6 degrees of freedom. Here we restrict ourselves to methods that use directly the intensity images as features, thus avoiding the preliminary extraction of corresponding features in the two images.C. Brain SegmentationThe next step of preprocessing consists of brain segmentation. Several methods have been proposed to perform this operation and some of them are available in software’s such as Brain-Visa, FSL and Brain suite. Unfortunately most of them fail in the case of the presence of a tumor in the brain, especially if located on the border of the brain (Figure 5). To solve this problem, we propose to perform a symmetry analysis, based on the assumption that tumors are generally not symmetrically placed in both hemispheres, while the whole brain is approximately symmetrical.First we segment the brain using histogram analysis and morphological operations. This leads to a partial segmentation, where a part corresponding to the tumor may be missing. The algorithm is applied on the gray level image of the head to compute the approximate symmetry plane, because the segmented brain is not symmetric. The computed symmetry planes of the head and of the segmented brain in normal cases are approximately equal and this approximation is acceptable in pathological cases for tumor detectionpurpose.(a)(b)(c) (d)Figure 5: Pathological brain segmentation using existing methods. (a) One slice of the original image on two examples. (b) Segmented brain by histogram analysis and morphological operations using Brain Visa.(c) Segmented brain by BET using FSL. (d) Segmented brain by BSE using Brain suite.D. Structure SegmentationThe proposed method for internal brain structures segmentation, such as for tumors, has two phases: initialization and refinement. In other words, we first segment the brain tissues (consequently the internal structures of the brain) and since this segmentation for internal brain structures is not fine enough, we then refine them one by one using prior information. Toperform these two phases, the segmentation procedure consists of the following steps [6]:Global segmentation of the brain,Retrieving spatial relations,Selecting the valid spatial relations,Fuzzification and fusion of relations and providing the ROI,Searching the initial segmentation of structure,Refining the initial segmentation,Repeating from step 2 for other structures.Global segmentation of the brain to segment the brain tissues and its structures we use two methods, the first one is the MPFCM method and the second one is the multiphase level sets.VIII. PERFORMANCE MEASURES Characterizing the performance of image segmentation methods is a challenge in image analysis. An important difficulty we have to face in developing segmentation methods is the lack of a gold standard for their evaluation. Accuracy of a segmentation technique refers to the degree to which the segmentation results agree with the true segmentation. Although physical or digital phantoms can provide a level of known “ground truth”, they are still una ble to reproduce the full range of imaging characteristics and normal and abnormal anatomical variability observed in clinical data.Manual segmentation of desired objects by domain experts can be considered as an acceptable approach (it still suffers from inter-expert and intra-expert variability). The result of an automated method is then compared to the manually segmented object by an expert or a group of experts, and if the algorithm generates segmentations sufficiently similar to the ones provided by the experts, it is accepted. A number of metrics have been proposed to measure the similarity between the segmentations, including volume measures and surface measures.IX. EXPERIMENTAL RESULTSIn order to implement this 3D brain tumors and internal brain structures segmentation in MR images efficiently, program is used. This program could speed up the development of this system because it has facilities to draw forms and to add library easily [6].The proposed method was applied to 10 clinical MRI datasets of various origins and types. We illustrate the results on four cases, for which manual segmentation of several structures was available, and which exhibit tumors with different shapes, locations, sizes, intensities and contrasts. Evaluation of the segmentation results was performed through quantitative comparisons with manual segmentations, using volume and surface measures. Segmentation results are quantitative evaluations are high accuracy. The voxel size is typically 1 × 1 × 1.3 mm3, so that the average error is less than one voxel. The Hausdorff distance represents the error for the worst point, which explains its higher values. Although the segmented structures are relatively small (about 4000 m3), the volume metrics shows good results. For the similarity index measures, values above 70% are satisfactory. The results show that the segmentation of caudate nuclei is better than thalamus due to their well defined borders. The comparison of the results obtained using the initial segmentation of MPFCM and multiphase level sets illustrates that there is not a large difference between them. But the MPFCM method is faster than the multiphase level sets method.X. CONCLUSIONIn this paper we deal with 3D MR images in order to segment brain tumors and internal brain structures for the applications such as treatment and follow-up, surgery, individual modeling, etc. We first review the discussed topics and the contributions and following this we discuss possible future directions. We proposed a new method for segmentation of pathological brain structures. This method combines prior information of structures and image information (region and edge) for segmentation. To represent the prior information weused ontological engineering tools. We also proposed a simple ontology for a specific classification of tumors and it can be extended for other classification of tumors (such as tumor grading). Brain tumor segmentation method uses contrast enhanced T1- weighted and FLAIR images for segmentation and it consists of two steps: preprocessing and segmentation. In the preprocessing step, in addition to use the classical methods for reducing the noise and inhomogeneity and registration, we proposed a new adapted method for correct and robust brain segmentation. The brain is segmented by a combination of histogram analysis, morphological operations and symmetry analysis. A new symmetry-based histogram analysis was proposed that is able to detect automatically the tumor type and the pathological hemisphere.The segmentation of the pathological brain structures is a difficult task due to the different effects of the different tumors. Using prior information such as an atlas or adapting it to guide the segmentation is also difficult because of these different effects. We proposed a new method that in addition to region and edge information uses a type of prior information which is more consistent in pathological cases. The spatial relations between structures are the prior information used in this method. Here we deal with three main problems: explicit representation of spatial relations for each structure, adaptation of spatial relations for pathological cases and segmentation method and volume considered an important parameter such as, storage, transmission, visualization, and quantitative analysis.REFERENCES[1] Algorri, M. E. and Flores-Mangas, F. (2004). Classification ofanatomical structures in MR brain images using fuzzy parameters. IEEE Transactions on Biomedical Engineering, 51(9):1599–1608. [2] Dou, W., Ruan, S., Chen, Y., Bloyet, D., and Constans, J. M.(2007). A framework of fuzzy information fusion for segmentation of brain tumor tissues on MR images. Image and Vision Computing, 25:164–171.[3] Fletcher-Heath, L. M., Hall, L. O., Goldgof, D. B., and Murtagh, F.(2001). Automatic segmentation of non-enhancing brain tumor in magnetic resonance images. Artificial Intelligence in Medicine, 21:43–63.[4] Gering, D. T. (2003). Recognizing Deviations from Normalcy forBrain Tumor Segmentation. PhD thesis, Massachusetts Institute of Technology.[5] Hata, N., Muragaki, Y., Inomata, T., Maruyama, T., Iseki, H., Hori,T., and Dohi, T. (2005). Interaoperative tumor segmentation and volume measurement in MRI guided glioma surgery for tumor resection rate control. Academic Radiology, 12:116–122.[6] Hu, S. and Collins, D. L. (2007). Joint level-set shape modelingand appearance modeling for brain structure segmentation. NeuroImage, 36:672–683.[7] K. M., Jia, W., and Marsh, R. (2003). Fractal analysis of tumor inbrain MR images. Machine Vision and Applications, 13:352–362. [8] Khotanlou, H., Colliot, O., and Bloch, I. (2007). Automatic BrainTumor Segmentation using Symmetry Analysis and Deformable Models. In International Conference on Advances in Pattern Recognition (ICAPR), pages 198–202, Kolkata, India.[9] Lefohn, A., Cates, J., and Whitaker, R. (2003). Interactive, GPU-based level sets for 3D brain tumor segmentation. Technical report, University of Utah.[10] Maintz, J. and Viergever, M. (1998). A survey of medical imageregistration. Medical Image Analysis, 2(1):1–36.First Author Profile:Mr. P.NARENDRAN M.Sc., PGDCA, M.Phil., B.Ed., is the Head&AssociateProfessor,Department of Computer Science, Gobi Arts & Science College (Autonomous), Gobichettipalayam –638 453, Erode District, Tamil Nadu, India. He received his M. Phil., degree in Computer Science in 1995. His research interests include medical image processing.Second Author Profile:Mr.V.K.NARENDIRAKUMARM.C.A.,M.Phil.,Assistant Professor, Department of Information Technology, Gobi Arts&ScienceCollege(Autonomous), Gobichettipalayam – 638 453, Erode District, TamilNadu, India. He received his M.Phil. Degree in Computer Science from Bharathiar University in 2007. He has authored or co-authored more than 38 technical papers and conference presentations. He is a reviewer for several scientific journals. His research interests are focused on advanced network, image processing, video processing, visual human-computer interaction, and multimodal biometrics technologies.Third Author Profile:Dr. K. SOMASUNDARAM, isHead & Professor, Department ofComputer Science & Applications,Gandhigram Rural University,Gandhigram –624 302, TamilNadu, India. He received his Ph.D. in the year 1984 from Indian Institute of Science, Bangalore. He has published 32 papers and 3 books. His research interests include Medical Imaging, Image Compression, Image Enhancement, Multimedia for Teaching, PC-Based instrumentation and Theoretical and Computational Plasma Physics.。
2012年考研英语二真题(全部答案解析完整版)

英语二真题:Section 1 Use of EninglishDirections :Millions of Americans and foreigners see GI.Joe as a mindless war toy ,the symbol of American military adventurism, but that’s not how it used to be .To the men and women who 1 )in World War II and the people they liberated ,the GI.was the 2) man grown into hero ,the pool farm kid torn away from his home ,the guy who 3) all the burdens of battle ,who slept in cold foxholes,who went without the 4) of food and shelter ,who stuck it out and drove back the Nazi reign of murder .this was not a volunteer soldier ,not someone well paid ,5) an average guy ,up 6 )the best trained ,best equipped ,fiercest ,most brutal enemies seen in centuries.His name is not much.GI. is just a military abbreviation 7) Government Issue ,and it was on all of the article 8) to soldiers .And Joe? A common name for a guy who neve r 9) it to the top .Joe Blow ,Joe Magrac …a working class name.The United States has 10) had a president or vicepresident or secretary of state Joe.GI .joe had a (11)career fighting German ,Japanese , and Korean troops . He appers as a character ,or a (12 ) of american personalities, in the 1945 movie The Story of GI. Joe, based on the last days of war correspondent Ernie Pyle. Some of the soldiers Pyle(13)portrayde themselves in the film. Pyle was famous for covering the (14)side of the warl, writing about the dirt-snow –and-mud soldiers, not how many miles were(15)or what towns were captured or liberated, His reports(16)the “willie” cartoons of famed Stars and Stripes artist Bill Maulden. Both men(17)the dirt and exhaustion of war, the (18)of civilization that the soldiers shared with each other and the civilians: coffee, tobacco, whiskey, shelter, sleep.(19)Egypt, France, and a dozen more countries, G.I. Joe was any American soldier,(20)the most important person in their lives.1.[A] performed [B]served [C]rebelled [D]betrayed2.[A] actual [B]common [C]special [D]normal3.[A]bore [B]cased [C]removed [D]loaded4.[A]necessities [B]facilitice [C]commodities [D]propertoes5.[A]and [B]nor [C]but [D]hence6.[A]for [B]into [C] form [D]against7.[A]meaning [B]implying [C]symbolizing [D]claiming8.[A]handed out [B]turn over [C]brought back [D]passed down9.[A]pushed [B]got [C]made [D]managed10.[A]ever [B]never [C]either [D]neither11.[A]disguised [B]disturbed [C]disputed [D]distinguished12.[A]company [B]collection [C]community [D]colony13.[A]employed [B]appointed [C]interviewed [D]questioned14.[A]ethical [B]military [C]political [D]human15.[A]ruined [B]commuted [C]patrolled [D]gained16.[A]paralleled [B]counteracted [C]duplicated [D]contradicted17.[A]neglected [B]avoided [C]emphasized [D]admired18.[A]stages [B]illusions [C]fragments [D]advancea19.[A]With [B]To [C]Among [D]Beyond20.[A]on the contrary [B] by this means [C]from the outset [D]at thatpointSection II Resdiong ComprehensionPart ADirections:Read the following four texts. answer the question after each text by choosing A,B,C or D. Mark your answers on ANSWER SHEET 1.(40 points)Text 1Homework has never been terribly popular with students and even many parents, but in recent years it has been particularly scorned. School districts across the country, most recently Los Angeles Unified, are revising their thinking on his educational ritual. Unfortunately, L.A. Unified has produced an inflexible policy which mandates that with the exception of some advanced courses, homework may no longer count for more than 10% of a student’s academic grade.This rule is meant to address the difficulty that students from impoverished or chaotic homes might have in completing their homework. But the policy is unclear and contradictory. Certainly, no homework should be assigned that students cannot do without expensive equipment. But if the district is essentially giving a pass to students who do not do their homework because of complicated family lives, it is going riskily close to the implication that standards need to be lowered for poor children.District administrators say that homework will still be a pat of schooling: teachers are allowed to assign as much of it as they want. But with homework counting for no more than 10% of their grades, students can easily skip half their homework and see vey little difference on their report cards. Some students might do well on state tests without completing their homework, but what about the students who performed wellon the tests and did their homework? It is quite possible that the homework helped. Yet rather than empowering teachers to find what works best for their students, the policy imposes a flat, across-the-board rule.At the same time, the policy addresses none of the truly thorny questions about homework.If the district finds homework to be unimportant to its students’ academic achievement, it should move to reduce or eliminate the assignments, not make them count for almost nothing. Conversely, if homework does nothing to ensure that the homework students are not assigning more than they are willing to review and correct.The homework rules should be put on hold while the school board, which is responsible for setting educational policy, looks into the matter and conducts public hearings. It is not too late for L.A. Unified to do homework right.21.It is implied in paragraph 1 that nowadays homework_____.[A]is receiving more criticism[B]is no longer an educational rit ual[C]is not required for advanced courses[D]is gaining more preferences22.L.A.Unified has made the rule about homework mainly because poor students_____.[A]tend to have moderate expectations for their education[B]have asked for a different educational standard[C]may have problems finishing their homewo rk[D]have voiced their complaints about homework23.According to Paragraph 3,one problem with the policy is that it may____.[A]discourage students from doing homewor k[B]result in students' indifference to their report cards[C]undermine the authority of state tests[D]restrict teachers' power in education24. As mentioned in Paragraph 4, a key question unanswered about homework is whether______. [A] it should be eliminat ed[B]it counts much in schooling[C]it places extra burdens on teachers[D]it is important for grades25.A suitable title for this text could be______.[A]Wrong Interpretation of an Educational Polic y[B]A Welcomed Policy for Poor Students[C]Thorny Questions about HomeworkA Faulty Approach to HomeworkText2Pretty in pink: adult women do not rememer being so obsessed with the colour, yet it is per vasive in our young girls’ lives. Tt is not that pink is intrinsically bad, but it is such a tiny slice of the rainbow and, though it may celebrate girlhood in one way, it also repeatedly and firmly fuses girls’ identity to appearance. Then it presents tha t connection, even among two-year-olds, between girls as not only innocent but as evidence of innocence. Looking around, I despaired at the singular lack of imagination about girls’ lives and interests.Girls’ attraction to pink may seem unavoidable, s omehow encoded in their DNA, but according to Jo Paoletti, an associate professor of American Studies, it is not. Children were not colour-coded at all until the early 20th century: in the era before domestic washing machines all babies wore white as a practical matter, since the only way of getting clothes clean was to boil them. What’s more, both boys and girls wore what were thought of as gender-neutral dresses.When nursery colours were introduced, pink was actually considered the more masculine colour, a pastel version of red, which was associated with strength. Blue, with its intimations of the Virgin Mary, constancy and faithfulness, symbolised femininity. It was not until the mid-1980s, when amplifying age and sex differences became a dominant childre n’s marketing strategy, that pink fully came into its own,when it began to seem inherently attractive to girls, part of what defined them as female, at least for the first few critical years.I had not realised how profoundly marketing trends dictated our perception of what is natural to kins, including our core beliefs about their psychological development. Take the toddler. I assumed that phase was something experts developed after years of research into child ren’s behavior: wrong. Turns out, acdording to Daniel Cook, a historian of childhood consumerism, it was popularised as a marketing trick by clothing manufacturers in the 1930s.Trade publications counselled department stores that, in order to increase sales, they should create a “third stepping stone” between infant wear and older kids’ clothes. Tt was only after “toddler”became a common shoppers’ term that it evolved into a broadly accepted developmental stage. Splitting kids, or adults,into ever-tinier categories has proved a sure-fire way to boost profits. And one of the easiest ways to segment a market is to magnify gender differences – or invent them where they did not previously exist.26.By saying "it is...the rainbow"(Line 3, Para.1),the author means pink______.[A]should not be the sole representation of girlho od[B]should not be associated with girls' innocence[C]cannot explain girls' lack of imagination[D]cannot influence girls' lives and interests27.According to Paragraph 2, which of the following is true of colours?[A]Colours are encoded in girls' DNA.[B]Blue used to be regarded as the colour for girl s.[C]Pink used to be a neutral colour in symbolising genders.[D]White is prefered by babies.28.The author suggests that our perception of children's psychological development was much influenced by_____.[A]the marketing of products for childr en[B]the observation of children's nature[C]researches into children's behavior[D]studies of childhood consumption29.We may learn from Paragraph 4 that department stores were advised to_____.[A]focus on infant wear and older kids' clothes[B]attach equal importance to different genders[C]classify consumers into smaller group s[D]create some common shoppers' terms30.It can be concluded that girls' attraction to pink seems to be____.[A] clearly explained by their inborn tendency[B]fully understood by clothing manufacturers[C] mainly imposed by profit-driven businessm en[D]well interpreted by psychological expertsText3In2010.afederaljudgeshookAmerica'paniesh adwonpatentsforisolatedDNAfordecades-by2005some20%ofhumangeneswereparented.ButinMarch2010ajudgeruledthatgeneswereunpa tentable.Executiveswereviolentlyagitated.TheBiotechnologyIndustryOrga nisation(BIO),atradegroup,assuredmembersthatthiswasjusta“preliminary step”inalongerbattle.OnJuly29ththeywererelieved,atleasttemporarily.Afederalappealscourtoverturnedthepriordecision,rulingthatMyriadGeneticscouldindeedholbpatent stotwogenssthathelpforecastawoman'sriskofbreastcancer.Thechiefexecuti veofMyriad,acompanyinUtah,saidtherulingwasablessingtofirmsandpatients alike.Butascompaniescontinuetheirattemptsatpersonalisedmedicine,thecourtswi llremainratherbusy.TheMyriadcaseitselfisprobablynotoverCriticsmakethr eemainargumentsagainstgenepatents:ageneisaproductofnature,soitmaynotb epatented;genepatentssuppressinnovationratherthanrewardit;andpatents' monopoliesrestrictaccesstogenetictestssuchasMyriad's.Agrowingnumberse styearafederaltask-forceurgedreformforpatentsrelatedtogen etictests.InOctobertheDepartmentofJusticefiledabriefintheMyriadcase,a rguingthatanisolatedDNAmolecule“isnolessaproductofnature...thanareco ttonfibresthathavebeenseparatedfromc ottonseeds.”Despitetheappealscourt'sdecision,bigquestionsremainunanswered.Forexam ple,itisunclearwhetherthesequencingofawholegenomeviolatesthepatentsof individualgeneswithinit.ThecasemayyetreachtheSupremeCourt.AS the industry advances ,however,other suits may have an even greater panies are unlikely to file many more patents for human DNA molecules-most are already patented or in the public domain .firms are now studying how genes intcract,looking for correlations that might be used to deter mine the causes of disease or predict a drug’s efficacy,companies are eager to win patents for ‘connecting the dits’,expaainshanssauer,alawyer for the BIO.Their success may be determined by a suit related to this issue, brought by the Mayo Clinic, which the Supreme Court will hear in its next term. The BIO rtcently held a convention which included seddions to coach lawyers on the shifting landscape for patents. Each meeting was packed.31.itcanbe learned from paragraph I that the biotech companies would like-----A.their executives to be activeB.judges to rule out gene patentingC.genes to be patcntablcD.the BIO to issue a warning32.those who are against gene patents believe that----A.genetic tests are not reliableB.only man-made products are patentableC.patents on genes depend much on innovatiaonD.courts should restrict access to gene tic tests33.according to hanssauer ,companies are eager to win patents for----A.establishing disease comelationsB.discovering gene interactionsC.drawing pictures of genesD.identifying human DNA34.By saying “each meeting was packed”(line4,para6)the author means that -----A.thesupreme court was authoritativeB.the BIO was a powerful organizationC.gene patenting was a great concernwyers were keen to attend conventiongs35.generally speaking ,the author’s attitude toward gene patenting is----A.criticalB.supportiveC.scornfulD.objectiveText 4The great recession may be over, but this era of high joblessness is probably beginning. Before it ends,it will likely change the life course and character of a generation of young adults. And ultimately, it is likely to reshape our politics,our culture, and the character of our society for years.No one tries harder than the jobless to find silver linings in this national economic disaster. Many said that unemployment, while extremely painful, had improved them in some ways; they had become less materialistic and more financially prudent; they were more aware of the struggles of others. In limited respects, perhaps the recession will leave society better off. At the very least, it has awoken us from our national fever dream of easy riches and bigger houses, and put a necessary end to an era of reckless personal spending.But for the most part, these benefits seem thin, uncertain, and far off. In The Moral Consequences of Economic Growth, the economic historian Benjamin Friedman argues that both inside and outside the U.S. ,lengthy periods of economic stagnation or decline have almost always left society more mean-spirited and less inclusive, and have usually stopped or reversed the advance of rights and freedoms. Anti-immigrant sentiment typically increases, as does conflict between races and classes.Income inequality usually falls during a recession, but it has not shrunk in this one,. Indeed, this period of economic weakness may reinforce class divides, and decrease opportunities to cross them--- especially for young people. The research of Till Von Wachter, the economist in Columbia University, suggests that not all people graduating into a recession see their life chances dimmed: those with degrees from elite universities catch up fairly quickly to where they otherwise would have been if they had graduated in better times; it is the masses beneath them that are left behind.In the internet age, it is particularly easy to see the resentment that has always been hidden winthin American society. More difficult, in the moment , is discerning precisely how these lean times are affecting society’s character. In many respects, the U.S. was more socially tolerant entering this resession than at any time in its history, and a variety of national polls on social conflict since then have shown mixed results. We will have to wait and see exactly how these hard times will reshape our social fabric. But they certainly it, and all the more so the longer they extend.36.By saying “to find silver linings”(Line 1,Para.2)the author suggest that the jobless try to___.[A]seek subsidies from the govemment[B]explore reasons for the unermployment[C]make profits from the troubled economy[D]look on the bright side of the recession37.According to Paragraph 2,the recession has made people_____.[A]realize the national dream[B]struggle against each other[C]challenge their lifestyle[D]reconsider their lifestyle38.Benjamin Friedman believe that economic recessions may_____.[A]impose a heavier burden on immigrants[B]bring out more evils of human nature[C]Promote the advance of rights and freedoms[D]ease conflicts between races and classes39.The research of Till Von Wachther suggests that in recession graduates from elite universities tend to _____.[A]lag behind the others due to decreased opportunities[B]catch up quickly with experienced employees[C]see their life chances as dimmed as the others’[D]recover more quickly than the others40.The author thinks that the influence of hard times on society is____.[A]certain[B]positive[C]trivial[D]destructivePart BDirections:Read the following text and answer the questions by finding information from the left column that corresponds to each of the marked details given in the right column. There are two extra choices in the right column. Mark your answers on ANSWER SHEERT 1.(10 points)“Universal history, the history of what man has accomplished in this world, is at bottom the History of the Great Men who have worked here,” wrote the Victorian sage Thomas Carlyle. Well, not any more it is not.Suddenly, Britain looks to have fallen out with its favourite historical form. This could be no more than a passing literary craze, but it also points to a broader truth about how we now approach the past: less concerned with learning from forefathers and more interested in feeling their pain. Today, we want empathy, not inspiration.From the earliest days of the Renaissance, the writing of history meant recounting the exemplary lives of great men. In 1337, Petrarch began work on his rambling writing De VirisIllustribus –On Famous Men, highlighting the virtus (or virtue) of classical heroes. Petrarch celebrated their greatness in conquering fortune and rising to the top. This was the biographical tradition which Niccolo Machiavelli turned on its head. In The Prince, the championed cunning, ruthlessness, and boldness, rather than virtue, mercy and justice, as the skills of successful leaders.Over time, the attributes of greatness shifted. The Romantics commemorated the leading painters and authors of their day, stressing the uniqueness of the artist's personal experience rather than public glory. By contrast, the Victorian author Samual Smiles wrote Self-Help as a catalogue of the worthy lives of engineers , industrialists and explores . "The valuable examples which they furnish of the power of self-help, ifpatient purpose, resolute working and steadfast integrity, issuing in the formulation of truly noble and many character, exhibit,"wroteSmiles."what it is in the power of each to accomplish for himself"His biographies of James Walt, Richard Arkwright and Josiah Wedgwood were held up as beacons to guide the working man through his difficult life.This was all a bit bourgeois for Thomas Carlyle, who focused his biographies on the truly heroic lives of Martin Luther, Oliver Cromwell and Napoleon Bonaparte. These epochal figures represented lives hard to imitate, but to be acknowledged as possessing higher authority than mere mortals.Communist Manifesto. For them, history did nothing, it possessed no immense wealth nor w aged battles:“It is man, real, living man who does all that.” And history should be the story of the masses and their record of struggle. As such, it needed to appreciate the economic realities, the social contexts and power relations in which each epoch s tood. For:“Men make their own history, but they do not make it just as they please; they do not make it under circumstances chosen by themselves, but under circumstances directly found, given and transmitted from the past.”This was the tradition which revolutionized our appreciation of the past. In place of Thomas Carlyle, Britain nurtured Christopher Hill, EP Thompson and Eric Hobsbawm. History from below stood alongside biographies of great men. Whole new realms of understanding —from gender to race to cultural studies — were opened up as scholars unpicked the multiplicity of lost societies. And it transformed public history too: downstairs became just as fascinating as upstairs.Section III Translation46.Directions:Translate the following text from English into Chinese.Write your translation on ANSWER SHEET2.(15 points)When people in developing countries worry about migration,they are usually concerned at the prospect of ther best and brightest departure to Silicon Valley or to hospitals and universities in the developed world ,These are the kind of workers that countries like Britian ,Canada and Australia try to attract by using immigration rules that privilege college graduates .Lots of studies have found that well-educated people from developing countries are particularly likely to emigrate .A big survey of Indian households in 2004 found that nearly 40%of emigrants had more than a high-school education,compared with around 3.3%of all Indians over the age of 25.This "brain drain "has long bothered policymakers in poor countries ,They fear that it hurts their economies ,depriving them of much-needed skilled workers who could have taught at their universities ,worked in their hospitals and come up with clever new products for their factories to make .Section IV WritingPart A47.DirectionsSuppose you have found something wrong with the electronic dictionary that you bought from an onlin store the other day ,Write an email to the customer service center to1)make a complaint and2)demand a prompt solutionYou should write about 100words on ANSERE SHEET 2Do not sign your own name at the end of the letter ,Use "zhangwei "instead .48、write an essay based on the following table .In your writing you should1)describe the table ,and2)give your commentsYou should write at least 150 words(15points)英语二答案:完形填空:1.B2.B3.A4.A5.C6.B7.C8.A9.D 10.B11.D 12.B 13.C 14.D 15.B16.A 17.C 18.B 19.B 20.DTEXT1:21. A 22.C 23.A 24.B 25.DTEXT2:26.A 27.B 28.A 29.C 30.CTEXT3:31.C 32.B 33.A 34.D 35.DTEXT4:36.D 37.D 38.B 39.D 40.A翻译:而发展中国家担心移民,则通常考虑的是,他们最优秀的人才流入了硅谷,或是发达国家的一些医院和大学。
Ambady_et_al-2012-genesis

ARTICLE Identification of Novel MicroRNAs in Xenopus laevis Metaphase II Arrested EggsSakthikumar Ambady,1*Zheyang Wu,2and Tanja Dominko31Department of Biomedical Engineering,Worcester Polytechnic Institute,Worcester,Massachusetts2Department of Mathematical Sciences,Worcester Polytechnic Institute,Worcester,Massachusetts3Department of Biology and Biotechnology,Worcester Polytechnic Institute,Worcester,MassachusettsReceived13September2011;Revised26December2011;Accepted29December2011Summary:Using a combination of deep sequencingand bioinformatics approach,we for thefirst time iden-tify miRNAs and their relative abundance in mature,metaphase II arrested eggs in Xenopus laevis.We char-acterize115miRNAs that have been described either in Xenopus tropicalis(85),evis(9),or other vertebrate species(21)that also map to known Xenopus pre-miRNAs and to the X.tropicalis genome.In addition,72new evis putative candidate miRNAs are identifiedbased on mapping to X.tropicalis genome withinregions that have the propensity to form hairpin loops.These data expand on the availability of genetic infor-mation in evis and identify target miRNAs for futurefunctional studies.genesis50:286–299,2012.V C2012 Wiley Periodicals,Inc.Key words:deep sequencing;bioinformatics;miRBase; genetics;amphibianINTRODUCTIONIn recent years,microRNAs(miRNAs)have emerged as an important class of gene regulators.They mediate complex post-transcriptional regulatory activities by interacting with coding and noncoding regions of mes-senger RNAs(Abdelmohsen et al.,2008;Tay et al., 2008).While they recognize their targets with incom-plete complementarity,the hallmark of these short sequences is perfect base pairing of nucleotides2–7at the5’end of a mature miRNA,referred to as‘‘seed’’sequence,to its target mRNA.This seed-mediated bind-ing is required for modulation of mRNA expression (Doench and Sharp,2004;Grimson et al.,2007).Their initial role in post-transcriptional repression was more recently expanded to include post-transcriptional acti-vation/derepression(Cordes et al.,2009;Mortensen et al.,2011)and mRNA stability(Valencia-Sanchez et al.,2006).Biogenesis of miRNAs is quite elaborate.Primary miR transcripts are produced mostly by RNA Pol II as kilobase long transcripts from either intronic sequen-ces of protein coding genes or their own transcrip-tion units that are not associated with protein coding genes.The primary transcripts(pri-miRNAs)are proc-essed in the nucleus by RNase-III enzyme Drosha and cleaved into60–100nt long pre-miRNAs that form a secondary hairpin structure.The pre-miRNAs are exported into the cytoplasm and processed to double stranded stretches of miRNAs by another RNase-III enzyme,Dicer.The passenger strands are removed and degraded and the remaining guide strands are loaded onto RNA-induced silencing complex(RISC) where they become single stranded mature miRNAs (reviewed by(Kim et al.,2009).The location of their interaction within the target mRNA as well as the degree of complementarity determines their activity as translational repressors,translational activators,or mRNA stabilizers.Additional Supporting Information may be found in the online version of this article.*Correspondence to:Sakthikumar Ambady,Worcester Polytechnic Insti-tute,100Institute Road,Worcester,MA01609.E-mail:sambady@Contract grant sponsor:NIH;Contract grant number:R01GM085456. Published online5January2012inWiley Online Library().DOI:10.1002/dvg.22010'2012Wiley Periodicals,Inc.genesis50:286–299(2012)MicroRNAs participate in various cellular functionsand are increasingly being identified as diagnosticmarkers in several diseases and cancers(Brase et al.,2010;Chen et al.,2011;Cortez and Calin,2009;Kroh et al.,2010;Lawrie et al.,2008;Mahn et al.,2011;Shah et al.,2009;Weigel and Dowsett,2010;Wittmann and Ja¨ck,2010;Zen and Zhang,2010).The role of miRNAsin modification of chromatin remodeling complexesand their ability to promote trans-differentiation of cellphenotype have been described more recently(Cardi-nali et al.,2009;Chen et al.,2006;Fineberg et al.,2009;Yoo et al.,2009,2011).MicroRNAs exhibit a high levelof conservation across species(Altuvia et al.,2005;Liu et al.,2008).Computational prediction of miRNAs sug-gests thousands of miRNA molecules for human and other species(Mazie`re and Enright,2007;Olena and Patton,2010).The most current version of miRBase contains16,772entries representing hairpin precursor miRNAs,expressing19,724mature miRNA products in 153species(miRBase Registry,release17;Kozomara and Griffiths-Jones,2011).The African clawed frog Xenopus laevis is an im-portant model organism that has been used in devel-opmental biology research for evis eggextracts have been invaluable in studying biologicalprocesses such as chromatin remodeling and acquisi-tion of transcriptional competence(Blow and Laskey,1986;Dimitrov and Wolffe,1996;Kikyo et al.,2000;Lohka and Masui,1983a,b),cell cycle(Lohka,1989),and DNA replication(Blow,1993).One of the mostcompelling abilities of evis metaphase II arrestedegg extracts is its ability to reprogram differentiatedsomatic cells into stem cell gene expressing cells(Alberio et al.,2005;Byrne et al.,2003;Gurdon et al.,1958;2005;;Hansis et al.,2004;Miyamoto et al.,2007).While an expanding database of e-vis genomic and genetic information is emerging, sequencing of the evis genome has not yet been completed.Similarly,the evis transcriptome, small RNAome and proteome remain incomplete com-pared to other species.For example,1,902mature miRNAs have been published for human,207for X. tropicalis,and only22for evis(miRBase,version 17).All the miRNA sequences in evis are derived from a single published study(Watanabe et al.,2005) and from unpublished data from Biasci et al.(2008) (miRBase).Traditionally,miRNAs have been discovered by clon-ing of small RNAs(Watanabe et al.,2005)followingthe commonly used Sanger sequencing methods.How-ever,this methodology has limited application in dis-covering low abundant miRNAs.Another avenue ofmiRNA discovery is computational analysis based onRNA secondary structure predictions and sequenceconservation across species.Even though computa-tional methods are useful in predicting miRNAs,expression of these transcripts still needs to be verifiedby experimentation.Recent advances in high-through-put sequencing technologies are paving the way fordiscovering new and rare species of expressed smallRNAs.Illumina sequencing(formerly Solexa)is an idealplatform for miRNA discovery due to the35bp readlengths and the level of sequencing depth afforded bythis platform(Szittya et al.,2008).Therefore,a combi-nation of deep sequencing and computational analysisis an attractive proposition for identifying new andrarely expressed,nonconserved,and species-specificmiRNAs.Here,we undertook a deep sequencing approach toidentify miRNAs in metaphase II arrested evis eggs.Combined with bioinformatics and interrogation ofgenomic sequences available for X.tropicalis,we char-acterize populations of miRNAs in metaphase II eggextracts,describe their likely precursor sequences(pre-miRNAs),identify putative new miRNAs,map their loca-tions to the genomic scaffolds of X.tropicalis,and iden-tify their mRNA targets.RESULTSSmall RNA Reads in evis Metaphase II Arrested EggsA total of12,526,420raw reads were obtained fromsequencing short RNAs from evis metaphase IIarrested eggs.Reads werefiltered to11,302,087map-pable reads using the criteria described in Table1andassigned to groups described in detail in Figure 1.Only reads between15and24nucleotides,corre-sponding to conventionally accepted miRNA length,and mapping perfectly to the available X.tropicalis ge-nome scaffolds were included in the dataset.All identi-fied sequences were able to fold into the hairpin-loopstructure characteristic of a folded pre-miRNA.As X. laevis genome sequence data becomes available,addi-tional sequences identified(but not presented)in thisstudy may be revisited in the future.The comprehen-sive dataset is included in Supporting Information TableS1and available at / tdominko/XenopusProject/.A total of115unique reads in the15–24nucleotide size range mapped to mature miR-NAs and pre-miRNAs of evis,X.tropicalis,andother vertebrate species in the current version of miR-Base and pre-miRNAs further mapped to the X.tropi-calis genome(Figure2,Groups1a,b,c and Group2). Additional72putative candidate—PC miRNAs were identified that did not map to any known pre-miRNAs, but mapped to X.tropicalis genome within sequences with propensity to form hairpin structures(Figure2, Group4).Distribution of small reads is presented in Figure3.287MICRORNAS IN EVIS EGGSReads Mapping to Known miRNAs in evis or X.tropicalis(Group1a)A total of94reads mapped to previously described X. laevis(Table2)or X.tropicalis(Table3)miRNAs/pre-miRNAs in miRBase(miRBase,Release17)and these pre-miRNAs further mapped to X.tropicalis genome (JGI v4.1,).Thirty-nine of the94Xenopus miRNA identified here (4out of9laevis and35out of85tropicalis)showed a perfect match,while the remaining reads contained changes when compared to Xenopus miRNAs in miR-Base.These changes most frequently included single nu-cleotide additions or deletions to either the5’or the3’end of mature reads,base substitutions,or a combina-tion of these changes.The majority of the reads contain-ing changes had a single or double base addition or dele-tion at their3’end(5in laevis and39in tropicalis). Three reads contained a single nucleotide substitution. None of these changes affected the seed sequence, located at the5’end of the miRNA(5’nucleotides2 through8).Four reads contained base additions or dele-tions at their5’ends and four reads contained changes at both ends;these changes consequently altered the seed sequence.Eight reads showed a5’or3’location shift on known Xenopus pre-miRNAs(Supporting Infor-mation Table S2).Reads Mapping to Other Known Vertebrate (Other Than Xenopus)miRNAs and Pre-miRNAs (Group1b)Three of21reads that mapped to X.tropicalis ge-nome matched sequences described in zebrafish(dre-mir-24)and human(hsa-mir-98and hsa-miR-129). Eleven reads matched miRNAs in other species and mapped to new regions within previously described pre-miRNAs;and seven reads matched miRNAs in other species but mapped to novel regions in X.tropicalis ge-nome that have not been previously identified as pre-miRNAs(Table4).Hence,these last seven represent potentially new miRNAs and new pre-miRNAs for X.tro-picalis and evis.The genomic sequencesflanking the reads could form stable hairpin structures(Sup-porting Information Table S2).Reads Mapping to X.tropicalis Genomic Regions—‘‘Putative Candidate’’miRNAs(Group4)A third category of72unique reads did not map to any known vertebrate miRNAs or pre-miRNAs in miR-Base(Table5).However,they all mapped to X.tropica-lis genome in regions that showed propensity to form thermodynamically stable hairpin structures.These are considered Putative Candidate—PC—miRNAs for X. laevis and X.tropicalis.Importantly,none of these mapped to Xenopus genomic repeats(repbase).Fifty-eight reads mapped to a single locus and15mapped to multiple loci on X.tropicalis scaffolds(Supporting In-formation Table S3).Genomic Clustering and Expression of evis miRNAsThe location of the reads was analyzed within X.tro-picalis genomic scaffolds.Clustering was examined for multiple copies of the same read or single copies of multiple reads.Any reads that localized within approxi-mately15kb within the same genome ID were assigned to a cluster(Table6and Supporting Information Table S3).About half of all the known miRNAs localized to one of the clusters described previously(Tang and Max-well,2008).The size of clusters ranged from142bp (cluster#2)to14,946bp(cluster#1).The largest clus-ter(#5)contained eight known miRNAs,three of which have not been previously described for evis(Sup-porting Information Table S3).As observed previously, some miRNA localized to multiple genomic loci,andTable1Criteria Used for miRNA Annotation and Hairpin StructureDeterminationmiR annotation presented in Supporting Information TablesmiRNA_name is the name of detected miRNA sequence.The miR_name is composed of the1st known miR name in acluster,an underscore,and a matching annotation,such as:L-n means the miRNA_seq(detected)is n bases less than knownrep_miRSeq in the left sideR-n means the miRNA_seq(detected)is n bases less than knownrep_miRSeq in the right sideL1n means the miRNA_seq(detected)is n bases more than knownrep_miRSeq in the left sideR1n means the miRNA_seq(detected)is n bases more thanknown rep_miRSeq in the right side2ss5TC13TA means2‘‘sequence substitutions’’(ss),which areT>C at position5and T>A at position13of the representativemiRNA.Hairpin determination in Supporting Information TablesDefinition of MFEI:MFEI52dG*100/mirLen/CG%.Reference:CellMol Life Sci63(2006)246-254.Definition of#base_in Loop:This is the maximum number of bases appearing in hairpin loop region.This number is only for gp1cand gp2.Criteria:1.Number of allowed errors in one bulge in stem: 122.Number of basepairs(bp)in stem region: 163.Free energy(dG in kcal/mol): 2154.Length of hairpin(up and down stem1terminal loop): 505.Length of terminal loop: 206.Number of allowed errors in one bulge in mature region: 87.Number of allowed biased errors in one bulge in mature region:48.Number of allowed biased bulges in mature region: 29.Number of basepairs(bp)in mature or mature*region: 1210.Percentage of small RNA in stem region(pm): 80%11.Number of allowed errors in mature region: 7288AMBADY ET AL.the rest of the miRNAs appeared to be single copy genes.Expression of clustered miRNAs varied significantly between different clusters.Some clustered miRNAs exhibited similar expression pattern (e.g.,xtr-miR-130b/130c in cluster #11and miR-23a/27a in cluster #8),consistent with co-transcription of a common pri-miRNA (Baskerville and Bartel,2005;Tang and Maxwell,2008).Examination of other evis clustered miR-NAs,however,indicated their post-transcriptional regu-lation.One of the most striking examples was expres-sion of seven miRNAs from the 724bp cluster #5(miR-363/363-3p/92a/19b/20/20b/18/106).The number of copies for individual miRNAs in this cluster ranged from 1to 461(Supporting Information Table S3).If these miRNAs are expressed from a single pri-miRNA,then differential processing of the pri-miRNA may be respon-sible for the observed variation.Alternatively,different miRNAs in the cluster could be expressed under control of different promoters.We observed otherexpressionFIG.1.Data analysis flowchart.289MICRORNAS IN EVIS EGGSimbalances including miR-98-p5and miR-let-7f in cluster #14(2and 2,162copies,respectively).Interestingly,both miRNAs are co-expressed from the same pri-miRNA and demonstrate significant quantitative differ-ence in all evis tissues examined but could not be detected in evis oocytes (Tang and Maxwell,2008).The expression of miRNA from a specific cluster could reflect developmental stage-specific transcription or dif-ferential pre-miRNA post-transcriptional processing.Only five PC miRNAs localized to four clusters,each containing multiple copies of the same PC miRNA (clus-ters #26,27,and 29)or two copies of two PC miRNAs (cluster #28).Expression of these PC miRNAs was low and differential expression from different loci could not be determined by sequencing alone.Cluster #25con-tained one known miRNAs and one PC miRNA (Table 6).Some miRNAs Are Expressed from the Same Pre-miRNA SequenceMapping of reads to the genome identified sequences of 17pre-miRNAs that expressed two miRNAs each.Mature and star miRNAs (*)and those with the suffix 5p,3p and p5,p3represent miRNAs derived from different regions of the same pre-miRNA (Supporting Information Table S3).We observed differential expression of miRNA pairs derived from the same pre-miRNA.Among the known miRNAs,the expression ratio between the pairs varied widely.For example,xtr-let-7a and hsa-let-7a*were expressed at a ratio of 1,055to 1while xtr-miR-132and newly identified hsa-miR-132*exhibited similar copy numbers (Supporting Information Table S3).Expression of the majority of PC miRNA pairs expressed from the same pre-miRNA showed much less variation,possibly due to their relatively low levels of expression.Target Predictions for Putative Candidate miRNAs and Seed-Shifted miRNAsTarget prediction was performed using custom Target-Scan (Release 5.2,June 2011)based oncomplementarityFIG.2.Criteria used to group miRNAs identified in evis metaphase II arrestedeggs.FIG.3.Length distribution of sequencing reads between 15and 25nucleotides.290AMBADY ET AL .between the seed sequences of mature miRNAs (posi-tions 2–8)and frog genes for 72PC miRNAs identified in the study.Four reads contained seed sequences perfectly matched to seed sequences of known miRNAs (miR-106/93a/302;miR-199/199-5p;miR-139-5p;miR-1/206).Seed sequences of 68PC miRNAs did not match previously identified miRNA seed sequences but all recognized frog transcript 3’UTR sequences.The number of target genes for individual reads ranged between 1and 142(Supporting Information Table S4).Target prediction was also performed for 12known miRNAs that showed ‘‘seed shift’’occurring as a result of base changes (addition or deletion)at the 5’end of the reads (Supporting Information Table S5).Shift in the seed sequence for any miRNA resulted in loss of some target genes,in acquisition of some new target genes,and in maintenance of some of the same target genes.For example,comparison of target genes for xtr-miR-124_L-1R 11(242targets)and its representative xtr-miR-124(174targets)reveals that 117targets remain the same even though the seed sequence has shifted.As a diametrically opposite example,seed sequence of hsa-miR-132*does not have any reported targets in Xenopus genome,but its seed-shifted variant hsa-miR-132*_L-1,detected in Xenopus eggs,recognizes three gene targets.DISCUSSIONevis represents an important model system in cell and developmental biology.In particular,the availability of its large size oocytes and eggs has established e-vis as a unique source of cytoplasmic extracts.While much has been learned about its smaller cousin X.tropi-calis ,genomic and genetic information for evis remains largely unavailable.Here we report the first complete analysis of miRNA populations in evis metaphase II arrested eggs using deep sequencing and bioinformatics analysis.As deep sequencing approach is independent of known genome sequence,our data will remain suitable for further analysis when the genome in-formation for evis becomes available.Available bioinformatics tools and approaches for analysis of deep sequencing data in any species rely on comparison of identified sequences to an existing ge-nome sequence database.As sequencing of the genome of evis is still in progress,our analysis included only sequences matching to the X.tropicalis .The ge-nome assembly for X.tropicalis is available (JGI v4.1,)with an estimated coverage of 90%on 19,759scaffolds.While still not assembled into chro-mosomes,this resource represents the only tool for sequence comparison and analysis for evis .Even though they share the same genus,significant differen-ces between the two genomes exist (Cannatella and de S,1993)evis has a genome size of 3.1Gb distrib-uted over 18pairs of chromosomes,while X.tropicalis is diploid with a genome size of 1.7Gb distributed over 10pairs of chromosomes.Due to the differences in ploidy and chromosome number,it is reasonable to expect that comparison between evis and X.tropicalis will identify sequence variations.The majority of Xenopus miRNA sequences in miRBase have been deposited based on similarity to vertebrate miRNAs.Recent reports on deep sequencing data for other species indicate that variations have been observed compared to miRBase sequences (Fernandez-Valverde et al.,2010;Lee et al.,2010).Such variations are attributed to RNA degradation,to sequenc-ing errors,and to differences in miRNA processing in a cell-and tissue-dependent manner.In an allopolyploid species such as evis ,in which genomic sequence in-formation is scant,it is difficult to tease out the exact causes of observed sequence variations.The variations observed in this study possibly represent evis spe-cific variations.It is also possible that factors such as han-dling of small RNAs,depth of sequencing,and default fac-tors set for specific algorithms used for analysis can impact sequence analysis results.Alternatively,dynamic expression of miRNAs can change during development and the identity of small RNA species captured at a par-ticular developmental stage will vary.Twenty-five miR-NAs we found in common with Armisen et al .(2009)contained the same sequence variations reported here.Further probing into Xenopus miRNA sequences in miRBase revealed that a majority of the sequences were deposited purely based on sequence similarity.Table 2Nine Known evis Mature miRNAs Identified in evis Metaphase II Arrested Eggs (Group 1a)#ReferencemiR/pre-miR_name miR_seqAccession #1xla-miR-15c TAGCAGCACATCATGGTTTGTA JN7950182xla-miR-18TAAGGTGCATCTAGTGCAGTTA JN7950223xla-miR-19b TGTGCAAATCCATGCAAAACTGA JN7950254xla-miR-20CAAAGTGCTCATAGTGCAGGTAG JN7950265xla-miR-23a ATCACATTGCCAGGGATTTCCA JN7950316xla-miR-92a TATTGCACTTGTCCCGGCCTGT JN7950517xla-miR-205TCCTTCATTCCACCGGAGTCTGT JN7951028xla-miR-363CGGGTGGATCACGATGCAATTT JN7951159xla-miR-427AAAGTGCTTTCTGTTTTGGGCGTJN795119291MICRORNAS IN EVIS EGGS292AMBADY ET AL.Table3Eighty-five miRNAs Identified in evis Metaphase II Arrested Eggs that Correspond To Known X.tropicalis Mature miRNAs/pre-miRNAs(Group1a)#Reference miR/pre-miR_name miR_seq Suggested name Accession# 1xtr-let-7a TGAGGTAGTAGGTTGTATAGTT xla-let-7a JN795004 2xtr-let-7b TGAGGTAGTAGTTTGTGTAGTT xla-let-7b JN795006 3xtr-let-7c TGAGGTAGTAGGTTGTATGGTT xla-let-7c JN795007 4xtr-let-7e TGAGGTAGTAGGTTGTTTAGTT xla-let-7e JN795008 5xtr-let-7f TGAGGTAGTAGATTGTATAGTT xla-let-7f JN795009 6xtr-let-7g TGAGGTAGTAGTTTGTACAGTT xla-let-7g JN795010 7xtr-let-7i TGAGGTAGTAGTTTGTGCTGTT xla-let-7i JN795011 8xtr-miR-1a TGGAATGTAAAGAAGTATGTAT xla-miR-1a JN795012 9xtr-miR-7TGGAAGACTAGTGATTTTGTTGT xla-miR-7JN795013 10xtr-miR-10a TACCCTGTAGATCCGAATTTGT xla-miR-10a JN795014 11xtr-miR-10b TACCCTGTAGAACCGAATTTGT xla-miR-10b JN795015 12xtr-miR-15a TAGCAGCACATAATGGTTTGTGA xla-miR-15a JN795016 13xtr-miR-15b TAGCAGCACATCATGATTTGCA xla-miR-15b JN795017 14xtr-miR-16b TAGCAGCACGTAAATATTGGGT xla-miR-16b JN795019 15xtr-miR-16c TAGCAGCACGTAAATACTGGAG xla-miR-16c JN795020 16xtr-miR-17-5p CAAAGTGCTTACAGTGCAGGTAG xla-miR-17-5p JN795021 17xtr-miR-18a TAAGGTGCATCTAGTGCAGATA xla-miR-18a JN795023 18xtr-miR-20a TAAAGTGCTTATAGTGCAGGTAG xla-miR-20a JN795027 19xtr-miR-22AAGCTGCCAGTTGAAGAACTGT xla-miR-22JN795029 20xtr-miR-22*AGTTCTTCAGTGGCAAGCTTT xla-miR-22*JN795030 21xtr-miR-23b ATCACATTGCCAGGGATT xla-miR-23b JN795032 22xtr-miR-24a TGGCTCAGTTCAGCAGGAACAG xla-miR-24a JN795034 23xtr-miR-25CATTGCACTTGTCTCGGTCTGA xla-miR-25JN795035 24xtr-miR-26TTCAAGTAATCCAGGATAGGCT xla-miR-26JN795036 25xtr-miR-27a TTCACAGTGGCTAAGTTCCG xla-miR-27a JN795038 26xtr-miR-27b TTCACAGTGGCTAAGTTCTGC xla-miR-27b JN795039 27xtr-miR-27c TTCACAGTGGCTAAGTTC xla-miR-27c JN795040 28xtr-miR-29c TAGCACCATTTGAAATCGGTTA xla-miR-29c JN795041 29xtr-miR-30a-3p CTTTCAGTCAGATGTTTGCAGC xla-miR-30a-3p JN795042 30xtr-miR-30a-5p TGTAAACATCCTCGACTGGAAGC xla-miR-30a-5p JN795043 31xtr-miR-30b TGTAAACATCCTACACTCAGCT xla-miR-30b JN795044 32xtr-miR-30c TGTAAACATCCTACACTCTCAGCT xla-miR-30c JN795045 33xtr-mir-30c-1GTGAACATAAGGTGGCTGGGAGAA xla-mir-30c-1-3p JN795046 34xtr-miR-30d TGTAAACATCCCCGACTGGAAGCT xla-miR-30d JN795047 35xtr-miR-30e TGTAAACATCCTTGACTGGAAGCT xla-miR-30e JN795048 36xtr-miR-34a TGGCAGTGTCTTAGCTGGTTGT xla-miR-34a JN795049 37xtr-miR-34b AGGCAGTGTAGTTAGCTGATTG xla-miR-34b JN795050 38xtr-miR-92b TATTGCACTCGTCCCGGCCTCC xla-miR-92b JN795053 39xtr-miR-93a CAAAGTGCTGTTCGTGCAGGTAG xla-miR-93a JN795054 40xtr-miR-99AACCCGTAGATCCGATCTTGTG xla-miR-99JN795057 41xtr-miR-100AACCCGTAGATCCGAACTTGTG xla-miR-100JN795058 42xtr-miR-101a TACAGTACTGTGATAACTGAA xla-miR-101a JN795059 43xtr-miR-106AAAAGTGCTTATAGTGCAGGTAG xla-miR-106JN795060 44xtr-miR-107AGCAGCATTGTACAGGGCTAT xla-miR-107JN795061 45xtr-miR-122TGGAGTGTGACAATGGTGTTT xla-miR-122JN795062 46xtr-miR-124TAAGGCACGCGGTGAATGCCAA xla-miR-124JN795064 47xtr-miR-125a TCCCTGAGACCCTTAACCTGTGA xla-miR-125a JN795065 48xtr-miR-125b TCCCTGAGACCCTAACTTGTGA xla-miR-125b JN795066 49xtr-miR-128TCACAGTGAACCGGTCTCTTT xla-miR-128JN795067 50xtr-miR-130a CAGTGCAATGTTAAAAGGGCAT xla-miR-130a JN795069 51xtr-miR-130b CAGTGCAATGATGAAAGGGCAT xla-miR-130b JN795070 52xtr-miR-130c CAGTGCAATATTAAAAGGGCAT xla-miR-130c JN795071 53xtr-miR-132TAACAGTCTACAGCCATGGTC xla-miR-132JN795072 54xtr-miR-135TATGGCTTTTTATTCCTATGT xla-miR-135JN795074 55xtr-miR-140CAGTGGTTTTACCCTATGGTA xla-miR-140JN795075 56xtr-miR-143TGAGATGAAGCACTGTAGCTC xla-miR-143JN795077 57xtr-miR-145GTCCAGTTTTCCCAGGAATCCCT xla-miR-145JN795079 58xtr-miR-146TGAGAACTGAATTCC xla-miR-146JN795080 59xtr-miR-146b TGAGAACTGAATTCCATGGACT xla-miR-146b JN795081 60xtr-miR-148a TCAGTGCACTACAGAACTTTGT xla-miR-148a JN795082 61xtr-miR-148b TCAGTGCATCACAGAACTTTGT xla-miR-148b JN795084 62xtr-miR-153TTGCATAGTCACAAAAGTGATT xla-miR-153JN795086 63xtr-miR-184TGGACGGAGAACTGATAAGGG xla-miR-184JN795089 64xtr-miR-184TGGACGGAGAACTGATAAGG xla-miR-184-2JN795090We confirmed expression of many miRNAs described previously in adult evis tissues (Michalak and Malone,2008;Tang and Maxwell,2008),but describe expression of the majority of these miRNAs now also for the metaphase II arrested evis eggs.Some previ-ous studies failed to detect a number of miRNAs in evis oocytes and eggs by Northern blotting (Tang and Maxwell,2008).This is not surprising as the method may not be sensitive enough for low expressing miR-NAs.Among the 22known evis miRNAs in miR-Base,only 7have been verified by cloning (Watanabe et al.,2005)and the remaining 15were presented based on similarity (Biasci et al .,2008,unpublished).Of the 191X.tropicalis miR/pre-miRNAs in miRBase,only 2sequences have been verified by sequencing (Armisen et al.,2009),41miRNAs were verified by Northern blot-ting (Tang and Maxwell,2008),and 148were deposited based on sequence similarity.Therefore miRBase sequences may not be a true representation of expressed Xenopus miRNAs.Identical sequence observed in the present study and the study of Armisen et al .(2009),particularly of those reads with sequence changes points to the importance of verifying miRNAs by sequencing before miRBase submission.Of the 94reads only 9correspond to evis miRs in miRBase.The remaining 85are therefore new xla-miRs.Due to high conservation of miRNAs across different species,it is not surprising that additional expressed reads in our study matched miRNA sequences described in other vertebrates.Metaphase II arrested eggs repre-sent a very unique developmental stage during oogene-sis,and these miRNAs may be specific to this develop-mental stage.Conversely,we identified expression of miRNAs in metaphase II eggs that were believed to be testis specific, e.g.,miR-100(Michalak and Malone,2008),or associated with central nervous system miR-124and miR-23b/miR-24a/miR27b cluster (Walker and Harland,2008).The 72putative candidate miRNA reads identified in this study are genuine expressed sequences with strong secondary structure prediction which makes them puta-tive candidate miRNAs for both evis and X.tropica-lis .Because they are unique to Xenopus they are most likely genus-specific.It is unlikely these reads belong to piRNAs,the predominant species of small RNAs in oocytes (Armisen et al.,2009).None of the reads map to genomic repeats (Okamura and Lai,2008),they are shorter than 25nucleotides (Grimson et al.,2007),and their precursor genomic sequences have the propensity to form hairpin structures.Our data indicate the presence of a repertoire of miR-NAs that show high level of conservation with X.tropi-calis and a subset that show sequence differences at the RNA level.Some of the identified miRNAs show differ-ences with X.tropicalis genomic sequences suggesting that these sequences may be derived from evis spe-cific loci.Therefore it is possible that miRNAs are expressed from one or more loci from multiple paralogs present in evis genome.Redundant genes,present in allopolyploid evis,could exhibit preferential silencing or differential expression in a cell,tissue or stage-specific manner for the duplicate paralogs,as well as exhibit heterochrony (Adams et al.,2003;Evans,2007;Hellsten et al.,2007;Morin et al.,2006,2008;Se´mon and Wolfe,2008;Yanai et al.,2011).Contrary to the gene silencing theory,there is evidence to suggestTable 3(Continued)#Reference miR/pre-miR_name miR_seqSuggested name Accession #65xtr-miR-191CAACGGAATCCCAAAAGCAGCTGT xla-miR-191JN79509166xtr-miR-192ATGACCTATGAATTGACAGCCA xla-miR-192JN79509267xtr-mir-194-2CCGGTGGAGATGCTGTTATCTT xla-mir-194-2-3p JN79509468xtr-miR-199a*ACAGTAGTCTGCACATTGGTT xla-miR-199a*JN79509569xtr-miR-200a TAACACTGTCTGGTAACGATGTT xla-miR-200a JN79509670xtr-miR-200b AATACTGCCTGGTAATGATGATT xla-miR-200b JN79509771xtr-miR-202*TTCCTATGCATATACCTCTTT xla-miR-202*JN79509872xtr-miR-203GTGAAATGTTTAGGACCACTTG xla-miR-203JN79509973xtr-miR-204TTCCCTTTGTCATCCTATGCCT xla-miR-204JN79510074xtr-miR-206TGGAATGTAAGGAAGTGTGTGG xla-miR-206JN79510375xtr-miR-210CTGTGCGTGTGACAGCGGCTAA xla-miR-210JN79510476xtr-miR-215ATGACCTATGAAATGACAGCCA xla-miR-215JN79510777xtr-miR-216TAATCTCAGCTGGCAACTGTGA xla-miR-216JN79510878xtr-miR-217ATACTGCATCAGGAACTGATTG xla-miR-217JN79510979xtr-miR-221AGCTACATTGTCTGCTGGGTTT xla-miR-221JN79511080xtr-miR-222AGCTACATCTGGCTACTGGGTCTC xla-miR-222JN79511181xtr-miR-338TCCAGCATCAGTGATTTTGTTG xla-miR-338JN79511382xtr-miR-363-3p AATTGCACGGTATCCATCTGTAA xla-miR-363-3p JN79511683xtr-miR-375TTTGTTCGTTCGGCTCGCGTTA xla-miR-375JN79511884xtr-miR-455TATGTGCCCTTGGACTACATCG xla-miR-455JN79512085xtr-miR-499TTAAGACTTGCAGTGATGTTTAxla-miR-499JN795124293MICRORNAS IN EVIS EGGS。
2012版自考英语二课文中英文对照

第1课Text AText A Critical ReadingCritical reading applies to non-fiction writing in which the author puts forth a position or seeks to make a statement. Critical reading is active reading. It involves more than just understanding what an author is saying. Critical reading involves questioning and evaluating what the author is saying, and forming your own opinions about what the author is saying. Here are the things you should do to be a critical reader.批判性地阅读批判性阅读适合于那种作者提出一个观点或试图陈述一个说法的纪实类写作。
批判性阅读是积极阅读。
它不仅仅包括理解作者说了些什么,还包括质疑和评价作者的话,并对此形成自己的观点。
成为一名批判性阅读者需要做到以下几点。
Consider the context of what is written. You may be reading something that was written by an author from a different cultural context than (=from) yours. Or, you may be reading something written some time ago in a different time context than yours. In either case, you must recognize and take into account any differences between your values and attitudes and those represented by the author.考虑写作背景。
将军澳官立中学

普23.通動感話普通話校本版 中一
音
24.
音(附樂樂學新習世光界碟及1 (學20習06錦年囊版連)光碟)
黃詠芝 王令
培培 生生
布裕民 李孝聰等 啟 思 布裕民 李孝聰等 啟 思
朗文
張陳志錦義輝 文永光 張陳志錦義輝 文永光
齡記 齡記
王仙瀛Байду номын сангаас全玉莉 精 工
Longman Longman
156.00 156.00 129.00 121.00 90.00 65.00 298.00 238.00
188.00
188.00
168.00 168.00
HISTORY
12. Issue – Enquiry Series Section 22 – Life in the Main Nelson Y.Y. Kan
Roald Dahl L.Frank Baum
Aristo
Puffin O.U.P. Longman
O.U.P.
MATHEMATICS
8. Mathematics in Action (Second Edition) Book 1A P.F. Man, C.M. Longman
(Modular Binding) (with Learning CD-ROM 1A, Yeung, K.H. Yeung, Revision Handbook 1A (Enhanced Version) , Bridge Y.F. Kwok, Programme P6 to S1 & Supplementary Exercise 1A) H.Y. Cheung
8. Mathematics in Action (Second Edition) Book 2B (Modular Binding) (with Learning CD-ROM 2B, Revision Handbook 2B (Enhanced Version) & Supplementary Exercise 2B)
- 1、下载文档前请自行甄别文档内容的完整性,平台不提供额外的编辑、内容补充、找答案等附加服务。
- 2、"仅部分预览"的文档,不可在线预览部分如存在完整性等问题,可反馈申请退款(可完整预览的文档不适用该条件!)。
- 3、如文档侵犯您的权益,请联系客服反馈,我们会尽快为您处理(人工客服工作时间:9:00-18:30)。
2012-DSE-BIO
2~2
2
l.(b) In the study described in (a), the participants perfonned exercise in a room maintained at 34°C and 60-70% relative humidity.
Start each question
(3) (4) (5)
The diagrams in this paper are NOT necessarily drawn to scale .
•
Hong Kong Examinations and Assessment Authority All Rights Reserved 2012
..a
S
~
~ 200 .§
\ • • \ ..
..
\
5
-.;
.0 '"0
3
500
0 0
~
O~~~--~~To.~.--.---··~-~··~--
-
---. - - - ._.
•• ••
...•........
....
a
0
150 100 50 0 0
~
. \ ...
0
00
;g
.8
•••
"
-500
20 I2-DSE-BIO 2-1
Not to be taken away before the end of the examination session
SECTION A
Human Physiology: Regulation and Control
Answer ALL parts of the question.
5
6
(i)
Describe the general patterns of the urine output after consuming drinks with different sodium concentrations. (4 marks) Account for the high urine output of the participants who consumed sports drink with 0 mmol / L sodium. (4 marks)
Figure 2B
18
16
14
(!)
-+- AR site ·····0···· Sandy seabed area
<Il
'0 (!)
.d
12 10 8
Cl.. <Il
t;:::
<Il
"-' 0
(!)
....
.I:J
Z
~
6
4 2
........,...,.,.0.,.,..........0 ................. ............. 0·· ..........0 ............... ................0 ..........'.....0 ...........0 0 0
(i)
State two major ways in which participants lost water during exercise.
(2 marks)
(ii)
If the temperature and relative humidity in the room had been set higher, it would have been dangerous to the participants. Explain why this is so. (4 marks) Explain the (3 marks)
2012·DSE
810
PAPER 2
HONG KONG EXAMINATIONS AND ASSESSMENT AUTHORITY HONG KONG DIPLOMA OF SECONDARY EDUCATION EXAMINATION 2012
BIOLOGY PAPER 2
11.45 am - 12.45 pm (1 hour)
(ii)
(iii) As soil nutrients are depleted in Stage I, the leaves of the crops become yellow. nutrient is probably lacking? Explain your answer. (iv) State two ways in which soil nutrients are lost from land.
0 0
2
3
4
5
6
7
8
9
10
Year after the deployment ofAR
0)
(ii)
Compare the results obtained in terms of the number of fish species at the AR site and the sandy seabed area. (3 marks) Explain why AR has an effect on the number of fish species at the AR site. (3 marks)
" " -"
•• •
~
~-l 000
-1500
' ; ' - . ' : ' : . II
Intake of sports drink
i
2 4 3 Time (hour)
5
6
t End of
exercise
0
Intake of
sports drink
i
2 4 3 Time (hour)
(iii) After the exercise, the breathing rate of the participants remained at a fairly high level. significance of this.
•
•
2012-DSE-BIO 2-3
3
I Go on to the next page>
Figure 2A
20
o
Stage I: actively growing crop
Stage II: abandoned land
Stage III: vegetation restored
(i)
Briefly describe the process in Stage II that leads to the restoration of the natural vegetation in Stage III. (3 marks) Suggest two agricultural activities which could account for the difference in the annual runoff between Stage I and Stage III. (5 marks) Which soil (2 marks) (2 marks)
(ii)
(iii) In terms of the replenishment of water, which sports drink would you recommend for athletes to consume after exercise? Explain your answer. (3 marks)
(2)
Write your answers in the Answer Book DSE (C) provided. (not part of a question) on a new page. Present your answers in paragraphs wherever appropriate. Illustrate your answers with diagrams wherever appropriate.
2012-DSE-BIO 2-4
4
2.(b) An artificial reef (AR) is a man-made structure deployed on the seabed to enhance biodiversity. A ten-year study was carried out on a sandy seabed to investigate the effect of an AR on the number of fish species. The number of fish species at the AR site (around and within AR) was compared with another similar sandy seabed area nearby. The results are shown in Figure 2B below:
This paper must be answered in English
INSTRUCTIONS
(1)
There are FOUR sections, A, B, C and 0 in this Paper. in any TWO sections.
பைடு நூலகம்
Attempt ALL questions
- -_ _ 0 mmol / L sodium
•••• ••• • • •• 50 mmol / L sodium
