Inference of Genetic Regulatory Networks by Evolutionary Algorithm and H ∞ Filtering
《人工智能》测试题答案

《人工智能》测试题答案测试题——人工智能原理一、填空题1.人工智能作为一门学科,它研究的对象是______,而研究的近期目标是____________ _______;远期目标是___________________。
2.人工智能应用的主要领域有_________,_________,_________,_________,_______和__________。
3.知识表示的方法主要有_________,_________,_________,_________和________。
4.产生式系统由三个部分所组成,即___________,___________和___________。
5.用归结反演方法进行定理证明时,可采取的归结策略有___________、___________、_________、_________、_________和_________。
6.宽度优先搜索对应的数据结构是___________________;深度优先搜索是________________。
7.不确定知识处理的基本方法有__________、__________、__________和__________。
8.AI研究的主要途径有三大学派,它们是________学派、________学派和________学派。
9.专家系统的瓶颈是________________________;它来自于两个阶段,第一阶段是,第二阶段是。
10.确定因子法中函数MB是描述________________________、而函数MD是描述________________________。
11.人工智能研究的主要领域有_________、_________、_________、_________、_______和__________。
12.一阶谓词逻辑可以使用的连接词有______、_______、_______和_______。
24188874_青海牛苦头矿区两期岩浆岩及其矽卡岩型成矿作用

1000 0569/2021/037(05) 1567 86ActaPetrologicaSinica 岩石学报doi:10 18654/1000 0569/2021 05 14青海牛苦头矿区两期岩浆岩及其矽卡岩型成矿作用王新雨1 祝新友1 李加多2 王玉往1 蒋斌斌1 吴锦荣2 黄行凯1 赵子烨1WANGXinYu1,ZHUXinYou1 ,LIJiaDuo2,WANGYuWang1,JIANGBinbin1,WUJinRong2,HUANGXingKai1andZHAOZiYe11 北京矿产地质研究院有限责任公司,北京 1000122 青海鸿鑫矿业有限公司,格尔木 8160991 BeijingInstituteofGeologyforMineralResourcesCo ,Ltd,Beijing100012,China2 TheQinghaiHongxinMiningCo ,Ltd ,Golmud816099,China2020 08 25收稿,2021 04 20改回WangXY,ZhuXY,LiJD,WangYW,JiangBB,WuJR,HuangXKandZhaoZY 2021 Twostagemagmatismsandtheirskarn typemineralizationintheNiukutouoredistrict,QinghaiProvince ActaPetrologicaSinica,37(5):1567-1586,doi:10 18654/1000 0569/2021 05 14Abstract TheNiukutoudeposit,locatedinthecentral westernpartofQimantagharea,EastKunlunorogenicbelt,isoneofthemostpotentialPb ZnskarndepositsinQimantaghareawithaprovenPb+Znresourceof1 15milliontons(inferenceorabove).M1,M4andM2magneticanomalyareas(oreblocks)arethemainoreblocks(points)inthisdistrict LargevolumeofgraniticrocksarecloselyrelatedtothemineralizationofPb Znpolymetallicdeposit Basedonthestudyofgeologicalcharacteristics,alterationandmineralizationzonationoftypicaldepositsinM1,M4andM2oreblocks,itispreliminarilyconcludedthatgranodioriteandmonzoniticgraniteatthebottomofdrillholesaretheoreformingrockbodies LA ICP MSanalysisshowsthatthegraniticrocksinNiukutouoredistrictcouldbedividedintotwostages:(1)thegranodioriteinM1andM4oreblocks,withagesof375Maand353Ma,correspondingtoHercynian;and(2)themonzoniticgraniteatthebottomofboreholeinM2oreblockis216Mato212Ma,correspondingtoIndosinian ThewholerockgeochemicalstudiesshowthattheHercyniangranodioritesinM1andM4oreblocksbelongtocalcalkalineseries high KcalcalkalineseriesI typegranites,withrelativelyhighMg#(average=44 23),alargenumberofdarkenclavesandweakδEunegativeanomaly(0 68~0 87).Theabovecharacteristicssuggestthatmantlematerialparticipatedintheformationofthesourcearea,similartometa basalticamphibolitesource TheyareenrichedinLREEandlargeionlithophileelements,depletedinHREEandhighfieldstrengthelements,showingthegeochemicalfeaturesofpost collisionalmagmaticrocks TheIndosinianmonzoniticgraniteatthebottomofdrillholeinM2oreblockbelongstohigh Kcalc alkalinetoshoshoniteI typegranites,withrelativelylowMg#value(average=27 05),nodarkenclaves,obviousnegativeδEuanomaly(0 11~0 56),andΣREEvaryingfrom113×10-6to512×10-6 ItisrichinLREEandlargeionlithophileelements,depletedinHREEandhighfieldstrengthelements,andhighRb/Srvalues,representingahighlydifferentiatedrock Itssourceareaissimilartothatofmeta sandstonesource Basedonregionaltectonic magmaticevolutionarygeologicalbackgrounds,wesuggestthattheNiukutouHercyniangranodioriteswereformedinthepostcollisionalsettingaftertheclosureoftheEo TethyanOceaninLateDevonian,whenthemantleunderplatedtheancientcontinentalcrust,andtheoredepositwasformedbythemixedmineralizationofmantlederivedmagmaandcrustderivedmagma TheLateIndosinianmonzogranitewasformedinthepostcollisionalenvironmentaftertheclosureofthePaleo TethysintheLatePaleozoic,similartothepartialmeltingofmetamorphicsandstonelithosphereandexperiencedstrongdifferentiationandevolution Combinedwiththegeologicalandmineralogicalcharacteristicsofthedeposit,theNiukutouoredistrictispreliminarilydividedintotwometallogenicsystems:theM1andM4HercynianmetallogenicsystemandtheM2Indosinianmetallogenicsystem,correspondingtoHercynianandIndosinianmagmaticore formingprocessesrespectivelyKeywords Geochronology;Two stagemagmaticrocks;Skarntypemineralization;Niukutou;Qimantagh本文受国家重点研发计划项目(2017YFC0602403)、中国地质调查局国家二级项目(DD20190815、DD20160072)和中国铜业重点科技项目(QHHXKCZYB007、QHHX KZ JF2020 001)联合资助.第一作者简介:王新雨,男,1991年生,博士后,从事矽卡岩铅锌矿成矿理论研究,E mail:wxyu1991@126.com通讯作者:祝新友,男,1965年生,教授级高级工程师,长期从事矿床地质、地球化学研究,E mail:Zhuxinyou@outlook.com摘 要 牛苦头矿床位于东昆仑造山带祁漫塔格地区中段,目前已探明Pb+Zn资源量116万吨(推断及以上),为祁漫塔格地区探明的最大矽卡岩型铅锌矿床之一。
细胞信号转导通路

Chromatin/Epigenetics Resources
Overview of Chromatin / Epigenetics
Chromatin regulation refers to the events affecting chromatin structure and therefore, transcriptional control of gene expression patterns. Epigenetics, specifically, refers to the heritable modifications which result in altered gene expression and are not known to be encoded in DNA. The nucleosome, made up of four histone proteins (H2A, H2B, H3, and H4), is the primary building block of chromatin. Originally thought to function as a static scaffold for DNA packaging, histones have more recently been shown to be dynamic proteins, undergoing multiple types of post-translational modifications (PTMs) and interacting with regulatory proteins to control gene expression. Protein acetylation plays a crucial role in regulating chromatin structure and transcriptional activity. Histone hyperacetylation by histone acetyltransferases (HATs) is associated with transcriptional activation, whereas histone deacetylation by histone deacetylases (HDACs) is associated with transcriptional repression. Hyperacetylation can directly affect chromatin structure by neutralizing the positive charge on histone tails and disrupting nucleosome-nucleosome and nucleosomeDNA interactions. In addition, acetylation creates binding sites for bromodomain-containing chromatin regulatory proteins (histone modification readers). Unlike acetylation, methylation does not alter the charge of arginine and lysine residues and is unlikely to directly modulate nucleosomal interactions required for chromatin folding. Methylated arginine and lysine residues are major determinants for formation of active and inactive regions of the genome. Methylation facilitates binding of chromatin regulatory proteins/histone modification readers that contain various methyl-lysine or methyl-arginine binding domains (PHD, chromo, WD40, Tudor, MBT, Ankyrin repeats, PWWP domains). Recruitment of co-activator and co-repressor proteins is dependent on the specific lysine residue that is modified. The modulation of chromatin structure is an essential component in the regulation of transcriptional activation and repression. One strategy by which chromatin structure can be modulated is through disruption of histone-DNA contacts by ATP-dependent chromatin remodelers, such as the NuRD, Polycomb, and SWI/SNF complexes, which have been shown to regulate gene activation/repression, cell growth, the cell cycle, and differentiation. Chromatin structure is also modulated through other PTMs such as phosphorylation of histone proteins, which affects association with DNA-interacting proteins and has been recently identified to play a role in coordinating other histone modifications. Furthermore, methylation of DNA at cytosine residues in mammalian cells affects chromatin folding and is a heritable, epigenetic modification that is critical for proper regulation of gene silencing, genomic imprinting, and development. Three families of mammalian DNA methyl-transferases have been identified, DNMT1/2/3, that play distinct roles in embryonic stem cells and adult somatic cells. In addition to the core histone proteins, a number of histone variants exist that confer different structural properties to nucleosomes and play a number of specific functions such as DNA repair, proper kinetochore assembly and chromosome segregation during mitosis, and regulation of transcription. Chromatin and epigenetic regulation is crucial for proper programming of the genome during development and under stress conditions, as the misregulation of gene expression can lead to diseased states such as cancer.
State of charge estimation of lithium-ion batteries using the open-circuit
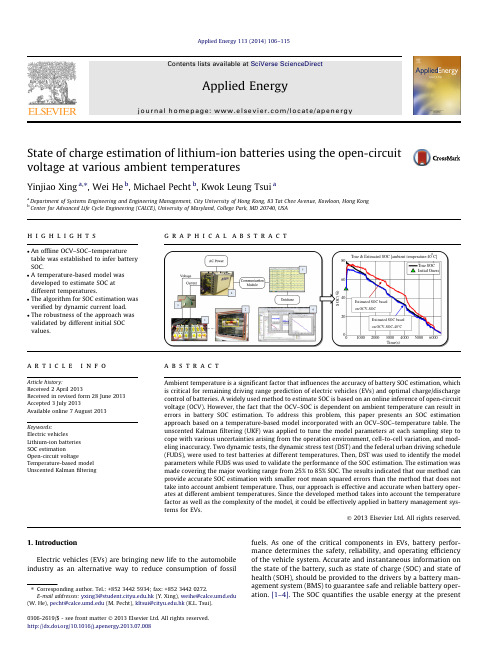
State of charge estimation of lithium-ion batteries using the open-circuit voltage at various ambienttemperaturesYinjiao Xing a ,⇑,Wei He b ,Michael Pecht b ,Kwok Leung Tsui aa Department of Systems Engineering and Engineering Management,City University of Hong Kong,83Tat Chee Avenue,Kowloon,Hong Kong bCenter for Advanced Life Cycle Engineering (CALCE),University of Maryland,College Park,MD 20740,USAh i g h l i g h t sg r a p h i c a l a b s t r a c t100020003000400050006000020406080Time(s)S O C (%)True & Estimated SOC [ambient temperature:40oC]True SOC Initial GuessEstimated SOC based on OCV-SOCEstimated SOC based on OCV-SOC-40°Ca r t i c l e i n f o Article history:Received 2April 2013Received in revised form 28June 2013Accepted 3July 2013Available online 7August 2013Keywords:Electric vehiclesLithium-ion batteries SOC estimationOpen-circuit voltageTemperature-based model Unscented Kalman filteringa b s t r a c tAmbient temperature is a significant factor that influences the accuracy of battery SOC estimation,which is critical for remaining driving range prediction of electric vehicles (EVs)and optimal charge/discharge control of batteries.A widely used method to estimate SOC is based on an online inference of open-circuit voltage (OCV).However,the fact that the OCV–SOC is dependent on ambient temperature can result in errors in battery SOC estimation.To address this problem,this paper presents an SOC estimation approach based on a temperature-based model incorporated with an OCV–SOC–temperature table.The unscented Kalman filtering (UKF)was applied to tune the model parameters at each sampling step to cope with various uncertainties arising from the operation environment,cell-to-cell variation,and mod-eling inaccuracy.Two dynamic tests,the dynamic stress test (DST)and the federal urban driving schedule (FUDS),were used to test batteries at different temperatures.Then,DST was used to identify the model parameters while FUDS was used to validate the performance of the SOC estimation.The estimation was made covering the major working range from 25%to 85%SOC.The results indicated that our method can provide accurate SOC estimation with smaller root mean squared errors than the method that does not take into account ambient temperature.Thus,our approach is effective and accurate when battery oper-ates at different ambient temperatures.Since the developed method takes into account the temperature factor as well as the complexity of the model,it could be effectively applied in battery management sys-tems for EVs.Ó2013Elsevier Ltd.All rights reserved.1.IntroductionElectric vehicles (EVs)are bringing new life to the automobile industry as an alternative way to reduce consumption of fossil fuels.As one of the critical components in EVs,battery perfor-mance determines the safety,reliability,and operating efficiency of the vehicle system.Accurate and instantaneous information on the state of the battery,such as state of charge (SOC)and state of health (SOH),should be provided to the drivers by a battery man-agement system (BMS)toguarantee safe and reliable battery oper-ation.[1–4].The SOC quantifies the usable energy at the present0306-2619/$-see front matter Ó2013Elsevier Ltd.All rights reserved./10.1016/j.apenergy.2013.07.008⇑Corresponding author.Tel.:+852********;fax:+852********.E-mail addresses:yxing3@.hk (Y.Xing),weihe@ (W.He),pecht@ (M.Pecht),kltsui@.hk (K.L.Tsui).cycle,while the SOH denotes the remaining performance of the battery over its whole life cycle[5].Battery SOC is a direct and immediate look at the remaining charge of the battery,which re-flects residual range of an EV.This has gained more attention due to drivers’range anxiety i.e.running out of power on the road. Additionally,an accurate SOC is an indicator of how to improve a battery’s operational reliability,extend its lifespan,and optimize the power management of the vehicle[1,2,6].However,SOC can-not be measured directly but must be estimated according to mea-surable parameters such as current and voltage.Moreover, ambient temperature is a critical factor that affects the accuracy of SOC estimation[7–12].There are three main types of methods for SOC estimation:cou-lomb counting,machine learning methods,and their combination using a model-based estimation approach.These three types of methods are described below.Coulomb counting is a straightforward method for estimating SOC that accumulates the net charge at the last time period in units of ampere-hours(Ah).Its performance is highly reliant on the pre-cision of current sensors and the accurate estimation of the initial SOC[3,13].However,coulomb counting is an open-loop estimator that does not eliminate the accumulation of measurement errors and uncertain disturbances.In addition,it is not able to determine the initial SOC,and address the variation of the initial SOC caused by self-discharging.Without the knowledge of the initial SOC,this method will cause accumulating errors on SOC estimation.Taking into account these factors,regular recalibration is recommended and widely used by methods such as fully discharging the battery, or referring to other measurements such as open-circuit voltage (OCV),as suggested in[3,6,7,14].Machine learning approaches,including artificial neural net-works,fuzzy logic–based models,and support vector machines, have been used to estimate SOC online.Li et al.[15]designed a 12-input-2-level merged fuzzy neural network(FNN)that was fused with a reduced-form generic algorithm(RGA)to estimate SOC.Bo et al.[16]developed parallel chaos immune evolutionary programming(PICEP)to train a neural network model in which five input variables were selected.This approach was used to esti-mate the SOC of nickel–metal hydride(Ni/MH)batteries.The per-formance of the kind of black-box models is reliant on the reliability of the training data,i.e.whether it is sufficient to cover the entire loading conditions.Once the battery operated at the un-known loading conditions,the robustness of these models was subject to challenge.Wang[17]employed a support vector ma-chine to model the dynamic behavior of a Ni/MH battery under dy-namic current loading.However,model training is time consuming and requires a large amount of data.Also,the estimation based on this model causes a large prediction error due to the uncertainty of the new data set.A model-basedfiltering estimation approach is being widely ap-plied due to its close-loop nature and concerning various uncer-tainties.Both electrochemical models and equivalent circuit models aim to capture the dynamic behavior of the battery.The former are usually presented in the form of partial differential equations with many unknown parameters.They are accurate but not desirable in practice because of a high requirement for memory and computation.To guarantee the accuracy of the model and the feasibility,equivalent circuit models have been imple-mented in BMSs such as the enhanced self-correcting(ESC)model and the hysteresis model,as found in[10,18,19],and one or two-order resistance–capacitance(RC)network models[1,2,10,11,20]. OCV is a vital element in the above-mentioned battery equivalent models and is a function of SOC in nature.The premise of utilizing OCV–SOC is that the battery needs to rest a long time and terminal voltage approaches the OCV.However,in real life,a long resting time may not be possible.To make up for theflaws of OCV meth-ods,nonlinearfiltering techniques based on state-space models have been developed to enhance SOC estimation through combin-ing coulomb counting and OCV[7].Plett applied extended Kalman filters(EKF)into BMS to implement SOC estimation of a lithium polymer battery(LiPB)using different battery models in [10,21,22].Plett later proposed the use of two sigma-point Kalman filtering(SPKF)estimators,including the unscented Kalmanfilter (UKF)and central difference Kalmanfilter(CDKF),in[18,23].Sub-sequently,adaptive EKF[7,20],dual EKF[11],and adaptive UKF[3] were developed to improve the accuracy of the SOC estimation based on their own sample sets and some common equivalent cir-cuit models.Charkhgard and Farrokhi[13]also proposed the com-bination of NN and EKF to estimate SOC.NN was employed to train a lithium-ion battery model using some charging data from the battery.The effectiveness of this method was not verified under the dynamic discharging data,which would lead to a larger uncer-tainty on estimating SOC.However,several existing issues are seldom addressed in the literature.Firstly,the temperature dependence of the OCV–SOC lookup table is seldom discussed with regard to battery SOC esti-mation.Instead,a single OCV–SOC table constructed at a certain temperature(e.g.,room temperature)is widely employed.It will cause a large error in inferring SOC when the battery is operating at other ambient temperatures(not room temperature) [1,8,10,11].Secondly,lithium-ion batteries have a relativelyflat OCV curve over the SOC,especially for lithium iron phosphate (LiFePO4)batteries,which are widely used in the electric vehicle market[24].That means a small error on the inferred OCV will produce a larger deviation in SOC.Thirdly,different models were adopted by individuals based on their own experimental data. Although a sophisticated model with more parameters might be able to provide a smaller modeling error,it would run the risk of adding more uncertainties,such as over-fitting problems and the introduction of unnecessary noises,especially concerning temperature.Therefore,it makes more sense to investigate a gen-eric but accurate temperature-based model with fewer parame-ters for real-time applications.Kim et al.[25]considered temperature as an input variable into afirst-order RC circuit model.However,the effect of temperature on the OCV–SOC was ignored due to a slight difference between OCV curves from 30%SOC to80%at different temperatures.Moreover,their samples have an obvious linear slope of OCV–SOC that is prone to infer SOC accurately.Nevertheless,for a relativelyflat OCV curve dependent on ambient temperature,it is significant not only to develop an accurate and generic model considering ambient temperature,but also to enhance the capability of online estimation due to the uncertainty,including unit-to-unit varia-tion,measurement noise,operational uncertainties,and model inaccuracy[26].In this paper,a temperature-based internal resistance(R int)bat-tery model combined with a nonlinearfiltering method was put forward to improve the SOC estimation of lithium-ion batteries un-der dynamic loading conditions at different ambient temperatures. The research proceeds as follows.Three tests at different tempera-tures are introduced in Section2.The dynamic stress test(DST)and federal urban driving schedule(FUDS)are two kinds of dynamic loading conditions tested at different temperatures to identify the model parameters and verify the estimated performance, respectively.The purpose of the OCV–SOC–temperature(OCV–SOC–T)test is to extend the OCV–SOC behavior to temperature field.Due to various uncertainties of the system,UKF-based SOC estimator is proposed due to its superiority of reaching to the 3rd order of any nonlinearity over the EKF.The implemented procedure for our battery study is followed by Section3.The experimental results are presented in Section4to compare our developed method based on OCV–SOC–T with the originalY.Xing et al./Applied Energy113(2014)106–115107estimation using a single OCV–SOC table.The robustness is vali-dated and compared under the different initial true values and dif-ferent initial guesses of SOC.2.ExperimentsThe experiment setup is shown in Fig.1.It consisted of (1)lith-ium-ion cells (LiFePO4)of the 18650cylindrical type (the key spec-ifications are shown in Table 1);(2)Vötsch temperature test chamber (The cells were placed in cell holders in the chamber);(3)a current and voltage sampling cable for loading and sampling;(4)a battery test system (Arbin BT2000tester);(5)a PC with Ar-bins’Mits Pro Software (v4.27)for battery charging/discharging control;(6)Matlab R2009b for data analysis.During battery oper-ation,the sampling time of current,voltage was 1s.Three separate test schedules were conducted on the battery test bench for model identification,OCV–SOC–T table construction,and method valida-tion,respectively.2.1.Model identification testFor model identification,the dynamic stress test (DST)was run from 0°C to 50°C at an interval of 10°C.DST is employed to investigate the dynamic electric behavior of the battery.It is designed by US Advanced Battery Consortium (USABC)to simu-late a variable-power discharge regime that represents the expected demands of an EV battery [27].A completed DST cycle is 360s long and can be scaled down to any desired maximum demand regarding the specifications of the test samples.There-fore,in our study,DST was run continuously from 100%SOC at 3.6V to empty at 2V over several cycles in a discharge process.The positive current responds to discharging while the negative denotes charging.The measured current and voltage profile at 20°C is shown in Fig.2.2.2.The OCV–SOC–T testOCV is a function of SOC for the cells.If the cell is able to rest for a long period until the terminal voltage approached the true OCV,OCV can be used to infer SOC accurately.However,this method is not practical for dynamic SOC estimation.To address this issue,the SOC can be estimated by combining the online identification of the OCV with the predetermined offline OCV–SOC lookup table.Taking into account the temperature dependence of the OCV–SOC table,the OCV–SOC test was conducted from 0°C to 50°C at an interval of 10°C.The test procedure at each temperature is the same as fol-lows.Firstly,the cell was fully charged using a constant current of 1C-rate (1C-rate means that a full discharge of the battery takes approximately 1h)until the voltage reached to the cut-off voltage of 3.6V and the current was 0.01C.Secondly,the cell was fully dis-charged at a constant rate of C/20until the voltage reached 2.0V,which corresponds to 0%SOC.Finally,the cell was fully charged at a constant rate of C/20to 3.6V,which corresponds to 100%SOC.The terminal voltage of the cell is considered as a close approximation to the real equilibrium potential [6,10].AsshownFig.1.Schematic of the battery test bench.Table 1The key specifications of the test samples.Type Nominal voltage Nominal capacity Upper/lower cut-off voltage Maximum continuous discharge current LiFePO 43.3V1.1Ah3.6V/2.0V30A010002000300040005000-2024Time (s)C u r r e n t (A )in Fig.3,the equilibrium potential during the charging process is higher than that during discharging process.It accounts for a hys-teresis phenomenon of the OCV during the charging/discharging.In our paper,the OCV curve was defined as the average value of the charge and discharge equilibrium potentials.The effect of the hys-teresis was ignored.In addition,referring to[28],when SOC is normalized relative to the specific cell capacity,the OCV–SOC curve can be referred to as being unique for the same type at the same testing condition.Fig.3shows the average OCV at20°C.A flat OCV slope between25%and80%SOC is emphasized in another small plot in Fig.3.Its effect will be discussed in Section3.1.2.3.Method validation testA validation test with a more sophisticated dynamic current profile,the federal urban driving schedule(FUDS),was conducted to verify the estimation algorithm based on the developed model. FUDS is a dynamic electric vehicle performance test based on a time–velocity profile from an automobile industry standard vehi-cle[27].In the laboratory test,a dynamic current sequence was transferred from the time–velocity profile,programmed to charge or discharge the battery and applied to battery performance test [3,17,20,29].Similar to the DST test,the current sequence is scaled tofit the specification of the test battery and the limitation of the testing system of Fig.1.The current profile of FUDS causes varia-tion of the SOC from fully charged at3.6V to empty at2V.The FUDS test was also run form0°C to50°C at an interval of10°C. The measured current,voltage profile,and the cumulative SOC at 20°C are shown in Fig.4.A completed FUDS current profile over 1372s is emphasized in another graph in Fig.4(a).3.Battery modelingFor lithium-ion batteries,the internal resistance(R int)model is generic and straightforward to characterize a battery’s dynamics with one estimated parameter.Although a sophisticated model with more parameters would possibly show a well-fitting result, such as an equivalent circuit model with several amounts of paral-lel resistance–capacitance(RC)networks,it would also pose a risk of over-fitting and introducing more uncertainties for online esti-mation at the same time.Especially taking into account tempera-ture factor,more complexity should be imposed on battery modeling.Therefore,we would prefer a simple model to a sophis-ticated model if the former had generalization ability and provided sufficiently good results.In this paper,model modification based on the original R int model is proposed to balance the model com-plexity and the accuracy of battery SOC estimation.The schematic of the original R int model is shown in Fig.5.U term;k¼U OCVÀI kÂRð1ÞU OCV/fðSOC kÞð2ÞIn Eqs.(1)and(2),U term,k is the measured terminal voltage of the battery under a normal dynamic current load at time k,and I k is the dynamic current at the same time.The positive current re-sponds to discharging while the negative value means charging.R is the simplified total internal resistance of the battery.U OCV is a function of SOC of the battery that should be tested following the procedure as presented in Section2.2.The battery model Eq.(1) can be used to infer OCV directly according to the measured termi-Schematic of the internal resistance(R int)modelY.Xing et al./Applied Energy113(2014)106–1151093.1.Model parameter identificationThe DST was run on the LiFePO4batteries to identify the model parameter R in Eq.(1).Taking the current and voltage profile of DST at20°C as an example,the voltage and current are measured and recorded from fully charged to empty with a sampling period of1s based on our battery test bench.The accumulative charge(exper-imental SOC)is calculated synchronously from100%SOC.Thus, the parameter R can befitted using a sequence of the current,volt-age,and the offline OCV–SOC by the least square algorithm.In terms of thefitted R value,the model performance can be evalu-ated based on the measured terminal voltage(U term,k)and the esti-mated voltage(U term;k).Fig.6shows the measured and the estimated voltage response on the DST profile at20°C based on the original model.In statistics,the mean absolute error(MAE)and the root mean squared(RMS)error can be used together to evaluate the good-ness-of-fit of the model.These two indicators are given by the fol-lowing equations respectively.MAE¼1X nk¼1j e k jð3ÞRMS error¼ffiffiffiffiffiffiffiffiffiffiffiffiffiffiffiffiffiffiffiffiffiffiffiffiffiffiffi1nX nk¼1ðe kÞ2rð4ÞHere,e k is the modeling error(U term,kÀU term;k)at time k.The MAE measures how close forecasts are to the corresponding out-comes without considering the direction.The RMS error is more sensitive to large errors than the MAE.It is able to characterize the variation in errors.The statistics list of the model is shown in Table2.According to the small graph in Fig.3,the OCV slope,namely, dOCV/dSOC is approximately equal to0.0014between25%and 80%SOC.This means that the deviation on the OCV inference will cause an estimated deviation of SOC up to21%when there is a MAE of0.0288V based on the current model.Additionally,a large mean error was plotted over time in Fig.6.Thus,the residuals should be reduced to improve the model adequacy with smaller MAE and RMS error values.3.2.Model improvement and validation3.2.1.The OCV–SOC–T table for model improvementAccording to the test in Section2.2,six OCV curves were ob-tained from0°C to50°C at an interval of10°C.Fig.7(a)empha-sizes the differences of OCV–SOC curves between30%and80% SOC at different temperatures.It can be seen that SOC0°C is much larger than other SOC values at higher temperatures when the OCV inference is the same,i.e.,3.3V.It makes sense that the releas-able capability of the charge is reduced at low temperatures. Fig.7(b)shows the SOC values if the OCV inference was equal to the specific values from3.28V to3.32V at intervals of0.01V at three temperatures:0°C,20°C,40°C.One issue of interest can be seen in Fig.7(b).That is,the same OCV inference at different temperatures corresponds to different SOC values.For example,the SOC difference between0°C and 40°C reaches approximately22%at an OCV of3.30V.Therefore, we propose adding the OCV–SOC–T to the battery model to im-prove the model accuracy.The improved battery model is as follows:U term;k¼U OCVðSOC k;TÞÀI kÂRðTÞþCðTÞð5Þwhere U OCV is a function of SOC and ambient temperature(T).C(T)is a function of temperature that facilitates the reduction of the offset due to model inaccuracy and environmental conditions.Fig.8 shows the measured and the estimated voltage response on the DST profile at20°C based on the proposed model.It can be found that the mean error of the new model is reduced with small varia-tions as compared to the original model in Fig.6.Another issue of interest in Fig.7(b)is that a small deviation of 0.01V in OCV inference will lead to a large difference in SOC at the same temperature condition.It is the same issue as shown in Fig.3. Therefore,if the SOC estimation were directly inferred from a bat-tery model,it would have a high requirement on the model and measurement accuracy.To address this issue and improve the accuracy of the SOC estimation,the model-based unscented Kal-manfiltering approach was employed and introduced in Section4.3.2.2.The validation of the proposed modelBased on the developed model in Eq.(5),it is noted that the spe-cific OCV–SOC look-up table should be selected in terms of the ambient temperature(here it is viewed as an average value).Least squarefitting was also used to identify model parameters,R and C. Thefitted model parameter list and the statistics list of the pro-posed model are shown in Table3.In comparison to thefitted results at20°C of Table2,here the MAE is one order of magnitude smaller than that of the original model and the RMS modeling error is also reduced.In addition, the correlation coefficient(Corrcoef)was calculated for residual analysis.The Corrcoef(e k,I k)values close to zero indicate that the residuals and the input variable hardly have linear relationship. Thus,the corrected model can be betterfitted on the dynamic cur-rent load.Onefinding of interest is C values that can befitted over the ambient temperature(T)using a regression curve,as Fig.9 shows.Referring to the paper[30],the exponential function can be selected tofit C values over T because the internal elements of the battery,i.e.,battery resistance follow the Arrhenius equation, which has exponential dependency on the temperature.In our study,five C values at0°C,10°C,20°C,25°C,30°C,40°C wereTable2Model parameter and statistics list of thefitted error of the model.R(X)Mean absolute error RMS modeling error0.24450.0288V0.0301110Y.Xing et al./Applied Energy113(2014)106–115used for curvefitting while C(50°C)was used to test thefitted per-formance of this exponential function.The95%prediction bounds are shown in Fig.9based on C values andfitted curve.Apparently, C(50°C)drops within the95%prediction bounds.It can be seen that the function of C(T)in Fig.9can be used to estimate C when the corresponding temperature test has not been run.4.Algorithm implementation for online estimationThe online SOC estimation has strong nonlinearity.This point can be seen from any battery model in which OCV has a nonlinear relationship with SOC.Additionally,the uncertainties due to the model inaccuracy,measurement noise,and operating conditions will cause a large variation in the estimation.The model-based nonlinearfiltering approach has been developed to implement dy-namic SOC estimation.The objective is to estimate the hidden sys-tem state,estimate the model parameters for system identification, or both.Thus,an error-feedback-based unscented Kalmanfiltering approach is proposed by shifting the system noise to improve the accuracy of the estimation.4.1.Unscented KalmanfilteringThe extended Kalmanfiltering(EKF)technique has become a popular technique for addressing the issue of state or parameter estimation for nonlinear systems.The rationale behind EKF is still the KF approach based on state space modeling.It aims to utilize the error between the current measurement and the model output to adjust the model state by virtue of a Kalman gain.Its principle and implementation can be found in[31].Since KF is only available for linear systems,extended Kalmanfiltering(EKF)used a lineari-zation process at each time step to approximate a nonlinear system through thefirst-order Taylor series expansion[31,32].However, thefirst order approximation will probably lead to large errors inTable3Fitted model parameter list and statistics list of modelfitting.T(°C)R(X)C Mean absoluteerrors(V)RMSmodelingerrorsCorrcoef(e k,I k)00.2780À0.05520.01530.0188 1.36eÀ13100.2396À0.04360.01120.01348.45eÀ14200.2249À0.03600.00870.0105 1.09eÀ13250.2020À0.03260.00800.0095 1.02eÀ13300.1838À0.02890.00730.0085À7.62eÀ13400.1565À0.02370.00600.0071 2.85eÀ13500.1816À0.02010.00990.0131 3.15eÀ14Y.Xing et al./Applied Energy113(2014)106–115111the true posterior mean and covariance of the noise and could even result in divergence of thefilter.Under this situation,unscented Kalmanfiltering(UKF)based on unscented transformation was suggested to avoid the weakness that comes from using Taylor ser-ies expansion.The core idea of UKF is easier to approximate the state distribu-tion that is represented by a minimal set of chosen sample points called sigma points,which can capture the mean and covariance of Gaussian random variable when propagated through a nonlinear system.The state-space model of a nonlinear system is represented as follows:x k¼fðx kÀ1;u kÀ1Þþw kÀ1yk¼hðx k;u kÞþm kð6Þwhere x k is the system state vector and y k is the measurement vec-tor at time k.Correspondingly,f(Á)and h(Á)are the state function and the measurement function,respectively;u k is the known input vector;w k$Nð0;PwÞis the Gaussian process noise;and v k$Nð0;PVÞis the Gaussian measurement noise.Assume the state x has mean x and covariance P ing the unscented transformation (UT),the state will be transformed as a matrix of2n+1sigma vec-tors v i with corresponding weights w i.These sigma points are shown in the following equation:vkÀ1¼½ xð0ÞkÀ1; xð1:nÞkÀ1; xðnþ1:2nÞkÀ1¼½ x kÀ1; x kÀ1þffiffiffiffiffiffiffiffiffiffiffiffiffiffiffiffiffiffiffiffiffiffiffiffiffiðnþkÞP kÀ1p; x kÀ1ÀffiffiffiffiffiffiffiffiffiffiffiffiffiffiffiffiffiffiffiffiffiffiffiffiffiðnþkÞP kÀ1pð7Þwhere n is the dimension of the state and k is a scaling parameter. These sigma points are propagated through a nonlinear function,re-estimated,and then they are used to capture the posterior mean and covariance to the3rd Taylor expansion,as shown in Eqs.(9) and(10).yi¼hðv iÞ;i¼0;...;2nð8Þ y%X2ni¼0wðiÞmyið9ÞP y%X2ni¼0wðiÞcf y iÀ y gf y iÀ yg Tð10Þwhere wðiÞm and wðiÞc are the weights of the corresponding sigma points that can be calculated in[33–35]for details.4.2.SOC estimation based on proposed methodIn our battery study,the state vector is x=[SOC,R]T.Thefirst state equation in Eq.(11)follows the coulomb counting method mentioned in Section1.Peukert effect and capacity aging could be partially compensated when introducing the process noise x1,kÀ1.A random walk is applied to the model parameter R regard-ing the cell-to-cell variation and operation uncertainties.Tuning the R will also be able to compensate for the variation of the C in our proposed model.The terminal voltage of the battery is the measured vector y=U term,that is,the proposed battery model as shown in Eq.(5).State function:SOC k¼SOC kÀ1ÀI kÀ1ÂD t=C nþx1;kÀ1R k¼R kÀ1þx2;kÀ1ð11ÞMeasurement function:U term;k¼U OCVðSOC k;TÞÀI kÂR kðTÞþCðTÞþm kð12Þwhere I k is the current as the input(u k in Eq.(6))at time k;D t is the sampling interval,which is1s according to the sampling rate;and C n is the rated capacity.The rated capacity of the test samples is1.1 Ah.And x1,k,x2,k and m k are zero-mean white stochastic processes with covariancesPx1,Px2andPm,respectively.According to the proposed state-space model in Eqs.(11)and(12),the procedure of SOC estimation based on UKF is summarized in Table4.Table4Summary of the UKF approach for SOC estimation.Initialize:–Measure ambient temperature,prepare U OCV(SOC,T)and R0,C0–Initial guess:S0,–Covariance matrix:P o–Process and measurement noise covariance:Pw0;PVGenerate sigma points at time kÀ1,(k2½l;...;1 ):vkÀ1¼S kÀ1R kÀ1¼½ v kÀ1; v kÀ1þffiffiffiffiffiffiffiffiffiffiffiffiffiffiffiffiffiffiffiffiffiffiffiffiffiðnþkÞP kÀ1p; v kÀ1ÀffiffiffiffiffiffiffiffiffiffiffiffiffiffiffiffiffiffiffiffiffiffiffiffiffiðnþkÞP kÀ1pPredict the prior state mean and covariance–Calculate sigma points through state function:v ik j kÀ1¼S i k j kÀ1R ik j kÀ1"#¼S i kÀ1ÀI kÀ1ÂD tC nR i kÀ1"#;i¼1; (2)–Calculate the prior mean and covariance:^xÀk ¼P2ni¼0w imv ik j kÀ1;PÀk¼P2ni¼0w ichv ik j kÀ1À^xÀkihv ik j kÀ1À^xÀki TþPwUpdate using the measurement function–Calculate sigma points y k j kÀ1¼U OCVðS k j kÀ1;TÞÀI kÂRðTÞk j kÀ1þCðTÞ–Calculate the propagated mean:^yÀk ¼P2ni¼0w i m y ik j kÀ1–Calculate the covariance of the measurement:P yÀk ;yÀk¼P2ni¼0w j c½y ik j kÀ1À^yÀkTþPm–Calculate the cross-covariance and the state and measurement:of battery SOC estimation by UKF112Y.Xing et al./Applied Energy113(2014)106–115。
support-vector-machine
1
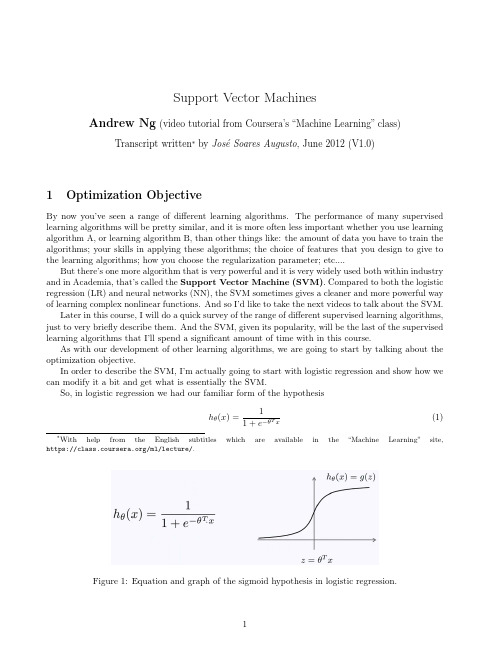
Figure 2: The two partial cost terms belonging to the cost function J (θ) for logistic regression: in the left, the positive case for y = 1 is − log 1+1 ; in the right, the negative case for y = 0, is e−z − log 1 −
m
y (i) log hθ x(i) + 1 − y (i) log 1 − hθ x(i)
i=1
+
λ 2m
n 2 θj j =1
(2)
you find that each example, (x, y ), contributes the term (forgetting averaging with the ห้องสมุดไป่ตู้/m weight) − (y log(hθ (x)) + (1 − y ) log(1 − hθ (x))) to the overall cost function, J (θ). If I take the definition of my hypothesis (1), and plug it in the above cost term, what I get is that each training example contributes with the quantity −y log 1 1 + e−θT x − (1 − y ) log 1 − 1 1 + e−θT x (3)
in the objective
Recall that z = θT x. If we plot − log
某理工大学《生物化学》考试试卷(107)

某理工大学《生物化学》课程试卷(含答案)__________学年第___学期考试类型:(闭卷)考试考试时间:90 分钟年级专业_____________学号_____________ 姓名_____________1、判断题(145分,每题5分)1. 凡是与茚三酮反应不产生特有的蓝紫色的氨基酸,即可认为不是蛋白质氨基酸。
()答案:错误解析:有的氨基酸与茚三酮反应不产生特有的蓝紫色,如脯氨酸、羟脯氨酸与茚三酮反应生成的是黄色产物。
2. 脂肪的皂化价高表示含低相对分子质量的脂肪酸少。
()[山东大学2017研]答案:错误解析:皂化值(价)是指皂化1g脂肪所需的KOH的毫克数。
通常从皂化值的数值即可略知混合脂酸或混合脂肪的平均相对分子质量。
皂化值与脂肪(或脂酸)的相对分子质量成反比,脂肪的皂化值高表示含低相对分子质量的脂酸较多,因为相同质量的低级脂酸皂化时所需的KOH数量比高级脂酸为多。
3. 处于等电点时氨基酸的溶解度最小。
()答案:正确解析:4. 有n个不对称C原子单糖的旋光异构体为2n-1。
()[四川大学2017研]答案:错误解析:糖分子的旋光异构体数目取决于其所含不对称碳原子数目,用n表示不对称碳原子数目,C表示不对称碳原子。
含n个C的单糖有2n个旋光异构体,组成2n-1对不同的对映体。
5. 复性后DNA分子中的两条链并不一定是变性前该分子原先的两条链。
()答案:正确解析:6. 所谓肽单位就是指组成蛋白质的氨基酸残基。
()答案:错误解析:肽单位又称肽基,是肽链主链上的重复结构,是由参与肽键形成的氮原子、碳原子和它们的4个取代成分羰基氧原子、酰胺氢原子和两个相邻的α碳原子组成的一个平面单位。
7. 蛋白质构象是蛋白质分子中的原子绕单键旋转而产生的蛋白质分子中的各原子的空间排布。
因此,构象并不是一种可以分离的单一立体结构形式。
答案:正确解析:8. Km是酶的特征常数,只与酶的性质有关,与酶浓度无关。
()。
matlab工具箱安装教程
1.1 如果是Matlab安装光盘上的工具箱,重新执行安装程序,选中即可;1.2 如果是单独下载的工具箱,一般情况下仅需要把新的工具箱解压到某个目录。
2 在matlab的file下面的set path把它加上。
3 把路径加进去后在file→Preferences→General的Toolbox Path Caching里点击update Toolbox Path Cache更新一下。
4 用which newtoolbox_command.m来检验是否可以访问。
如果能够显示新设置的路径,则表明该工具箱可以使用了。
把你的工具箱文件夹放到安装目录中“toolbox”文件夹中,然后单击“file”菜单中的“setpath”命令,打开“setpath”对话框,单击左边的“ADDFolder”命令,然后选择你的那个文件夹,最后单击“SAVE”命令就OK了。
MATLAB Toolboxes============================================/zsmcode.htmlBinaural-modeling software for MATLAB/Windows/home/Michael_Akeroyd/download2.htmlStatistical Parametric Mapping (SPM)/spm/ext/BOOTSTRAP MATLAB TOOLBOX.au/downloads/bootstrap_toolbox.htmlThe DSS package for MATLABDSS Matlab package contains algorithms for performing linear, deflation and symmetric DSS. http://www.cis.hut.fi/projects/dss/package/Psychtoolbox/download.htmlMultisurface Method Tree with MATLAB/~olvi/uwmp/msmt.htmlA Matlab Toolbox for every single topic !/~baum/toolboxes.htmleg. BrainStorm - MEG and EEG data visualization and processingCLAWPACK is a software package designed to compute numerical solutions to hyperbolic partial differential equations using a wave propagation approach/~claw/DIPimage - Image Processing ToolboxPRTools - Pattern Recognition Toolbox (+ Neural Networks)NetLab - Neural Network ToolboxFSTB - Fuzzy Systems ToolboxFusetool - Image Fusion Toolboxhttp://www.metapix.de/toolbox.htmWAVEKIT - Wavelet ToolboxGat - Genetic Algorithm ToolboxTSTOOL is a MATLAB software package for nonlinear time series analysis.TSTOOL can be used for computing: Time-delay reconstruction, Lyapunov exponents, Fractal dimensions, Mutual information, Surrogate data tests, Nearest neighbor statistics, Return times, Poincare sections, Nonlinear predictionhttp://www.physik3.gwdg.de/tstool/MATLAB / Data description toolboxA Matlab toolbox for data description, outlier and novelty detectionMarch 26, 2004 - D.M.J. Taxhttp://www-ict.ewi.tudelft.nl/~davidt/dd_tools/dd_manual.htmlMBEhttp://www.pmarneffei.hku.hk/mbetoolbox/Betabolic network toolbox for Matlabhttp://www.molgen.mpg.de/~lieberme/pages/network_matlab.htmlPharmacokinetics toolbox for Matlabhttp://page.inf.fu-berlin.de/~lieber/seiten/pbpk_toolbox.htmlThe SpiderThe spider is intended to be a complete object orientated environment for machine learning in Matlab. Aside from easy use of base learning algorithms, algorithms can be plugged together and can be compared with, e.g model selection, statistical tests and visual plots. This gives all the power of objects (reusability, plug together, share code) but also all the power of Matlab for machine learning research.http://www.kyb.tuebingen.mpg.de/bs/people/spider/index.htmlSchwarz-Christoffel Toolbox/matlabcentral/fileexchange/loadFile.do?objectId=1316&objectT ype=file#XML Toolbox/matlabcentral/fileexchange/loadFile.do?objectId=4278&object Type=fileFIR/TDNN Toolbox for MATLABBeta version of a toolbox for FIR (Finite Impulse Response) and TD (Time Delay) NeuralNetworks./interval-comp/dagstuhl.03/oish.pdfMisc.http://www.dcsc.tudelft.nl/Research/Software/index.htmlAstronomySaturn and Titan trajectories ... MALTAB astronomy/~abrecht/Matlab-codes/AudioMA Toolbox for Matlab Implementing Similarity Measures for Audiohttp://www.oefai.at/~elias/ma/index.htmlMAD - Matlab Auditory Demonstrations/~martin/MAD/docs/mad.htmMusic Analysis - Toolbox for Matlab : Feature Extraction from Raw Audio Signals for Content-Based Music Retrihttp://www.ai.univie.ac.at/~elias/ma/WarpTB - Matlab Toolbox for Warped DSPBy Aki Härmä and Matti Karjalainenhttp://www.acoustics.hut.fi/software/warp/MATLAB-related Softwarehttp://www.dpmi.tu-graz.ac.at/~schloegl/matlab/Biomedical Signal data formats (EEG machine specific file formats with Matlab import routines)http://www.dpmi.tu-graz.ac.at/~schloegl/matlab/eeg/MPEG Encoding library for MATLAB Movies (Created by David Foti)It enables MATLAB users to read (MPGREAD) or write (MPGWRITE) MPEG movies. That should help Video Quality project.Filter Design packagehttp://www.ee.ryerson.ca:8080/~mzeytin/dfp/index.htmlOctave by Christophe COUVREUR (Generates normalized A-weigthing, C-weighting, octave and one-third-octave digital filters)/matlabcentral/fileexchange/loadFile.do?objectType=file&object Id=69Source Coding MATLAB Toolbox/users/kieffer/programs.htmlBio Medical Informatics (Top)CGH-Plotter: MATLAB Toolbox for CGH-data AnalysisCode: http://sigwww.cs.tut.fi/TICSP/CGH-Plotter/Poster: http://sigwww.cs.tut.fi/TICSP/CSB2003/Posteri_CGH_Plotter.pdfThe Brain Imaging Software Toolboxhttp://www.bic.mni.mcgill.ca/software/MRI Brain Segmentation/matlabcentral/fileexchange/loadFile.do?objectId=4879Chemometrics (providing PCA) (Top)Matlab Molecular Biology & Evolution Toolbox(Toolbox Enables Evolutionary Biologists to Analyze and View DNA and Protein Sequences) James J. Caihttp://www.pmarneffei.hku.hk/mbetoolbox/Toolbox provided by Prof. Massart research grouphttp://minf.vub.ac.be/~fabi/publiek/Useful collection of routines from Prof age smilde research grouphttp://www-its.chem.uva.nl/research/pacMultivariate Toolbox written by Rune Mathisen/~mvartools/index.htmlMatlab code and datasetshttp://www.acc.umu.se/~tnkjtg/chemometrics/dataset.htmlChaos (Top)Chaotic Systems Toolbox/matlabcentral/fileexchange/loadFile.do?objectId=1597&objectT ype=file#HOSA Toolboxhttp://www.mathworks.nl/matlabcentral/fileexchange/loadFile.do?objectId=3013&objectTy pe=fileChemistry (Top)MetMAP - (Metabolical Modeling, Analysis and oPtimization alias Met. M. A. P.)http://webpages.ull.es/users/sympbst/pag_ing/pag_metmap/index.htmDoseLab - A set of software programs for quantitative comparison of measured and computed radiation dose distributions/GenBank Overview/Genbank/GenbankOverview.htmlMatlab: /matlabcentral/fileexchange/loadFile.do?objectId=1139CodingCode for the estimation of Scaling Exponentshttp://www.cubinlab.ee.mu.oz.au/~darryl/secondorder_code.htmlControl (Top)Control Tutorial for Matlab/group/ctm/AnotherCommunications (Top)Channel Learning Architecture toolbox(This Matlab toolbox is a supplement to the article "HiperLearn: A High Performance Learning Architecture")http://www.isy.liu.se/cvl/Projects/hiperlearn/Source Coding MATLAB Toolbox/users/kieffer/programs.htmlTCP/UDP/IP Toolbox 2.0.4/matlabcentral/fileexchange/loadFile.do?objectId=345&objectT ype=fileHome Networking Basis: Transmission Environments and Wired/Wireless Protocols Walter Y. Chen/support/books/book5295.jsp?category=new&language=-1MATLAB M-files and Simulink models/matlabcentral/fileexchange/loadFile.do?objectId=3834&object Type=file•OPNML/MATLAB Facilities/OPNML_Matlab/Mesh Generation/home/vavasis/qmg-home.htmlOpenFEM : An Open-Source Finite Element Toolbox/CALFEM is an interactive computer program for teaching the finite element method (FEM)http://www.byggmek.lth.se/Calfem/frinfo.htmThe Engineering Vibration Toolbox/people/faculty/jslater/vtoolbox/vtoolbox.htmlSaGA - Spatial and Geometric Analysis Toolboxby Kirill K. Pankratov/~glenn/kirill/saga.htmlMexCDF and NetCDF Toolbox For Matlab-5&6/staffpages/cdenham/public_html/MexCDF/nc4ml5.htmlCUEDSID: Cambridge University System Identification Toolbox/jmm/cuedsid/Kriging Toolbox/software/Geostats_software/MATLAB_KRIGING_TOOLBOX.htmMonte Carlo (Dr Nando)http://www.cs.ubc.ca/~nando/software.htmlRIOTS - The Most Powerful Optimal Control Problem Solver/~adam/RIOTS/ExcelMATLAB xlsheets/matlabcentral/fileexchange/loadFile.do?objectId=4474&objectTy pe=filewrite2excel/matlabcentral/fileexchange/loadFile.do?objectId=4414&objectTy pe=fileFinite Element Modeling (FEM) (Top)OpenFEM - An Open-Source Finite Element Toolbox/NLFET - nonlinear finite element toolbox for MATLAB ( framework for setting up, solving, and interpreting results for nonlinear static and dynamic finite element analysis.)/GetFEM - C++ library for finite element methods elementary computations with a Matlabinterfacehttp://www.gmm.insa-tlse.fr/getfem/FELIPE - FEA package to view results ( contains neat interface to MATLA/~blstmbr/felipe/Finance (Top)A NEW MATLAB-BASED TOOLBOX FOR COMPUTER AIDED DYNAMIC TECHNICAL TRADINGStephanos Papadamou and George StephanidesDepartment of Applied Informatics, University Of Macedonia Economic & Social Sciences, Thessaloniki, Greece/fen31/one_time_articles/dynamic_tech_trade_matlab6.htm Paper: :8089/eps/prog/papers/0201/0201001.pdfCompEcon Toolbox for Matlab/~pfackler/compecon/toolbox.htmlGenetic Algorithms (Top)The Genetic Algorithm Optimization Toolbox (GAOT) for Matlab 5/mirage/GAToolBox/gaot/Genetic Algorithm ToolboxWritten & distributed by Andy Chipperfield (Sheffield University, UK)/uni/projects/gaipp/gatbx.htmlManual: /~gaipp/ga-toolbox/manual.pdfGenetic and Evolutionary Algorithm Toolbox (GEATbx)/Evolutionary Algorithms for MATLAB/links/ea_matlab.htmlGenetic/Evolutionary Algorithms for MATLABhttp://www.systemtechnik.tu-ilmenau.de/~pohlheim/EA_Matlab/ea_matlab.html GraphicsVideoToolbox (C routines for visual psychophysics on Macs by Denis Pelli)/VideoToolbox/Paper: /pelli/pubs/pelli1997videotoolbox.pdf4D toolbox/~daniel/links/matlab/4DToolbox.htmlImages (Top)Eyelink Toolbox/eyelinktoolbox/Paper: /eyelinktoolbox/EyelinkToolbox.pdfCellStats: Automated statistical analysis of color-stained cell images in Matlabhttp://sigwww.cs.tut.fi/TICSP/CellStats/SDC Morphology Toolbox for MATLAB (powerful collection of latest state-of-the-art gray-scale morphological tools that can be applied to image segmentation, non-linear filtering, pattern recognition and image analysis)/Image Acquisition Toolbox/products/imaq/Halftoning Toolbox for MATLAB/~bevans/projects/halftoning/toolbox/index.htmlDIPimage - A Scientific Image Processing Toolbox for MATLABhttp://www.ph.tn.tudelft.nl/DIPlib/dipimage_1.htmlPNM Toolboxhttp://home.online.no/~pjacklam/matlab/software/pnm/index.htmlAnotherICA / KICA and KPCA (Top)ICA TU Toolboxhttp://mole.imm.dtu.dk/toolbox/menu.htmlMISEP Linear and Nonlinear ICA Toolboxhttp://neural.inesc-id.pt/~lba/ica/mitoolbox.htmlKernel Independant Component Analysis/~fbach/kernel-ica/index.htmMatlab: kernel-ica version 1.2KPCA- Please check the software section of kernel machines.KernelStatistical Pattern Recognition Toolboxhttp://cmp.felk.cvut.cz/~xfrancv/stprtool/MATLABArsenal A MATLAB Wrapper for Classification/tmp/MATLABArsenal.htmMarkov (Top)MapHMMBOX 1.1 - Matlab toolbox for Hidden Markov Modelling using Max. Aposteriori EM Prerequisites: Matlab 5.0, Netlab. Last Updated: 18 March 2002./~parg/software/maphmmbox_1_1.tarHMMBOX 4.1 - Matlab toolbox for Hidden Markov Modelling using Variational Bayes Prerequisites: Matlab 5.0,Netlab. Last Updated: 15 February 2002../~parg/software/hmmbox_3_2.tar/~parg/software/hmmbox_4_1.tarMarkov Decision Process (MDP) Toolbox for MatlabKevin Murphy, 1999/~murphyk/Software/MDP/MDP.zipMarkov Decision Process (MDP) Toolbox v1.0 for MATLABhttp://www.inra.fr/bia/T/MDPtoolbox/Hidden Markov Model (HMM) Toolbox for Matlab/~murphyk/Software/HMM/hmm.htmlBayes Net Toolbox for Matlab/~murphyk/Software/BNT/bnt.htmlMedical (Top)EEGLAB Open Source Matlab Toolbox for Physiological Research (formerly ICA/EEG Matlabtoolbox)/~scott/ica.htmlMATLAB Biomedical Signal Processing Toolbox/Toolbox/Powerful package for neurophysiological data analysis ( Igor Kagan webpage)/Matlab/Unitret.htmlEEG / MRI Matlab Toolbox/Microarray data analysis toolbox (MDAT): for normalization, adjustment and analysis of gene expression_r data.Knowlton N, Dozmorov IM, Centola M. Department of Arthritis and Immunology, Oklahoma Medical Research Foundation, Oklahoma City, OK, USA 73104. We introduce a novel Matlab toolbox for microarray data analysis. This toolbox uses normalization based upon a normally distributed background and differential gene expression_r based on 5 statistical measures. The objects in this toolbox are open source and can be implemented to suit your application. AVAILABILITY: MDAT v1.0 is a Matlab toolbox and requires Matlab to run. MDAT is freely available at:/publications/2004/knowlton/MDAT.zipMIDI (Top)MIDI Toolbox version 1.0 (GNU General Public License)http://www.jyu.fi/musica/miditoolbox/Misc. (Top)MATLAB-The Graphing Tool/~abrecht/matlab.html3-D Circuits The Circuit Animation Toolbox for MATLAB/other/3Dcircuits/SendMailhttp://carol.wins.uva.nl/~portegie/matlab/sendmail/Coolplothttp://www.reimeika.ca/marco/matlab/coolplots.htmlMPI (Matlab Parallel Interface)Cornell Multitask Toolbox for MATLAB/Services/Software/CMTM/Beolab Toolbox for v6.5Thomas Abrahamsson (Professor, Chalmers University of Technology, Applied Mechanics,Göteborg, Sweden)http://www.mathworks.nl/matlabcentral/fileexchange/loadFile.do?objectId=1216&objectType =filePARMATLABNeural Networks (Top)SOM Toolboxhttp://www.cis.hut.fi/projects/somtoolbox/Bayes Net Toolbox for Matlab/~murphyk/Software/BNT/bnt.htmlNetLab/netlab/Random Neural Networks/~ahossam/rnnsimv2/ftp: ftp:///pub/contrib/v5/nnet/rnnsimv2/NNSYSID Toolbox (tools for neural network based identification of nonlinear dynamic systems) http://www.iau.dtu.dk/research/control/nnsysid.htmlOceanography (Top)WAFO. Wave Analysis for Fatigue and Oceanographyhttp://www.maths.lth.se/matstat/wafo/ADCP toolbox for MATLAB (USGS, USA)Presented at the Hydroacoustics Workshop in Tampa and at ADCP's in Action in San Diego /operations/stg/pubs/ADCPtoolsSEA-MAT - Matlab Tools for Oceanographic AnalysisA collaborative effort to organize and distribute Matlab tools for the Oceanographic Community /Ocean Toolboxhttp://www.mar.dfo-mpo.gc.ca/science/ocean/epsonde/programming.htmlEUGENE D. GALLAGHER(Associate Professor, Environmental, Coastal & Ocean Sciences)/edgwebp.htmOptimization (Top)MODCONS - a MATLAB Toolbox for Multi-Objective Control System Design/mecheng/jfw/modcons.htmlLazy Learning Packagehttp://iridia.ulb.ac.be/~lazy/SDPT3 version 3.02 -- a MATLAB software for semidefinite-quadratic-linear programming .sg/~mattohkc/sdpt3.htmlMinimum Enclosing Balls: Matlab Code/meb/SOSTOOLS Sum of Squares Optimi zation Toolbox for MATLAB User’s guide/sostools/sostools.pdfPSOt - a Particle Swarm Optimization Toolbox for use with MatlabBy Brian Birge ... A Particle Swarm Optimization Toolbox (PSOt) for use with the Matlab scientific programming environment has been developed. PSO isintroduced briefly and then the use of the toolbox is explained with some examples. A link to downloadable code is provided.Plot/software/plotting/gbplot/Signal Processing (Top)Filter Design with Motorola DSP56Khttp://www.ee.ryerson.ca:8080/~mzeytin/dfp/index.htmlChange Detection and Adaptive Filtering Toolboxhttp://www.sigmoid.se/Signal Processing Toolbox/products/signal/ICA TU Toolboxhttp://mole.imm.dtu.dk/toolbox/menu.htmlTime-Frequency Toolbox for Matlabhttp://crttsn.univ-nantes.fr/~auger/tftb.htmlVoiceBox - Speech Processing Toolbox/hp/staff/dmb/voicebox/voicebox.htmlLeast Squared - Support Vector Machines (LS-SVM)http://www.esat.kuleuven.ac.be/sista/lssvmlab/WaveLab802 : the Wavelet ToolboxBy David Donoho, Mark Reynold Duncan, Xiaoming Huo, Ofer Levi /~wavelab/Time-series Matlab scriptshttp://wise-obs.tau.ac.il/~eran/MATLAB/TimeseriesCon.htmlUvi_Wave Wavelet Toolbox Home Pagehttp://www.gts.tsc.uvigo.es/~wavelets/index.htmlAnotherSupport Vector Machine (Top)MATLAB Support Vector Machine ToolboxDr Gavin CawleySchool of Information Systems, University of East Anglia/~gcc/svm/toolbox/LS-SVM - SISTASVM toolboxes/dmi/svm/LSVM Lagrangian Support Vector Machine/dmi/lsvm/Statistics (Top)Logistic regression/SAGA/software/saga/Multi-Parametric Toolbox (MPT) A tool (not only) for multi-parametric optimization. http://control.ee.ethz.ch/~mpt/ARfit: A Matlab package for the estimation of parameters and eigenmodes of multivariate autoregressive modelshttp://www.mat.univie.ac.at/~neum/software/arfit/The Dimensional Analysis Toolbox for MATLABHome: http://www.sbrs.de/Paper: http://www.isd.uni-stuttgart.de/~brueckner/Papers/similarity2002.pdfFATHOM for Matlab/personal/djones/PLS-toolbox/Multivariate analysis toolbox (N-way Toolbox - paper)http://www.models.kvl.dk/source/nwaytoolbox/index.aspClassification Toolbox for Matlabhttp://tiger.technion.ac.il/~eladyt/classification/index.htmMatlab toolbox for Robust Calibrationhttp://www.wis.kuleuven.ac.be/stat/robust/toolbox.htmlStatistical Parametric Mapping/spm/spm2.htmlEVIM: A Software Package for Extreme Value Analysis in Matlabby Ramazan Gençay, Faruk Selcuk and Abdurrahman Ulugulyagci, 2001.Manual (pdf file) evim.pdf - Software (zip file) evim.zipTime Series Analysishttp://www.dpmi.tu-graz.ac.at/~schloegl/matlab/tsa/Bayes Net Toolbox for MatlabWritten by Kevin Murphy/~murphyk/Software/BNT/bnt.htmlOther: /information/toolboxes.htmlARfit: A Matlab package for the estimation of parameters and eigenmodes of multivariate autoregressive models/~tapio/arfit/M-Fithttp://www.ill.fr/tas/matlab/doc/mfit4/mfit.htmlDimensional Analysis Toolbox for Matlab/The NaN-toolbox: A statistic-toolbox for Octave and Matlab®... handles data with and without MISSING VALUES.http://www-dpmi.tu-graz.ac.at/~schloegl/matlab/NaN/Iterative Methods for Optimization: Matlab Codes/~ctk/matlab_darts.htmlMultiscale Shape Analysis (MSA) Matlab Toolbox 2000p.br/~cesar/projects/multiscale/Multivariate Ecological & Oceanographic Data Analysis (FATHOM)From David Jones/personal/djones/glmlab (Generalized Linear Models in MATLA.au/staff/dunn/glmlab/glmlab.htmlSpacial and Geometric Analysis (SaGA) toolboxInteresting audio links with FAQ, VC++, on the topic机器学习网站北京大学视觉与听觉信息处理实验室北京邮电大学模式识别与智能系统学科复旦大学智能信息处理开放实验室IEEE Computer Society北京映象站点计算机科学论坛机器人足球赛模式识别国家重点实验室南京航空航天大学模式识别与神经计算实验室- PARNEC南京大学机器学习与数据挖掘研究所- LAMDA南京大学人工智能实验室南京大学软件新技术国家重点实验室人工生命之园数据挖掘研究院微软亚洲研究院中国科技大学人工智能中心中科院计算所中科院计算所生物信息学实验室中科院软件所中科院自动化所中科院自动化所人工智能实验室ACL Special Interest Group on Natural Language Learning (SIGNLL)ACMACM Digital LibraryACM SIGARTACM SIGIRACM SIGKDDACM SIGMODAdaptive Computation Group at University of New MexicoAI at Johns HopkinsAI BibliographiesAI Topics: A dynamic online library of introductory information about artificial intelligence Ant Colony OptimizationARIES Laboratory: Advanced Research in Intelligent Educational SystemsArtificial Intelligence Research in Environmental Sciences (AIRIES)Austrian Research Institute for AI (OFAI)Back Issues of Neuron DigestBibFinder: a computer science bibliography search engine integrating many other engines BioAPI ConsortiumBiological and Computational Learning Center at MITBiometrics ConsortiumBoosting siteBrain-Style Information Systems Research Group at RIKEN Brain Science Institute, Japan British Computer Society Specialist Group on Expert SystemsCanadian Society for Computational Studies of Intelligence (CSCSI)CI Collection of BibTex DatabasesCITE, the first-stop source for computational intelligence information and services on the web Classification Society of North AmericaCMU Advanced Multimedia Processing GroupCMU Web->KB ProjectCognitive and Neural Systems Department of Boston UniversityCognitive Sciences Eprint Archive (CogPrints)COLT: Computational Learning TheoryComputational Neural Engineering Laboratory at the University of FloridaComputational Neurobiology Lab at California, USAComputer Science Department of National University of SingaporeData Mining Server Online held by Rudjer Boskovic InstituteDatabase Group at Simon Frazer University, CanadaDBLP: Computer Science BibliographyDigital Biology: about creating artificial lifeDistributed AI Unit at Queen Mary & Westfield College, University of LondonDistributed Artificial Intelligence at HUJIDSI Neural Networks group at the Université di Firenze, ItalyEA-related literature at the EvALife research group at DAIMI, University of Aarhus, Denmark Electronic Research Group at Aberdeen UniversityElsevierComputerScienceEuropean Coordinating Committee for Artificial Intelligence (ECCAI)European Network of Excellence in ML (MLnet)European Neural Network Society (ENNS)Evolutionary Computing Group at University of the West of EnglandEvolutionary Multi-Objective Optimization RepositoryExplanation-Based Learning at University of Illinoise at Urbana-ChampaignFace Detection HomepageFace Recognition Vendor TestFace Recognition HomepageFace Recognition Research CommunityFingerpassftp of Jude Shavlik's Machine Learning Group (University of Wisconsin-Madison)GA-List Searchable DatabaseGenetic Algorithms Digest ArchiveGenetic Programming BibliographyGesture Recognition HomepageHCI Bibliography Project contain extended bibliographic information (abstract, key words, table of contents, section headings) for most publications Human-Computer Interaction dating back to 1980 and selected publications before 1980IBM ResearchIEEEIEEE Computer SocietyIEEE Neural Networks SocietyIllinois Genetic Algorithms Laboratory (IlliGAL)ILP Network of ExcellenceInductive Learning at University of Illinoise at Urbana-ChampaignIntelligent Agents RepositoryIntellimedia Project at North Carolina State UniversityInteractive Artificial Intelligence ResourcesInternational Association of Pattern RecognitionInternational Biometric Industry AssociationInternational Joint Conference on Artificial Intelligence (IJCAI)International Machine Learning Society (IMLS)International Neural Network Society (INNS)Internet Softbot Research at University of WashingtonJapanese Neural Network Society (JNNS)Java Agents for Meta-Learning Group (JAM) at Computer Science Department, Columbia University, for Fraud and Intrusion Detection Using Meta-Learning AgentsKernel MachinesKnowledge Discovery MineLaboratory for Natural and Simulated Cognition at McGill University, CanadaLearning Laboratory at Carnegie Mellon UniversityLearning Robots Laboratory at Carnegie Mellon UniversityLaboratoire d'Informatique et d'Intelligence Artificielle (IIA-ENSAIS)Machine Learning Group of Sydney University, AustraliaMammographic Image Analysis SocietyMDL Research on the WebMirek's Cellebration: 1D and 2D Cellular Automata explorerMIT Artificial Intelligence LaboratoryMIT Media LaboratoryMIT Media Laboratory Vision and Modeling GroupMLNET: a European network of excellence in Machine Learning, Case-based Reasoning and Knowledge AcquisitionMLnet Machine Learning Archive at GMD includes papers, software, and data sets MIRALab at University of Geneva: leading research on virtual human simulationNeural Adaptive Control Technology (NACT)Neural Computing Research Group at Aston University, UKNeural Information Processing Group at Technical University of BerlinNIPSNIPS OnlineNeural Network Benchmarks, Technical Reports,and Source Code maintained by Scott Fahlman at CMU; source code includes Quickprop, Cascade-Correlation, Aspirin/Migraines Neural Networks FAQ by Lutz PrecheltNeural Networks FAQ by Warren S. SarleNeural Networks: Freeware and Shareware ToolsNeural Network Group at Department of Medical Physics and Biophysics, University ofNeural Network Group at Université Catholique de LouvainNeural Network Group at Eindhoven University of TechnologyNeural Network Hyperplane Animator program that allows easy visualization of training data and weights in a back-propagation neural networkNeural Networks Research at TUT/ELENeural Networks Research Centre at Helsinki University of Technology, FinlandNeural Network Speech Group at Carnegie Mellon UniversityNeural Text Classification with Neural NetworksNonlinearity and Complexity HomepageOFAI and IMKAI library information system, provided by the Department of Medical Cybernetics and Artificial Intelligence at the University of Vienna (IMKAI) and the Austrian Research Institute for Artificial Intelligence (OFAI). It contains over 36,000 items (books, research papers, conference papers, journal articles) from many subareas of AI OntoWeb: Ontology-based information exchange for knowledge management and electronic commercePortal on Neural Network ForecastingPRAG: Pattern Recognition and Application Group at University of CagliariQuest Project at IBM Almaden Research Center: an academic website focusing on classification and regression trees. Maintained by Tjen-Sien LimReinforcement Learning at Carnegie Mellon UniversityResearchIndex: NECI Scientific Literature Digital Library, indexing over 200,000 computer science articlesReVision: Reviewing Vision in the Web!RIKEN: The Institute of Physical and Chemical Research, JapanSalford SystemsSANS Studies of Artificial Neural Systems, at the Royal Institute of Technology, Sweden Santa-Fe InstituteScirus: a search engine locating scientific information on the InternetSecond Moment: The News and Business Resource for Applied AnalyticsSEL-HPC Article Archive has sections for neural networks, distributed AI, theorem proving, and a variety of other computer science topicsSOAR Project at University of Southern CaliforniaSociety for AI and StatisticsSVM of ANU CanberraSVM of Bell LabsSVM of GMD-First BerlinSVM of MITSVM of Royal Holloway CollegeSVM of University of SouthamptonSVM-workshop at NIPS97TechOnLine: TechOnLine University offers free online courses and lecturesUCI Machine Learning GroupUMASS Distributed Artificial Intelligence LaboratoryUTCS Neural Networks Research Group of Artificial Intelligence Lab, Computer Science Department, University of Texas at AustinVivisimo Document Clustering: a powerful search engine which returns clustered results Worcester Polytechnic Institute Artificial Intelligence Research Group (AIRG)Xerion neural network simulator developed and used by the connectionist group at the University of TorontoYale's CTAN Advanced Technology Center for Theoretical and Applied Neuroscience ZooLand: Artificial Life Resource。
C.parvum全基因组序列
DOI: 10.1126/science.1094786, 441 (2004);304Science et al.Mitchell S. Abrahamsen,Cryptosporidium parvum Complete Genome Sequence of the Apicomplexan, (this information is current as of October 7, 2009 ):The following resources related to this article are available online at/cgi/content/full/304/5669/441version of this article at:including high-resolution figures, can be found in the online Updated information and services,/cgi/content/full/1094786/DC1 can be found at:Supporting Online Material/cgi/content/full/304/5669/441#otherarticles , 9 of which can be accessed for free: cites 25 articles This article 239 article(s) on the ISI Web of Science. cited by This article has been /cgi/content/full/304/5669/441#otherarticles 53 articles hosted by HighWire Press; see: cited by This article has been/cgi/collection/genetics Genetics: subject collections This article appears in the following/about/permissions.dtl in whole or in part can be found at: this article permission to reproduce of this article or about obtaining reprints Information about obtaining registered trademark of AAAS.is a Science 2004 by the American Association for the Advancement of Science; all rights reserved. The title Copyright American Association for the Advancement of Science, 1200 New York Avenue NW, Washington, DC 20005. (print ISSN 0036-8075; online ISSN 1095-9203) is published weekly, except the last week in December, by the Science o n O c t o b e r 7, 2009w w w .s c i e n c e m a g .o r g D o w n l o a d e d f r o m3.R.Jackendoff,Foundations of Language:Brain,Gram-mar,Evolution(Oxford Univ.Press,Oxford,2003).4.Although for Frege(1),reference was established rela-tive to objects in the world,here we follow Jackendoff’s suggestion(3)that this is done relative to objects and the state of affairs as mentally represented.5.S.Zola-Morgan,L.R.Squire,in The Development andNeural Bases of Higher Cognitive Functions(New York Academy of Sciences,New York,1990),pp.434–456.6.N.Chomsky,Reflections on Language(Pantheon,New York,1975).7.J.Katz,Semantic Theory(Harper&Row,New York,1972).8.D.Sperber,D.Wilson,Relevance(Harvard Univ.Press,Cambridge,MA,1986).9.K.I.Forster,in Sentence Processing,W.E.Cooper,C.T.Walker,Eds.(Erlbaum,Hillsdale,NJ,1989),pp.27–85.10.H.H.Clark,Using Language(Cambridge Univ.Press,Cambridge,1996).11.Often word meanings can only be fully determined byinvokingworld knowledg e.For instance,the meaningof “flat”in a“flat road”implies the absence of holes.However,in the expression“aflat tire,”it indicates the presence of a hole.The meaningof“finish”in the phrase “Billfinished the book”implies that Bill completed readingthe book.However,the phrase“the g oatfin-ished the book”can only be interpreted as the goat eatingor destroyingthe book.The examples illustrate that word meaningis often underdetermined and nec-essarily intertwined with general world knowledge.In such cases,it is hard to see how the integration of lexical meaning and general world knowledge could be strictly separated(3,31).12.W.Marslen-Wilson,C.M.Brown,L.K.Tyler,Lang.Cognit.Process.3,1(1988).13.ERPs for30subjects were averaged time-locked to theonset of the critical words,with40items per condition.Sentences were presented word by word on the centerof a computer screen,with a stimulus onset asynchronyof600ms.While subjects were readingthe sentences,their EEG was recorded and amplified with a high-cut-off frequency of70Hz,a time constant of8s,and asamplingfrequency of200Hz.14.Materials and methods are available as supportingmaterial on Science Online.15.M.Kutas,S.A.Hillyard,Science207,203(1980).16.C.Brown,P.Hagoort,J.Cognit.Neurosci.5,34(1993).17.C.M.Brown,P.Hagoort,in Architectures and Mech-anisms for Language Processing,M.W.Crocker,M.Pickering,C.Clifton Jr.,Eds.(Cambridge Univ.Press,Cambridge,1999),pp.213–237.18.F.Varela et al.,Nature Rev.Neurosci.2,229(2001).19.We obtained TFRs of the single-trial EEG data by con-volvingcomplex Morlet wavelets with the EEG data andcomputingthe squared norm for the result of theconvolution.We used wavelets with a7-cycle width,with frequencies ranging from1to70Hz,in1-Hz steps.Power values thus obtained were expressed as a per-centage change relative to the power in a baselineinterval,which was taken from150to0ms before theonset of the critical word.This was done in order tonormalize for individual differences in EEG power anddifferences in baseline power between different fre-quency bands.Two relevant time-frequency compo-nents were identified:(i)a theta component,rangingfrom4to7Hz and from300to800ms after wordonset,and(ii)a gamma component,ranging from35to45Hz and from400to600ms after word onset.20.C.Tallon-Baudry,O.Bertrand,Trends Cognit.Sci.3,151(1999).tner et al.,Nature397,434(1999).22.M.Bastiaansen,P.Hagoort,Cortex39(2003).23.O.Jensen,C.D.Tesche,Eur.J.Neurosci.15,1395(2002).24.Whole brain T2*-weighted echo planar imaging bloodoxygen level–dependent(EPI-BOLD)fMRI data wereacquired with a Siemens Sonata1.5-T magnetic reso-nance scanner with interleaved slice ordering,a volumerepetition time of2.48s,an echo time of40ms,a90°flip angle,31horizontal slices,a64ϫ64slice matrix,and isotropic voxel size of3.5ϫ3.5ϫ3.5mm.For thestructural magnetic resonance image,we used a high-resolution(isotropic voxels of1mm3)T1-weightedmagnetization-prepared rapid gradient-echo pulse se-quence.The fMRI data were preprocessed and analyzedby statistical parametric mappingwith SPM99software(http://www.fi/spm99).25.S.E.Petersen et al.,Nature331,585(1988).26.B.T.Gold,R.L.Buckner,Neuron35,803(2002).27.E.Halgren et al.,J.Psychophysiol.88,1(1994).28.E.Halgren et al.,Neuroimage17,1101(2002).29.M.K.Tanenhaus et al.,Science268,1632(1995).30.J.J.A.van Berkum et al.,J.Cognit.Neurosci.11,657(1999).31.P.A.M.Seuren,Discourse Semantics(Basil Blackwell,Oxford,1985).32.We thank P.Indefrey,P.Fries,P.A.M.Seuren,and M.van Turennout for helpful discussions.Supported bythe Netherlands Organization for Scientific Research,grant no.400-56-384(P.H.).Supporting Online Material/cgi/content/full/1095455/DC1Materials and MethodsFig.S1References and Notes8January2004;accepted9March2004Published online18March2004;10.1126/science.1095455Include this information when citingthis paper.Complete Genome Sequence ofthe Apicomplexan,Cryptosporidium parvumMitchell S.Abrahamsen,1,2*†Thomas J.Templeton,3†Shinichiro Enomoto,1Juan E.Abrahante,1Guan Zhu,4 Cheryl ncto,1Mingqi Deng,1Chang Liu,1‡Giovanni Widmer,5Saul Tzipori,5GregoryA.Buck,6Ping Xu,6 Alan T.Bankier,7Paul H.Dear,7Bernard A.Konfortov,7 Helen F.Spriggs,7Lakshminarayan Iyer,8Vivek Anantharaman,8L.Aravind,8Vivek Kapur2,9The apicomplexan Cryptosporidium parvum is an intestinal parasite that affects healthy humans and animals,and causes an unrelenting infection in immuno-compromised individuals such as AIDS patients.We report the complete ge-nome sequence of C.parvum,type II isolate.Genome analysis identifies ex-tremely streamlined metabolic pathways and a reliance on the host for nu-trients.In contrast to Plasmodium and Toxoplasma,the parasite lacks an api-coplast and its genome,and possesses a degenerate mitochondrion that has lost its genome.Several novel classes of cell-surface and secreted proteins with a potential role in host interactions and pathogenesis were also detected.Elu-cidation of the core metabolism,including enzymes with high similarities to bacterial and plant counterparts,opens new avenues for drug development.Cryptosporidium parvum is a globally impor-tant intracellular pathogen of humans and animals.The duration of infection and patho-genesis of cryptosporidiosis depends on host immune status,ranging from a severe but self-limiting diarrhea in immunocompetent individuals to a life-threatening,prolonged infection in immunocompromised patients.Asubstantial degree of morbidity and mortalityis associated with infections in AIDS pa-tients.Despite intensive efforts over the past20years,there is currently no effective ther-apy for treating or preventing C.parvuminfection in humans.Cryptosporidium belongs to the phylumApicomplexa,whose members share a com-mon apical secretory apparatus mediating lo-comotion and tissue or cellular invasion.Many apicomplexans are of medical or vet-erinary importance,including Plasmodium,Babesia,Toxoplasma,Neosprora,Sarcocys-tis,Cyclospora,and Eimeria.The life cycle ofC.parvum is similar to that of other cyst-forming apicomplexans(e.g.,Eimeria and Tox-oplasma),resulting in the formation of oocysts1Department of Veterinary and Biomedical Science,College of Veterinary Medicine,2Biomedical Genom-ics Center,University of Minnesota,St.Paul,MN55108,USA.3Department of Microbiology and Immu-nology,Weill Medical College and Program in Immu-nology,Weill Graduate School of Medical Sciences ofCornell University,New York,NY10021,USA.4De-partment of Veterinary Pathobiology,College of Vet-erinary Medicine,Texas A&M University,College Sta-tion,TX77843,USA.5Division of Infectious Diseases,Tufts University School of Veterinary Medicine,NorthGrafton,MA01536,USA.6Center for the Study ofBiological Complexity and Department of Microbiol-ogy and Immunology,Virginia Commonwealth Uni-versity,Richmond,VA23198,USA.7MRC Laboratoryof Molecular Biology,Hills Road,Cambridge CB22QH,UK.8National Center for Biotechnology Infor-mation,National Library of Medicine,National Insti-tutes of Health,Bethesda,MD20894,USA.9Depart-ment of Microbiology,University of Minnesota,Min-neapolis,MN55455,USA.*To whom correspondence should be addressed.E-mail:abe@†These authors contributed equally to this work.‡Present address:Bioinformatics Division,Genetic Re-search,GlaxoSmithKline Pharmaceuticals,5MooreDrive,Research Triangle Park,NC27009,USA.R E P O R T S SCIENCE VOL30416APRIL2004441o n O c t o b e r 7 , 2 0 0 9 w w w . s c i e n c e m a g . o r g D o w n l o a d e d f r o mthat are shed in the feces of infected hosts.C.parvum oocysts are highly resistant to environ-mental stresses,including chlorine treatment of community water supplies;hence,the parasite is an important water-and food-borne pathogen (1).The obligate intracellular nature of the par-asite ’s life cycle and the inability to culture the parasite continuously in vitro greatly impair researchers ’ability to obtain purified samples of the different developmental stages.The par-asite cannot be genetically manipulated,and transformation methodologies are currently un-available.To begin to address these limitations,we have obtained the complete C.parvum ge-nome sequence and its predicted protein com-plement.(This whole-genome shotgun project has been deposited at DDBJ/EMBL/GenBank under the project accession AAEE00000000.The version described in this paper is the first version,AAEE01000000.)The random shotgun approach was used to obtain the complete DNA sequence (2)of the Iowa “type II ”isolate of C.parvum .This isolate readily transmits disease among numerous mammals,including humans.The resulting ge-nome sequence has roughly 13ϫgenome cov-erage containing five gaps and 9.1Mb of totalDNA sequence within eight chromosomes.The C.parvum genome is thus quite compact rela-tive to the 23-Mb,14-chromosome genome of Plasmodium falciparum (3);this size difference is predominantly the result of shorter intergenic regions,fewer introns,and a smaller number of genes (Table 1).Comparison of the assembled sequence of chromosome VI to that of the recently published sequence of chromosome VI (4)revealed that our assembly contains an ad-ditional 160kb of sequence and a single gap versus two,with the common sequences dis-playing a 99.993%sequence identity (2).The relative paucity of introns greatly simplified gene predictions and facilitated an-notation (2)of predicted open reading frames (ORFs).These analyses provided an estimate of 3807protein-encoding genes for the C.parvum genome,far fewer than the estimated 5300genes predicted for the Plasmodium genome (3).This difference is primarily due to the absence of an apicoplast and mitochondrial genome,as well as the pres-ence of fewer genes encoding metabolic functions and variant surface proteins,such as the P.falciparum var and rifin molecules (Table 2).An analysis of the encoded pro-tein sequences with the program SEG (5)shows that these protein-encoding genes are not enriched in low-complexity se-quences (34%)to the extent observed in the proteins from Plasmodium (70%).Our sequence analysis indicates that Cryptosporidium ,unlike Plasmodium and Toxoplasma ,lacks both mitochondrion and apicoplast genomes.The overall complete-ness of the genome sequence,together with the fact that similar DNA extraction proce-dures used to isolate total genomic DNA from C.parvum efficiently yielded mito-chondrion and apicoplast genomes from Ei-meria sp.and Toxoplasma (6,7),indicates that the absence of organellar genomes was unlikely to have been the result of method-ological error.These conclusions are con-sistent with the absence of nuclear genes for the DNA replication and translation machinery characteristic of mitochondria and apicoplasts,and with the lack of mito-chondrial or apicoplast targeting signals for tRNA synthetases.A number of putative mitochondrial pro-teins were identified,including components of a mitochondrial protein import apparatus,chaperones,uncoupling proteins,and solute translocators (table S1).However,the ge-nome does not encode any Krebs cycle en-zymes,nor the components constituting the mitochondrial complexes I to IV;this finding indicates that the parasite does not rely on complete oxidation and respiratory chains for synthesizing adenosine triphosphate (ATP).Similar to Plasmodium ,no orthologs for the ␥,␦,or εsubunits or the c subunit of the F 0proton channel were detected (whereas all subunits were found for a V-type ATPase).Cryptosporidium ,like Eimeria (8)and Plas-modium ,possesses a pyridine nucleotide tran-shydrogenase integral membrane protein that may couple reduced nicotinamide adenine dinucleotide (NADH)and reduced nico-tinamide adenine dinucleotide phosphate (NADPH)redox to proton translocation across the inner mitochondrial membrane.Unlike Plasmodium ,the parasite has two copies of the pyridine nucleotide transhydrogenase gene.Also present is a likely mitochondrial membrane –associated,cyanide-resistant alter-native oxidase (AOX )that catalyzes the reduction of molecular oxygen by ubiquinol to produce H 2O,but not superoxide or H 2O 2.Several genes were identified as involved in biogenesis of iron-sulfur [Fe-S]complexes with potential mitochondrial targeting signals (e.g.,nifS,nifU,frataxin,and ferredoxin),supporting the presence of a limited electron flux in the mitochondrial remnant (table S2).Our sequence analysis confirms the absence of a plastid genome (7)and,additionally,the loss of plastid-associated metabolic pathways including the type II fatty acid synthases (FASs)and isoprenoid synthetic enzymes thatTable 1.General features of the C.parvum genome and comparison with other single-celled eukaryotes.Values are derived from respective genome project summaries (3,26–28).ND,not determined.FeatureC.parvum P.falciparum S.pombe S.cerevisiae E.cuniculiSize (Mbp)9.122.912.512.5 2.5(G ϩC)content (%)3019.43638.347No.of genes 38075268492957701997Mean gene length (bp)excluding introns 1795228314261424ND Gene density (bp per gene)23824338252820881256Percent coding75.352.657.570.590Genes with introns (%)553.9435ND Intergenic regions (G ϩC)content %23.913.632.435.145Mean length (bp)5661694952515129RNAsNo.of tRNA genes 454317429944No.of 5S rRNA genes 6330100–2003No.of 5.8S ,18S ,and 28S rRNA units 57200–400100–20022Table parison between predicted C.parvum and P.falciparum proteins.FeatureC.parvum P.falciparum *Common †Total predicted proteins380752681883Mitochondrial targeted/encoded 17(0.45%)246(4.7%)15Apicoplast targeted/encoded 0581(11.0%)0var/rif/stevor ‡0236(4.5%)0Annotated as protease §50(1.3%)31(0.59%)27Annotated as transporter 69(1.8%)34(0.65%)34Assigned EC function ¶167(4.4%)389(7.4%)113Hypothetical proteins925(24.3%)3208(60.9%)126*Values indicated for P.falciparum are as reported (3)with the exception of those for proteins annotated as protease or transporter.†TBLASTN hits (e Ͻ–5)between C.parvum and P.falciparum .‡As reported in (3).§Pre-dicted proteins annotated as “protease or peptidase”for C.parvum (CryptoGenome database,)and P.falciparum (PlasmoDB database,).Predicted proteins annotated as “trans-porter,permease of P-type ATPase”for C.parvum (CryptoGenome)and P.falciparum (PlasmoDB).¶Bidirectional BLAST hit (e Ͻ–15)to orthologs with assigned Enzyme Commission (EC)numbers.Does not include EC assignment numbers for protein kinases or protein phosphatases (due to inconsistent annotation across genomes),or DNA polymerases or RNA polymerases,as a result of issues related to subunit inclusion.(For consistency,46proteins were excluded from the reported P.falciparum values.)R E P O R T S16APRIL 2004VOL 304SCIENCE 442 o n O c t o b e r 7, 2009w w w .s c i e n c e m a g .o r g D o w n l o a d e d f r o mare otherwise localized to the plastid in other apicomplexans.C.parvum fatty acid biosynthe-sis appears to be cytoplasmic,conducted by a large(8252amino acids)modular type I FAS (9)and possibly by another large enzyme that is related to the multidomain bacterial polyketide synthase(10).Comprehensive screening of the C.parvum genome sequence also did not detect orthologs of Plasmodium nuclear-encoded genes that contain apicoplast-targeting and transit sequences(11).C.parvum metabolism is greatly stream-lined relative to that of Plasmodium,and in certain ways it is reminiscent of that of another obligate eukaryotic parasite,the microsporidian Encephalitozoon.The degeneration of the mi-tochondrion and associated metabolic capabili-ties suggests that the parasite largely relies on glycolysis for energy production.The parasite is capable of uptake and catabolism of mono-sugars(e.g.,glucose and fructose)as well as synthesis,storage,and catabolism of polysac-charides such as trehalose and amylopectin. Like many anaerobic organisms,it economizes ATP through the use of pyrophosphate-dependent phosphofructokinases.The conver-sion of pyruvate to acetyl–coenzyme A(CoA) is catalyzed by an atypical pyruvate-NADPH oxidoreductase(Cp PNO)that contains an N-terminal pyruvate–ferredoxin oxidoreductase (PFO)domain fused with a C-terminal NADPH–cytochrome P450reductase domain (CPR).Such a PFO-CPR fusion has previously been observed only in the euglenozoan protist Euglena gracilis(12).Acetyl-CoA can be con-verted to malonyl-CoA,an important precursor for fatty acid and polyketide biosynthesis.Gly-colysis leads to several possible organic end products,including lactate,acetate,and ethanol. The production of acetate from acetyl-CoA may be economically beneficial to the parasite via coupling with ATP production.Ethanol is potentially produced via two in-dependent pathways:(i)from the combination of pyruvate decarboxylase and alcohol dehy-drogenase,or(ii)from acetyl-CoA by means of a bifunctional dehydrogenase(adhE)with ac-etaldehyde and alcohol dehydrogenase activi-ties;adhE first converts acetyl-CoA to acetal-dehyde and then reduces the latter to ethanol. AdhE predominantly occurs in bacteria but has recently been identified in several protozoans, including vertebrate gut parasites such as Enta-moeba and Giardia(13,14).Adjacent to the adhE gene resides a second gene encoding only the AdhE C-terminal Fe-dependent alcohol de-hydrogenase domain.This gene product may form a multisubunit complex with AdhE,or it may function as an alternative alcohol dehydro-genase that is specific to certain growth condi-tions.C.parvum has a glycerol3-phosphate dehydrogenase similar to those of plants,fungi, and the kinetoplastid Trypanosoma,but(unlike trypanosomes)the parasite lacks an ortholog of glycerol kinase and thus this pathway does not yield glycerol production.In addition to themodular fatty acid synthase(Cp FAS1)andpolyketide synthase homolog(Cp PKS1), C.parvum possesses several fatty acyl–CoA syn-thases and a fatty acyl elongase that may partici-pate in fatty acid metabolism.Further,enzymesfor the metabolism of complex lipids(e.g.,glyc-erolipid and inositol phosphate)were identified inthe genome.Fatty acids are apparently not anenergy source,because enzymes of the fatty acidoxidative pathway are absent,with the exceptionof a3-hydroxyacyl-CoA dehydrogenase.C.parvum purine metabolism is greatlysimplified,retaining only an adenosine ki-nase and enzymes catalyzing conversionsof adenosine5Ј-monophosphate(AMP)toinosine,xanthosine,and guanosine5Ј-monophosphates(IMP,XMP,and GMP).Among these enzymes,IMP dehydrogenase(IMPDH)is phylogenetically related toε-proteobacterial IMPDH and is strikinglydifferent from its counterparts in both thehost and other apicomplexans(15).In con-trast to other apicomplexans such as Toxo-plasma gondii and P.falciparum,no geneencoding hypoxanthine-xanthineguaninephosphoribosyltransferase(HXGPRT)is de-tected,in contrast to a previous report on theactivity of this enzyme in C.parvum sporo-zoites(16).The absence of HXGPRT sug-gests that the parasite may rely solely on asingle enzyme system including IMPDH toproduce GMP from AMP.In contrast to otherapicomplexans,the parasite appears to relyon adenosine for purine salvage,a modelsupported by the identification of an adeno-sine transporter.Unlike other apicomplexansand many parasitic protists that can synthe-size pyrimidines de novo,C.parvum relies onpyrimidine salvage and retains the ability forinterconversions among uridine and cytidine5Ј-monophosphates(UMP and CMP),theirdeoxy forms(dUMP and dCMP),and dAMP,as well as their corresponding di-and triphos-phonucleotides.The parasite has also largelyshed the ability to synthesize amino acids denovo,although it retains the ability to convertselect amino acids,and instead appears torely on amino acid uptake from the host bymeans of a set of at least11amino acidtransporters(table S2).Most of the Cryptosporidium core pro-cesses involved in DNA replication,repair,transcription,and translation conform to thebasic eukaryotic blueprint(2).The transcrip-tional apparatus resembles Plasmodium interms of basal transcription machinery.How-ever,a striking numerical difference is seenin the complements of two RNA bindingdomains,Sm and RRM,between P.falcipa-rum(17and71domains,respectively)and C.parvum(9and51domains).This reductionresults in part from the loss of conservedproteins belonging to the spliceosomal ma-chinery,including all genes encoding Smdomain proteins belonging to the U6spliceo-somal particle,which suggests that this par-ticle activity is degenerate or entirely lost.This reduction in spliceosomal machinery isconsistent with the reduced number of pre-dicted introns in Cryptosporidium(5%)rela-tive to Plasmodium(Ͼ50%).In addition,keycomponents of the small RNA–mediatedposttranscriptional gene silencing system aremissing,such as the RNA-dependent RNApolymerase,Argonaute,and Dicer orthologs;hence,RNA interference–related technolo-gies are unlikely to be of much value intargeted disruption of genes in C.parvum.Cryptosporidium invasion of columnarbrush border epithelial cells has been de-scribed as“intracellular,but extracytoplas-mic,”as the parasite resides on the surface ofthe intestinal epithelium but lies underneaththe host cell membrane.This niche may al-low the parasite to evade immune surveil-lance but take advantage of solute transportacross the host microvillus membrane or theextensively convoluted parasitophorous vac-uole.Indeed,Cryptosporidium has numerousgenes(table S2)encoding families of putativesugar transporters(up to9genes)and aminoacid transporters(11genes).This is in starkcontrast to Plasmodium,which has fewersugar transporters and only one putative ami-no acid transporter(GenBank identificationnumber23612372).As a first step toward identification ofmulti–drug-resistant pumps,the genome se-quence was analyzed for all occurrences ofgenes encoding multitransmembrane proteins.Notable are a set of four paralogous proteinsthat belong to the sbmA family(table S2)thatare involved in the transport of peptide antibi-otics in bacteria.A putative ortholog of thePlasmodium chloroquine resistance–linkedgene Pf CRT(17)was also identified,althoughthe parasite does not possess a food vacuole likethe one seen in Plasmodium.Unlike Plasmodium,C.parvum does notpossess extensive subtelomeric clusters of anti-genically variant proteins(exemplified by thelarge families of var and rif/stevor genes)thatare involved in immune evasion.In contrast,more than20genes were identified that encodemucin-like proteins(18,19)having hallmarksof extensive Thr or Ser stretches suggestive ofglycosylation and signal peptide sequences sug-gesting secretion(table S2).One notable exam-ple is an11,700–amino acid protein with anuninterrupted stretch of308Thr residues(cgd3_720).Although large families of secretedproteins analogous to the Plasmodium multi-gene families were not found,several smallermultigene clusters were observed that encodepredicted secreted proteins,with no detectablesimilarity to proteins from other organisms(Fig.1,A and B).Within this group,at leastfour distinct families appear to have emergedthrough gene expansions specific to the Cryp-R E P O R T S SCIENCE VOL30416APRIL2004443o n O c t o b e r 7 , 2 0 0 9 w w w . s c i e n c e m a g . o r g D o w n l o a d e d f r o mtosporidium clade.These families —SKSR,MEDLE,WYLE,FGLN,and GGC —were named after well-conserved sequence motifs (table S2).Reverse transcription polymerase chain reaction (RT-PCR)expression analysis (20)of one cluster,a locus of seven adjacent CpLSP genes (Fig.1B),shows coexpression during the course of in vitro development (Fig.1C).An additional eight genes were identified that encode proteins having a periodic cysteine structure similar to the Cryptosporidium oocyst wall protein;these eight genes are similarly expressed during the onset of oocyst formation and likely participate in the formation of the coccidian rigid oocyst wall in both Cryptospo-ridium and Toxoplasma (21).Whereas the extracellular proteins described above are of apparent apicomplexan or lineage-specific in-vention,Cryptosporidium possesses many genesencodingsecretedproteinshavinglineage-specific multidomain architectures composed of animal-and bacterial-like extracellular adhe-sive domains (fig.S1).Lineage-specific expansions were ob-served for several proteases (table S2),in-cluding an aspartyl protease (six genes),a subtilisin-like protease,a cryptopain-like cys-teine protease (five genes),and a Plas-modium falcilysin-like (insulin degrading enzyme –like)protease (19genes).Nine of the Cryptosporidium falcilysin genes lack the Zn-chelating “HXXEH ”active site motif and are likely to be catalytically inactive copies that may have been reused for specific protein-protein interactions on the cell sur-face.In contrast to the Plasmodium falcilysin,the Cryptosporidium genes possess signal peptide sequences and are likely trafficked to a secretory pathway.The expansion of this family suggests either that the proteins have distinct cleavage specificities or that their diversity may be related to evasion of a host immune response.Completion of the C.parvum genome se-quence has highlighted the lack of conven-tional drug targets currently pursued for the control and treatment of other parasitic protists.On the basis of molecular and bio-chemical studies and drug screening of other apicomplexans,several putative Cryptospo-ridium metabolic pathways or enzymes have been erroneously proposed to be potential drug targets (22),including the apicoplast and its associated metabolic pathways,the shikimate pathway,the mannitol cycle,the electron transport chain,and HXGPRT.Nonetheless,complete genome sequence analysis identifies a number of classic and novel molecular candidates for drug explora-tion,including numerous plant-like and bacterial-like enzymes (tables S3and S4).Although the C.parvum genome lacks HXGPRT,a potent drug target in other api-complexans,it has only the single pathway dependent on IMPDH to convert AMP to GMP.The bacterial-type IMPDH may be a promising target because it differs substan-tially from that of eukaryotic enzymes (15).Because of the lack of de novo biosynthetic capacity for purines,pyrimidines,and amino acids,C.parvum relies solely on scavenge from the host via a series of transporters,which may be exploited for chemotherapy.C.parvum possesses a bacterial-type thymidine kinase,and the role of this enzyme in pyrim-idine metabolism and its drug target candida-cy should be pursued.The presence of an alternative oxidase,likely targeted to the remnant mitochondrion,gives promise to the study of salicylhydroxamic acid (SHAM),as-cofuranone,and their analogs as inhibitors of energy metabolism in the parasite (23).Cryptosporidium possesses at least 15“plant-like ”enzymes that are either absent in or highly divergent from those typically found in mammals (table S3).Within the glycolytic pathway,the plant-like PPi-PFK has been shown to be a potential target in other parasites including T.gondii ,and PEPCL and PGI ap-pear to be plant-type enzymes in C.parvum .Another example is a trehalose-6-phosphate synthase/phosphatase catalyzing trehalose bio-synthesis from glucose-6-phosphate and uridine diphosphate –glucose.Trehalose may serve as a sugar storage source or may function as an antidesiccant,antioxidant,or protein stability agent in oocysts,playing a role similar to that of mannitol in Eimeria oocysts (24).Orthologs of putative Eimeria mannitol synthesis enzymes were not found.However,two oxidoreductases (table S2)were identified in C.parvum ,one of which belongs to the same families as the plant mannose dehydrogenases (25)and the other to the plant cinnamyl alcohol dehydrogenases.In principle,these enzymes could synthesize protective polyol compounds,and the former enzyme could use host-derived mannose to syn-thesize mannitol.References and Notes1.D.G.Korich et al .,Appl.Environ.Microbiol.56,1423(1990).2.See supportingdata on Science Online.3.M.J.Gardner et al .,Nature 419,498(2002).4.A.T.Bankier et al .,Genome Res.13,1787(2003).5.J.C.Wootton,Comput.Chem.18,269(1994).Fig.1.(A )Schematic showing the chromosomal locations of clusters of potentially secreted proteins.Numbers of adjacent genes are indicated in paren-theses.Arrows indicate direc-tion of clusters containinguni-directional genes (encoded on the same strand);squares indi-cate clusters containingg enes encoded on both strands.Non-paralogous genes are indicated by solid gray squares or direc-tional triangles;SKSR (green triangles),FGLN (red trian-gles),and MEDLE (blue trian-gles)indicate three C.parvum –specific families of paralogous genes predominantly located at telomeres.Insl (yellow tri-angles)indicates an insulinase/falcilysin-like paralogous gene family.Cp LSP (white square)indicates the location of a clus-ter of adjacent large secreted proteins (table S2)that are cotranscriptionally regulated.Identified anchored telomeric repeat sequences are indicated by circles.(B )Schematic show-inga select locus containinga cluster of coexpressed large secreted proteins (Cp LSP).Genes and intergenic regions (regions between identified genes)are drawn to scale at the nucleotide level.The length of the intergenic re-gions is indicated above or be-low the locus.(C )Relative ex-pression levels of CpLSP (red lines)and,as a control,C.parvum Hedgehog-type HINT domain gene (blue line)duringin vitro development,as determined by semiquantitative RT-PCR usingg ene-specific primers correspondingto the seven adjacent g enes within the CpLSP locus as shown in (B).Expression levels from three independent time-course experiments are represented as the ratio of the expression of each gene to that of C.parvum 18S rRNA present in each of the infected samples (20).R E P O R T S16APRIL 2004VOL 304SCIENCE 444 o n O c t o b e r 7, 2009w w w .s c i e n c e m a g .o r g D o w n l o a d e d f r o m。
2023年考研英语真题及解析
1.[A] selected [B] prepared [C] obliged [D] pleased2.[A] unique [B] particular [C] special [D] rare3.[A] of [B] with [C] in [D] against4.[A] subsequently [B] presently [C] previously [D] lately5.[A] Only [B] So [C] Even [D] Hence6.[A] thought [B] sight [C] cost [D] risk7.[A] advises [B] suggests [C] protests [D] objects8.[A] progress [B] fact [C] need [D] question9.[A] attaining [B] scoring [C] reaching [D] calculating10.[A] normal [B] common [C] mean [D] total11.[A] unconsciously[B] disproportionately[C] indefinitely[D] unaccountably12.[A] missions [B] fortunes [C] interests [D] careers13.[A] affirm [B] witness [C] observe [D] approve14.[A] moreover [B] therefore [C] however [D] meanwhile15.[A] given up [B] got over [C] carried on [D] put down16.[A] assessing [B] supervising [C] administering [D] valuing17.[A] development [B] origin [C] consequence [D] instrument18.[A] linked [B] integrated [C] woven [D] combined19.[A] limited [B] subjected [C] converted [D] directed20.[A] paradoxical [B] incompatible [C] inevitable [D] continuousSection II Reading ComprehensionPart ADirections:Read the following four texts. Answer the questions below each text by choosing [A], [B], [C] or [D]. Mark your answers on ANSWER SHEET 1. (40 points)Text 1While still catching up to men in some spheres of modern life, women appear to be way ahead in at least one undesirable category. “Women are particularly susceptible to developing depression and anxiety disorders in response to stress compared to men,” according to Dr. Yehuda, chief psychiatrist at New York’s Veteran’s Administration Hospital.Studies of both animals and humans have shown that sex hormones somehow affect the stress response, causing females under stress to produce more of the trigger chemicals than do males under the same conditions. In several of the studies, when stressed-out female rats had their ovaries (the female reproductive organs) removed, their chemical responsesbecame equal to those of the males.Adding to a woman’s increased dose of stress chemicals, are her increased “opportunities” for stress. “It’s not necessarily that women don’t cope as well. It’s just that they have so much more to cope with,” says Dr. Yehuda. “Their capacity for tolerating stress may even be greater than men’s,” she observes, “it’s just that they’re dealing with so many more things that they become worn out from it more visibly and sooner.”Dr. Yehuda notes another difference between the sexes. “I think that the kinds of things that women are exposed to tend to be in more of a chronic or repeated nature. Men go to war and are exposed to combat stress.Men are exposed to more acts of random physical violence. The kinds of interpersonal violence that women are exposed to tend to be in domestic situations, by, unfortunately, parents or other family members, and they tend not to be one-shot deals. The wear-and-tear that comes from these longer relationships can be quite devastating.”Adeline Alvarez married at 18 and gave birth to a son, but was determined to finish college. “I struggled a lot to get the college degree. I was living in so much frustration that that was my escape, to go to school, and get ahead and do better.” Later, her marriage ended and she became a single mother. “It’s the hardest thing to take care of a teenager, have a job, pay the rent, pay the car payment, and pay the debt.I lived from paycheck to paycheck.”Not everyone experiences the kinds of severe chronic stresses Alvarez describes. But most women today are coping with a lot of obligations, with few breaks, and feeling the strain. Alvarez’s experienc e demonstrates the importance of finding ways to diffuse stress before it threatens your health and your ability to function.21. Which of the following is true according to the first two paragraphs?[A] Women are biologically more vulnerable to stress.[B] Women are still suffering much stress caused by men.[C] Women are more experienced than men in coping with stress.[D] Men and women show different inclinations when faced with stress.22. Dr. Yehuda’s research suggests that women .[A] need extra doses of chemicals to handle stress[B] have limited capacity for tolerating stress[C] are more capable of avoiding stress[D] are exposed to more stress23. According to Paragraph 4, the stress women confront tends to be .[A] domestic and temporary[B] irregular and violent[C] durable and frequent[D] trivial and random24. The sentence “I lived from paycheck to paycheck.” (Line 5, Para. 5) shows that .[A] Alvarez cared about nothing but making money[B] Alvarez’s salary barely covered her household expense s[C] Alvarez got paychecks from different jobs[D] Alvarez paid practically everything by check25. Which of the following would be the best title for the text?[A] Strain of Stress: No Way Out?[B] Response to Stress: Gender Difference[C] Stress Analysis: What Chemicals Say?[D] Gender Inequality: Women Under StressText 2It used to be so straightforward. A team of researchers working together in the laboratory would submit the results of their research to a journal. A journal editor would then remove t he author’s names and affiliations from the paper and send it to their peers for review. Depending on the comments received, the editor would accept thepaper for publication or decline it. Copyright rested with the journal publisher, and researchers seeking knowledge of the results would have to subscribe to the journal.No longer. The Internet—and pressure from funding agencies, who are questioning why commercial publishers are making money fromgovernment–funded research by restricting access to it—is making access to scientific results a reality. The Organization for Economic Co-operation and Development (OECD) has just issued a report describing the far-reaching consequences of this. The report, by John Houghton of Victoria University in Australia and Graham Vickery of the OECD, makes heavy reading for publishers who have, so far, made handsome profits. But it goes further than that. It signals a change in what has, until now, been a key element of scientific endeavor.The value of knowledge and the return on the public investment in research depends, in part, upon wide distribution and ready access. It is big business. In America, the core scientific publishing market is estimated at between $7 billion and $11 billion. The International Association of Scientific, Technical and Medical Publishers says that there are more than 2,000 publishers worldwide specializing in these subjects. They publish more than 1.2 million articles each year in some 16,000 journals.This is now changing. According to the OECD report, some 75% of scholarly journals are now online. Entirely new business models are emerging; three main ones were identified by the report’s authors. There is the so-called big deal, where institutional subscribers pay for access to a collection of online journal titles through site-licensing agreements. There is open-access publishing, typically supported by asking the author (orhis employer) to pay for the paper to be published. Finally, there are open-access archives, where organizations such as universities or international laboratories support institutional repositories. Other models exist that are hybridsof these three, such as delayed open-access, where journals allow only subscribers to read a paper for the first six months, before making it freely available to everyone who wishes to see it. All this could change the traditional form of the peer-review process, at least for the publication of papers.26. In the first paragraph, the author discusses .[A] the background information of journal editing[B] the publication routine of laboratory reports[C] the relations of authors with journal publishers[D] the traditional process of journal publication27. Which of the following is true of the OECD report?[A] It criticizes government-funded research.[B] It introduces an effective means of publication.[C] It upsets profit-making journal publishers.[D] It benefits scientific research considerably.28. According to the text, online publication is significant in that .[A] it provides an easier access to scientific results[B] it brings huge profits to scientific researchers[C] it emphasizes the crucial role of scientific knowledge[D] it facilitates public investment in scientific research29. With the open-access publishing model, the author of a paper is required to .[A] cover the cost of its publication[B] subscribe to the journal publishing it[C] allow other online journals to use it freely[D] complete the peer-review before submission30. Which of the following best summarizes the text?[A] The Internet is posing a threat to publishers.[B] A new mode of publication is emerging.[C] Authors welcome the new channel for publication.[D] Publication is rendered easily by online service.Text 3In the early 1960s Wilt Chamberlain was one of the only three players in the National Basketball Association (NBA) listed at over seven feet. If he had played last season, however, he would have been one of 42. The bodies playing major professional sports have changed dramatically over the years, and managers have been more than willing to adjust team uniforms to fit the growing numbers of bigger, longer frames.The trend in sports, though, may be obscuring an unrecognized reality: Americans have generally stopped growing. Though typically about two inches ta ller now than 140 years ago, today’s people—especially those born to families who have lived in the U.S. for many generations—apparently reached their limit in the early 1960s.And they aren’t likely to get any taller. “In the general population today, at t his genetic, environmental level, we’ve pretty much gone as far as we can go,” says anthropologist WilliamCameron Chumlea of Wright State University. In the case of NBA players, their increase in height appears to result from the increasingly common practice of recruiting players from all over the world.Growth, which rarely continues beyond the age of 20, demands calories and nutrients—notably, protein—to feed expanding tissues. At the start of the 20th century, under-nutrition and childhood infections got in the way. But as diet and health improved, children and adolescents have, on average, increased in height by about an inch and a half every 20 years, a pattern known as the secular trend in height. Yet according to the Centers for Disease Control and Prevention, average height—5'9" for men, 5'4" for women—hasn’t really changed since 1960.Genetically speaking, there are advantages to avoiding substantial height. During childbirth, larger babies have more difficulty passing through the birth canal. Moreover, even though humans have been upright for millions of years, our feet and back continue to struggle with bipedal posture and cannot easily withstand repeated strain imposed by oversize limbs. “There are some real constraints that are set by the genetic architecture of the individual organism,” says anthropologist William Leonard of Northwestern University.Genetic maximums can change, but don’t expect this to happen soon. Claire C. Gordon, senior anthropologist at the Army Research Center in Natick, Mass., ensures that 90 percent of the uniforms and workstations fit recruits without alteration. She says that, unlike those for basketball, the length of military uniforms has not changed for some time. And if you need to predict human height in the near future to design a piece of equipment, Gordon says that by and large, “you could use today's data and feel fairly confident.”31. Wilt Chamberlain is cited as an example to .[A] illustrate the change of height of NBA players[B] show the popularity of NBA players in the U.S.[C] compare different generations of NBA players[D] assess the achievements of famous NBA players32. Which of the following plays a key role in body growth according to the text?[A] Genetic modification.[B] Natural environment.[C] Living standards.[D] Daily exercise.33. On which of the following statements would the author most probably agree?[A] Non-Americans add to the average height of the nation.[B] Human height is conditioned by the upright posture.[C] Americans are the tallest on average in the world.[D] Larger babies tend to become taller in adulthood.34. We learn from the last paragraph that in the near future .[A] the garment industry will reconsider the uniform size[B] the design of military uniforms will remain unchanged[C] genetic testing will be employed in selecting sportsmen[D] the existing data of human height will still be applicable35. The text intends to tell us that .[A] the change of human height follows a cyclic pattern[B] human height is becoming even more predictable[C] Americans have reached their genetic growth limit[D] the genetic pattern of Americans has alteredText 4In 1784, five years before he became president of the United States, George Washington, 52, was nearly toothless. So he hired a dentist to transplant nine teeth into his jaw—having extracted them from the mouths of his slaves.That’s a far different image from the cherry-tree-chopping George most people remember from their history books. But recently,many historians have begun to focus on the role slavery played in the lives of the founding generation. They have been spurred in part by DNA evidence made available in 1998, which almost certainly proved Thomas Jefferson had fathered at least one child with his slave Sally Hemings. And only over the past 30 years have scholars examined history from the bottom up. Works of several historians reveal the moral compromises made by the nation’s early leaders and the fragile nature of the country’s infancy. More significant, they argue that many of the Founding Fathers knew slavery was wrong—and yet most did little to fight it.More than anything, the historians say, the founders were hampered by the culture of their time. While Washington and Jefferson privately expressed distaste for slavery, they also understood that it was part of the political and economic bedrock of the country they helped to create.For one thing, the South could not afford to part with its slaves. Owning slaves was “like having a large bank account,” says Wiencek, auth or of An Imperfect God: George Washington, His Slaves, and the Creation of America. The southern states would not have signed the Constitution without protections for the “peculiar institution,” including a clause that counted a slave as three fifths of a man for purposes of congressional representation.And the statesmen’s political lives depended on slavery. The three-fifths formula handed Jefferson his narrow victory in the presidential election of 1800 by inflating the votes of the southern states in the Electoral College. Once in office, Jefferson extended slavery with the Louisiana Purchase in 1803; the new land was carved into 13 states, including three slave states.Still, Jefferson freed Hemings’s children—though not Hemings herself or his approximately 150 other slaves. Washington, who had begun to believe that all men were created equal after observing the bravary of the black soldiers during the Revolutionary War, overcame the strong opposition of his relatives to grant his slaves their freedom in his will. Only a decade earlier, such an act would have required legislative approval in Virginia.36. George Washington’s dental surgery is mentioned to .[A] show the primitive medical practice in the past.[B] demonstrate the cruelty of slavery in his days.[C] stress the role of slaves in the U.S. history.[D] reveal some unknown aspect of his life.37. We may infer from the second paragraph that .[A] DNA technology has been widely applied to history research.[B] in its early days the U.S. was confronted with delicate situations.[C] historians deliberately made up some stories of Jefferson’s life.[D] political compromises are easily found throughout the U.S. history.38. What do we learn about Thomas Jefferson?[A] His political view changed his attitude towards slavery.[B] His status as a father made him free the child slaves.[C] His attitude towards slavery was complex.[D] His affair with a slave stained his prestige.39. Which of the following is true according to the text?[A] Some Founding Fathers benefit politically from slavery.[B] Slaves in the old days did not have the right to vote.[C] Slave owners usually had large savings accounts.[D] Slavery was regarded as a peculiar institution.40. Washington’s decision to free slaves originated from his .[A] moral considerations.[B] military experience.[C] financial conditions.[D] political stand.Part BDirections:In the following text, some segments have been removed. For Questions 41-45, choose the most suitable one from the list A-G to fit into each ofthe numbered blanks. There are two extra choices, which do not fit in any of the blanks. Mark your answers on ANSWER SHEET 1. (10 points)The time for sharpening pencils, arranging your desk, and doing almost anything else instead of writing has ended. The first draft will appear on the page only if you stop avoiding the inevitable and sit, stand up, or lie down to write. (41)_______________.Be flexible. Your outline should smoothly conduct you from one point to the next, but do not permit it to railroad you. If a relevant and important idea occurs to you now, work it into the draft. (42) _______________. Grammar, punctuation, and spelling can wait until you revise. Concentrate on what you are saying. Good writing most often occurs when you are in hot pursuit of an idea rather than in a nervous search for errors.(43) _______________. Your pages will be easier to keep track of that way, and, if you have to clip a paragraph to place it elsewhere, you will not lose any writing on either side.If you are working on a word processor, you can take advantage of its capacity to make additions and deletions as well as move entire paragraphs by making just a few simple keyboard commands. Some software programs can also check spelling and certain grammatical elements in your writing. (44) _______________. These printouts are also easier to read than the screen when you work on revisions.Once you have a first draft on paper, you can delete material that is unrelated to your thesis and add material necessa ry to illustrate your points and make your paper convincing. The student who wrote “The A&P as a Stateof Mind” wisely dropped a paragraph that questioned whether Sammy displays chauvinistic attitudes toward women. (45) _______________.Remember that your initial draft is only that. You should go through the paper many times—and then again—working to substantiate and clarify your ideas. You may even end up with several entire versions of the paper. Rewrite. The sentences within each paragraph should be related to a single topic. Transitions should connect one paragraph to the next so that there are no abrupt or confusing shifts. Awkward or wordy phrasing or unclear sentences and paragraphs should be mercilessly poked and prodded into shape.[A] To make revising easier, leave wide margins and extra space between lines so that you can easily add words, sentences andcorrections. Write on only one side of the paper.[B] After you have already and adequately developed the body of your paper, pay particular attention to the introductory and concluding paragraphs. It’s probably best to write the introduction last, after you know precisely what you are introducing. Concluding paragraphs demand equal attention because they leave the reader with a final impression.[C] It’s worth remembering, however, that though a clean copy fresh off a printer may look terrible, it will read only as well as the thinking and writing that have gone into it. Many writers prudently store their data on disks and print their pages each time they finish a draft to avoid losing any material because of power failures or other problems.[D] It makes no difference how you write, just so you do. Now that you have developed a topic into a tentative thesis, you can assemble your notes and begin to flesh out whatever outline you have made.[E] Although this is an interesting issue, it has nothing to do with the thesis, which explains how the setting influences Sammy’s decision to quit his job. Instead of including that paragraph, she added one that d escribed Lengel’s crabbed response to the girls so that she could lead up to the A & P “policy” he enforces.[F] In the final paragraph about the significance of the setting in “A&P” the student brings together the reasons Sammy quit his job by referring t o his refusal to accept Lengel’s store policies.[G] By using the first draft as a means of thinking about what you want to say, you will very likely discover more than your notes originally suggested. Plenty of good writers don’t use outlines at all but discover ordering principles as they write. Do not attempt to compose a perfectly correct draft the first time around.Part CDirections:Read the following text carefully and then translate the underlined segments into Chinese. Your translation should be written neatly on ANSWER SHEET 2. (10 points)In his autobiography,Darwin himself speaks of his intellectualpowers with extraordinary modesty. He points out that he always experienced much difficulty in expressing himself clearly and concisely, but (46)he believes that this very difficulty may have had the compensating advantage of forcing him to think long and intently about every sentence, and thus enabling him to detect errors in reasoning and in his ownPart A51. Directions:You have just come back from Canada and found a music CDin your luggage that you forgot to return to Bob, your landlord there. Write him a letter to1) make an apology, and2) suggest a solution.You should write about 100 words on ANSWER SHEET 2.Do not sign your own name at the end of the letter. Use “Li Ming” instead.Do not write the address. (10 points)Part B52. Directions:Write an essay of 160-200 words based on the following drawing. In your essay, you should1) describe the drawing briefly,2) explain its intended meaning, and then3) give your comments.You should write neatly on ANSHWER SHEET 2. (20 points)2023年全国硕士硕士招生考试英语(一)答案详解Section I Use of English一、文章总体分析这是一篇议论文。
mTOR通路
Seminars in Cell &Developmental Biology 36(2014)79–90Contents lists available at ScienceDirectSeminars in Cell &DevelopmentalBiologyj o u r n a l h o m e p a g e :w w w.e l s e v i e r.c o m /l o c a t e /s e m c dbGrowing knowledge of the mTOR signaling networkKezhen Huang a ,Diane C.Fingar a ,b ,∗aDepartment of Cell and Developmental Biology,University of Michigan Medical School,Ann Arbor,MI 48109-2200,United StatesbDivision of Metabolism,Endocrinology,and Diabetes (MEND),Department of Internal Medicine,University of Michigan Medical School,Ann Arbor,MI 48109-2200,United Statesa r t i c l ei n f oArticle history:Available online 19September 2014Keywords:mTOR mTORC1mTORC2InsulinAmino acids Energya b s t r a c tThe kinase mTOR (mechanistic target of rapamycin)integrates diverse environmental signals and trans-lates these cues into appropriate cellular responses.mTOR forms the catalytic core of at least two functionally distinct signaling complexes,mTOR complex 1(mTORC1)and mTOR complex 2(mTORC2).mTORC1promotes anabolic cellular metabolism in response to growth factors,nutrients,and energy and functions as a master controller of cell growth.While significantly less well understood than mTORC1,mTORC2responds to growth factors and controls cell metabolism,cell survival,and the organization of the actin cytoskeleton.mTOR plays critical roles in cellular processes related to tumorigenesis,metabolism,immune function,and aging.Consequently,aberrant mTOR signaling contributes to myriad disease states,and physicians employ mTORC1inhibitors (rapamycin and analogs)for several pathological conditions.The clinical utility of mTOR inhibition underscores the important role of mTOR in organismal physiology.Here we review our growing knowledge of cellular mTOR regulation by diverse upstream signals (e.g.growth factors;amino acids;energy)and how mTORC1integrates these signals to effect appropriate downstream signaling,with a greater emphasis on mTORC1over mTORC2.We highlight dynamic sub-cellular localization of mTORC1and associated factors as an important mechanism for control of mTORC1activity and function.We will cover major cellular functions controlled by mTORC1broadly.While signif-icant advances have been made in the last decade regarding the regulation and function of mTOR within complex cell signaling networks,many important findings remain to be discovered.©2014Elsevier Ltd.All rights reserved.Contents 1.Introduction ..........................................................................................................................................802.Growth factor sensing by the TSC–Rheb axis ........................................................................................................812.1.Insulin-PI3K-Akt signaling ....................................................................................................................812.2.EGF-Ras-MAPK signaling......................................................................................................................822.3.mTORC2regulation ...........................................................................................................................822.4.Negative feedback signaling ..................................................................................................................823.Amino acid sensing by the Rag–Ragulator axis and other emerging factors ........................................................................823.1.The Rag GTPases ..............................................................................................................................833.2.The Ragulator complex .......................................................................................................................833.3.The v-ATPase ..................................................................................................................................833.4.Emerging amino acid sensing factors .........................................................................................................844.The lysosome as a critical platform for upstream signal integration and mTORC1activation ......................................................855.Energy and stress sensing ............................................................................................................................856.Major cellular functions controlled by mTORC1.....................................................................................................86∗Corresponding author at:Department of Cell and Developmental Biology,University of Michigan Medical School,Ann Arbor,MI 48109-2200,United States.Tel.:+17347637541.E-mail address:dfingar@ (D.C.Fingar)./10.1016/j.semcdb.2014.09.0111084-9521/©2014Elsevier Ltd.All rights reserved.80K.Huang,D.C.Fingar/Seminars in Cell&Developmental Biology36(2014)79–907.Future directions (87)Acknowledgments (87)References (87)1.IntroductionAll cells from single-celled organisms to those comprising mul-ticellular organisms sense and respond rapidly tofluctuations in their nutritional and energetic environments in order to modulate cell metabolism appropriately and maintain cellular homeostasis. Consequently,cells coordinate nutritional and energetic supply and demand tightly to prevent engagement in ATP-consuming anabolic processes when environmental resources become limited. During evolution,multicellular organisms acquired the additional ability to sense and respond to long-range systemic signals(i.e. hormones;growth factors;mitogens;cytokines)(referred to col-lectively as“growth factors”here)to enable communication between tissues and organ systems.mTOR,the mechanistic tar-get of rapamycin,functions as a critical integrator of these diverse environmental cues by integrating them into appropriate cellu-lar responses.mTOR,an evolutionarily conserved serine/threonine protein kinase,belongs to the phosphatidylinositol-3kinase(PI3K)-related kinase(PIKK)superfamily.mTOR represents the functional target of a natural macrolide antibiotic called rapamycin(clini-cally known as sirolimus).Rapamycin,produced by the bacterium Streptomyces hygroscopicus,was discovered in soil samples from Easter Island(known as Rapa Nui to the native population)in the 1970s[1,2].Rapamycin reduces eukaryotic cell proliferation to var-ious degrees,with immune cells showing strong sensitivity.To identify the target of rapamycin,Hall and colleagues performed an elegant genetic screen in1991in the budding yeast Saccha-romyces cerevisiae.Mutations in three genes,Fpr1(an orthologue of FKBP12[FK506-binding protein12]),Tor1and Tor2,conferred resis-tance to rapamycin[3](and reviewed in[2]).Today we understand that rapamycin associates with an endogenous cellular protein, FKBP12,and this complex docks to the FRB(FKBP12rapamycin binding)domain located immediately N-terminal to the C-terminal mTOR kinase domain(see the accompanying article for greater detail regarding mTOR structure),resulting in allosteric inhibi-tion of mTOR activity and signaling[4–6].By affinity purification of FKPB12-rapamycin binding proteins,several groups identified the mammalian orthologue of budding yeast Tor1/2in1994–1995 [7–9].mTOR constitutes the catalytic core of two known signal-ing complexes,mTOR complex1(mTORC1)and mTOR complex2 (mTORC2)[10,11].These mTOR complexes(mTORCs)possess dis-tinct substrates,cellular functions,and sensitivity to rapamycin. Acute rapamycin treatment inhibits cellular mTORC1but not mTORC2signaling while longer-term rapamycin treatment sup-presses mTORC2function by compromising complex integrity to variable degrees depending on cell type[12].Rapamycin fails to inhibit the phosphorylation of all mTORC1substrates equally [13,14].It completely inhibits phosphorylation of S6K1(ribosomal protein S6kinase1)but only partially inhibits phosphorylation of 4EBP1(eukaryotic initiation factor4E binding protein1).The devel-opment of ATP-competitive mTOR catalytic inhibitors(i.e.Torin1; Ku-0063794)revealed that mTORC1phosphorylates substrates in both rapamycin-sensitive and-insensitive manners[15–18],pos-sibly due to differential substrate access to the kinase active site controlled by the mTOR FRB domain[19]and/or due to differen-tial substrate quality conferred by phosphorylation site consensus sequence[20,21].The exclusive mTOR partner raptor(regulatory-associated pro-tein of mTOR)defines mTORC1[22,23]while the exclusive mTOR partner rictor(rapamycin-insensitive companion of mTOR)defines mTORC2[24,25](Fig.1).In addition to raptor,mTORC1contains mLST8/GL(mammalian lethal with Sec13protein8/G-protein -protein subunit like)[26],PRAS40(Akt/PKB substrate40kDa) [27,28],and deptor(DEP-domain-containing mTOR interacting protein)[29].Raptor serves a scaffolding role,functioning to recruit substrates to the mTOR kinase through their TOS(TOR signal-ing)motifs[30,31].Global deletion of raptor in mice results in early embryonic lethality(e5.5)[32],similar to the global knock-out of mTOR[33].PRAS40and deptor function as both suppressors and targets of mTORC1,likely by acting as competitive sub-strates[29,34],while mLST8/GL is not essential for mTORC1 function[32].In addition to rictor[24,25],mTORC2contains mSIN1 (mammalian stress-activated protein kinase interacting protein1) [35,36],protor1/2(protein observed with rictor1/2)(aka PRR5) [37],mLST8/GL[24,25],and deptor[29].Thus,mTORC1and mTORC2contain distinct and shared partner proteins.Similar to raptor within mTORC1,rictor and mSin1serve as critical scaffolds that control mTORC2integrity,regulation by upstream signals, and substrate choice[24,25,35,36].Unlike mTORC1,mLST8/GL is required for mTORC2function;like rictor,its deletion in mice causes embryonic lethality(e10.5)[32].The role of protor remains unclear.It is important to note that mTOR also assembles into relatively homologous TORC1and TORC2complexes in bud-ding andfission yeast,underscoring the ancestral origin of the TORCs.While TORC1in yeast responds simply to environmental nutri-ents and energy,mTORC1in higher eukaryotes responds to a broader array of upstream signals,integrating cues from growth factors(i.e.insulin;IGF;EGF;cytokines)to modulate cellular func-tions appropriately(Fig.1)[10,11].mTORC1function absolutely requires sufficient levels of amino acids such that their withdrawal inactivates mTORC1signaling rapidly and renders mTORC1acti-vation refractory to virtually all other inputs,including growth factors.To limit cellular engagement in energy costly anabolic pro-cesses,nutrient and growth factor withdrawal as well as diverse types of cell stress(i.e.low energy;hypoxia;ER stress;ROS(reac-tive oxygen species))downregulate mTORC1signaling[38].Upon activation,mTORC1signaling drives cap-dependent protein syn-thesis,cell growth,and cell proliferation through phosphorylation of the ribosomal protein S6kinases(S6K1/2)and the eukaryotic ini-tiation factor4E(eIF4E)binding proteins1–3(4EBP1-3)at least in part[10,11,39].While the current set of direct mTORC1substrates remains somewhat limited(e.g.S6Ks;4EBPs;IRS-1;ULK1;Lipin1; TFEB;Grb10),quantitative phosphoproteomic screens identified a large number of downstream mTORC1effectors,many of which likely represent bonafide mTORC1substrates[40,41](Fig.1). mTORC1promotes other anabolic processes including lipid and nucleotide synthesis and suppresses autophagy,a degradative pro-cess in which autophagosomes break down macromolecules and organelles during nutrient and energy starvation.Thus,mTORC1 drives anabolic and suppresses catabolic cellular processes.Our understanding of the regulation and function of mTORC2lags far behind that of mTORC1due to its more recent discovery[24,25] and the lack of mTORC2-specific inhibitors.Growth factors activate mTORC2,which phosphorylates a limited set of known substrates including Akt(aka PKB),PKC␣(protein kinase C␣),and SGK1(serum and glucocorticoid-induced protein kinase).mTORC2modulates cell metabolism and the organization of the actin cytoskeleton and enhances cell survival,due to its activation of the survival kinase Akt[42,43](Fig.1).K.Huang,D.C.Fingar /Seminars in Cell &Developmental Biology 36(2014)79–9081Fig.1.Regulation of the mTORC1and mTORC2signaling network by diverse upstream inputs.Growth factors such as insulin or EGF activate mTORC1through either the PI3K-Akt or the Ras-MAPK (ERK)-RSK axes,respectively.Growth factor-mediated activation of mTORC1absolutely requires sufficient levels of amino acids,which are sensed through a variety of factors,as indicated.mTORC1action also requires sufficient levels of energy (i.e.ATP)and/or oxygen,which are sensed by AMPK,REDD1,and TCA cycle metabolites (i.e.␣KG).The TSC complex integrates diverse upstream signals to regulate mTORC1action.TSC suppresses the conversion of Rheb-GDP to Rheb-GTP,a small GTPase that activates mTORC1.mTORC1phosphorylates a limited known set of bona fide substrates to drive anabolic and suppress catabolic cellular processes and to mediate negative feedback toward PI3K.Growth factors (i.e.insulin)also activate mTORC2through poorly defined signaling intermediates.Aberrant mTORC1function contributes to myriad patho-logic conditions including cancer and benign tumor syndromes,metabolic disorders (e.g.type II diabetes;obesity),cardiovascu-lar disorders,inflammatory disorders,and neurological disorders [10,11].Consequently,clinicians employ rapamycin (aka sirolimus)and rapamycin analogs (rapalogs)(i.e.everolimus;temsirolimus)for immunosuppression following renal transplantation and for treatment of renal cell carcinoma,neuroendocrine tumors of pan-creatic origin,tuberous sclerosis complex (TSC,a benign tumor syndrome),and cardiac restenosis following angioplasty [6].The role of mTOR in pathophysiology of disease combined with the utility of mTORC1inhibitors in clinical medicine underscores the importance of elucidating the regulation and function of mTORC1at the cellular level [44].2.Growth factor sensing by the TSC–Rheb axisGrowth factors,in particular insulin/IGF (insulin-like growth factor)and EGF (epidermal growth factor),represent the best understood inputs that lead to activation of mTORC1upon converg-ing on the TSC (tuberous sclerosis complex)/Rheb axis [45].TSC,composed of TSC1(aka hamartin),TSC2(aka tuberin),and a morerecently discovered third subunit,TBC1D7(TBC [Tre2-Bub2-Cdc16]1domain family member 7),functions as a tumor suppressor that inhibits mTORC1[46](Fig.1).Inactivating mutations in either TSC1or TSC2increases mTORC1signaling and causes an autosomal dominant disease in which benign tumors form in various organs including brain,kidney,and heart [47].TSC inhibits mTORC1by inhibiting Rheb (Ras homolog enriched in brain),a small Ras-like GTPase essential for mTORC1activation by all upstream signals.2.1.Insulin-PI3K-Akt signalingInsulin/IGF binding to its cognate cell surface tyrosine kinase receptor leads to tyrosine phosphorylation of IRS (insulin receptor substrate)proteins,which recruits and activates PI3K (phos-phatidylinositol 3-kinase)[48](Fig.1).Increased production of the phospho-lipid PI(3,4,5)P3on lipid membranes by PI3K recruits Akt via its PH (pleckstrin homology)domain,leading to PDK1-mediated phosphorylation of Akt on its activation loop (T308)[49].It is important to note that additional phosphorylation of Akt on its hydrophobic motif (S473)by PI3K-controlled mTORC2boosts Akt activity several fold further [49,50].Activated Akt then phosphorylates TSC2on several sites (S939;T1462)to82K.Huang,D.C.Fingar/Seminars in Cell&Developmental Biology36(2014)79–90suppress the inhibitory effect of the TSC complex toward mTORC1, thus leading to increased mTORC1signaling[51,52].TSC2pos-sesses a GAP(GTPase activating protein)domain that hydrolyzes active Rheb-GTP to inactive Rheb-GDP.Thus,in response to insulin/PI3K signaling,Akt phosphorylates and inactivates TSC2, which increases Rheb-GTP loading and mTORC1kinase activity [27].While Rheb-GTP interacts with the mTOR kinase domain [27,53],the underlying molecular mechanism by which Rheb acti-vates mTORC1remains unclear.Other parallel mechanisms contribute to activation of mTORC1 by insulin/IGF.Akt and mTORC1phosphorylate PRAS40,an mTORC1inhibitory partner(on T246and S183/S212/S221,respec-tively),inducing the dissociation of PRAS40from mTORC1and thus relieving PRAS40-mediated substrate competition[27,28,34,54]. Insulin/PI3K signaling also leads to mTORC1-mediated phospho-rylation of raptor(on S863)to promote mTORC1signaling[55,56]. Moreover,phosphorylation of mTOR itself(on S1261,S2159,and T2164)by unknown kinases[57,58]and on S1415by IKK␣[59] contributes to increased mTORC1signaling.While mTOR autophos-phorylation on S2481plays no known role in mTORC1function, it serves as a biomarker for mTORC1and mTORC2catalytic activ-ity in intact cells[5].Many phospho-proteomic studies agree that mTOR and its partner proteins undergo phosphorylation on many sites[40,41,60].Consequently,a challenge for the future will be to identify the kinases that act on these sites directly and to deci-pher the regulation and functional significance of complex mTORC phosphorylation.2.2.EGF-Ras-MAPK signalingEGF activates mTORC1signaling independently of the PI3K/Akt axis.EGF binding to its cell surface tyrosine kinase receptor acti-vates the Ras GTPase,which leads to activation of c-Raf,MEK (MAPK/ERK kinase),MAPK(mitogen activated protein kinase) (aka ERK)and RSK(p90ribosomal S6kinase(Fig.1).Similar to Akt,MAPK and RSK phosphorylate TSC2on different sites (S540/S644and S1798,respectively)to suppress the inhibitory action of TSC2toward Rheb[61,62].By a parallel pathway,the Ras/MAPK pathway converges on raptor.MAPK phosphorylates raptor(on S8/S696/S863)and RSK phosphorylates raptor(on S719/S721/S722)[63,64]to promote mTORC1signaling.2.3.mTORC2regulationInsulin/PI3K signaling leads to mTORC2-mediated phosphory-lation of Akt on its hydrophobic motif(HM)site,S473(Fig.1)as well as the HM sites of other AGC kinases,PKC␣(on S657),and SGK1(on S422)[25,50,65].Insulin increases the kinase activity of mTORC2in vitro in a manner sensitive to cellular treatment with the PI3K inhibitor wortmannin[66].Thus,insulin/PI3K signaling activates mTORC2;it is important to note,however,that the sig-naling intermediates that link PI3K to mTORC2remain virtually unknown.Interestingly,while TSC inhibits mTORC1,TSC activates mTORC2[66].MEFs lacking TSC2display reduced mTORC2kinase activity toward Akt in vitro and decreased Akt S473phosphoryla-tion in intact cells in a manner independent of the well-known mTORC1-mediated negative feedback loop that attenuates PI3K signaling(discussed below)[66].In addition,TSC associates with mTORC2.These data suggest quite different regulation of mTORC2 compared to mTORC1.On the other hand,the Rac1GTPase interacts with mTOR and provides an activating signal to both mTORC1and mTORC2in response to growth factors in a PI3K/Akt independent manner,suggesting that common upstream inputs co-regulate both mTORCs[67].There is no question that important discoveries await regarding mTORC2regulation.As mTORC2mediates Akt S473phosphorylation,it would seem that mTORC2lies upstream of mTORC1.While such epistasis may hold true in certain cellular contexts,genetic knockout or knockdown of core mTORC2components(i.e.rictor;mSin1)in many cultured cell types has no effect on TSC2phosphoryla-tion and mTORC1signaling[32,68].Thus,mTORC2function is not required for mTORC1action.On the other hand,mTORC2function is required for Akt-mediated phosphorylation of other substrates (i.e.FoxO1/3a)[32,68].These data can be explained by the known essential requirement for activation loop site(T308)but not HM site(S473)phosphorylation for Akt kinase activity;Akt S473phos-phorylation boosts Akt activity further and may modulate substrate specificity[32,49,68].Thus,in many cellular contexts Akt phospho-rylation on its activation loop without HM-site phosphorylation provides sufficient activity to mediate downstream signaling to mTORC1.Interestingly,mTORC2associates with ribosomes in a growth factor sensitive manner[69,70].Structurally intact ribosomes,but not protein synthesis itself,are required for mTORC2kinase activity in vitro and signaling in intact cells[70].Thus,a direct interaction of mTORC2with ribosomes may play a role in insulin/PI3K-meditated mTORC2activation.mTORC2also promotes turn-motif (TM)site phosphorylation of Akt(T450)and several conventional PKCs(PKC␣T638and PKCT641)in a co-translational manner independently of growth factor status,functioning to increase protein stability and folding[71,72].In addition to interacting with ribosomes(likely those associated with ER engaged in pro-tein translation),mTORC2associates with an ER sub-compartment called MAM(mitochondrial-associated ER membrane)in a growth factor stimulated manner.mTORC2inactivation decreases MAM integrity,mitochondrial metabolism,and cell survival[73].2.4.Negative feedback signalingSeveral negative feedback mechanisms modulate the mTOR signaling network,as signal attenuation limits signal amplitude and duration critical for homeostatic control of complex biolog-ical systems.Cellular TSC loss leads to elevated and constitutive mTORC1signaling independent of growth factor status and atten-uates PI3K signaling,thus producing a state of cellular insulin resistance[74,75].S6K1and mTORC1phosphorylate IRS-1directly to induce its degradation,thus uncoupling the insulin receptor from PI3K.mTORC2also limits PI3K signaling by inducing IRS-1 degradation[76].mTORC2phosphorylates and stabilizes Fbw7,an ubiquitin ligase subunit that targets IRS-1for degradation[76]. Grb10was identified as a direct mTORC1substrate in phospho-proteomic screens[40,76].mTORC1-mediated phosphorylation of Grb10,a growth factor receptor-bound adaptor that limits growth factor signaling,stabilizes Grb10and attenuates both PI3K and MAPK/ERK signaling.Depending on cellular context,either S6K1 or Akt phosphorylate mSin1directly(on T86and T398),a crit-ical mTORC2partner,dissociating mSin1from the complex and decreasing mTORC2signaling[40].Along similar lines,several groups reported that S6K1phosphorylates rictor directly(T1135) [77–80],which may reduce mTORC2signaling to Akt[78,79].These data reveal that both mTORC1and mTORC2engage in negative feedback to maintain proper signaling by growth factor receptors and the mTORCs.3.Amino acid sensing by the Rag–Ragulator axis and other emerging factorsSufficient levels of amino acids,particularly leucine,are essen-tial for basal mTORC1signaling from yeast to mammals and for robust activation of mTORC1in response to growth factor signalsK.Huang,D.C.Fingar/Seminars in Cell&Developmental Biology36(2014)79–9083Fig. 2.Rag heterodimers recruit mTORC1to lysosomal membranes for Rheb-mediated activation.Activation of mTORC1by amino acids through Rag GTPase heterodimers involves the v-ATPase,Ragulator complex,and Rag regulatory factors. The Ragulator complex,which acts as a GEF toward RagA/B GTPases,induces forma-tion of active RagA/B GTP–RagC/D GDP heterodimers.The GATOR1complex functions as a GAP(GTPase activating protein)for RagA/B(thus inhibiting Rag heterodimers) while folliculin(FLCN)and its associated proteins(FNIP1/2)functions as a GAP for Rag C/D(thus promoting a Rag heterodimer active state).The GATOR2complex sup-presses GATOR1.Active RagA/B GTP–RagC/D GDP heterodimers bind mTORC1through raptor to recruit mTORC1to the lysosomal surface where Rheb resides.When loaded with GTP,Rheb activates mTORC1through a poorly defined mechanism.An“inside-out”model proposes that the v-ATPase and Ragulator respond to amino acid levels inside the lysosomal lumen to control Rag nucleotide binding state.in higher eukaryotes[81,82].How cells sense amino acid levels remains poorly defined,but great progress has been made in recent years identifying the machinery that propagates amino acid sens-ing proximal to mTORC1.While several signaling molecules that link amino acid sensing to mTORC1have been identified(see text below),the Rag GTPases,the Ragulator complex,and the v-ATPase represent the best-characterized links between amino acid sensing and mTORC1.3.1.The Rag GTPasesThe evolutionarily conserved family of Rag GTPases function as obligate heterodimers in which Rag A or Rag B dimerizes with RagC or RagD(Fig.2).Upon amino acid stimulation,RagA/B loads with GTP and binds raptor while RagC/D loads with GDP[83,84]. Expression of dominant-active RagA/B mutants(loaded consti-tutively with GTP)promote mTORC1signaling in the absence of cellular amino acids while expression of dominant-negative RagA/B mutants(nucleotide-free)suppress mTORC1signaling in the presence of amino acids.Thus,heterodimers composed of RagA/B GTP–RagC/D GDP form during amino acid sufficiency to promote mTORC1signaling and heterodimers composed of RagA/B GDP–RagC/D GTP form during amino acid withdrawal[83,84]. As exogenous expression of RagA/B GTP–RagC/D GDP heterodimers more strongly activate mTORC1in amino acid deprived cells than exogenous expression of dominant-active RagA/B alone,these data suggest that the nucleotide-bound state of RagC/D as well as that of RagA/B indeed contributes to mTORC1signaling in response to amino acids.How do Rags control amino acid-mediated activation of mTORC1?While Rheb-GTP provided in vitro increases mTORC1 kinase activity directly[27],active RagA/B GTP–RagC/D GDP het-erodimers provided in vitro are insufficient[83].Cellular amino acid stimulation induces the translocation of mTOR and raptor from a poorly defined cytoplasmic compartment to LAMP1/2-positive lysosomal membranes,a site to which the Rags and Rheb also localize[83,85].Importantly,mTORC1translocation requires the Rags[83].These data suggest a model in which the amino acid-Rag axis activates mTORC1by controlling mTORC1subcel-lular localization:amino acid signaling drives the formation of active RagA/B GTP–RagC/D GDP heterodimers,which bind raptor to recruit mTORC1to the lysosomal surface where mTORC1receives an essential activating input from Rheb(and possibly from other inputs)(Fig.3).While Rheb docks to internal membranes by a farnesyl lipid moiety,Rag GTPases do not possess lipid-anchoring motifs.3.2.The Ragulator complexA pentameric complex called the Ragulator,consisting of p18 (LAMTOR1),p14(LAMTOR2),MP1(LAMTOR3),c7orf59(LAMTOR4), and HBXIP(hepatitisB virus X interacting protein)(LAMTOR5),was found to anchor the Rags to lysosomal membranes through p18 myristoylation[85,86](Fig.3).The Ragulator complex not only tethers the Rags to lysosomal membranes,it possesses GEF activity toward RagA/B to enable exchange of GDP for GTP,thus converting Rag heterodimers to an active state[85].Conversely,a complex of proteins termed GATOR1,which contains proteins DEPDC5,Nprl2, and Nprl3,binds Rag heterodimers and possesses GAP activity for RagA/B,thus converting Rag heterodimers to an inactive state[87].A complex called GATOR2,which contains proteins Mios,Wdr24, Wdr59,Seh1L,and Sec13,suppresses GATOR1.Indeed,inactivation of GATOR1subunits renders mTORC1resistant to amino acid depri-vation while inactivation of GATOR2subunits suppresses mTORC1 signaling[87].RagC/D not only function as obligate binding partners for RagA/B,their nucleotide binding state also participates in amino acid controlled mTORC1function.Folliculin(FLCN)and its inter-acting partners FNIP1/2are required for amino acid-stimulated translocation of mTORC1to lysosomes,where they dock to Rag GTPases in the absence of amino acids,poised to convert RagC/D GTP to RagC/D GDP upon docking of mTORC1to the lysosomal sur-face upon amino acid addition[88,89](Fig.2).FLCN possess GAP activity toward RagC/D but not RagA/B and thus converts Rag heterodimers from an inactive RagA/B GDP–RagC/D GTP to an active RagA/B GTP–RagC/D GDP nucleotide-bound state that stabilizes mTORC1docking[89].The discovery that a spatial-temporal mech-anism governs amino acid-mediated mTORC1signaling explained a long-standing mystery in thefield regarding why growth factors fail to activate mTORC1in the absence of amino acids:if mTORC1local-izes within the cell to the wrong place at the wrong time,mTORC1 cannot be activated by upstream inputs.3.3.The v-ATPaseHow do cells sense amino acid levels?The discovery that the v-ATPase(vacuolar H+-adenosine triphosphatase)interacts with the Ragulator complex and Rag GTPases on the lysosomal surface and senses amino acids levels,possibly from within the lysoso-mal lumen,begins to elucidate these important but poorly resolved questions[90].v-ATPase subunits and its ATP hydrolyzing catalytic activity are required for generation of active RagA/B GTP–RagC/D GDP heterodimers,for amino acid-mediated localization of mTORC1to lysosomes,and for activation of mTORC1(Fig.2).Its classical func-tion as a proton pump that lowers luminal pH of lysosomes is not required for mTORC1activation,however[90].Amino acids modulate interactions between the v-ATPase,Ragulator,and Rags [90],and amino acid accumulation within the lysosomal lumen correlates with recruitment of mTORC1to lysosomal membranes. Thus,mTORC1appears to sense intra-lysosomal amino acids by an“inside-out”mechanism.It is important to note,however, that the overall importance of intra-lysosomal amino acids for mTORC1recruitment remains unclear.In this proposed model,。
- 1、下载文档前请自行甄别文档内容的完整性,平台不提供额外的编辑、内容补充、找答案等附加服务。
- 2、"仅部分预览"的文档,不可在线预览部分如存在完整性等问题,可反馈申请退款(可完整预览的文档不适用该条件!)。
- 3、如文档侵犯您的权益,请联系客服反馈,我们会尽快为您处理(人工客服工作时间:9:00-18:30)。
InferenceofGeneticRegulatoryNetworksbyEvolutionaryAlgorithmandH∞Filtering
LijunQianandHaixinWangDepartmentofElectricalandComputerEngineeringPrairieViewA&MUniversityPrairieView,Texas77446Email:LiQian,HWang@pvamu.edu
Abstract—Thecorrectinferenceofgeneticregulatorynetworksplaysacriticalroleinunderstandingbiologicalregulationinphenotypicdeterminationanditcanaffectadvancedgenome-basedtherapeutics.Inthisstudy,weproposeajointevolutionaryalgorithmandH∞filteringapproachtoinfergeneticregulatorynetworksusingnoisytimeseriesdatafrommicroarraymea-surements.Specifically,aniterativealgorithmisproposedwheregeneticprogrammingisappliedtoidentifythestructureofthemodelandH∞filteringisusedtoestimatetheparametersineach
iteration.Theproposedmethodcanobtainaccuratedynamicnonlinearordinarydifferentialequation(ODE)modelofgeneticregulatorynetworksevenwhenthenoisestatisticsisunknown.Bothsyntheticdataandexperimentaldatafrommicroarraymeasurementsareusedtodemonstratetheeffectivenessoftheproposedmethod.Withtheincreasingavailabilityoftimeseriesmicroarraydata,thealgorithmdevelopedinthispapercouldbeappliedtoconstructmodelstocharacterizecancerevolutionandserveasthebasisfordevelopingnewregulatorytherapies.
I.INTRODUCTION
Ageneticregulatorynetwork(GRN)isacollectionofDNAsegmentsinacellwhichinteractwitheachotherandwithothersubstancesinthecell,therebygoverningthegenetranscriptions.Thecorrectinferenceofgeneticregulatorynetworksplaysacriticalroleinunderstandingbiologicalreg-ulationinphenotypicdeterminationanditcanaffectadvancedgenome-basedtherapeutics.Inlightoftherecentdevelopmentofhigh-throughputDNAmicroarraytechnology,itbecomespossibletodiscoverGRNs,whicharecomplexandnonlinearinnature.Specifically,theincreasingexistenceofmicroar-raytime-seriesdatamakespossiblethecharacterizationofdynamicnonlinearregulatoryinteractionsamonggenes.Themodeling,analysisandcontrolofGRNsarecriticalforstudy-ingcancerevolutionandmayserveasthebasisfordevelopingnewregulatorytherapies.BecauseGRNmodelsaredifficulttodeducesolelybymeansofexperimentaltechniques,computationalandmath-ematicalmethodsareindispensable.MuchresearchhasbeendoneonGRNmodelingbylineardifferential/differenceequa-tionsusingtime-seriesdata,forexample,[1-8],justtonameafew.Thebasicideaistoapproximatethecombinedeffectsofdifferentgenesbymeansofaweightedsumoftheirexpressionlevels.In[5],aconnectionistmodelisusedtomodelsmallgenenetworksoperatingintheblastodermofDrosophila.In[1],theconcentrationsofmRNAandproteinaremodeledbylineardifferentialequations.Asimpleformof
linearadditivefunctionsissuggestedby[2],wheredxi/dt=
n
j=1wijx
j.Thedegradationrateofgenei’smRNAand
environmentaleffectsareassumedtobeincorporatedintheparameterswijandtheirinfluenceongenei’sexpressionlevel
xiisassumedtobelinear.Amethodtoobtainacontinuous
lineardifferentialequationmodelfromsampledtime-seriesdataisproposedin[7].Foraddedbiologicalrealism(allconcentrationsgetsaturatedatsomepointintime),asigmoid(squashing)functionmaybeincludedintotheequation.Ithasbeenshownthatthissortofquasi-linearmodelcanbesolvedbyfirstapplyingtheinverseofthesquashingfunction[3].Inourstudy,aGRNismodeledbycontinuousnonlinearOrdinaryDifferentialEquations(ODEs).Comparedtolinearmodels,identificationofthenonlineardifferentialequationmodeliscomputationallymoreintensiveandcanrequiremoredata;however,therangeofnonlinearbehaviorsex-hibitedbyGRNscanbemorethoroughlyunderstoodwithnonlineardifferentialequations.Inaddition,wellestablisheddynamicalsystemstheoryisavailabletocharacterizethedynamicsproducedbythesemodels.Whenmoretime-seriesdatabecomeavailableowingtoadvancesinmicroarrayorothertechnologies,andassumingcontinuedimprovementincomputationalcapability,itcanbeexpectedthatcontinuousnonlineardynamicmodelswillplayacriticalroleinrevealingcomplicatedgenebehavior.Ingeneral,modelinggeneregulatorynetworksisanonlinearidentificationproblem.AssumingthereareNgenesofinterestandxidenotesthestate(suchasthemicroarrayreading)of
theithgene,thenthedynamicsoftheGRNmaybemodeled
as
dxi
dt=
211-4244-1198-X/07/$25.00 ©2007 IEEESSP 2007
