细菌群体感应
QS

信息素的抗生素活性
有些细菌产生的信息素兼有抗生素的双重活性。如 Lactococcus lactis(乳酸乳球菌 )产生的乳链菌素 nisin不但作为信息素调节细胞生物合成和免疫基因的表 达,也作为抗生素拮抗其它微生物。
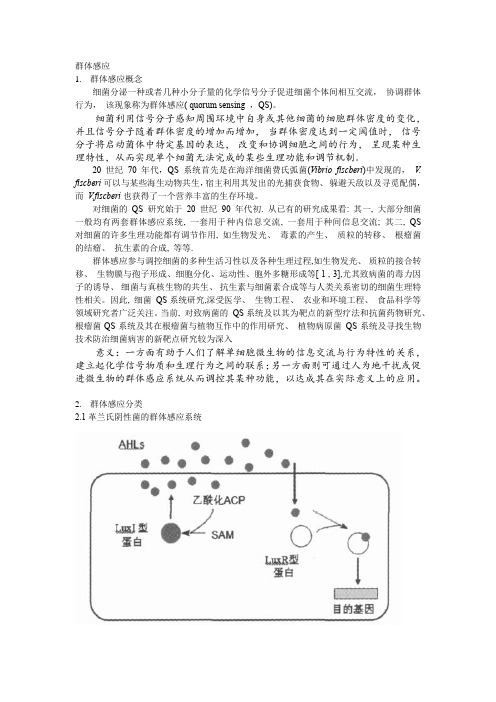
三、细菌信号分子的调大多数G-菌有着相似的调节机制,通常以V. fischeri 的生物发光调节系统LuxI-LuxR为基本模型, 其中LuxI是信息素合成酶,LuxR是相应的调节蛋白, 已知的许多G-菌的QS系统与LuxI-LuxR系统同源。 此外,还有:LuxM型合成酶,与LuxI的同源性不是 很高,可以利用同样的底物合成AHLs,以及从荧光假 单胞菌F113中鉴定出的Hdts合成酶,与LuxI和LuxM 均无同源性。
'Quorum Sensing' (QS) describe the phenomenon whereby the accumulation of signalling molecules enable a single cell to sense the number of bacteria (cell density).
2. 革兰氏阴性细菌的信号分子---AI-1
革兰氏阴性细菌的信号分子大多是酰 基高丝氨酸内酯(acyl-homoserine lactones,AHLs)衍生物,是一类水 溶性、膜透过性分子, 可自由出入细胞 内, 故胞内胞外浓度一致。这些小分子 被称为第一类自身诱导物, AI-1。 AHL 由LuxI 类蛋白酶催化脂肪酸代 谢途径中的酰基-酰基载体蛋白的酰基 侧链与S-腺苷甲硫氨酸中高丝氨酸部 分的结合, 并进一步内酯化而生成的。 AHL 类自诱导剂都是以高丝氨酸为母 体, 之间的差别只是酰基侧链的有无及 侧链的长短不同。
细菌群体感应系统功能

细菌群体感应系统功能
细菌群体感应系统是一种细菌激发细胞间相互作用的机制,通过该系统细菌能够感知并响应外界刺激,调节自身生长和行为,实现一种集体行为。
细菌群体感应系统包含以下功能:
1. 信息传递:细菌通过释放化学信号物质(自动诱导物质、群体感应激素等),使周围细菌感知到外界环境的变化。
这些信号物质可以通过扩散或分泌到周围环境中,也可以直接通过细胞间连接的纤毛或细胞间通道传递。
2. 群体行为:细菌感知到外界环境的变化后,能够通过群体行为来响应和适应。
例如,一些细菌在感知到相对高密度的环境后会进行群体聚集,形成生物膜或菌落。
这种群体行为可以提供保护、资源共享和传递信号等功能。
3. 调控基因表达:细菌群体感应系统能够影响细菌内部的基因表达,通过调节特定基因的转录和翻译过程来实现对环境的适应。
这些基因可能与细菌的生长、生存、毒力等相关。
4. 抗生素生产和耐药性:一些细菌群体感应系统能够诱导或抑制细菌对抗生素的产生。
此外,一些感应系统还能够调节细菌对抗生素的敏感性,从而实现对抗生素的耐药性。
细菌群体感应系统的功能使细菌能够在群体中实现一种高效的信息传递、协作和适应性,为它们在复杂的生态环境中生存和繁衍提供了竞争优势。
这种系统在医药、环境保护、生物工程等领域都有重要的应用潜力。
e42-1群体感应

e42-1 群体感应
群体感应是指某个菌体能够感应到周围环境中同种细菌的其他成员的存在并做出反应的现象。
在上个世纪60年代后期,J.Woodland Hastings等人发现,某些海洋发光细菌只有在达到临界数量后才会发光,而在细菌数量不足时就保持黯淡。
对此他们认为,细菌释放了一种叫自诱导物(autoinducer)的信号分子,来对生物荧光进行调控,同时用它来监测同种细菌的密度。
直到1981年,他们才首次纯化并确定自诱导物是一种脂酰高丝氨酰内酯(acylated homoserine lactone,AHL)。
目前已知具有群体效应的菌体会持续地释放出自诱导物,随着群体扩展,更多自诱导物被增殖的细菌制造,并释放到菌体周围,其浓度也因此渐渐上升。
一旦自诱导物浓度达到一个临界值,细菌便可感应到群体数目的变化,一些细胞行为也会因此改变,如生物荧光、接合作用、转化作用、孢子生成、生物薄膜(biofilm)形成、抗生素和毒素的合成。
迄今为止,具有群体感应的菌种已达数十种,其中,革兰氏阴性菌有两类自诱导物——AHL和呋喃糖硼酸二酯(furanosyl borate diester),革兰氏阳性菌则以寡肽为自诱导物。
以AHL为自诱导物的系统是由luxI和luxR两个结构基因组成,分别编码AHL合酶和AHL反应调节蛋白。
产生的自诱导物分子需要和反应调节蛋白结合,才能控制与群体感应有关的多个基因的表达(图e42-1),例如Vibrio fischeri及V. harveyi的生物荧光基因。
图e42-1 细菌群体感应的LuxI/LuxR系统。
群体感应
群体感应1.群体感应概念细菌分泌一种或者几种小分子量的化学信号分子促进细菌个体间相互交流,协调群体行为,该现象称为群体感应( quorum sensing ,QS)。
细菌利用信号分子感知周围环境中自身或其他细菌的细胞群体密度的变化,并且信号分子随着群体密度的增加而增加,当群体密度达到一定阈值时,信号分子将启动菌体中特定基因的表达,改变和协调细胞之间的行为,呈现某种生理特性,从而实现单个细菌无法完成的某些生理功能和调节机制。
20世纪70年代,QS系统首先是在海洋细菌费氏弧菌(Vibrio fiscberi)中发现的,V. fiscberi 可以与某些海生动物共生,宿主利用其发出的光捕获食物、躲避天敌以及寻觅配偶,而V.fiscberi也获得了一个营养丰富的生存环境。
对细菌的QS 研究始于20 世纪90 年代初. 从已有的研究成果看: 其一, 大部分细菌一般均有两套群体感应系统, 一套用于种内信息交流, 一套用于种间信息交流; 其二, QS 对细菌的许多生理功能都有调节作用, 如生物发光、毒素的产生、质粒的转移、根瘤菌的结瘤、抗生素的合成, 等等.群体感应参与调控细菌的多种生活习性以及各种生理过程,如生物发光、质粒的接合转移、生物膜与孢子形成、细胞分化、运动性、胞外多糖形成等[ 1 , 3],尤其致病菌的毒力因子的诱导、细菌与真核生物的共生、抗生素与细菌素合成等与人类关系密切的细菌生理特性相关。
因此, 细菌QS系统研究,深受医学、生物工程、农业和环境工程、食品科学等领域研究者广泛关注。
当前, 对致病菌的QS系统及以其为靶点的新型疗法和抗菌药物研究、根瘤菌QS系统及其在根瘤菌与植物互作中的作用研究、植物病原菌QS系统及寻找生物技术防治细菌病害的新靶点研究较为深入意义:一方面有助于人们了解单细胞微生物的信息交流与行为特性的关系,建立起化学信号物质和生理行为之间的联系;另一方面则可通过人为地干扰或促进微生物的群体感应系统从而调控其某种功能,以达成其在实际意义上的应用。
群体感应.

2.另外,群体感应系统也在于真菌中,比如白色 念珠菌、新生隐球菌等,但人们是对真菌中的群体 感应系统研究还比较浅,尤其是对真菌群体感应系 统的效应分子、效应分子受体、靶蛋白、相关信号 转导通路以及靶基因的调控等方面的研究有待进一 步深入。
3.最近,一种被称为LED209的分子被发现能够抑制 QseC介导的致病基因激活及诸如EHEC、鼠伤寒沙门 菌和土拉弗朗西斯菌等细菌在活体哺乳动物体内所产 生的不良反应,而且其对哺乳动物的不良反应很小, 对这种分子的研究也许会有一个很好的前景。总之, 不久的将来,随着研究人员的不断探索,人们将可能 通过各种渠道来抑制群体感应系统中的各个环节,从 而达到治疗一些细菌性疾病的目的。
感谢您的关注
3. QS系统的特点 多样性
(1)信号分子的多样性 (2)分布的多样性
细菌种内、 种间,细菌与植物、 动物间
(3)信号分子产生机制的多样性
G-菌——信号分子合成酶,G+菌——前体,经蛋白酶切割
(4)信号分子运输的多样性
G+菌——ABC转运系统,G-菌——直接透过细胞膜
(5)信号响应的多样性
G+菌——双组分信号转导系统; G-菌——受体蛋白
群体感应概述
目录
1 群体感应的发现及其概念
2
群体感应的分类及机制
3
群体反应的特点
4 群体反应的应用与研究前景
1.1 发现
20世纪70年代
海洋细菌费氏弧菌(Vibrio fiscberi)和哈氏弧菌(V . harveyi) 生物发光现象
与海生动物共生,宿主利用其发出的光捕获食物、 躲避天敌以及寻觅配偶,而 V. fiscberi也获得了一个 营养丰富的生存环境
(3)不同 QS系统之间关系的复杂性
群体感应系统

细菌能自发产生、释放一些特定的信号分子,并能感知其浓度变化,调节微生物的群体行为,这一调控系统称为群体感应。
细茵群体感应参与包括人类、动植物病原茵致病力在内的多种生物学功能的调节。
简介群体感应(Quorum-Sensing):近年来的研究证明细菌之间存在信息交流,许多细菌都能合成并释放一种被称为自诱导物质(autoinducer,AI)的信号分子,胞外的AI 浓度能随细菌密度的增加而增加,达到一个临界浓度时,AI能启动菌体中相关基因的表达,调控细菌的生物行为。
如产生毒素、形成生物膜、产生抗生素、生成孢子、产生荧光等,以适应环境的变化,我们将这一现象称为群体感应调节(quorum sensing.QS)。
这一感应现象只有在细菌密度达到一定阈值后才会发生,所以也有人将这一现象称为细胞密度依赖的基因表达(cell density de- pendent control of gene expression)。
[1]自身诱导物质AI细菌可以合成一种被称为自身诱导物质( auto-inducer .AI ) 的信号分子,细菌根据特定的信号分子的浓度可以监测周围环境中自身或其它细菌的数量变化,当信号达到一定的浓度阈值时,能启动菌体中相关基因的表达来适应环境的变化,如芽胞杆菌中感受态与芽胞形成、病原细菌胞外酶与毒素产生、生物膜形成、菌体发光、色素产生、抗生素形成等等。
根据细菌合成的信号分子和感应机制不同,QS系统基本可分为三个代表性的类型:革兰氏阴性细菌一般利用酰基高丝氨酸内酯( AHL) 类分子作为AI ,革兰氏阳性细菌—般利用寡肽类分子(Al P) 作为信号因子,另外许多革兰氏阴性和阳性细菌都可以产生一种AI - 2的信号因子,一般认为AI - 2是种间细胞交流的通用信号分子,另外最近研究发现,有些细菌利用两种甚至三种不同信号分子调节自身群体行为,这说明群体感应机制是极为复杂的。
细菌信息素的特点1,分子量小:细菌信息素都是一些小分子物质,如酰基-高丝氨酸内酯(AHL)衍生物、寡肽、伽马一丁内酯等,能自由进出细胞或通过寡肽通透酶分泌到环境中,在环境中积累。
细菌群体感应系统及其应用

种内交流:G+ 的QS系统
AIP不能自由穿透细胞 壁,需要ABC(ATPbinding-cassette)转运 系统或其它膜通道蛋 白作用到达胞外行使 功能 AIP浓度在胞外达到某 一阈值 膜上激酶识别信号分 子,并促进激酶中组 氨酸残基磷酸化 经过天冬氨酸残基的 传递,把磷酸基团传 递给受体蛋白 AIP前体肽经转录 后的一系列修饰加 工,在不同细菌内 形成长短不同、稳 定、特异的AIP 磷酸化的受体蛋白与 DNA 特定的靶位点结 合,调控基因表达
酶。AiiA蛋白能打开胡萝卜软腐欧文氏菌产生的AHL的内酯键,使软腐
菌的QS系统失灵,由其调控的致病基因与碳青烯抗生素基因不能表达, 从而大大削弱了该菌的致病力
群体感应的抑制
2.产生病原菌信号分子的类似物与信号分子受体蛋 白竞争结合,从而阻断病原菌的QS系统
海洋红藻(Delisea pulchra)产生的卤化呋喃酮结构和AHL相似,用 该卤化呋喃酮处理V. fiscberi后,其QS系统被竞争性的抑制。另外吡 咯酮类化合物、某些取代的HSL化合物、二酮哌嗪类化合物等也能够起 到相类似的作用。在G+菌中,尽管AIP分子调控许多致病基因的表达, 但目前还没有专门针对其QS系统的防病策略。仅在金黄色葡萄球菌发现 其产生不同种类的AIP之间可以相互抑制。因此可以通过设计与病菌AIP 分子相似的物质来破坏其QS统,从而增强植物等的抗病性
小结与展望:
群体感应现象的发现被视为近 20 年来微生物研究领 域中最重大的进展之一。细菌利用 QS 调控系统以群体协 作的方式对种群的社会行为产生影响,赋予细菌类似多细
胞群体行为的能力,使之更好地适应不断变化的环境。QS
在农业、生物技术和医学等诸多领域展示了广阔的应用前 景。
细菌群体感应在微生物生态系统中的作用研究

细菌群体感应在微生物生态系统中的作用研究细菌群体感应是一种自协调的细菌行为,细菌通过分泌信号分子来与它们周围的同种细菌进行通信,并协同地做出响应。
这种协作行为有助于建立细菌社区,并有助于它们在复杂的微生物生态系统中生存和繁殖。
本文将讨论细菌群体感应在微生物生态系统中的作用,并探讨该领域目前的研究进展。
1. 细菌群体感应的基本原理细菌群体感应是一种通过细菌间分泌的信号分子进行交流的行为,这些分子可以传递不同的信息,例如细胞密度、群体方向、环境变化等。
在感应过程中,当一定数量的信号分子被积累到足够数量时,细菌将协调做出共同的行为。
例如,一些细菌会通过群体感应来形成生物膜,从而形成细菌社区,或者来协同合成一些生物活性物质,如光合色素、激素、抗生素等。
这些共同的行为有助于细菌在微生物生态系统中生存和繁殖。
2. 细菌群体感应在微生物生态系统中的作用细菌群体感应在微生物生态系统中起着至关重要的作用。
首先,它有助于细菌建立稳定的细菌社区,并与其他细菌、微生物甚至宿主紧密相连。
这些细菌社区有时会形成生物膜,从而能够更好地抵御环境压力。
其次,它有助于细菌在微生物生态系统中发挥“分工协作”的作用,不同种类的细菌能够通过群体感应来分布不同的环境和角色,以最大化资源利用率并优化生态系统。
另外,细菌群体感应还发挥着各种生态学角色。
例如,在土壤微生物系统中,细菌群体感应可以促进植物生长和根际土壤释放养分。
一些细菌群体感应所产生的代谢产物还被发现对宿主免疫反应和免疫功能具有重要意义。
此外,细菌群体感应还被认为是生态系统中细菌和其他生物之间相互作用的重要媒介,它能够帮助生物维持相互联系并参与生态系统的稳定性。
3. 细菌群体感应的研究进展目前,细菌群体感应的研究进展日新月异。
这是因为细菌群体感应在医学、环境保护、农业等领域都有重要应用价值。
例如,在医学中,对细菌群体感应的深入研究能够有助于探索新型抗生素的生产和应用;环境保护中,它可以帮助减少有毒物质的生产和释放,改善微生物生态环境;在农业中,它能够协助控制农业害虫和植物病害。
- 1、下载文档前请自行甄别文档内容的完整性,平台不提供额外的编辑、内容补充、找答案等附加服务。
- 2、"仅部分预览"的文档,不可在线预览部分如存在完整性等问题,可反馈申请退款(可完整预览的文档不适用该条件!)。
- 3、如文档侵犯您的权益,请联系客服反馈,我们会尽快为您处理(人工客服工作时间:9:00-18:30)。
Antimicrobial resistance,respiratory tract infections and role of bio films in lung infections in cystic fibrosis patients ☆Oana Ciofu a ,Tim Tolker-Nielsen a ,Peter Østrup Jensen a ,Hengzhuang Wang a ,b ,Niels Høiby a ,b ,⁎a Institute of International Health,Immunology and Microbiology,Costerton Bio film Centre,University of Copenhagen,Copenhagen,Denmark bDepartment of Clinical Microbiology,Rigshospitalet,Copenhagen,Denmarka b s t r a c ta r t i c l e i n f o Available online 2December 2014Keywords:Pseudomonas aeruginosa Bio filmCystic fibrosisLung infection is the main cause of morbidity and mortality in patients with cystic fibrosis and is mainly dominat-ed by Pseudomonas aeruginosa .The bio film mode of growth makes eradication of the infection impossible,and it causes a chronic in flammation in the airways.The general mechanisms of bio film formation and antimicrobial tolerance and resistance are reviewed.Potential anti-bio film therapeutic targets such as weakening of bio films by quorum-sensing inhibitors or antibiotic killing guided by pharmacokinetics and pharmacodynamics of antibiotics are presented.The vicious circle of adaptive evolution of the persisting bacteria imposes important therapeutic challenges and requires development of new drug delivery systems able to reach the different niches occupied by the bacteria in the lung of cystic fibrosis patients.©2014Elsevier B.V.All rights reserved.Contents1.Cystic fibrosis and chronic lung infection ..................................................82.The occurrence and architecture of bacterial bio films –general aspects ....................................83.High cell density and quorum-sensing in bio films ..............................................104.Tolerance of bio films to antimicrobials and immune system .........................................114.1.Restricted penetration .......................................................114.2.Differential physiological activity ..................................................114.3.Differential expression of speci fic genes in bio film ..........................................124.4.Persister cells ...........................................................125.Hydroxyl radical formation during antibiotic treatment of bio films ......................................126.Antibiotic resistance and mutability in bio films ...............................................137.Bio film treatment strategies ........................................................137.1.Quorum-sensing inhibitors .....................................................137.2.Antibiotic killing in bio films:pharmacokinetic and pharmacodynamic issues in Pseudomonas aeruginosa bio film lung infections.......147.2.1.PK/PD of colistin on P .aeruginosa bio films ..........................................147.2.2.PK/PD of β-lactam antibiotics on P .aeruginosa bio films ....................................158.Actual treatment of the P .aeruginosa infection in CF patients .........................................159.Challenges of the treatment of P .aeruginosa airways infection in CF ......................................179.1.P.aeruginosa in the airways of CF patients:sites,niches and bio film ..................................179.2.P .aeruginosa adaptation to the CF lung:bacterial heterogeneity ....................................1810.Therapeutic strategies to circumvent these challenges ............................................1811.Conclusions ...............................................................19Acknowledgement ..............................................................19References (19)Advanced Drug Delivery Reviews 85(2015)7–23☆This review is part of the Advanced Drug Delivery Reviews theme issue on "Inhaled antimicrobial chemotherapy for respiratory tract infections:Successes,challenges and the road ahead".⁎Corresponding author at:Department of Clinical Microbiology,9301,University Hospital Rigshospitalet,Juliane Mariesvej 28,2100Copenhagen Ø,Denmark.Tel.:+4535457778;fax:+4535456412.E-mail address:hoiby@hoibyniels.dk (N.Høiby)./10.1016/j.addr.2014.11.0170169-409X/©2014Elsevier B.V.All rightsreserved.Contents lists available at ScienceDirectAdvanced Drug Delivery Reviewsj o u r na l h om e p a g e :w w w.e l s e v i e r.c o m /l o c a t e /a d d r1.Cysticfibrosis and chronic lung infectionCysticfibrosis(CF)is a congenital,recessively inherited disorder which affects one out of2000newborns,in Caucasian populations and affects around70,000people around the world.There are considerable variations through Europe from as high as1in1400live births in Ireland,to1in25,000in Finland[1].The genetic background is one of N2000mutations in the cysticfibrosis transmembrane conductance regulator gene(CFTR)in each of the two chromosomes7which gives malfunction of the chloride channel in CF patients().If intensively treated,the mean expected lifetime of CF patients is N35 years and in some centres N50years(http://www.cysticfibrosis.ca/). Any progress in treatment is therefore important for CF patients.CF is a multiorgans disease affecting the airways,pancreas,small intestine,liver,reproductive tract and the sweat glands.The clinical symptoms from the lungs are viscid mucus and respiratory infections leading to chronic bronchial disease and bronchiectasis.The malfunction of the chloride channel in CF patients leads to decreased volume of the paraciliaryfluid in the lower respiratory tract and to impaired mucus detachment from the submucosal gland ducts in the conducting airways[2]and that in turn leads to impaired mucociliary clearance of inhaled microbes[3].This impairment of the non-inflammatory defense mechanism of the respiratory tract leads to early recruitment of the inflammatory defense mechanisms notably polymorphonuclear leukocytes(PMN)and antibodies[4–6].CF patients,therefore,from early childhood suffer from recurrent and chronic respiratory tract infections characterized by PMN inflammation. In spite of the inflammatory response and intensive antibiotic therapy, however,infections caused by P.aeruginosa,the Burkholderia cepacia complex(mostly B.multivorans and B.cenocepacia)and Achromobacter xylosoxidans,persist and lead to respiratory failure and lung transplanta-tion or death of the patients[7].Several other species including Staphylo-coccus aureus,Haemophilus influenzae,Stenotrophomonas maltophilia and Mycobacteria other than tuberculosis and Aspergillus fumigatus also contribute to the morbidity and mortality[7].Chronic P.aeruginosa lung infection is the cause of much of the morbidity and most of the mortality in CF patients.About80%of CF adults suffer from chronic P.aeruginosa infection.Previously50%of CF patients would succumb within5years after onset of the chronic P.aeruginosa infection.This prognosis has been changed completely by the introduction of intensive early eradication therapy and many patients will therefore not contract the chronic infection during childhood anymore[8].Adaptive mechanisms of P.aeruginosa exists which explain why this pathogen is able to survive and persist for several decades in the respi-ratory tract of CF patients in spite of the defense mechanisms of the host and intensive antibiotic therapy.P.aeruginosa is able to survive by switching to the biofilm mode of growth which provides tolerance to the inflammatory defense mechanism,to the aerobic respiratory zone and to the conductive zone of the lungs which contain anaerobic spu-tum,and to the antibiotic therapy[9–12].The conductive and respirato-ry zones of the lung are presented in Fig.1.During the adaptation mucoid(biofilm mode of growth)and non-mucoid phenotypes are split off due to mutations(Table1).Microscopy pictures of P.aeruginosa biofilms from the lung of CF patients are presented in Fig.2A,B,C.The biofilm strategy is also used by Burkholderia,sp A.xylosoxidans and Stenotrophomonas species[13](Fig.2D.E,F)although this has been questioned by some authors suggesting that Burkholderia species may exist predominantly as single cells in CF lungs[14].2.The occurrence and architecture of bacterialbiofilms–general aspectsMicrobial biofilms are ubiquitous in nature and of importance for a number of environmental processes.However,biofilms are also the un-derlying cause of persistent infections in various parts of the human body,including the teeth[15],urinary tract[16],heart valves[17],CF lungs[11],middle ears[18],nasal passages and sinus cavities[19], bones[20],prostate[21],and chronic wounds[22].In addition,biofilm formation on implants and catheters gives rise to problematic infections in connection with the use of medical devices including urinary cathe-ters,central venous catheters,fracturefixation devices,dental implants, joint prostheses,vascular grafts,cardiac pacemakers,breast implants, mechanical heart valves,penile implants,and heart assist devices[23].The formation and maintenance of biofilms depends critically on the presence of bacteria-to-bacteria interconnecting extracellular substances that serve as a biofilm matrix[24].Many kinds of exopolymers, e.g.polysaccharide,protein,and DNA,may be part of the matrix of biofilms.In addition,outer membrane proteins and a variety of bacterial cell appendages such asfimbriae,pili,andflagella may also have bacteria-to-bacteria interconnecting functions,and can therefore be con-sidered part of the biofilm matrix.The components of the biofilm matrix are usually,but not always,produced by the bacteria themselves.A single bacterial species can produce several different biofilm matrix com-ponents,and it appears that not all of these components are expressed during biofilm formation in a particular environment.It is anticipated that the capacity of bacteria to produce different biofilmmatrix Fig.1.The conductive and respiratory zones of the lungs.Inhalation antibiotic therapy is mainly targeting the conductive zone where sputum is located,whereas systemic antibiotic therapy is mainly targeting the respiratory zones with no sputum[12,75]. Table1Important properties of mucoid and non-mucoid phenotypes of Pseudomonas aeruginosa in the respiratory tract of cysticfibrosis[9,10].Property Mucoid phenotype Nonmucoidphenotype Location in the lungs Respiratory zone andconductive zone in sputumConductivezone in sputum Biofilm formation in vitro Yes YesBiofilm formation in vivo Yes NoMultiply antibiotic resistance dueto conventional mechanismsSeldom FrequentResistance due to biofilm properties Yes No Responsible for lung tissue damage Yes NoInduces pronounced antibody response Yes No8O.Ciofu et al./Advanced Drug Delivery Reviews85(2015)7–23components allows colonization of different niches through different bio film formation pathways.The extracellular bio film matrix is believed to offer protection against various adverse factors including immune responses and antibiotic treatment [25–28].Studies using confocal laser scanning microscopy of bio films formed in laboratory experimental systems by fluorescence-tagged bacteria have provided detailed knowledge about structural bio film develop-ment (e.g.[29–33]).Bio film formation can initiate when bacteria attach to a surface or to each other and form aggregates.The bio film developmental cycle is believed to include the processes:i)transport of solitary microbes to a surface or each other,ii)initial attachment of the microbes to the surface or each other,iii)formation of microcolonies,iv)maturation of the bio film,and v)dispersal of the bio-film.In many bacteria,the initiation of bio film formation and the pro-duction of bio film matrix components occur in response to high intrabacterial levels of the second messenger molecule c-di-GMP,whereas dispersal of the bio film occurs in response to low levels of c-di-GMP [34].Synthesis and degradation of c-di-GMP in the bacteriaisFig.2.Bio films of CF-related pathogensin the lungs of patients with cystic fibrosis.A.Mucoid bio film of P .aeruginosa in an alveolar sac (lower arrow)surrounded by severely in flammed tissue (PMNs,pneumonia)(upper arrow).Autopsy of a CF girl who died due to chronic P .aeruginosa lung infection.HE stain ×100(photo by N.Høiby).B.Pseudomonas aeruginosa bio film in a detached lung alveole in sputum from a CF patient suffering from chronic lung infection.Gram stained smear,×1000magni fication (photo by N.Høiby).C.Pseudomonas aeruginosa bio film in sputum form a cystic fibrosis patient suffering from chronic lung infection.Gram stained smear,×1000magni fication (photo by N.Høiby).D.Achromobacter xylodoxidans bio film in sputum from a CF patient suffering from chronic lung infection.Gram stained smear,×400magni fication (photo by N.Høiby).E.Burkholderia multivorans bio film in sputum from a CF patient suffering from chronic lung infection.Gram stained smear,×1000magni fication (photo by N.Høiby).F.Stenotrophomonas maltophilia bio film in sputum from a CF patient suffering from chronic lung infection.Gram stained smear,×1000magni fication (photo by N.Høiby).9O.Ciofu et al./Advanced Drug Delivery Reviews 85(2015)7–23accomplished by diguanylate cyclases and phosphodiesterases that con-tain sensory domains,enabling translation of diverse environmental cues into synthesis or degradation of c-di-GMP,which bind to down-stream effector molecules and modulates their function,resulting in regulation of the production of different adhesins and bio film matrix products.In between initiation and termination of bio film formation we have de fined speci fic bio film stages,but the currently available evi-dence suggests that these transitions are mainly governed by adaptive responses,and not by speci fic genetic programs.In the case of P .aeruginosa a number of microarray analyses have been performed in order to monitor genes that are expressed during bio film formation [35–38].However,the transcriptomic analyses performed by the differ-ent research groups have failed to consistently identify speci fic bio film regulons,in support of the suggestion that bio film formation is mainly governed by adaptive responses.Bio film formation can occur through multiple pathways,and the spatial structure of the bio films is species dependent as well as dependent on the environmental conditions.For example,P .aeruginosa forms bio films with mushroom shaped struc-tures in flow-chambers perfused with glucose-medium,whereas it forms flat bio films in flow-chambers perfused with citrate medium [29](Fig.3).The bacteria are still susceptible to antibiotics at the earliest stage of bio film formation,and this is in accordance with the success of peroperative antibiotic prophylaxis for e.g.alloplastic surgery,however more developed bio films display recalcitrance to antibiotic treatment (as addressed below).When dispersal occurs at focal areas of bio films the liberated bacteria can spread to other locations where new bio films can be formed.The difference in the properties of bio film-growing and planktonic bacteria has signi ficant diagnostic and therapeutic consequences.In clinical specimens (e.g.biopsies,pus,sputum)bio films can often be rec-ognized by light microscopy,but precise identi fication of the bacteria within a bio film can only be done by the use of nucleic acid hybridiza-tion techniques,and identi fication of the components of the bio film ma-trix requires specialized staining techniques [11].Traditional sampling techniques may not be suf ficient to culture bacteria from bio films unless the bacteria are released from the bio film by ultasonic pre-treatment [39].The ordinary culture techniques,however,reveals only the proper-ties of planktonic bacteria,and antibiotic susceptibility testing therefore does not indicate the susceptibility of the bio film-growing bacteria.Methods to test antibiotic susceptibility of bio film-growing bacteria have been developed,but their clinical relevance awaits con firmation [25,40,41].3.High cell density and quorum-sensing in bio filmsMany bacterial species use cell-to-cell signaling,also referred to as quorum-sensing,to coordinate gene expression in relation to population density.Production by the bacteria of extracellular signal molecules gives rise to a high concentration of signal molecule in high-density bacterial populations.The signal molecules are sensed by the bacteria via speci fic quorum-sensing receptors,and the bacteria can therefore induce target genes when the concentration of quorum-sensing signal molecule is high,and restrict expression of speci fic genes to conditions of high cell density.P .aeruginosa utilizes the three quorum-sensing signal molecules 3-oxododecanoyl-L-homoserine lactone (3-oxo-C12-HSL),N-butanoyl homoserine lactone (C4-HSL),and 2-heptyl-3-hydroxy-4-quinolone (Pseudomonas quinolone signal,denoted PQS)[42].The 3-oxo-C12-HSL molecule is synthesized via the LasI synthase and sensed by the LasR re-ceptor,whereas C4-HSL is synthesized via the RhlI synthase and sensed by the RhlR receptor [43],and PQS is synthesized via the pqsABCDE and pqsH gene products and sensed by the PqsR receptor [44].In many P .aeruginosa strains,the quorum-sensing systems are interconnected and work in a hierarchical manner with the Las system affecting expres-sion of the Rhl system [45,43],and the Pqs system intertwined between the Las and Rhl systems [46],but exceptions to this hierarchical organiza-tion have been found [47].Transcriptomic analysis of P .aeruginosa bio films has con firmed that quorum-sensing controls gene expression in these high density populations [35].Quorum-sensing primarily regulates the expression of virulence factors in P .aeruginosa [48],but in addition quorum-sensing appears to regulate the production of several factors that play roles in P .aeruginosa bio film formation.Davies et al.[49]found that a P .aeruginosa lasI quorum-sensing mu-tant formed flat and undifferentiated bio films in flow-chambers,where-as the wild-type strain formed bio films with large mushroom-shaped structures.The flat bio films formed by the lasI mutant strain were vulnerable to treatment with the detergent SDS,while the structured bio films formed by the wild-type strain resisted SDS treatment.Quorum-sensing plays a role in the generation of extracellular DNA in P .aeruginosa bio films [32],and the extracellular DNA functions as a matrix component in the bio films [50].In addition to a small amount of extracellular DNA present in the initial phase in P .aeruginosa bio films,release of a large amount of extracellular DNA occurs at a later stage of P .aeruginosa bio film formation,regulated via the PQS-based quorum-sensing system,and evidently as a consequence of lysis of a small sub-population of the bacteria [32].In agreement with a role ofPQS-basedFig.3.Confocal laser scanning microscopy (CLSM)micrographs acquired in 5day-old P .aeruginosa -Gfp bio films grown in flow-chambers on glucose minimal medium (A)or citrate min-imal medium (B).The central pictures show-top down fluorescence projections and the flanking pictures show vertical sections.Bars,20μm.Adapted from Mol Microbiol .48:1511–1524with permission from Wiley-Blackwell publishing.10O.Ciofu et al./Advanced Drug Delivery Reviews 85(2015)7–23QS in the generation of extracellular DNA that functions as a biofilm ma-trix component,P.aeruginosa pqs mutants were found to have defects in biofilm formation,and theflat biofilms that were formed by the pqs mu-tants contained little extracellular DNA[51,32,33,52].The mechanism involved in PQS-mediated DNA-release is unknown,but evidence has been provided that PQS can act as a cell-sensitizing pro-oxidant[53], which may cause lysis of a subpopulation of P.aeruginosa bacteria.In ad-dition to extracellular DNA,quorum-sensing signaling controls the pro-duction of the biosurfactant rhamnolipid,which was shown to be important for biofilm formation by P.aeruginosa[54–56],and to play a role in the tolerance of P.aeruginosa biofilms towards immune cells [57].Furthermore,the production of the P.aeruginosa LecA and LecB lectins was shown to be regulated by quorum-sensing[58,59],and the lectins were shown to play a role in P.aeruginosa biofilm formation [60,61].Moreover,transcriptome analysis has indicated that genes encoding CupA and CupBfimbriae are regulated by quorum-sensing in P.aeruginosa biofilms[35],and thefimbriae were shown to function as biofilm matrix components[62].Quorum-sensing signaling in P.aeruginosa also controls the production of siderophores such as pyoverdine and pyochelin,which are also of importance for biofilm for-mation[63].Furthermore,evidence has been presented that the Rhl quorum-sensing system is necessary for preventing accumulation of toxic nitric oxide during anaerobic nitrate respiration which may play an important role in P.aeruginosa biofilms in clinical settings[64–66]. In agreement,genes required for respiratory nitrate reduction,e.g. nirCMSQ and napEF,were found to be strongly up-regulated in P.aeruginosa during biofilm growth[35],and genes in the nar operon, encoding the denitrification pathway,were found to be upregulated in some P.aeruginosa CF lung isolates[67].4.Tolerance of biofilms to antimicrobials and immune systemMicrobial biofilms can tolerate activities of the host immune system and treatment with antimicrobial compounds[26–28]The minimal in-hibitory concentration(MIC)and minimal bactericidal concentration (MBC)of antibiotics to biofilm-growing bacteria may be100–1000 fold higher than to planktonic bacteria[25,28,41].The tolerance of biofilms to immune defenses and antimicrobials is connected to the biofilm mode of growth,and if bacteria originating from a biofilm are grown in planktonic culture they will usually display susceptibility to the antimicrobial activities[68].Biofilm-associated antimicrobial toler-ance is therefore fundamentally different from antimicrobial resistance, which can be displayed by bacteria in planktonic culture and is not connected to the biofilm mode of growth per se.4.1.Restricted penetrationRestricted penetration of antimicrobials through the biofilm matrix can in some cases contribute to the antimicrobial tolerance of biofilms. Although biofilm matrices do not inhibit diffusion of antibiotics in gen-eral,restricted penetration of antibiotics through biofilms may occur in cases where the antibiotics bind to components of the biofilm matrix or the bacterial membranes e.g.;[69,70].For example,alginate and extra-cellular DNA in the matrix of P.aeruginosa biofilms may bind aminogly-coside antibiotics and thereby play a role in the antimicrobial tolerance of P.aeruginosa biofilms to antibiotics such as tobramycin[71].demon-strated that biofilms formed by an alginate over-producing P.aeruginosa strain displayed enhanced tolerance to tobramycin in comparison to a biofilm formed by an isogenic nonmucoid P.aeruginosa strain.Much of the extracellular DNA that is present in P.aeruginosa biofilms is released by the bacteria via a quorum-sensing controlled process(as described above),and[70]provided evidence that the fragile biofilms formed by a DNA-release deficient P.aeruginosa quorum-sensing mu-tant are susceptible to tobramycin treatment,but become tobramycin tolerant if they were supplied with exogenous DNA.Furthermore, it was demonstrated that addition of lysed polymorphonuclear leukocytes,which are thought to be a source of extracellular DNA at sites of infection,increased the tolerance of P.aeruginosa biofilms to-wards tobramycin treatment[70].In accordance with these studies [72],demonstrated that co-administration of DNase and alginate lyase led to increased killing activity of tobramycin in P.aeruginosa biofilms.4.2.Differential physiological activityDifferential physiological activity of the bacteria in biofilms can also be an underlying cause of biofilm-associated antimicrobial tolerance. Studies of P.aeruginosa biofilms grown in various in vitro laboratory setups have provided evidence that the metabolic activity of the bacte-ria is high in the outer part of the biofilm whereas it is low in the inner part of the biofilm[69,73,74].The available evidence suggests that the differential physiological activity seen in biofilms is caused by limited oxygen and nutrient penetration through the biofilm due to bacterial consumption(e.g.[69].Because many antibiotics targets processes that occur in growing bacteria(e.g.replication,transcription,transla-tion,and cell wall synthesis),biofilm bacteria with low metabolic activity may display increased antimicrobial tolerance.Studies of P.aeruginosa biofilms grown inflow-chambers have provided evidence that the antibiotics tobramycin,ciprofloxacin,and tetracycline preferen-tially kill the metabolically active bacteria located in the outer part of the biofilm,whereas the non-growing bacteria in the inner part of the bio-film survives treatment with these antibiotics[75–77,33,78,74,79] (Fig.4).However,some antimicrobials such as colistin,SDS,EDTA,and chlorhexidine preferentially kill the non-growing bacteria located in the inner part of the biofilm[74,79,80](see Fig.4).Colistin,SDS,EDTA, and chlorhexidin,on the contrary,do not kill the actively growing biofilm bacteria as they are able to induce the expression of defense mechanisms such as efflux pumps and the pmr genes,the latter encoding enzymes that adds amino arabinose to LPS and thereby abolish binding of the antimicrobials to the bacterial surface[74,80].Fig.4.Visualization of live and dead bacteria inflow-chamber-grown P.aeruginosa-Gfp biofilms treated with antibiotics.P.aeruginosa-Gfp biofilms were grown for4days and then continuously exposed to no antibiotic(A),25μg/ml colistin(B),60μg/ml ciproflox-acin(C),or200μg/ml tetracycline(D)for24hours.The central pictures show horizontal CLSM optical sections,and theflanking pictures show vertical CLSM optical sections.Live cells appear green due to expression of Gfp and dead cells appear red due to staining with the dead-cell indicator propidium iodide.Adapted from Mol.Microbiol.68:223–240with permission from Wiley-Blackwell publishing.11O.Ciofu et al./Advanced Drug Delivery Reviews85(2015)7–234.3.Differential expression of specific genes in biofilmAs indicated above,biofilms can display antimicrobial tolerance mechanisms that can be related to the expression of a few specific genes in the bacteria.Another example of this is the ndvB gene in P.aeruginosa PA14which encodes an enzyme that is involved in the syn-thesis of periplasmic glucans that binds tobramycin and prevents cell death by sequestering the antibiotic[81].Evidence was provided that the ndvB gene is expressed specifically in P.aeruginosa PA14biofilms but not in planktonic cells,and biofilms formed by a P.aeruginosa ndvB mutant were demonstrated to be more sensitive to tobramycin than wild type biofilms,whereas the ndvB mutant and wild type strain showed no difference in tobramycin sensitivity in planktonic culture [81].Another biofilm-specific resistance mechanism was discovered by Zhang and Mah[82]who reported that the PA1874-1877genes in P. aeruginosa encode a putative efflux pump that is expressed only during the biofilm mode of growth,and mediates tolerance to tobramycin,gen-tamicin,and ciprofloxacin.Moreover,Liao and Sauer[83]reported that the transcriptional regulator BrlR is expressed in P.aeruginosa biofilms but not in planktonic cells,and plays a role in the antimicrobial tolerance of P.aeruginosa biofilms.Inactivation of the brlR gene rendered biofilms, but not planktonic cells,significantly more susceptible to hydrogen per-oxide andfive different classes of antibiotics.Subsequently,evidence was provided that BrlR is required for maximal expression in P.aeruginosa biofilms of genes encoding the MexAB-OprM and MexEF-OprN efflux pumps[84].Earlier studies have suggested that efflux pumps such as MexAB-OprM,MexCD-OprJ,MexEF-OprN,and MexXY-oprM do not play a role in the tolerance of P.aeruginosa biofilms to antibiotics[85]. However,more recent studies have provided evidence that these efflux pumps are involved in the tolerance of P.aeruginosa biofilms to azithromycin[86],colistin[74,80],tobramycin,tetracycline,trimetho-prim,and norfloxacin[84].Resistance of biofilms to tobramycin mediat-ed by efflux-pump has also been reported to occur in B.cenocepacia[87]. Efflux pumps might also have a general detoxifying role in P.aeruginosa biofilms.The P.aeruginosa MexEF-OprN and MexXY-oprM efflux sys-tems were shown to be up-regulated in response to reactive oxygen spe-cies,and it was proposed that these efflux systems export cellular constituents damaged by reactive oxygen species[88].Because bacteria in biofilms experience increased oxidative stress due to accumulation of waste products[89],efflux pumps might have a detoxifying role in P.aeruginosa and Burkholderia sp.biofilms[90].In line with this,Kvist et al.[91]reported that efflux pumps involved in removal of toxic sub-stances were highly up-regulated in E.coli biofilms.In addition,it was found that mutant E.coli strains which were unable to produce efflux pumps showed defects in biofilm formation[91].Evidence has been provided that quorum-sensing is involved in the tolerance of P.aeruginosa biofilms to tobramycin,kanamycin, and hydrogen peroxide[92,93,75].There is currently no evidence that quorum-sensing promotes antibiotic tolerance in planktonic P. aeruginosa cells,and the involvement of quorum-sensing in antimicro-bial tolerance therefore appears to be biofilm-specific.The involvement of quorum-sensing in antimicrobial tolerance of P.aeruginosa biofilms may in part be explained by its role in the production of extracellular DNA which inhibits penetration of some antibiotics into the biofilm. The extracellular DNA that is produced in P.aeruginosa biofilms also has a stabilizing effect[32].Because biofilms formed by P.aeruginosa quorum-sensing mutants are more fragile than wild type biofilms they may be more vulnerable to for example shear forces and phagocy-tosis,and because of the high bacterial turnover a large proportion of the bacteria will be growing actively and may therefore show increased sensitivity to some antibiotics.4.4.Persister cellsIn addition to the mechanism described above,slowly-dividing or non-dividing bacteria that show diminished susceptibility to antibiotics also contribute to the antimicrobial tolerance of biofilms[94–96].The fraction of these so-called persister cells is usually low(b0.1%)and they should be distinguished from the bulk subpopulation of metaboli-cally inactive bacteria that are present in biofilms as described above. Persister cells are believed to be the result of bacterial differentiation into a dormant state.The reduced metabolism exhibited by persister cells evidently enable them to escape the activity of antibiotics that tar-get fundamental cellular processes such as replication,translation,and cell wall synthesis,but persister cells also in some cases show tolerance to antibiotics that kill non-growing cells[94,95].Substantial evidence suggests that the formation of persister cells is connected to bacterial toxin/antitoxin systems[97,96,98].Increased expression of toxins was shown to block cell metabolism,indicating that randomfluctuations in the expression of toxin and antitoxin genes can lead to the formation of persister cells.However,mutant screens have suggested that a number of other genes(e.g.rpoS,spoT,relA,dksA,dinG,spuC,algR,pilH,ycgM,pheA) than those encoding toxin/antitoxins are involved in persister formation in P.aeruginosa[99,100,95],suggesting that multiple pathways can lead to the persister cell phenotype.Exogenous addition of quorum-sensing molecules has been shown to increase the formation of planktonic per-sister cells for P.aeruginosa PAO1,but had no such effect for P.aeruginosa PA14[101].However,there is currently no evidence that physiological levels of quorum-sensing molecules promote persister cell formation.Be-sides the persister cells,other phenotypic variants can also contribute to the antimicrobial tolerance of biofilms Drenkard and Ausubel[102],for example,identified a class of antibiotic tolerant P.aeruginosa variants in which the regulatory protein PvrR,was shown to control the conversion between the parental form and the tolerant variant.5.Hydroxyl radical formation during antibiotic treatmentof biofilmsThe involvement of hydroxyl radical(OH·)formation in the bacteri-cidal activity of major classes of antibiotics[103]may be attenuated in biofilm resulting in enhanced tolerance.As comprehensively reviewed [104,105],bactericidal treatment with antibiotics may cause hyper-oxidation of NADH produced in the tricarboxylic acid(TCA)cycle resulting in leakage of electrons from the respiratory electron transport chain.The leaking electrons reduce molecular oxygen(O2)to produce su-peroxide(O2-),which subsequently dismutates into hydrogenperoxide (H2O2)and liberates ferrous iron by disintegrating iron-sulphur clusters thereby providing the necessary substrates for generation of lethal amounts of highly reactive OH·through the Fenton reaction.The actual positions of the electron leak in the respiratory electron transport chain has not yet been determined,however the outflow of electrons is likely to occur upstream of the terminal oxidases,since electrons were mainly leaking from complex I and III in mammalian cells treated with antibiotics [106].Thus,the hyper-oxidation of NADH leads to oxidative stress involv-ing formation of both O2-,H2O2and OH·.But while O2-and H2O2can be enzymatically eradicated by superoxide dismutases,catalases and perox-idases,no known enzyme is able to catalyse the cellular detoxification of OH·.Despite the fact that OH·may impose toxic oxidative lesions on pro-teins,lipids and DNA[107],the main cause of lethality during treatment of E.coli with beta-lactams,quinolones and aminoglycosides was insuffi-cient repair of incorporated oxidized guanine in DNA[108].In addition to being dependent on the activity of the TCA cycle generation of lethal amounts of OH·is reliant on the available electron acceptors as both the bactericidal activity and the formation of OH·was decreased when O2was replaced by nitrate(NO3-)during ciproflox-acin treatment of growing P.aeruginosa[109].Thus,the lethal effect me-diated by formation of OH during bactericidal treatment with antibiotics depends on aerobic respiration and metabolic activity in order to devel-op the required reactive oxygen species(ROS).In biofilms,the contribution of OH·generation to the bactericidal ac-tivity during treatment with antibiotics has so far only been demon-strated for Pseudomonas aeruginosa treated with ciprofloxacin[110].12O.Ciofu et al./Advanced Drug Delivery Reviews85(2015)7–23。
