TaKaRa TaqTM DR100AM
TaKaRa一步法RT-PCR提取法

*2 用以校正孔与孔之间产生的荧光信号误差。ABI PRISM® 7000/7700 和 Applied
Biosystems® 7300 Real-Time PCR System 使用 ROX Reference Dye,7500
Real-Time PCR System 使用 ROX Reference Dye II,Thermal Cycler Dice Real
的荧光探针,游离荧光物质发出荧光。通过检测
反应体系中的荧光强度,可以达到检测 PCR 产物
扩增量的目的。具体原理见右图。
-2-
● 制品内容(50 μl 反应×100 次)
1. 2×One Step RT-PCR Buffer Ⅲ*1(2×)
840 μl ×3 支
2. TaKaRa Ex Taq HS(5 U/μl)
RNase Free dH2O
7.5 μl
Total
25 μl
*1 通常引物终浓度为 0.2 μM 可以得到较好结果。反应性能较差时,可以在 0.1~1.0 μM 范围 内调整引物浓度。
*2 探针浓度与所使用的 Real Time PCR 扩增仪、探针种类、荧光标记物质种类有关,使用时 请参考仪器说明书,或各荧光探针的具体使用要求进行。使用 Smart Cycler® System 时, 通常探针终浓度在 0.1~0.5 μM 范围内进行调整。
Stage 2:PCR 反应 Repeat:40 times 95℃ 5 sec 60℃ 20 sec
◆特别提示: 本制品中使用的 TaKaRa Ex Taq HS 是利用抗 Taq 抗体的 Hot Start 用 DNA 聚合酶,与其他公司的化 学修饰型 Hot Start 用 DNA 聚合酶相比,不需要 PCR 反应前的 95℃、5~15 分钟的酶的活性化反应。 如果高温处理时间过长,会使酶的活性下降,其 PCR 的扩增效率、定量精度等都会受到影响。PCR 反应前的反转录酶的热变性失活通常设定为 95℃、10 sec。
Takara公司的Taq酶比较
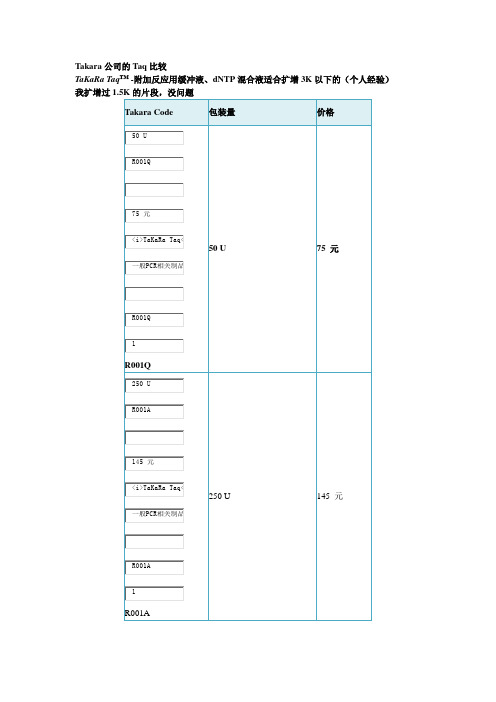
Takara公司的Taq比较
TaKaRa Taq™ -附加反应用缓冲液、dNTP混合液适合扩增3K以下的(个人经验)我扩增过1.5K的片段,没问题
说明:PCR反应性能
以λDNA为模板,可以很好地扩增8 kb的DNA片段。
TaKaRa Ex Taq®-附加反应用缓冲液、dNTP混合液适合扩增3-4Kb片段
说明:PCR反应性能
1. 以λDNA为模板,可以很好地扩增20 kb的DNA片段。
2. 以人基因组DNA为模板,可很好地扩增17.5 kb (β-Globin gene) 的DNA片段。
TaKaRa LA Taq®-附加反应用缓冲液、dNTP混合液大于4K的片段
我扩增过约9.5K的片段,没问题,但是该酶扩增3K的却不行
说明:用途PCR法扩增DNA。
尤其适用于15 kb以上DNA片段的扩增。
PCR反应性能
1. 以λDNA为模板,可以很好地扩增35 kb的DNA片段。
2. 以人基因组DNA为模板,可以很好地扩增17.5 kb (β-Globin gene) 的DNA片段。
阪崎肠杆菌实时荧光双重TaqMan PCR快速检测体系的建立

2 1, 2 ( 0) 01 7 1
Chi s ou n l o on e ne e J r a f Zo os s 8 7 5
文 章 编 号 :0 2 6 4 2 1 )0 8 7 4 1 0 —2 9 ( 0 1 1 —0 5 —0
阪崎肠杆 菌实时荧 光双重 T q nP R a Ma C 快速检测体 系的建立 *
s e i c a l ia i n r r s n e e h 0 o h rc mmo n e a h g n c b c e i a d s me io a e a sn o o o a p cf mp i c t s we ep e e t d wh n t e 5 t e o i f o n e t rp t o e i a t ra n o l t sc u ig n s c mil s
中 图分 类 号 : 3 8 2 文 献 标 识 码 : R 7 . A
No e u l x r a - i e Ta M a v ld p e e ltm q n PCR s a o he d t c i n a s y f r t e e to
o fE r 易 c r s a a i D n ak z k i
阪崎 肠杆 菌是 一种 革 兰 阴性 、 周生 鞭 毛 、 运 有 能 动 、 性厌 氧 的无芽 孢杆 菌 , 兼 起初 被认 为是 阴 沟肠杆 菌 的生物 变 型菌 。后来 F r r 通 过 DNA 杂交 实 ame
作 为一 种重 要 的条 件 致 病 菌 , 崎肠 杆 菌感 染 阪
的大多 数病 例都 是 婴儿 , 特别 是早 产儿 、 出生 体 重偏 低 等身 体 状 况 较 差 的 新 生 儿 。感 染 主要 引 起 脑 膜 炎 、 血症 和坏 死 性 小 肠 结 肠 炎p 在 阪崎 肠杆 菌 的致 病 过程 中发 挥 o A)
TAKARA公司的5' full RACE说明书

400 μl 100 μl 100 μl
For Control Reactions(5 次量):
Control HL60 Total RNA(1 μg /μl)*2 5′RACE Control Outer Primer(10 μM)*2 5′RACE Control Inner Primer(10 μM)*2
引物序列(5’→ 3’) CATGGCTACATGCTGACAGCCTA CGCGGATCCACAGCCTACTGATGATCAGTCGATG AGGTAGGTGATGTTCCGAGAGCGT TTGGAGTCGCCCTCAGCAGAGAT
长度 23 mers 34 mers 24 mers 23 mers
团。 3. 使用 T4 RNA Ligase 将 5′RACE Adaptor 连接到去帽后的 mRNA 上。 4. 以上述 3 的mRNA作为模板,使用Reverse Transcriptase M-MLV(RNase H-),Random 9 mers
进行反转录反应合成cDNA。 5. 使用下游外侧特异性引物 GSP1 和 5′RACE Outer Primer 进行 Outer PCR 反应,再使用下游内侧
行 5′RACE 实验。 3. 高特异性的 5′RACE 扩增。套式 PCR 法的使用,大大提高了 5′RACE DNA 片段扩增的特异性。
4. 长片段 5′RACE扩增。试剂盒中使用了本公司特殊改良的M-MLV(RNase H-)反转录酶,反转录性
能良好,使长片段的RT-PCR扩增成为了可能。
●制品内容(10 次量)
以下为使用本试剂盒时的注意事项,使用前请一定认真阅读!
1. 进行 5′RACE 实验时,为提高 RACE 结果的可信度,应同时进行 TAP(-)和 M-MLV(-)的负对 照实验。TAP(-)即:RNA 经去磷酸化反应后不进行 TAP 处理,直接进行 5′RACE Adaptor 的 连接及后续实验,以验证 RNA 去磷酸化是否充分。如果充分,RT-PCR 将不能扩增。但是,真正进 行 TAP(-)负对照实验时,由于不能保证 100%的去磷酸化,往往会有 RT-PCR 扩增结果,但其扩 增片段长度一般不同于目的片段。M-MLV(-)即:5′RACE Adaptor 连接反应后进行 RT 反应时 不添加 M-MLV,以排除基因组 DNA 污染造成的假阳性结果。如果 M-MLV(-)的负对照实验没有 特异性扩增,说明 5′RACE 结果来源于 mRNA。
TaKaRa Premix Ex Taq

Code No. RR390Q 研究用Premix Ex Taq™(Probe qPCR)说明书目录内容页码●制品说明1●试剂盒原理 1 ●制品内容 2 ●试剂盒外必备主要试剂和仪器2●保存 3 ●使用注意 3 ●操作方法 3 ●关联产品12●制品说明本制品是采用探针法进行Real Time PCR(qPCR)反应的专用试剂,可用于快速PCR。
Premix Ex Taq 是一种2X 浓度的Premix Type试剂,进行实验时,PCR反应液的配制十分方便简单。
Mix中添加了Tli RNaseH(耐热性RNaseH),以cDNA作为模板进行PCR反应时,可以很好地抑制由于cDNA中残存mRNA对PCR反应造成的阻害作用。
制品中使用了添加了抗体的Hot Start PCR酶TaKaRa Ex Taq HS,与Takara精心研制的Real Time PCR用Buffer组合使用,可以有效抑制非特异性的PCR扩增,大大提高PCR的扩增效率,进行高灵敏度的Real Time PCR扩增反应。
本制品可以在宽广的定量区域内得到良好的标准曲线,对靶基因进行准确定量、检测,重复性好,可信度高。
特长1.适用于Real Time PCR反应,可以快速、准确地对目的基因进行检测、定量。
2.是一种Premix Type试剂,操作简单方便。
3.DNA聚合酶使用了TaKaRa Ex Taq HS,可以进行Hot Start法PCR反应,再与Takara特别开发的Buffer系统相结合,具有高扩增效率、高扩增灵敏度之特点。
4.在2X 浓度的Premix中,预先添加了耐热性RNaseH(Tli RNaseH),可以很好地抑制以cDNA作为模板进行PCR反应时,由于cDNA中残存mRNA对PCR反应造成的阻害作用。
适用的Real Time PCR扩增仪Thermal Cycler Dice™ Real Time System III(Code No. TP950/TP970/TP980/TP990)Thermal Cycler Dice Real Time System II(Code No. TP900/TP960:终卖)Thermal Cycler Dice Real Time System Lite (Code No. TP700/TP760:终卖)Applied Biosystems 7300/7500/7500 Fast Real-Time PCR System and StepOnePlus Real-Time PCR System (Thermo Fisher Scientific)LightCycler/LightCycler 480 System (Roche Diagnostics)CFX96 Real-Time PCR Detection System (Bio-Rad)Smart Cycler System/Smart Cycler II System (Cepheid)●试剂盒原理本制品利用TaKaRa Ex Taq HS进行PCR扩增反应,通过使用Probe对PCR扩增荧光信号强度进行检测。
TAKARA_ColdExpresSystem

v.050630Table of contentsI. Description (2)II. Components (2)III. Vector map (3)IV. Storage (3)V. Protocols (3)VI. Multiple cloning site (4)VII. Application (4)VIII. Q&A (5)IX. Appendix (6)X. Related Products (10)XI. References (11)Elucidation of protein structure and function maintains an important role in post-genomic sequencing I. Descriptionand analysis studies. An efficient protein production system is critical for obtaining large amounts ofcorrectly folded recombinant protein for study. E. coli expression systems, which are used exten-sively for the production of recombinant proteins, offer two major advantages over other types ofexpression systems: (1) ease of use, and (2) low cost. However, some recombinant proteins do notfold correctly during expression in E. coli, and result in deposits of inactive insoluble protein termed"inclusion bodies".In collaboration with Prof. Masayori Inouye (University of Medicine and Dentistry of New Jersey,USA), Takara Bio has developed the pCold DNA Vectors, a series of novel protein expressionvectors. The pCold Vectors provide increased in vivo protein yield, purity, and solubility for ex-pressed recombinant proteins using "cold shock" technology. More specifically, the csp A (coldshock protein A) promoter and related elements have been incorporated into these vectors to up-regulate target protein production at lowered incubation temperatures (37o C-15o C). This temperaturedrop also suppresses expression of other cellular proteins and temporarily halts overall cell growth.This process allows expression of target proteins at high yield, high purity (up to 60% of cellularprotein), and increased solubility as compared with conventional E .coli expression systems.Co-expression of one or more chaperone proteins during expression of a heterologous targetprotein has proven effective for obtaining increased amounts of soluble recombinant protein inE. coli (see Takara's Chaperone Plasmid Set [Cat. # 3340]). This procedure, though, lacks theconvenience of a single transformation step.Takara's pCold TF DNA Vector is a fusion cold shock expression vector that expresses TriggerFactor (TF) chaperone as a soluble tag. Trigger Factor is a prokaryotic ribosome-associatedchaperone protein (48 kDa) which facilitates co-translational folding of newly expressed polypep-tides. Because of its E. coli origin, TF is highly expressed in E. coli expression systems. The pColdTF DNA Vector consists of the csp A promoter plus additional downstream sequences including a 5'untranslated region (5' UTR), a translation enhancing element (TEE), a His-Tag sequence, and amulticloning site (MCS). A lac operator is inserted downstream of the csp A promoter to ensure strictregulation of expression. Additionally, recognition sites for HRV 3C Protease, Thrombin, and FactorXa are located between TF-Tag and the Multiple Cloning Site (MCS) and function to facilitate tagremoval from the expressed fusion protein. Most E. coli strains can serve as expression hosts. ThepCold TF DNA Vector provides cold shock technology for high yield protein expression combinedwith Trigger Factor (chaperone) expression to facilitate correct protein folding, thus enabling efficientsoluble protein production for otherwise intractable target proteins.II. ComponentspCold TF DNA Vector 25 µg< Available E. coli host strains >Most E. coli strains can be used as expression hosts for Takara's pCold DNA Vector series sincethese vectors utilize the E. coli csp A (cold shock protein gene) promoter.v.050630Fig.1 pCold TF D N A : Vector MapIII. Vector map:1M3G I pCold TF DNA5,769 bpbp C o l E 1o r i A p m A ’UTR lac csp a l c I csp A 3’UTRFactor Xa site Thrombin site HRV 3C Protease site Trigger Factor (TF)His s Tag TEE IV. Storage:-20°C (for shipping and storage)V. Protocol:How to express the target gene;The cultivation / induction conditions (culture medium, culture temperature, aeration, timing ofinduction, concentration of an inducer, cultivation time after induction) should be examined for eachtarget protein.The example of general method is shown below.1) Insert the target gene to the multicloning site of pCold DNA to construct the plasmid forexpression.2) Transform the E.coli host strain (e.g. BL21) with the plasmid of expression, and select thetransformants on the selection plate including ampicillin.3) Inoculate the transformant in the medium including 50 µg/ml of ampicillin, and culture at 37°C withshaking.4) At OD 600= 0.4 - 0.5, refrigerate the culture solution at 15°C and leave to stand for 30 minutes.5) Add IPTG at the final concentration of 0.1- 1.0 mM, and continue the culture with shaking at 15°Cfor 24 hours.6) Collect the cells, and confirm the expression of target protein with SDS-PAGE in soluble andinsoluble fractions or activity assay.By selection of the E.coli host strains for expression and optimization of cultivation / induction condi-tions (culture medium, culture temperature, aeration, timing of induction, concentration of an inducer,cultivation time after induction), the expression level and the degree of soluble expression areimprovable. The tag sequence at the N-termini can be cut and removed by Factor Xa, Thrombin, andHRV 3C Protease. GeneBank Accession No. AB213654VI. Multiple cloning site:pCold TF DNA (Code. 3365)VII. ApplicationProtein expression using pCold TF DNA was compared against expression using (1) the pColdDNA I Vector alone, (2) co-expression using the pCold DNA I Vector with Takara's ChaperonePlasmid pTf16, and (3) a T7 promoter expression system which included experiments using othertags for solubilization. pCold DNA I and pCold TF DNA Vectors were transformed separately intoE.coli BL21 cells, cells were then cultured, and expression was performed according to each oftheir protocols. Expression from T7 promoter-driven vectors was additionally conducted using ageneral procedure involving addition of IPTG and subsequent culturing at 37o C.(1) Example 1: Successful protein expression resulting in soluble formThe expression of enzyme protein A (estimated molecular weight 29 kDa) was not verified as an exact band around at the estimated molecular weight, 29 kDa, with the expression system utilizing T7 promoter or even with pCold I DNA (either individual expression or co-expression with chaper- one). On the other hand, the expression of target protein (29 kDa and 52 kDa) was verified in case of using pCold TF DNA, and most of the obtained protein was in soluble form. It was confirmed that the expressed enzyme protein A has the enzyme activity even in the form of a fusion protein.Fig.2 Expression of enzyme protein ApCold TF pCold I pCold+ T7 chaperonekDa 1 2 1 2 1 2 1 297-66-45-31-22-*1: cell extract solution2: solulde fraction @ : target protein * :Co-expressed trigger factor@@@@v.0506302) Example 2: Expression resulting in improved levels of soluble protein.Expression of soluble enzyme protein B (M.W: ~63 kDa) was not able to be obtained using eitherpCold DNA I alone or co-expressed with chaperone proteins, nor with a T7 expression vector thatincluded other tags for solubilization (Trx Tag [~12 kDa], Nus Tag [~55 kDa], and GST Tag [~26kDa]). Alternatively, when pCold TF DNA Vector was used, most of the expressed target proteinwas verifiable as a soluble fraction and present at an expression level much higher than with othertags. (Note: The molecular weight of the target protein was observed as larger than its real size dueto fused expression with each tag.)1: Cell extract solution2: Soluble fraction3: Insoluble fractionFig.3 Expression of enzyme protein BVIII. Q&A Q1: What parameters should be examined when the expressed protein is insoluble?A1: The optimal conditions for cultivation and induction vary depending on a kind of expressed protein. The cultivation and induction conditions should be determined by referring to thefollowing points:- Change the induction timing. It should be examined within early and late logarithmic growthphase.- Change the concentration of inducer (IPTG) within 0.1-1 mM.- Examine the cultivation time after induction (Generally 15°C, 24 hours is the mostappropriate.)- Change the conditions for aeration.Q2: What remedial procedures should be taken in case that no protein is expressed or the expres-sion level is low?A2: It is recommended to examine again the conditions for cultivation and induction (Refer to theabove Q1.), or change the host E.coli strain.Q3: What host strains have been confirmed to work with pCold Vectors?A3: BL21, Rosetta TM, Origami TM from Novagen, Inc. BL21 is most commonly used as host.Origami TM lacks the trx/gor gene and allows the formation of disulfide bond in cytoplasm at high level. Accordingly, the solubility and refolding of expressed protein are facilitated.Rosetta TM contains a plasmid which supplies tRNAs corresponding to the codons that are rarely used in E.coli. It enables the universal transcription of the genes which are restricted by thecodon usage of E.coli.Q4: Can E. Coli retaining a pCold TF vector that contains target gene be stored at 4°C on a plate?A4: We don't recommend the 4°C storage on a plate because it causes a possible leak of target protein in the cell. Pick the colony from the plate promptly, prepare a glycerol stock and store at-80°C.Q5: Is it possible to express a large gene in pCold TF DNA?A5: It was confirmed that human gene of 125 kDa can be expressed successfully in a soluble form.(The band of 125 + 52 kDa was was verified in CBB staining.)IX. AppendixExpression Plasmid Construction - Example using the thioredoxin gene1) Overview of pCold TF expression vector constructiona) Select a restriction enzyme site such that the DNA fragment to be inserted will have the sequenceof its target gene positioned in a continuous reading frame with that of the pCold TF DNA Vector.b) Prepare the DNA fragment to be inserted into the vector.c) Cut the vector with the desired restriction enzymes.d) After ligating the digested vector with the insert DNA, transform it into an appropriate E. coli strain.e) Prepare purified plasmid from the appropriate colonies containing the target insert.f) Purified plasmid may be used for protein expression experiments.There are several ways in which the insert DNA may be prepared, including PCR amplification,excision of a cloned gene by restriction enzyme digestion, and gene synthesis. Transfer of insertsalready cloned into Takara's pCold I-IV Vectors can be easily accomplished since the pCold TF DNAmulticloning site (MCS) is identical to the pCold I-IV MCS. Presented below is an example experi-ment which uses PCR amplification as the insert DNA preparation method.2) Example Plasmid preparation for expression of the E. coli thioredoxin genea) Guidelines for primer designProtocol and points to consider when designing primers:i) Select two restriction enzymes whose sites are contained within the MCS of pCold TF DNA thathave additionally been verified not to cut the insert DNA sequence.ii) Construct a primer for the target sequence, adding the selected restriction sites from Step 2a) i) tothe 5' terminus of each primer. Adjust the base number between the insert DNA sequence andN-terminal restriction sites such that the frame of the insert matches the reading frame ofpCold TF DNA. "Either a restriction site or a stop codon can be directly added to the C-terminusif required.").iii) Add four or more bases to sequences directly flanking the restriction sites. Most restrictionenzymes require that several bases lie outside of the recognition site for efficient digestion tooccur. Without the presence of this extra sequence, digestion efficiency will be lowered.v.050630Insertion of the thioredoxin gene into the pCold TF DNA Nde I/Xho I MCS restriction enzyme cloning sitesNde I site: Primer 1 (normal direction primer)Xho I site: Primer 2 (reverse direction primer)*1: When using Nde I site, adjust the position of the thioredoxin gene start codon (ATG) to correspond with the ATG site of Nde I*2: Complementary Thioredoxin sequence with stop codons[Example - Primer Design]5' -GCCGCATATGAGCGATAAAATTATTCACextra sequence thioredoxin-origin sequence *15' - GCCGCTCGAGTTAGGCCAGGTTAGCGTCextra sequence thioredoxin-origin sequence *2b) Insert DNA Preparation[Example - PCR amplification of the thioredoxin gene (~ 350 bp)]i) PCR amplification of the insert DNA.Prepare the reaction mixture by combing the following reagents. (use of a PCR Enzyme, such as TaKaRa Ex Taq TM (Cat. # RR001A) is recommended).Amplify the insert DNA using the following PCR cycling parameters (30 cycles):When using Takara Thermal Cycler Dice (Cat. # TP600)98o C, 10 sec.55o C, 30 sec.72o C, 1 min.*1 For plasmid DNA, use 1-10 ng; for cDNA or genomic DNA, use 50-500 ng.*210x Ex Taq TM Buffer and dNTP Mixture is supplied with TaKaRa Ex Taq TM(Cat. # RR001A).Template DNA (5 ng)*11 µl 10x Ex Taq TM Buffer*25 µl dNTP Mixture (2.5 mM each)*24 µl Primer 1 (10-50 pmol/µl) 1 µlPrimer 2 (10-50 pmol/µl) 1 µlTaKaRa Ex Taq TM (5 units/µl) 0.25 µlSterilized distilled water37.75 µl Total 50 µlNde IXho Iii) Verification of amplified productVerify that the amplified insert DNA fragment is a single band of the correct expected size by performing agarose gel electrophorsis using 5 µl of the PCR product.iii) PCR product purificationFor DNA which is amplified and appears as a single band, purification using SUPREC TM-02 (Cat. # 9041) is suggested. When multiple PCR products are generated, first isolate the band of interest from the agarose gel and then further purify using SUPREC TM-01 (Cat.#9040) or TaKaRa RECOCHIP (Cat.#9039) or other similar method.iv) Restriction enzyme digestion of amplified productsDigest the purified insert DNA with Nde I and Xho I restriction enzymes.1) Prepare the following restriction enzyme digest mixture:Insert DNA; 0.5-1 µg X µl10x K Buffer 3 µlNde I (10 units/ul) 1 µlXho I (10 units/ul) 1 µlSterilized distilled water X µlTotal 30 µl2) Incubate at 37o C for 1 hour.3) Ethanol precipitate the digested DNA to purify it.*). 4) Verify fragment purity using agarose gel electrophoresis or by measuring absorbance (OD260* Both Nde I and Xho I can be inactivated by ethanol precipitation. However, when restrictionenzymes which are not completely inactivated by ethanol precipitation are used, the digestionreaction should be treated with phenol. In addition, further purification and recovery of digestedDNA by agarose gel electrophoresis can completely remove all short fragments generated by the digestion.[Ethanol precipitation protocol]1) Add 3M sodium acetate, pH 5.2, to the restriction enzyme digest mixture in a 1:10 ratio (e.g. 3 µl 3M sodium acetate added to 30 µl digest mixture), and mix well.2) Add 2-2.5 times the volume of 100% cold ethanol to the above solution (e.g. add 66 µl 100%cold ethanol to 33 µl sodium acetate-digest mixture), and mix well. Chill at -20o C for 30 minutes.3) Centrifuge at 4o C, 12,000 rpm, for 10-15 minutes. Discard the supernatant.4) Add 70% cold ethanol and centrifuge again at 4o C, 12,000 rpm, for 5 minutes.5) Discard the supernatant and air dry.6) Dissolve the precipitate in 10-50 µl of TE buffer.v.050630b) Restriction Enzyme Digestion of pCold TF DNA Digest pCold TF DNA with the same restriction enzymes that were used for the digestion of ampli-fied insert DNA, and purify. Dissolve the purified DNA in TE buffer, and measure the DNA concen-tration by measuring absorbance.i) Prepare the following reaction mixture:ii) Incubate at 37o C for 1-2 hours.iii) Ethanol precipitate the digested vector DNA to purify.iv) Dissolve the precipitated vector DNA pellet in TE buffer.v) Measure the absorbance (OD260) and calculate the DNA concentration. For dsDNA (double- stranded DNA), calculate the DNA concentration assuming 1 OD 260 = 50 µg/ml.vi) Adjust the DNA concentration to100 ng/µl.* After digestion with restriction enzymes, the vector DNA may be de-phosphorylated with E. coli Alkaline Phosphatase (BAP)(Cat.# 2120A), or Calf Intestinal Alkaline Phosphase (CIAP (Cat.# 2250A). Note that de-phosphorylation is essential if only a single restriction enzyme was used for digestion. In addition, complete removal of short fragments generated by restriction enzymedigestion is recommended. Purify the vector from any resulting short fragments using agarose gel electrophoresis, then further isolate and purify the vector from the gel.pCold TF DNA1 µg 10 X K Buffer3 µl Nde I (10 units/µl) 1 µlXho I (10 units/µl) 1 µlSterilized distilled water X µlTotal 30 µlc) Ligation of the DNA fragment and pCold TF DNA vector and transformationi) Ligation reactionMix together the digested pCold TF DNA and the insert DNA fragment, and use this mixture for performing a ligation reaction using Takara's DNA Ligation Kit <Mighty Mix> Cat.# 6023). A 1:3-1:10 molar ratio of vector:insert DNA is recommended.Prepare the following ligation reaction mixture on ice:Incubate at 16o C for 1 hour.Digested pCold TF DNA ; 100 ng (- 0.03 pmol) 1 µlInsert DNA fragment (0.1-0.3 pmol) 4 µlLigation Mix (from DNA Ligation Kit <MightyMix>) 5 µlTotal 10 µlii) TransformationTransform 100 µl E. coli JM109 Competent Cells (Cat.# 9052) with 10 µl ligated DNA mixture. Platetransformed cells on LB-ampicillin agar (100 µg/ml ampicilin) and grow at 37o C overnight.1) Thaw E. coli JM109 competent cells on ice just before use.2) Add 10 ul ligated DNA mixture to 100 µl competent cells, and mix gently.3) Chill on ice for 30 minutes.4) Incubate at 42o C for 45 seconds.5) Chill on ice for 1-2 minutes.6) Add warm (37o C) SOC medium to a final volume of 1 ml.7) Shake at 37o C for 1 hour.8) Plate on LB-ampicillin agar (100 µg/ml ampicilin) and incubate at 37o C overnight .d) Plasmid preparation and verificationInoculate a colony obtained in Step 4 ii) above into LB-ampicillin broth (100 µg/ml ampicilin) andincubate with gentle shaking at 37o C overnight. Use the resulting culture for plasmid maxi- or mini-preps (Takara's Mini Prep DNA Purification Kit [Cat.# 9085] is recommended).After obtaining isolated plasmid DNA, digest the plasmid with the restriction enzymes Nde I andXho I. Verify insertion of the correct DNA fragment by checking insert DNA fragment size usingagarose gel electrophoresis.When the vector construct has been verified, confirm the sequence of the inserted DNA fragmentby sequencing analysis. This plasmid can be used as an expression plasmid for subsequentexperiments. Sequencing primers should be designed such that they lie upstream and downstreamof multicloning site by 50-100 bases. The following primer can be used as a downstream primer: Downstream primer: pCold-R 5'-GGCAGGGATCTTAGATTCTGX. Related Products<E.coli Competent Cells >E. coli DH5_ Competent Cells (TaKaRa Cat.#9057)E. coli HB101 Competent Cells (TaKaRa Cat.#9051)E. coli JM109 Competent Cells (TaKaRa Cat.#9052)E. coli DH5_ Electro-Cells (TaKaRa Cat.#9027)E. coli HB101 Electro-Cells (TaKaRa Cat.#9021)E. coli JM109 Electro-Cells (TaKaRa Cat.#9022)< Other >IPTG (Isopropyl-`-D-thiogalactopyranoside) (TaKaRa Cat.#9030)Chaperone Plasmid Set (TaKaRa Cat.# 3340)pCold DNA vector series (TaKaRa Cat.# 3360-3364)XI. References1) Qing, G. et al (2004) Nature Biotechnology22, 877-2004.2) Gerlined, S., et al (1995) EMBO J.14, 4939-4948.Cold Shock Expression System pCold TF DNA TAKARA BIO INC.v.050630Cat.# 3365URL: Phone: +81-77-543-7247 Fax: +81-77-543-9254NOTE: (1) For research use only. Not for use in therapeutic or diagnostic use.(2) Protein Purification Technology of His-Taq used in pCold I and pCold II DNA is licensed from Hoffmann-La Roche, Inc., Nutley,NJ and/or Hoffmann-La Roche Ltd., Basel, Switzerland and is provided only for the use in research. Information about licenses for commercial use if available from QIAGEN GmbH, Qiagen Strasse 1, D-40724 Hilden, Germany.。
TaKaRa定量PCR介绍
Gene Specific Primer
Oligo dT Primer
PCR F-Primer
PCR R-Primer
实时荧光定量PCR介绍
February 17, 2019
•
24
RT Primer的选择
Random
5’
5’
F
目的片段 Random 目的片段
R
F
1,500 base
目的片段
R
5’
F
R
目的片段在mRNA任何位 3’ 置都能使用。通常mRNA 表达量分析最适。 适用于mRNA表达量分析, AAA· · · 3’ 提升反应检测的灵敏度。 Oligo dT Primer Oligo dT Primer 目的片段在距Poly(A) Tail AAA· · · 3’ 1.5 kbp 以内适用,RT效率 较高。
exon
cDNA
exon
PCR Primer exon intron
Real Time PCR
PCR Primer exon
Real Time RT-PCR
Small PCR Product
融 解 曲 线 分 析
Small PCR Product
此种情况必须采用DNase I 处理,去除基因组DNA。
PCR Primer exon exon
Real Time RT-PCR
Real Time PCR
No Product
融 解 曲 线 分 析
Small PCR Product
实时荧光定量PCR介绍
February 17, 2019
•
28
Real Time PCR引物设计
山西褐马鸡种群的遗传多样性初探
山西省褐马鸡种群的遗传多样性初探1常江,吴爱平,张正旺北京师范大学生物多样性与生态工程教育部重点实验室,生命科学学院,北京(100875)E-mail:cjfloating@摘要:褐马鸡现存种群和数量都正在大幅度减少。
本文测定和比较褐马鸡主要分布区山西省的78只褐马鸡个体的线粒体控制区(mtDNA CR)序列(478bp),发现仅有1个单核苷酸突变位点,定义了2个单倍型(H1,H2)。
其中H1为主要单倍型,现行分布广泛,占研究个体总数的93.6%,另一个单倍型H2由H1发生突变形成,占研究个体总数的6.4%。
山西省褐马鸡种群的平均核苷酸多样性指数(π)和单倍型多样性指数(h)极低,分别为0.2168和0.0006。
结果提示,山西省现存褐马鸡种群的线粒体遗传多样性极低,单倍型数量极少是褐马鸡分化成种后经历过严重的瓶颈效应,母系数量锐减和现行人为破坏影响的结果。
关键词:褐马鸡,mtDNA,控制区,瓶颈效应1.引言褐马鸡(Crossoptilon mantchuricum)为我国Ⅰ级重点保护动物,隶属于鸡形目(Galliformes)雉科(Phasianidae)马鸡属(Crossoptilon),为我国特有种[1][2],分布于山西西北部、陕西省黄龙山、河北西北部及北京门头沟地区[3](图1)。
在各分布区内褐马鸡的栖息地环境破坏严重,尤其是在山西许多地区普遍存在开采煤矿、铁矿等人为干扰,对褐马鸡的生存极为不利[4][5],再加上森林退化加剧,褐马鸡的栖息地大幅度片段化,导致褐马鸡的种群数量锐减。
以往对褐马鸡的研究包括:分布现状[6]、种群数量调查[7][8]、窝卵数及其变异[9]、后肢肌肉的比较研究[10]、巢址选择[11]、繁殖生物学[12]、集群行为[13]、取食及梳羽等行为观察[14]、年活动规律[15]、笼养褐马鸡冬季社群等级[16]、核型及染色体G带带型[17]以及其分类地位的讨论[18][19]等。
抗Taq抗体说明书
Hot Start PCR用试剂Taq Antibody使用说明书TaKaRa Code:D9002A●包装量: 250 U●制品说明Taq Antibody是Hot Start PCR用抗Taq抗体,其与Taq 酶结合后抑制DNA聚合酶活性。
使用本制品进行PCR扩增时,高温变性前抗Taq抗体与Taq酶结合抑制DNA聚合酶活性,能够在低温条件下有效抑制引物的非特异性退火及引物二聚体引起的非特异性扩增。
抗Taq抗体在PCR反应最初的DNA变性步骤中变性,DNA聚合酶活性恢复,达到Hot Start PCR效果。
使用本制品无需特殊的抗Taq抗体失活处理,可以在常规PCR反应条件下使用。
●制品内容Taq Antibody(5 U/μl) 50 μl●保 存: -20℃。
●贮存溶液Tris-HCl(pH8.0) 22 mM KCl 100 mM NaCl15 mM EDTA 0.1 mM DTT 1 mM Tween® 20 0.5 % Nonidet P-40®0.5 % Glycerol 50 % ●活性定义Taq Antibody与Taq DNA Polymerase混合,25℃,10 min温浴后,在55℃,10 min条件下抑制90%以上的1 U 的Taq DNA Polymerase 活性的Taq Antibody量定义为1 U。
●纯 度1.10 U的Taq Antibody和1 μg的λ DNA-Hin d III在37℃或70℃下反应1小时,DNA的电泳谱带不发生变化。
2.10 U 的Taq Antibody和1 μg的Super-coiled pBR322 DNA在37℃或70℃下反应1时,DNA的电泳谱带不发生变化。
3.10 U的Taq Antibody和1 μg的λ DNA在37℃或70℃下反应1小时,DNA的电泳谱带不发生变化。
4.10 U的Taq Antibody和1 μg的16S,23SrRNA在37℃或70℃下反应1小时,RNA的电泳谱带不发生变化。
SYBR Premix Ex TaqTM II
TaKaRa Code:DRR081A SYBR®Premix Ex Taq TM II (Perfect Real Time)(200次量)目录内 容 页 码●制品说明 1●制品内容 1 ●适用的Real Time PCR扩增仪1●保 存 1 ●TaKaRa Ex Taq TM HS活性定义 1 ●纯 度 1●Real Time PCR性能检测 2 ●试剂盒原理 2 ●试剂盒特长 2 ●操作注意 2 ●Real Time PCR操作顺序 3 ●操作方法 3 ◆应用Thermal Cycler Dice Real Time System扩增仪的操作方法 3 ◆应用ABI PRISM 7000/7700/7900 HT,7300/7500/7500 Fast Real - Time PCR的操作方法 4 ◆应用Light Cycler Real Time PCR扩增仪的操作方法5●PCR反应条件说明 7 ●进行RT-PCR反应时的实验方法 7 ●反应例 9 ●引物设计说明 9 ●引物设计服务说明:本公司提供用于基因表达定量分析的引物设计及合成服务 10 ●问 答 10●制品说明本制品是采用SYBR® Green I嵌合荧光法进行Real Time PCR的专用试剂。
制品中已经将DNA聚合酶、反应用Buffer、dNTP、SYBR® Green I等试剂预混在一起,是一种2×浓度的Premix Type试剂,进行实验时,PCR反应液的配制十分方便简单。
本制品Buffer经过改良,使反应特异性比SYBR®Premix Ex Taq TM(Perfect Real Time)(TaKaRa Code:DRR041)更高。
抑制非特异性反应,能够在更广的范围内进行准确定量。
本Buffer和改良后的Hot Start 法用DNA聚合酶TaKaRa Ex Taq TM HS组合使用,可以进行再现性好、可信度高的Real Time PCR解析。
- 1、下载文档前请自行甄别文档内容的完整性,平台不提供额外的编辑、内容补充、找答案等附加服务。
- 2、"仅部分预览"的文档,不可在线预览部分如存在完整性等问题,可反馈申请退款(可完整预览的文档不适用该条件!)。
- 3、如文档侵犯您的权益,请联系客服反馈,我们会尽快为您处理(人工客服工作时间:9:00-18:30)。
V2010.01
3. 结果。
M1 2 M
2% Agarose gel 5 μl 电泳结果 M :DL2,000TM DNA Marker* 1 :1 kbp PCR 产物 2 :1 kbp PCR 产物
*(TaKaRa Code:D501A)
●注意事项
PCR 的反应液请在冰中配制,然后置于 PCR 反应 仪上进行 PCR 反应。这种冷启动法(Cool Start Method)可增强 PCR 扩增的特异性,减少 PCR 过程中的非特异性反应,能得到良好的 PCR 结果。
●保 存:-20℃
●10×PCR Buffer 的组成
10×PCR Buffer(Mg2+ Free) Tris-HCl(pH8.3) KCl
100 mM 500 mM
●活性定义
用活性化的大马哈鱼精子 DNA 作为模板/引物,在 74℃,30 分钟内,摄入 10 nmol 的全核苷酸为酸 性不溶物的活性定义为 1 个活性单位(U)。
●纯 度 1)10 U 的本酶和 0.6 μg 的λ-Hind III 在 74℃下
反应 1 小时,DNA 的电泳谱带不发生变化。 2)10 U 的 本 酶 和 0.6 μg 的 Supercoiled
pBR322 DNA 在 74℃下反应 1 小时,DNA 的 电泳谱带不发生变化。 3)10 U 的本酶和 0.6 μg 的λDNA 在 74℃下反应 1 小时,DNA 的电泳谱带不发生变化。
TaKaRa TaqTM
使用说明书
TaKaRa Code:DR100AM
●包装量:250 U
●制品说明
本制品是 94 kDa 的耐热性 DNA 聚合酶。是把 Thermus aquaticus DNA Polymerase 的基因 经过克隆转化到大肠杆菌中进行表达后,分离提取 而得到的。它与天然 Taq DNA 聚合酶具有相同的 功能。使用本制品扩增得到的 PCR 产物的 3′端附 有一个“A”碱基,因此可直接克隆于 T-Vector 中。
●制品内容
TaKaRa Taq(5 U/μl) 10×PCR Buffer(Mg2+ Free) MgCl2(25 mM) 6×Loading Buffer*
50 μl 1 ml 1 ml 1 ml
*① ②
电泳时,请按每 5 μl PCR 反应液中加入 1 μl 本制品的比例添加混合后进行电泳。 6×Loading Buffer 详细说明请见 TaKaRa 商品目录。
2. PCR 反应条件。 以λDNA 为模板,扩增 1 kbp 的 DNA 片段的 PCR 反应条件如下:
94℃ 55℃ 72℃
30 sec. 30 sec.
1 min.
30 Cycles
注) PCR 反应条件视模板、引物等的结构条件不同 而各异。在实际操作中需根据模板、目的片段 的大小、碱基序列和引物的长短等具体情况, 设定最佳的反应条件(温度、时间等)。
●用 途
1) PCR 法扩增 DNA。 2) DNA 序列测定。
●PCR 反应性能
1) 以λDNA 为模板,可以很好地扩增 8 kbp 的 DNA 片段。
2) 以人基因组 DNA 为模板,可很好地扩增 3.0 kbp(p53 基因)的 DNA 片段。
●应用例 以λDNA 为模板进行 PCR 扩增反应
1. 按下列组份配制 PCR 反应液。
TaKaRa Taq(5 U/μl)
0.25 μl
10×PCR Buffer(Mg2+ Free)
5 μl
MgCl2(25 mM)*1
3 μl
dNTP Mixture(各 2.5 mM)
4 μl
模板 DNA(λDNA)*2
2.5 ng
引物 1(20 μM)
1 μl
引物 2(20 μM)
1 μl
灭菌蒸馏水
up to 50 μl来自*1 应视具体情况加入适量的 MgCl2,一般情况 下加入的 MgCl2 量为终浓度 1.5 mM。
*2 【50 μl PCR 反应体系中模板 DNA 推荐使用量】
人基因组 DNA 大肠杆菌基因组 DNA λDNA 质粒 DNA
0.1 μg~1 μg 10 ng~100 ng 0.5 ng~5 ng 0.1 ng~10 ng
