Translating the Histone Code
心境障碍的表观遗传学研究进展

心境障碍的表观遗传学研究进展李磊张志珺张向荣表观遗传学(epigenetics)主要研究不涉及DNA序列突变的可逆性、可遗传性基因功能调控机制及其对疾病发生的影响[1]。
不断积累的证据显示,表观遗传学机制可能在心境障碍的发病及治疗过程中起着重要作用,已成为近年来心境障碍研究中颇受关注的热点领域。
一、表观遗传学概述基因组表观遗传学常见机制包括DNA甲基化、组蛋白修饰、染色体重塑、基因组印迹、X染色体失活等,它们动态可逆地控制基因表达的位点、时间以及表达水平,从而精确调控基因组功能,目前研究主要集中于DNA甲基化及组蛋白修饰。
DNA甲基化是在DNA甲基化转移酶(DNA methyltransferase, DNMTs)的作用下将S-腺苷甲硫氨酸(S-adenosyl-L-methionine, SAM)的1个甲基添加到DNA分子上,与胞嘧啶-鸟嘌呤(CpG)二核苷酸中胞嘧啶的第5位碳原子共价结合形成5甲基胞嘧啶。
哺乳动物DNA甲基化在基因启动子区CpG岛较为密集,通常认为DNA甲基化与基因沉默有关。
需要注意的是,DNA甲基化可能不是基因沉默的原因,而是结果。
组蛋白是构成染色质的关键性结构,组蛋白修饰的主要形式有组蛋白乙酰化和甲基化,可以通过改变染色质缠绕的致密程度,影响转录调控因子的可接近性,进而导致基因表达水平变化。
组蛋白乙酰化修饰与DNA甲基化过程密切相关,甲基胞嘧啶结合蛋白(methyl CpG binding protein 2, MeGP2)与甲基化的DNA结合后,可以募集组蛋白去乙酰化酶(histone deacetylases, HDACs),诱导组蛋白去乙酰化,染色质去乙酰化水平的提高通常抑制基因转录,而乙酰化增加则表示转录活性的增强。
组蛋白甲基化修饰是由组蛋白转甲基酶(histone methyltransferases, HMTs)介导的。
发生在组蛋白H3和H4的赖氨酸和精氨酸残基上的可逆性甲基化过程。
表观遗传与肝癌关系

表观遗传与肝癌关系摘要:近年来,表观遗传学成为了研究热点。
表观遗传学的调控涉及多方面的领域,比如衰老、先天性遗传、癌症、多因子疾病、个体差异多样性、种群分化和进化等。
本文仅就表观遗传学与肝癌的关系进行阐述,探讨在DNA甲基化、组蛋白修饰、非编码RNA和基因组印记等方面的研究成果,为揭示开发肝癌新的诊治手段提供了理论依据。
关键词:表观遗传肝癌一.肝癌肝细胞癌(HCC)在我国发病率很高,其死亡率在消化系统恶性肿瘤中居第2名,据流行病学统计,HCC发病率在城市居民中仅次于肺癌,农村仅次于胃癌[1]。
原发性肝癌的形成是由于正常肝细胞在多种致癌因素影响下,因遗传学和表观遗传学改变的累积效应而导致。
近年来,越来越多的证据表明,表观遗传学改变在肿瘤进展中扮演十分重要的角色。
二.表观遗传学指DNA序列不发生变化但基因表达却发生了可遗传的改变,也就是说基因型未变化而表型却发生了改变,这种变化是细胞内除了遗传信息以外的其他可遗传物质的改变,并且这种改变在发育和细胞增殖中能稳定遗传下去[2]。
表观遗传可通过DNA甲基化、组蛋白修饰、非编码RNA以及基因组印记等方式来实现对基因表达的调控[3]。
表观遗传学主要研究与DNA序列变异无关的基因表达,可遗传性现象的本质、功能、形成机制及其在疾病发生和发展过程中的作用。
三.表观遗传分子与肝癌的关系(一)DNA甲基化DNA甲基化是DNA化学修饰的一种形式,能够在不改变DNA序列的前提下,改变遗传表现。
DNA甲基化的主要机制是由甲基转移酶(DNMTs)催化S-腺苷甲硫氨酸作为甲基供体,DNA的CG两个核苷酸的胞嘧啶被选择性的添加甲基,形成5-甲基胞嘧啶,这常见于基因的5‘-CG-3序列。
甲基化位点可随DNA的复制而遗传,因为DNA复制后,甲基化转移酶可将新合成的未甲基化的位点进行甲基化。
已知DNMTs在哺乳动物中有活性的有四种,其中DNMT3A、DNMT3B、DNMT3L可甲基化CpG岛,使其半甲基化,继而全甲基化,并参与细胞生长和分化调控。
药物分析专业英语

(dissolution) vessel 溶出杯(FTIR) 傅里叶变换红外光谱仪13C-NMR spectrum,13CNMR 碳-13核磁共振谱1ength basis 长度基准1H-NMR 氢谱2D-NMR 二维核磁共振谱:2D-NMR3D-spectrochromatogram 三维光谱-波谱图Aa stream of nitrogen 氮气流a wide temperature range 宽的温度范围absolute detector response 检测器绝对响应(值)absolute entropy 绝对熵absolute error 绝对误差absolute reaction rate theory 绝对反应速率理论absolute temperature scale 绝对温标absorbance 吸光度,而不是吸收率(absorptance)。
当我们忽略反射光强时,透射率(T)与吸光度(A)满足如下关系式:A=lg(1/T)。
absorbance noise, absorbing noise 吸光度噪音。
也称光谱的稳定性,是指在确定的波长范围内对样品进行多次扫描,得到光谱的均方差。
吸光度噪音是体现仪器稳定性的重要指标。
将样品信号强度与吸光度噪音相比可计算出信噪比。
absorbed water 吸附水absorptance 吸收率absorptant 吸收剂absorption band 吸收带absorption cell 吸收池absorption curve 吸收光谱曲线/光吸收曲线absorption tube 吸收管abundance 丰度。
即具有某质荷比离子的数量accelerated solvent extraction(ASE) 加速溶剂萃取accelerated testing 加速试验accelerating decomposition 加速破坏acceptance limit,acceptance criterion 验收限度,合格标准accidental error 随机误差accuracy 准确度。
生物专业英语第一章

20. mega Megaspore Megabasse Megakaryocyte Megavolt Megalopolitan 21. macro macrophage macrogamete macroelement macromolecular
巨大,兆,百万 大孢子, 兆碱基 巨核细胞 兆伏 特大城市 大,巨大,多 巨噬细胞 大配子 常量元素 大分子
细胞中的亚器官
• Chloroplast['klɔ:rəuplæst] 叶绿体 A plastid in which photosynthesis is carried out. Chloroplasts occur in all photosynthetic organisms except photosynthetic bacteria and blue-green algae.
17. Nano nanosecond nanometer 18. demi,hemi,semi demibariel hemicerebrum semiopaque semi-allel 19. holo holoenzyme holoprotein holocrine
十亿分之一,毫微,纳 十亿分之一秒 纳米 半 半桶 大脑半球 半透明 半等位基因 全,整体,完全 全酶 全蛋白 全(质分)泌
10.-ic 加在外来词根的名词上,构成形容词specific特异 的,magnetic磁性的,aerobic需氧的,pubic耻骨的, oxytocic催产的,催产剂,therapeutic治疗的, dramatic戏剧性的 11.-ish 加在颜色的形容词上,表示略带...色reddish带 红色的,微红的,yellowish带黄色的, 12.-ive 由动词构成形容词relative有关的,相关的 congestive充血性 13.-less 表示没有...的useless无用的,lifeless无生命 的,hopeless绝望的,医治不好的,fruitless无 效的,无益的.
生物学中的基因调控机制

生物学中的基因调控机制引言:在生物学领域中,研究基因调控机制是了解生命本质和进化的关键。
基因调控是指通过一系列的分子机制,控制基因表达的过程。
这些机制可以使细胞在不同的环境条件下适应和响应,从而实现生物体的正常生长和发育。
本文将深入探讨生物学中的基因调控机制,包括转录调控、转录后调控和表观遗传调控。
一、转录调控转录调控是指通过调控基因转录的过程来控制基因表达。
这一过程涉及到转录因子、启动子和增强子等多个分子元件的相互作用。
转录因子是一类蛋白质,可以结合到基因的启动子或增强子上,通过与DNA相互作用,激活或抑制基因的转录。
启动子是位于基因上游的DNA序列,能够吸引转录因子与RNA聚合酶结合,启动基因转录。
增强子是位于基因上游或下游的DNA序列,可以增强或减弱基因的转录活性。
这些元件之间的相互作用形成了复杂的转录调控网络。
二、转录后调控转录后调控是指在基因转录完成后,通过调控mRNA的稳定性、剪接和翻译等过程来控制基因表达。
mRNA的稳定性是指mRNA分子在细胞内的寿命,通过RNA降解酶的作用可以调控mRNA的稳定性。
剪接是指在mRNA合成过程中,通过剪接酶的作用,将mRNA前体分子中的内含子剪切掉,形成成熟的mRNA分子。
翻译是指mRNA转化为蛋白质的过程,通过调控翻译起始子、翻译终止子和翻译调控因子等分子机制,可以控制蛋白质的合成速率和翻译后修饰。
三、表观遗传调控表观遗传调控是指通过改变染色体结构和化学修饰,来调控基因表达的过程。
这些化学修饰包括DNA甲基化、组蛋白修饰和非编码RNA等。
DNA甲基化是指在DNA分子上加上甲基基团,通过甲基化酶的作用,可以使基因沉默或激活。
组蛋白修饰是指组蛋白蛋白质上的化学修饰,包括乙酰化、甲基化和磷酸化等,通过这些修饰可以改变染色体的结构和可及性,从而调控基因的表达。
非编码RNA是指不具有编码蛋白质的功能,但可以通过与DNA和RNA相互作用,调控基因表达的RNA分子。
translation 翻译过程.ppt
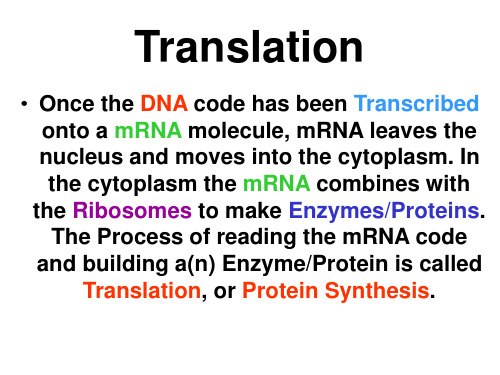
Summary of Translation
(or… so you want to make a Protein)
Let’s begin in the nucleus…… The DNA has the code to instruct the Ribosomes as to which Proteins to make. Ok, 2 problems here: 1.) the DNA
Now …… let’s say you are a ribosome….. you are just sitting around, in the cytoplasm, waiting for some amino acids to assemble into protein….you already have the instructions, remember the
The mRNA is made in the Nucleus through the process
of Transcription. mRNA is made up of RNA nucleotides. Each group of 3 nucleotides is called a Codon and codes for one amino acid of the newly
• tRNA is a small, folded RNA chain (74-93 nucleotides) that picks up and transfers specific amino acids to a growing polypeptide chain at the ribosomal site of protein synthesis during translation.
蛋白质的生物合成省名师优质课赛课获奖课件市赛课一等奖课件
Section 2 The Proce需要处理旳问题: 1. 原核生物和真核生物中,多肽链旳生物合成
涉及哪些主要旳环节? 2. 什么是核糖体循环?核糖体循环主要由哪些
反应过程所构成? 3. 多肽链合成时,延长一种氨基酸残基需要消
耗多少分子ATP?
factor,EF)。
EF-Tu bound with ribosome
原核生物中存在3种延长因子(EF-TU,EF-TS, EF-G),真核生物中存在2种(EF1,EF2)。 EF旳作用主要是促使氨基酰tRNA进入核蛋白旳 受位;并可增进移位过程,即具有转位酶活性, 可催化核糖体向mRNA 3’-端移动一种密码子旳 距离,使下一种密码子定位于A位。
摆动配对现象示意图
U
二、核糖体是多肽链生物合成旳场合
Ribosome is the place for peptide biosynthesis
核糖体(又称核蛋白体),是多肽链合成旳场合, 是由多种rRNA与蛋白质组装形成旳复合体。
核糖体旳构成
大肠杆菌核糖体旳空间构 造为一椭圆球体,其30S 亚基呈哑铃状,50S亚基 带有三角,中间凹陷形成 空穴,将30S小亚基抱住, 两亚基旳结合面为多肽链 生物合成旳场合。
(一)原核生物多肽链合成旳起始
涉及下列几种环节: ➢ 核糖体大、小亚基分离; ➢ mRNA在小亚基定位结合; ➢ 起始氨基酰-tRNA旳结合; ➢ 核糖体大亚基结合。
5. 摆动性(wobble): 转运氨基酸旳tRNA旳反密码需要经过碱基互 补与mRNA上旳遗传密码反平行配对结合。 但反密码与密码之间经常不严格遵守碱基配 对规律,称为摆动配对。
密码子与反密码子旳摆动配对
tRNA反密码子 第1位碱基
Histone Modification
K
Me
Me
K
Me Me Me
K
Histone Methylation Unlike acetylation, the consequence of methylation can be either positive or negative toward transcriptional expression, depending on the position of the residue within the histone.
Epigenetics: The study of reversible heritable changes in gene function that occur without a change in the sequence of nuclear DNA
Genetic code
What is the code in Epigenetics?
Type 1 and Type 2 have a related mechanism of deacetylation, which does not involve a cofactor, whereas the Sir2-related enzymes required the cofactor NAD as part of their catalytic mechanism.
+ 40 bp of DNA
+ 1 (H2A-H2B) dimer
+ 40 bp of DNA Folded Nucleosome
Nucleosome
The nucleosome is a complex between basic histone proteins and DNA. The histone proteins form an octamer composed of two subunits of each of the histones H2A, H2B, H3 and H4. DNA is wrapped around the octamer surface in helical turns (147bp).
Transliteration—音译
• ②Prefix+ Unit of measurement (由前缀加入计量单位构成复 合词,计量单位采用音译) • megavolt 百万伏特 microampere 微安培 • kilowatt 千瓦 decibel分贝
• ③Some compound words (某些复合词意音结合译) radar-man雷达手 valve-guide阀导 • ④Some terms containing people’s names :people’s names(sound)+the rest of the words (meaning) (有些由人名构 成的术语,人名音译,其余部分意译) Ohm law欧姆定律 Curie point居里点 Morse code莫尔斯电码 Monel metal蒙乃尔合金
George Bush 乔治.布什
Albert Einstein 阿尔伯特.爱因斯坦
Transcription table P326
Angela ???
Canada 加拿大
Australia 澳大利亚
The Alps 阿尔卑斯山
The Mississippi 密西西比河
France 法郎
Pound 镑
Classifications
Complete transliteration (纯音译) Combine sound and meaning (意音结合译)
Complete transliteration (纯音译)
1.words about unit of measurement (计量单位)
celluloid赛璐珞(硝纤象牙) sonar声纳(声波导航和测距设备) morphine吗啡
新标准大学英语 综合教程(外语教学与研究出版社)Unit2 重点单词
Unit2 Food,glorious food!Frown[fraun] 词形变化: frowningly frowner frowned frowned frowning frowns∙n. 皱眉,不悦v. 不同意,皱眉头∙vi.(因烦恼、焦虑或沉思而)皱眉、蹙额∙frown on 表示不满,不赞许,皱眉frown at 朝……皱眉头,对……表示不满,不赞许∙frown upon不赞成,不以为然frown down用皱眉蹙额压制住nasty['nɑ:sti]词形变化: nastily nastier nastiest nastiness nasties∙adj. (味道、气味、样子或感觉)令人作呕的,令人厌恶的;下流的,严重的,令人不快的,难懂的,危害的∙ a nasty piece of work n. 阴谋,下流的家伙cheap and nasty adj. 价廉物劣(中看不中用,金玉其外败絮其中)∙ a nasty one n. 责骂,使人一蹶不振的打击something nasty in the woodshed n.令人不愉快的经历∙video nasty n. 恐怖录像片nasty-nice adj. 笑里藏刀的∙leave a nasty taste in the mouth留下讨厌的气味Jack nasty鬼鬼祟祟的人∙have a nasty spill [口](被)摔得很重(指从马背或自行车上摔下)leavea nasty taste in someone’s mouth给(某人)留下坏印象∙sling a nasty foot跳舞跳得到家了nasty piece of work n. 令人讨厌的人,令人难以忍受的人∙nasty taste in the mouth不愉快的感觉Things look nasty.事态险恶,事情不妙,有恶化之势∙ a nasty proposition难对付的人a nasty piece of goods讨厌的家伙,卑鄙的人∙ a nasty quarter of an hour不愉快的短暂时刻cut up nasty [口]发怒,冒火;露出凶相;找人吵架∙ a nasty bit of goods讨厌的家伙,卑鄙的人a nasty bit of work讨厌的家伙。
- 1、下载文档前请自行甄别文档内容的完整性,平台不提供额外的编辑、内容补充、找答案等附加服务。
- 2、"仅部分预览"的文档,不可在线预览部分如存在完整性等问题,可反馈申请退款(可完整预览的文档不适用该条件!)。
- 3、如文档侵犯您的权益,请联系客服反馈,我们会尽快为您处理(人工客服工作时间:9:00-18:30)。
69.Y.Habu,T.Kakutani,J.Paszkowski,Curr.Opin.Genet.Dev.11,215(2001).70.M.Wassenegger,Plant Mol.Biol.43,203(2000).71.M.A.Matzke,A.J.Matzke,J.M.Kooter,Science293,1080(2001).72.J.Bender,Trends Biochem.Sci.23,252(1998).73.E.U.Selker,Cell97,157(1999).74.M.N.Raizada,M.I.Benito,V.Walbot,Plant J.25,79(2001).75.R.F.Ketting,T.H.Haverkamp,H.G.van Luenen,R.H.Plasterk,Cell99,133(1999).76.H.Tabara et al.,Cell99,123(1999).77.B.H.Ramsahoye et al.,Proc.Natl.Acad.Sci.U.S.A.97,5237(2000).78.P.Svoboda,P.Stein,H.Hayashi,R.M.Schultz,De-velopment127,4147(2000).79.C.Cogoni et al.,EMBO J.15,3153(1996).80.G.Faugeron,Curr.Opin.Microbiol.3,144(2000).81.L.Jackson-Grusby et al.,Nature Genet.27,31(2001).82.J.P.Vielle-Calzada,R.Baskar,U.Grossniklaus,Nature404,91(2000).83.P.S.Springer,D.R.Holding,A.Groover,C.Yordan,R.A.Martienssen,Development127,1815(2000).84.J.P.Vielle-Calzada et al.,Genes Dev.13,2971(1999).85.R.Vinkenoog et al.,Plant Cell12,2271(2000).86.S.Adams,R.Vinkenoog,M.Spielman,H.G.Dickinson,R.J.Scott,Development127,2493(2000).87.M.Byrne,M.Timmermans,C.Kidner,R.Martienssen,Curr.Opin.Plant Biol.4,38(2001).88.E.B.Cambareri,R.Aisner,J.Carbon,Mol.Cell.Biol.18,5465(1998).89.P.SanMiguel et al.,Science274,765(1996).90.R.Mauricio,Nature Rev.Genet.2,370(2001).91.P.Cubas,C.Vincent,E.Coen,Nature401,157(1999).92.R.Martienssen,Curr.Opin.Genet.Dev.8,240(1998).93.Z.J.Chen,C.S.Pikaard,Genes Dev.11,2124(1997).ai et al.,Plant Cell12,1551(2000).95.H.S.Lee,Z.J.Chen,Proc.Natl.Acad.Sci.U.S.A.98,6753(2001).96.We thank E.Selker,E.Richards,V.Chandler,S.Kaep-pler,S.Jacobsen,and J.Bender for communicatingresults prior to publication,two anonymous refereesfor suggestions for improvement,and our colleaguesfor many interesting discussions.R.M.and V.C.re-ceive grant support from the NSF(DBI1057338).R E V I E WTranslating the Histone CodeThomas Jenuwein1and C.David Allis2Chromatin,the physiological template of all eukaryotic genetic information,issubject to a diverse array of posttranslational modifications that largelyimpinge on histone amino termini,thereby regulating access to the underly-ing DNA.Distinct histone amino-terminal modifications can generate syner-gistic or antagonistic interaction affinities for chromatin-associated proteins,which in turn dictate dynamic transitions between transcriptionally active ortranscriptionally silent chromatin states.The combinatorial nature of histoneamino-terminal modifications thus reveals a“histone code”that considerablyextends the information potential of the genetic code.We propose that thisepigenetic marking system represents a fundamental regulatory mechanismthat has an impact on most,if not all,chromatin-templated processes,withfar-reaching consequences for cell fate decisions and both normal and patho-logical development.Genomic DNA is the ultimate template of our heredity.Yet despite the justifiable excitement over the human genome,many challenges re-main in understanding the regulation and trans-duction of genetic information(1).It is unclear, for example,why the number of protein-coding genes in humans,now estimated atϳ35,000, only doubles that of the fruit fly Drosophila melanogaster.Is DNA alone then responsible for generating the full range of information that ultimately results in a complex eukaryotic or-ganism,such as ourselves?We favor the view that epigenetics,im-posed at the level of DNA-packaging proteins (histones),is a critical feature of a genome-wide mechanism of information storage and retrieval that is only beginning to be under-stood.We propose that a“histone code”ex-ists that may considerably extend the infor-mation potential of the genetic(DNA)code. We review emerging evidence that histone proteins and their associated covalent modi-fications contribute to a mechanism that can alter chromatin structure,thereby leading to inherited differences in transcriptional“on-off”states or to the stable propagation of chromosomes by defining a specialized high-er order structure at centromeres.Under the assumption that a histone code exists,at least in some form,we discuss potential mecha-nisms for how such a code is“read”andtranslated into biological functions.Throughout this review,we have chosenepigenetic phenomena and underlying mecha-nisms in two general categories:chromatin-based events leading to either gene activation orgene silencing.In particular,we center our dis-cussion on examples where differences in“on-off”transcriptional states are reflected by dif-ferences in histone modifications that are either“euchromatic”(on)or“heterochromatic”(off)(Fig.1A).We also point out that,despite manyelegant genetic and biochemical insights intochromatin function and gene regulation in thebudding yeast Saccharomyces cerevisiae,someof the heterochromatic mechanisms(e.g.,HP1-based gene silencing)discussed here do notexist in an obvious form in this organism.Thus,we will need to pursue other model systems,such as Schizosaccharomyces pombe,Caeno-rhabditis elegans,Drosophila,and mice,to“crack”the histone code.Chromatin Template and HistoneCodeIn the nuclei of all eukaryotic cells,genomicDNA is highly folded,constrained,and com-pacted by histone and nonhistone proteins ina dynamic polymer called chromatin.Forexample,chromosomal regions that remaintranscriptionally inert are highly condensedin the interphase nucleus and remain cytolog-ically visible as heterochromatic foci or as the“Barr body,”which is the inactive X chromo-some in female mammalian cells(2).Thedistinct levels of chromatin organization aredependent on the dynamic higher order struc-turing of nucleosomes,which represent thebasic repeating unit of chromatin.In eachnucleosome,roughly two superhelical turnsof DNA wrap around an octamer of corehistone proteins formed by four histone part-ners:an H3-H4tetramer and two H2A-H2Bdimers(3).Histones are small basic proteinsconsisting of a globular domain and a moreflexible and charged NH2-terminus(histone“tail”)that protrudes from the nucleosome.Itremains unclear how nucleosomal arrays con-taining linker histone(H1)then twist and foldthis chromatin fiber into increasingly morecompacted filaments leading to defined high-er order structures.Central to our current thinking is thatchromatin structure plays an important regu-latory role and that multiple signaling path-ways converge on histones(4).Althoughhistone proteins themselves come in genericor specialized forms(5),exquisite variation isprovided by covalent modifications(acetyla-tion,phosphorylation,methylation)of the hi-stone tail domains,which allow regulatablecontacts with the underlying DNA.The en-zymes transducing these histone tail modifi-cations are highly specific for particular ami-no acid positions(6,7),thereby extendingthe information content of the genome pastthe genetic(DNA)code.This hypothesis pre-dicts that(i)distinct modifications of the1Research Institute of Molecular Pathology(IMP)atthe Vienna Biocenter,Dr.Bohrgasse7,A-1030Vi-enna,Austria.E-mail:jenuwein@nt.imp.univie.ac.at2Department of Biochemistry and Molecular Genetics,University of Virginia Health Science Center,Char-lottesville,VA22908,USA.E-mail:allis@Among the modifier genes identified in the above model systems,one subclass suppresses variegation[the Su(var)group]and comprises gene products such as histone deacetylases (HDACs),protein phosphatases(PPTases),and S-adenosylmethionine(SAM)synthetase(17), as well as chromatin-associated components that are best characterized by the heterochroma-tin protein HP1[Su(var)2-5](18).In addition to the Su(var)group of genes,an antagonizing class of PEV modifiers enhances variegation [E(var)group](12)and counteracts the Su(var)-induced silent chromatin state.Several E(var) gene products are components of adenosine triphosphate(ATP)–dependent nucleosome-re-modeling machines,such as the SWI/SNF and brahma complexes(19,20),which increase overall nucleosome mobility.Extending these parallels even further, Su(var)and E(var)gene products contain several conserved protein domains—the bro-mo-,chromo-,and SET domains—that are also shared with two other classes of antag-onizing chromatin regulators:the Polycomb (Pc-G)and trithorax(trx-G)groups.The Pc-G and trx-G genes are important for main-taining the expression boundaries of the ho-meotic selector genes and several other key developmental genes(21,22),presumably by modulating the chromatin structure of their target loci.The bromodomain(23)is found inSNF2,TAFII 250,and mammalian trithorax(HRX/Mll);the chromodomain(24,25)is shared between Polycomb and HP1;and the SET domain(26)is found in Su(var)3-9,in the Pc-G member E(z),and in trithorax. These modules have been widely used during evolution to generate a considerable function-al diversity among proteins specialized in modulating chromatin structure.Histone acetylation(27,28)and histone phosphorylation(29)modification systems have been characterized in detail.A further class of enzymatic activities that regulate the site-specific addition of methyl groups to hi-stones has recently been described.Original-ly identified as the PEV modifier Su(var)3-9 in Drosophila,homologs from fission yeast (Clr4)to human(SUV39H1)have been shown to encode histone methyltransferases (HMTases)that selectively methylate histone H3at Lys9(30).The HMTase function in the Su(var)3-9family maps to the highly con-served SET domain but also requires adjacent Cys-rich regions.Notably,generation of the H3-Lys9methyl epitope induces a hetero-chromatic affinity for HP1proteins that rec-ognize this epigenetic signal through their chromodomains(31,32).These results pro-vide a strong link among Su(var)function, gene-silencing activity,and the assembly of heterochromatin(31–35).By contrast,an enzymatic HMTase func-tion has not yet been demonstrated for Pc-G and trx-G proteins.Instead,E(z)has been associated with a Pc-G complex containingHDAC activity(36),and trx or HRX havebeen shown to interact with components ofchromatin-remodeling machines(37).In gen-eral terms,Su(var)and Pc-G gene functionwould be characterized by transducing theaddition of heterochromatic marks and theremoval of euchromatic marks on the chro-matin template.Conversely,the antagonizingactivity of E(var)and trx-G gene functionwould involve the establishment of euchro-matic signals(e.g.,increased nucleosomemobility)and destabilize or degrade(see be-low)heterochromatic“imprints”(Fig.1B).Translating the Histone CodeThe histone code hypothesis predicts that themodification marks on the histone tailsshould provide binding sites for effector pro-teins.In agreement with this notion,the bro-modomain has been the first protein moduleto be shown to selectively interact with acovalent mark(acetylated lysine)in the his-tone NH2-terminal tail(23,38,39).In addi-tion to the proteins discussed above,the bro-modomain is also present in many transcrip-tional regulators having intrinsic histoneacetyltransferase(HAT)activity(e.g.,GCN5,PCAF,TAFII250).Consistent with the sec-ond prediction of the histone code(that therebe combinatorial readout),TAFII250,whichitself harbors several histone-modifying ac-tivities,contains two tandem copies of thebromodomain.In this configuration it pref-erentially binds diacetylated histone pep-tides presenting acetyl-lysine moieties thatare appropriately spaced(40).Use of theSimple Modular Architectural ResearchTool(SMART;http://smart.embl-heidel-berg.de)indicates that there areϳ75bro-modomain-containing proteins in humans.Several of these proteins,such as humanpoly-bromodomain protein1,exhibit manycopies(six)of regularly spaced bromodo-mains,which could conceivably bind to aspecific combination of acetyl groups pre-sented on one or several histone tails.Chromodomains,on the other hand,ap-pear to be targeting modules for methylationmarks.The chromodomain of HP1is highlyselective for methylated H3at Lys9,and littleif any binding is observed with H3peptidescontaining a methylated Lys4position(32).Thus,although chromodomains are highlyconserved,it seems likely that not all chro-modomains—nor their methyl targets—be-have similarly.In support,chromodomainswapping experiments have not uniformlyindicated functional conservation in silencingassays(41,42).Interestingly,Su(var)3-9HMTase family members also contain a chro-modomain,whose integrity is critical for si-lencing in vivo(33,43).Several repressivechromatin-remodeling complexes comprisecomponents such as the Mi-2/CHD ATPasesubunit of the NuRD complex(44),whichharbors two chromodomains and might con-ceivably recognize dimethylated histone tailsin a manner analogous to double bromodo-mains.In this regard,we note that Lys9andLys27in the H3tail are embedded in similarsequence motifs,and both positions are“hotspots”for methylation by the SET domain–containing HMTase G9a(45).Finally,a hallmark property of all HP1proteins is the combination of a chromodo-main with a chromoshadow domain that areseparated by a short but variable hinge re-gion.Because the chromoshadow domain ofHP1appears to self-dimerize in solution(46,47),it is tempting to infer that full-lengthHP1may assemble intermolecular chromo-domains,thereby generating a bifunctionalcross-linker that is likely to stabilize the morerigid higher order structure of heterochroma-tin(35,48).Combinations and SwitchesThe above examples provide support for mod-ification-induced recruitment of chromatin-as-sociated proteins to acetylated and methylatedhistone NH2-termini(Fig.2A),and it is likelythat other modules exist that specifically recog-nize phosphorylation marks.Consistent withthe second prediction of the histone code hy-pothesis,all four NH2-termini of the core his-tones contain short“basic patches”that oftencomprise acetylation,phosphorylation,andmethylation marks in close proximity on oneindividual tail(4).All three of these modifica-tions can be found both in active or silencedchromatin regions,which raises the question ofhow combinatorial specificity is used in defin-ing an imprint for euchromatin or heterochro-matin(Fig.1,A and C).Some evidence is emerging about a pos-sible combinatorial code.For example,thehistone H3NH2-terminus appears to exist intwo distinct modification states that are likelyto be regulated by a“switch”between Lys9methylation and Ser10phosphorylation(Fig.1D).Ser10phosphorylation inhibits Lys9methylation(30)but is synergistically cou-pled with Lys9and/or Lys14acetylation dur-ing mitogenic and hormonal stimulation inmammalian cells(49–51).In this phos-phorylated-acetylated state,the modified H3tail marks transcriptional activation(Fig.1C).H3phosphorylation is also important for mi-totic chromosome condensation(52),whereit may be linked to other secondary signal(s)such as the nucleosomal incorporation of thepericentric H3analog Cenp-A(53).Con-versely,aberrant Lys9methylation antagoniz-es Ser10phosphorylation,leading to mitoticchromosome dysfunction(30,54).Further,deacetylation of Lys14in H3(33)is requiredto facilitate subsequent Lys9methylation bythe Clr4HMTase,again highlighting an or-dered interplay to establish distinct histoneImmortal ChromatinThe importance of chromatin in the informa-tion storage and decoding processes of the eukaryotic genome is reinforced by the growth in our knowledge about covalent modifications of histone proteins,and about the enzyme systems that transduce or remove these imprints.Moreover,histone modifica-tions may also be a“sensor”of the metabolic state of the cell.For example,the Sir2en-zyme uses an essential metabolic cofactor (nicotinamide adenine dinucleotide)to regu-late the activity of a family of silencing-associated HDACs(84).Will HDMases be uncovered only when the correct cofactor, itself possibly a direct product from interme-diary carbon metabolism,is added to the test reactions?The lessons learned from the Sir2 paradigm lead to an attractive new concept: Because chromatin is the physiological tem-plate of eukaryotic cells,are genomes pro-grammed to“open”and“close”on demand by enzyme complexes that evolved to re-spond directly to metabolic cues?If correct, we anticipate that further insights will be gained as we systematically investigate chro-matin changes during different physiological or pathological states.To what extent does a histone code link directly to our genetic code,or are these codes separate indexing mechanisms?Will we find evidence of interdependence between histone methylation and DNA methylation, similar to the interplay between histone deacetylation and DNA methylation(44)? Intriguingly,a“chromo-methylase”has re-cently been described in Arabidopsis that combines a chromodomain with a DNA methylating activity(85),and one member of the SET domain family contains a methyl CpG binding motif(35)(Fig.2C).Histone methylation may also help to explain poorly understood chromatin effects where deacety-lase inhibitors and/or5-aza-cytosine fail to cause reversal of previously silent genomic regions(86).Indeed,transcription of many genes is regulated by histone acetylation in organisms(e.g.,in yeast and flies)that exhib-it little DNA modification.Further,X chro-mosome inactivation in mammals correlates with hypoacetylation of histones,except for a few X-linked loci that escape this silencing mechanism(87).In addition,in some branch-es of mammalian evolution(e.g.,marsupials), no allele-specific DNA methylation has been observed.Could histone methylation be one of the conserved mechanisms substituting for the apparent absence of DNA methylation in these organisms,and to what extent is the inactive X chromosome hypoacetylated(88) because it may be hypermethylated at distincthistone NH2-termini?How far will epigenetics go past transcrip-tional effects?Emerging evidence indicates that programmed DNA rearrangements(89),imprinting phenomena(90),germ line si-lencing(57),developmentally cued stemcell divisions(91),and overall chromo-some stability and identity(52,92)are allinfluenced by epigenetic alterations of theunderlying chromatin structure.In keepingwith the distinct qualities of accessible andinaccessible nucleosomal states,could it bethat“open”(euchromatic)chromatin repre-sents the underlying principle that is syn-onymous for the character of progenitor,immortal,and young cells?Conversely,is“closed”(heterochromatic)chromatin thereflection of a developmental“memory”that stabilizes lineage commitment andgradually restricts the self-renewal poten-tial of our somatic cells?As pointed out byothers(93),epigenetics imparts a funda-mental regulatory system beyond the se-quence information of our genetic code andemphasizes that“Mendel’s gene is morethan just a DNA moiety.”References and Notes1.D.Baltimore,Nature409,814(2001).2.S.W.Brown,Science151,417(1966).3.K.Luger,A.W.Mader,R.K.Richmond,D.F.Sargent,T.J.Richmond,Nature389,251(1997).4.P.Cheung,C.D.Allis,P.Sassone-Corsi,Cell103,263(2000).5.A.P.Wolffe,D.Pruss,Trends Genet.12,58(1996).6.B.D.Strahl,C.D.Allis,Nature403,41(2000).7.B.M.Turner,Bioessays22,836(2000).8.R.Paro,Trends Genet.11,295(1995).9.Y.Dou,M.A.Gorovsky,Mol.Cell6,255(2000).10.A.P.Wolffe,D.Guschin,J.Struct.Biol.129,102(2000).11.H.J.Muller,Proc.Int.Congr.Genet.1,213(1932).12.G.Reuter,P.Spierer,Bioessays14,605(1992).13.G.Thon,A.J.S.Klar,Genetics131,287(1992).14.R.C.Allshire,J.-P.Javerzat,N.J.Readhead,G.Cran-ston,Cell76,157(1994).15.L.Pillus,M.Grunstein,in Chromatin Structure andGene Expression,S.C.R.Elgin,Ed.(IRL Press,NewYork,1995),pp.123–146.16.S.I.S.Grewal,A.J.S.Klar,Cell86,95(1996).17.L.L.Wallrath,Curr.Opin.Genet.Dev.8,147(1998).18.J.C.Eissenberg et al.,Proc.Natl.Acad.Sci.U.S.A.87,9923(1990).19.T.Tsukinama,C.Wu,Curr.Opin.Genet.Dev.7,182(1997).20.A.J.Kal,T.Mahmoudi,N.B.Zak,C.P.Verrijzer,GenesDev.14,1058(2000).21.R.Paro,P.Harte,in Epigenetic Mechanisms of GeneRegulation,V.E.A.Russo,R.A.Martienssen,A.D.Riggs,Eds.(Cold Spring Harbor Laboratory Press,ColdSpring Harbor,NY,1996),pp.507–528.22.M.van Lohuizen,Curr.Opin.Genet.Dev.9,355(1999).23.C.Dhalluin et al.,Nature399,491(1999).24.R.Paro,D.S.Hogness,Proc.Natl.Acad.Sci.U.S.A.88,263(1991).25.D.A.Jones,F.G.Cowell,P.B.Singh,Bioessays22,124(2000).26.B.Tschiersch et al.,EMBO J.13,3822(1994).27.S.Y.Roth,J.M.Denu,C.D.Allis,Annu.Rev.Biochem.70,81(2001).28.J.A.Johnson,B.M.Turner,Semin.Cell Dev.Biol.10,179(1999).29.J.-Y.Hsu et al.,Cell102,279(2000).30.S.Rea et al.,Nature406,593(2000).chner,D.O’Carroll,S.Rea,K.Mechtler,T.Jenu-wein,Nature410,116(2001).32.A.J.Bannister et al.,Nature410,120(2001).33.J.-I.Nakayama,J.C.Rice,B.D.Strahl,C.D.Allis,S.I.S.Grewal,Science292,110(2001);published online15March2001(10.1126/science.1060118).34.J.C.Rice,C.D.Allis,Curr.Opin.Cell Biol.13,263(2001).35.T.Jenuwein,Trends Cell Biol.11,266(2001).36.J.van der Vlag,A.P.Otte,Nature Genet.23,474(1999).37.O.Rozenblatt-Rosen et al.,Proc.Natl.Acad.Sci.U.S.A.95,4152(1998).38.F.Winston,C.D.Allis,Nature Struct.Biol.6,601(1999).39.D.J.Owen et al.,EMBO J.19,6141(2000).40.R.H.Jacobson,durner,D.S.King,R.Tjian,Science288,1422(2000).41.J.S.Platero,T.Harnett,J.C.Eissenberg,EMBO J.14,3977(1995).42.G.Wang et al.,Mol.Cell.Biol.20,6970(2000).43.A.V.Ivanova,M.J.Bonaduce,S.V.Ivanov,A.J.S.Klar,Nature Genet.19,192(1998).44.J.Ahringer,Trends Genet.16,351(2000).45.M.Tachibana,K.Sugimoto,T.Fukushima,Y.Shinkai,J.Biol.Chem.276,25309(2001).46.S.V.Brasher et al.,EMBO J.19,1587(2000).47.N.P.Cowieson,J.F.Partridge,R.C.Allshire,P.J.McLaughlin,Curr.Biol.10,517(2000).48.J.C.Eissenberg,S.C.R.Elgin,Curr.Opin.Genet.Dev.10,204(2000).49.P.Cheung et al.,Mol.Cell5,905(2000).50.W.S.Lo et al.,Mol.Cell5,917(2000).51.A.L.Clayton,S.Rose,M.J.Barratt,L.C.Mahadevan,EMBO J.19,3714(2000).52.Y.Wei,L.Yu,J.Bowen,M.A.Gorovsky,C.D.Allis,Cell97,99(1999).53.S.G.Zeitlin,C.M.Barber,C.D.Allis,K.E.Sullivan,J.Cell Sci.114,653(2001).54.M.Melcher et al.,Mol.Cell.Biol.20,3728(2000).55.B.M.Turner,A.J.Birley,vender,Cell69,375(1992).56.K.E.van Holde,Chromatin(Springer-Verlag,NewYork,1989).57.M.A.Jedrusik,E.Schulze,Development128,1069(2001).58.R.Holdeman,S.Nehrt,S.Strome,Development125,2457(1998).59.G.Reuter,personal communication.60.K.Stankunas et al.,Development125,4055(1998).61.T.Jenuwein,ible,R.Dorn,G.Reuter,Cell.Mol.Life Sci.54,80(1998).62.F.De Rubertis et al.,Nature384,589(1996).63.B.D.Strahl,R.Ohba,R.G.Cook,C.D.Allis,Proc.Natl.Acad.Sci.U.S.A.96,14967(1999).64.D.Chen et al.,Science284,2174(1999).65.M.R.Stallcup,Oncogene20,3014(2001).66.B.D.Strahl et al.,Curr.Biol,11,996(2001).67.H.Wang et al.,Science293,853(2001).68.S.Allard et al.,EMBO J.18,5108(1999).69.A.Eisen et al.,J.Biol.Chem.276,3484(2001).70.E.R.Smith et al.,Proc.Natl.Acad.Sci.U.S.A.95,3561(1998).71.A.S.Clarke,J.E.Lowell,S.J.Jacobson,L.Pillus,Mol.Cell.Biol.19,2515(1999).72.A.Akhtar,D.Zink,P.B.Becker,Nature407,405(2000).73.J.Taunton,C.A.Hassig,S.L.Schreiber,Science272,408(1996).74.C.D.Allis,J.K.Bowen,G.N.Abraham,C.V.Glover,M.A.Gorovsky,Cell20,55(1980).75.R.Lin,R.G.Cook,C.D.Allis,Genes Dev.5,1601(1991).76.W.M.Baarends et al.,Dev.Biol.207,322(1999).77.V.J.Palombella,O.J.Rando,A.L.Goldberg,T.Ma-niatis,Cell78,773(1994).78.K.Robzyk,J.Recht,M.A.Osley,Science287,501(2000).79.A.-D.Pham,F.Sauer,Science289,2357(2000).80.S.Henchoz,F.De Rubertis,D.Pauli,P.Spierer,Mol.Cell.Biol.16,5717(1996).81.J.Singh,V.Goel,A.J.S.Klar,Mol.Cell.Biol.18,5511(1998).82.D.Moazed,A.Johnson,Cell86,667(1996).83.R.Ballhorn,S.Weston,C.Thomas,A.J.Wyrobek,Exp.Cell Res.150,298(1984).84.S.Imai,C.M.Armstrong,M.Kaeberlein,L.Guarente,Nature403,795(2000).85.A.M.Lindroth et al.,Science292,2077(2001);published online10May2001(10.1126/sci-ence.1059745).86.A.El-Osta,A.P.Wolffe,Gene Expr.9,63(2000).87.C.M.Disteche,Trends Genet.11,17(1995).88.P.Jeppesen,B.M.Turner,Cell74,281(1993).89.D.R.Roth,S.Y.Roth,Cell103,699(2000).90.R.I.Gregory et al.,Mol.Cell.Biol,21,5426(2001).91.G.Cavalli,R.Paro,Science286,955(1999).92.A.H. F.M.Peters, D.O’Carroll,T.Jenuwein,unpublished data.93.A.S.J.Klar,Trends Genet.14,299(1998).94.We thank G.Reuter and members of our laborato-ries for allowing us to cite unpublished observa-tions.We are particularly grateful to S.Rea for hisassistance in preparing thefigures.Supported bythe IMP through Boehringer Ingelheim and bygrants from the Austrian Research Promotion Fundand the Vienna Economy Promotion Fund(T.J.),and by NIH grant GM53512and an NIH MERITaward(C.D.A.).This article is dedicated to the memory of AlanWolffe,an inspirational leader to all of us whohave pondered the mysteries of chromatin andgene regulation.V I E W P O I N TRNA:Guiding Gene SilencingMarjori Matzke,1*Antonius J.M.Matzke,1Jan M.Kooter2In diverse organisms,small RNAs derived from cleavage of double-strand-ed RNA can trigger epigenetic gene silencing in the cytoplasm and at thegenome level.Small RNAs can guide posttranscriptional degradation ofcomplementary messenger RNAs and,in plants,transcriptional gene si-lencing by methylation of homologous DNA sequences.RNA silencing is apotent means to counteract foreign sequences and could play an impor-tant role in plant and animal development.RNA silencing is a new field of research that has coalesced during the last decade from inde-pendent studies on various organisms.Scien-tists who study plants and fungi have known since the late1980s that interactions between homologous DNA and/or RNA sequences can silence genes and induce DNA methylation(1). The discovery of RNA interference(RNAi)in Caenorhabditis elegans in1998(2)focused attention on double-stranded RNA(dsRNA)as an elicitor of gene silencing,and indeed,many gene-silencing effects in plants are now known to be mediated by dsRNA(3).RNAi is usually described as a posttranscriptional gene-silenc-ing phenomenon in which dsRNA triggers deg-radation of homologous mRNA in the cyto-plasm(4).However,the potential for nuclear dsRNA to enter a pathway leading to epigenetic modifications of homologous DNA sequences and silencing at the transcriptional level should not be discounted.Although the nuclear aspects of RNA silencing have been studied primarily in plants,there are hints that similar RNA-directed DNA or chromatin modifications might occur in other organisms as well.Here we adopt a broad definition of RNA silencing that encompasses effects in the cytoplasm and the nucleus,and consider their possible devel-opmental roles and evolutionary origins. RNA Guiding Homologous RNA DegradationAlthough they may differ in detail,RNAi in animals and the related phenomena of post-transcriptional gene silencing(PTGS)in plants and quelling in Neurospora crassa re-sult from the same highly conserved mecha-nism,indicating an ancient origin(5–10).Thebasic process involves a dsRNA that is pro-cessed into shorter units that guide recogni-tion and targeted cleavage of homologousmRNA.dsRNAs that trigger PTGS/RNAican be made in the nucleus or cytoplasm in anumber of ways,including transcriptionthrough inverted DNA repeats,simultaneoussynthesis of sense and antisense RNAs,viralreplication,and the activity of cellular or viralRNA–dependent RNA polymerases(RdRP)on single-stranded RNA templates(Fig.1).InC.elegans,dsRNAs can be injected or intro-duced simply by soaking the worms in asolution containing dsRNA or feeding thembacteria expressing sense and antisense RNA(10).Genetic and biochemical approaches are be-ing used to dissect the mechanism of PTGS/RNAi.Putative RdRPs,putative helicases,andmembers of the PAZ/Piwi family are some ofthe common proteins identified in geneticscreens in N.crassa,C.elegans,and Arabidop-sis(3,5,8,10).Although these proteins provideclues about dsRNA synthesis and processing,the most detailed insight into the two-step RNAdegradation process has come from biochemi-cal experiments with cytoplasmic extracts fromDrosophila(11–15)(Fig.1).The first step in-volves a dsRNA endonuclease[ribonuclease III(RNase III)–like]activity that processes dsRNAinto sense and antisense RNAs21to25nucle-otides(nt)long.These small interfering RNAs(siRNAs),which were first described in a plantsystem(16),are generated in Drosophila by anRNase III–type protein termed Dicer.Orthologsof Dicer,which contains a helicase,dsRNAbinding domains,and a PAZ domain,havebeen identified in Arabidopsis, C.elegans,mammals,and Schizosaccharomyces pombe(15).In the second step,the antisense siRNAsproduced by Dicer serve as guides for a differ-ent ribonuclease complex,RISC(RNA-inducedsilencing complex),which cleaves the homolo-gous single-stranded mRNAs.RISC from Dro-sophila extracts cofractionates with siRNAsthat guide sequence-specific mRNA cleavage(12).RISC cuts the mRNA approximately inthe middle of the region paired with antisensesiRNA(14)(Fig.1),after which the mRNA isfurther degraded.Although most protein com-ponents of RISC have not yet been identified,they might include an endonuclease,an exonu-clease,a helicase,and a homology-searchingactivity(6,10).A candidate for a3Ј,5Ј-exonu-clease is C.elegans MUT7,an RNase D–likeprotein recovered in a screen for RNAi mutants(10).Another component of RISC is a proteinof the PAZ/Piwi family(17),which could in-teract with Dicer through their common PAZdomains(18)to incorporate the siRNA intoRISC(17).Genes encoding members of thePAZ/Piwi family(Arabidopsis:AGO1;N.crassa:QDE2;C.elegans:RDE1),which arehomologous to the translation factor eIF2C,have been shown to be required for PTGS/RNAi in several mutant screens(3,5,8,10).A putative RdRP was the first cellular pro-tein shown to be required for PTGS/RNAi ingenetic screens(N.crassa:QDE1;C.elegans:Ego1;Arabidopsis:SGS2/SDE1)(3,5,8,10),but its exact role is unclear and the predictedenzyme activity remains to be established.Thisprotein might be dispensible when largeamounts of dsRNA are produced from trans-genes or when viral RdRPs are present(5).RdRP might be needed only when dsRNA issynthesized to initiate silencing—for example,from“aberrant”sense RNAs that are prema-turely terminated or processed improperly(19).RISC-cleaved mRNAs may also be used astemplates and converted into dsRNA,increas-ing the level of siRNAs and enhancing PTGS/RNAi(Fig.1).Putative helicases are another class of en-zyme found repeatedly in mutant screens(N.crassa:QDE3;C.elegans:SMG-2;Chlamy-domas:MUT6;Arabidopsis:SDE3)(3,5,8,10).Those recovered so far are not highlyrelated and have not yet been characterizedbiochemically.A DNA helicase(QDE3)andmembers of two RNA helicase superfamilies(MUT6and SMG2/SDE3,respectively)have1Institute of Molecular Biology,Austrian Academy of Sciences,A-5020Salzburg,Austria.2Department of Developmental Genetics,Vrije Universiteit,Amster-dam,Netherlands.*To whom correspondence should be addressed.E-mail:mmatzke@imb.oeaw.ac.at。
