苹果-天冬氨酸穿梭
第十章电子传递与生物氧化

第十章电子传递与生物氧化一、单项选择题(在备选答案中只有一个是正确的)1.体内CO2来自:A.碳原子被氧原子氧化 B.呼吸链的氧化还原过程C.有机酸的脱羧 D.糖原的分解 E.真脂分解2.线粒体氧化磷酸化解偶联是意味着:A.线粒体氧化作用停止 B.线粒体膜ATP酶被抑制 C.线粒体三羧酸循环停止D.线粒体能利用氧,但不能生成ATP E.线粒体膜的钝化变性3.P/O比值是指:A.每消耗一分子氧所需消耗无机磷的分子数B.每消耗一分子氧所需消耗无机磷的克数C.每消耗一分子氧所需消耗无机磷的原子数4.各种细胞色素在呼吸链中传递电子的顺序是:A.a→a3→b→c1→c→1/2O2B.b→a→a3→c1→c→1/2 O2C.c1→c→b→a→a3→1/2 O2D.c→c1→a3→b→1/2 O2E.b→c1→c→a3→1/2 O25.细胞色素b,c1和c均含辅基:A.Fe3+ B.血红素C C.血红素A D.原卟啉 E.铁卟啉6.劳动或运动时ATP因消耗而大量减少,此时:A.ADP相应增加,ATP/ADP下降,呼吸随之加快B.ADP相应减少,以维持ATP/ADP恢复正常C.ADP大量减少,ATP/ADP增高,呼吸随之加快D.ADP大量磷酸化以维持ATP/ADP不变 E.以上都不对7.人体活动主要的直接供能物质是:A.葡萄糖 B.脂肪酸 C.磷酸肌酸 D.GTP E.ATP8.下列属呼吸链中递氢体的是:A.细胞色素 B.尼克酰胺 C.黄素蛋白 D.铁硫蛋白 E.细胞色素氧化酶9.氰化物中毒时,被抑制的是:A.Cyt b B.Cyt C1 C.Cyt C D.Cyt a E.Cyt aa310.下列物质中ATP的贮存形式是:A.磷酸烯醇式丙酮酸 B.磷脂酰肌醇 C.肌酸 D.磷酸肌酸 E.GTP二、多项选择题(在备选答案中有二个或二个以上是正确的,错选或未选全的均不给分)1.NAD+的性质包括:A.与酶蛋白结合牢固 B.尼克酰胺部份可进行可逆的加氢和脱氢C.每次接受一个氢原子和一个电子 D.为不需脱氢酶的辅酶2.铁硫蛋白的性质包括:A.由Fe-S构成活性中心 B.铁的氧化还原是可逆的C.每次传递一个电子 D.与辅酶Q形成复合物存在3.苹果酸天冬氨酸穿梭作用可以:A.生成3个ATP B.将线粒体外NADH所带的氢转运入线粒体C.苹果酸和草酰乙酸可自由穿过线粒体内膜D.谷氨酸和天冬氨酸可自由穿过线粒体膜4.氧化磷酸化的偶联部位是:A.复合体Ⅱ→泛醌 B.NADH→泛醌C.Cyt b→Cyt c D.复合体Ⅲ→1/2O25.抑制氧化磷酸化进行的因素有:A.CO B.氰化物 C.异戊巴比妥 D.二硝基酚6.下列关于解偶联剂的叙述正确的是A.可抑制氧化反应 B.使氧化反应和磷酸反应脱节C.使呼吸加快,耗氧增加 D.使ATP减少7.不能携带胞液中的NADH进入线粒体的物质是:A.肉碱 B.草酰乙酸 C.α-磷酸甘油 D.天冬氨酸三、填空题1.ATP的产生有两种方式,一种是__________,另一种是___________。
MDH1介导苹果酸-天冬氨酸NADH穿梭维持胎儿肝造血干细胞的活性水平。(Sonarsignal)
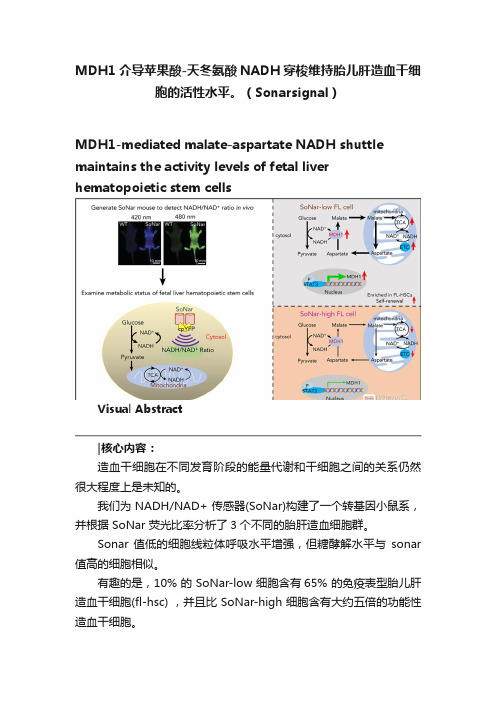
MDH1介导苹果酸-天冬氨酸NADH穿梭维持胎儿肝造血干细胞的活性水平。
(Sonarsignal)MDH1-mediated malate-aspartate NADH shuttle maintains the activity levels of fetal liver hematopoietic stem cellsVisual Abstract造血干细胞在不同发育阶段的能量代谢和干细胞之间的关系仍然很大程度上是未知的。
我们为 NADH/NAD+ 传感器(SoNar)构建了一个转基因小鼠系,并根据 SoNar 荧光比率分析了3个不同的胎肝造血细胞群。
Sonar 值低的细胞线粒体呼吸水平增强,但糖酵解水平与sonar 值高的细胞相似。
有趣的是,10% 的 SoNar-low 细胞含有65% 的免疫表型胎儿肝造血干细胞(fl-hsc) ,并且比 SoNar-high 细胞含有大约五倍的功能性造血干细胞。
Sonar 能够敏感地监测体内外造血干细胞能量代谢的动态变化。
STAT3通过反式激活 MDH1维持苹果酸-天冬氨酸 NADH 的穿梭活性和 HSC 的自我更新和分化。
我们揭示了一个意想不到的fl-hsc 代谢程序,为造血干细胞或其他类型干细胞的代谢研究提供了一个强有力的遗传工具。
原文摘要:The connections between energy metabolism and stemness of hematopoietic stem cells (HSCs) at different developmental stages remain largely unknown.We generated a transgenic mouse line for the genetically encoded NADH/NAD+ sensor (SoNar) and demonstrate that there are 3 distinct fetal liver hematopoietic cell populations according to the ratios of SoNar fluorescence.SoNar-low cells had an enhanced level of mitochondrial respiration but a glycolytic level similar to that of SoNar-high cells. Interestingly, 10% of SoNar-low cells were enriched for 65% of total immunophenotypic fetal liver HSCs (FL-HSCs) and contained approximately fivefold more functional HSCs than their SoNar-high counterparts.SoNar was able to monitor sensitively the dynamic changes of energy metabolism in HSCs both in vitro and in vivo.Mechanistically, STAT3 transactivated MDH1 to sustain the malate-aspartate NADH shuttle activity and HSC self-renewal and differentiation.We reveal an unexpected metabolic program of FL-HSCs and provide a powerful genetic tool for metabolic studies of HSCs or other types of stem cells.Key Points•FL-HSCs mainly use oxidative phosphorylation but with normal glycolysis, as indicated by a highly responsive NADH/NAD+ sensor.•FL-HSC activities are tightly regulated by the STAT3/MDH1-mediated malate-aspartate NADH shuttle.Subjects:Hematopoiesis and Stem CellsTopics:aspartate, fetus, fluorescence, liver, malates, mice, transgenic, mitochondria, nicotinamide adenine dinucleotide (nad), stat3 protein, hematopoietic stem cellsIntroductionHematopoietic stem cells (HSCs) originate from the aorta-gonad-mesonephros region1 and migrate into the fetal liver (FL) and undergo dramatic expansion,2,3 gradually localizing to and residing in the bone marrow niche after birth.4HSCs can self-renew to maintain the stem cell pool and generate all downstream progenitors and terminally differentiate into multiple lineages.5,6Increasing evidence indicates that the metabolic state is tightly connected to HSC activity.7-9Adult HSCs preferentially undergo glycolysis, rather than oxidative phosphorylation, in the hypoxic niche,7,10,11 which is extensively regulated by several signaling pathways, including HIF1A,12 MYC,13 PDK,14 DLK-GTL2,15 and vitamin A–retinoic acid signaling.16We have also shown that both murine and human HSCs adopt a glycolytic metabolic profile under certain conditions andthat this profile is fine-tuned by MEIS1/PBX1/HOXA9/HIF1A signaling pathways.17-19Interestingly, recent studies have suggested that adult HSCs also have high mitochondrial mass and enhanced dye efflux but possess limited respiratory and turnover capacity,20 which indicates that mitochondria are likely required for the function of adult HSCs, as evidenced by the fact that FOXO3 serves as a regulator to couple mitochondrial metabolism with HSC homeostasis.21The metabolic profiles of FL-HSCs and the effects of metabolism on HSC function, however, remain largely unknown.FL-HSCs undergo rapid division/expansion, conceivably through an increased demand on energy sources compared with that needed by adult HSCs, which are usually maintained in a relatively quiescent state.It is also possible that distinct microenvironments in different hematopoietic organs may affect the metabolism of HSCs.Interestingly, a recent report showed that loss of Rieske iron-sulfur protein, a mitochondrial complex III subunit, impairs the quiescent status of adult HSCs and the differentiation capacity of FL-HSCs.22FL-HSCs seem to have increased expression levels of many mitochondrial respiration–related genes, although whether metabolic status determines the cell fate of FL-HSCs remains unknown.23Results from previous studies indicate that mitochondrial activity may play a role in HSCs in the FL stage, although the detailed metabolic profiles and their underlying mechanisms await further investigation.Because of limitations in the availability of HSCs, moststudies related to the nutrient metabolism of HSCs have depended heavily on flow cytometric analysis with MitoTracker dyes, TMRE, and DCFDA to determine mitochondrial mass, membrane potential, and ROS level, respectively.Improved techniques have been used to measure several metabolic features of HSCs, such as oxygen consumption and lactate generation9,24 ; however, these studies may not directly reflect the true extent of glycolysis, oxidative phosphorylation, or other metabolic processes in HSCs.Recent studies have provided interesting evidence showing that it is feasible to perform a metabolomic analysis with fewer than 104 HSCs to explore the metabolic networks of different types of nutrients.25Nevertheless, it remains difficult to detect all of the metabolites sensitively with a limited number of HSCs using conventional metabolomic analysis.Few tools are available for real-time imaging of metabolic states in live HSCs, either in vitro or in vivo. Therefore, alternative approaches, such as metabolite biosensors, are required for the direct, precise, and real-time detection of subtle changes in nutrient metabolism in HSCs.Recently, we developed a highly responsive NADH/NAD+ sensor, called SoNar,26 which was designed by inserting cpYFP into the NAD(H)-binding domain of T-Rex.SoNar shows distinct fluorescence responses to NADH and NAD+.Inside the cell under physiological conditions, the total intracellular pool of NAD+ and NADH in the range of hundreds of micromolars27-30 far exceeds the dissociation constants of SoNar for NAD+ (K d, 5.0 μM) and NADH (K d, 0.2 μM);thus, the sensor would be occupied by either NAD+ or NADH molecules, and its steady-state fluorescence would report the NAD+/NADH ratio rather than the absolute concentrations of either of the 2 nucleotides according to equilibrium thermodynamics.26In addition, SoNar fluorescence is intrinsically ratiometric (比率计), with 2 excitation wavelengths, and its fluorescence excited at 420 (or 405) and 485 nm shows opposing responses to ligand binding.26,31This ratiometric property of a sensor is highly desired for quantitative imaging in live cells and in vivo,32,33 because it eliminates the differences in instrumental efficiency, environmental effects, and probe concentration, enabling it to be widely used in different biological samples.The SoNar sensor has a 15-fold (or 1500%) dynamic range, enabling us to measure the cytosolic NAD+/NADH ratio from 0.8 to 2000.26,31,34Interestingly, SoNar has many desirable properties that make it an ideal sensor; it has a rapid response, high sensitivity, intense fluorescence, and large dynamic range, and it is capable of reporting subtle perturbations in many pathways affecting energy metabolism, including glycolysis and mitochondrial respiration.We generated SoNar transgenic mice and examined the metabolic profiles of FL-HSCs and their contributions/connections to cell fate determinations, as well as the underlying mechanisms governing FL-HSC function.参考文献:/10.1182/blood.2019003940HEMATOPOIESIS AND STEM CELLS| JULY 30, 2020----------------------------------------------------------------------------------------------------------------------------MethodsMiceSoNar DNA consists of the sequence of cpYFP, truncated T-Rex (78-211), and the linkers between them.26 Its coding gene (1.2 kbp) is much smaller than those of the first-generation NADH sensors Peredox (2.8 kbp) and Frex35 (1.8 kbp). For transgenic studies, smaller is better for the expression of the sensor in cells and in vivo.31 To generate SoNar transgenic mice, SoNar DNA was cloned into the pCAG vector with chicken β-actinpromoter. The targeting construct was linearized, purified, and microinjected into FVB blastocysts. SoNar DNA was randomly incorporated into the genome and determined by polymerase chain reaction (PCR) assay. Messenger RNA (mRNA) and protein expressions of SoNar in different tissues of SoNar mice were further evaluated by both reverse transcription PCR (RT-PCR) and fluorescence microscopy. The resulting chimeric mice were bred with FVB mice to obtain germ line transmission. These mice were next backcrossed with a C57BL/6 CD45.2 background, and germ line transmission was checked by PCR and flow cytometry. Heterozygote transgenic SoNar mice were used for most of the experiments in the current study. C57BL/6 CD45.2 mice were purchased from the Shanghai SLAC Laboratory Animal Co., Ltd. CD45.1 mice were provided by Dr Jiang Zhu at Shanghai Jiao Tong University School of Medicine. All animal experiments were conducted according to the Guidelines for Animal Care at Shanghai Jiao Tong University School of Medicine. All these materials, including SoNar sensor, are available upon request.In vivo imaging of SoNar transgenic miceGenotyping, mRNA expression, and histology of SoNar transgenic miceCompetitive reconstitution assayMetabolic imaging and quantification of cytosolicNAD+/NADH ratio in living cellsReal-time metabolic imaging in the BM nicheFlow cytometryUltrahigh-performance LC–qTOF–MS analysisMicroarray and quantitative RT-PCRMetabolic analysisOxygen consumption rate (OCR) and extracellularacidification rate were determined in CD45.2+ SoNar-high and -low FL hematopoietic cells with the XF Cell Mito Stress Test Kit (Seahorse #103015-100) and XF Glycolysis Stress Test Kit (Seahorse #103020-100) according to the manufacturer’s instructions using a Seahorse XF96 analyzer. In brief, for the OCR analysis, 3 × 105 SoNar-high and -low FL hematopoietic cells were incubated in the 37°C carbon dioxide–free incubator in 175 μL of assay medium (XF Base Medium with 2 mM of glutamine, 1 mM of pyruvate, and 10 mM of glucose [pH, 7.4]; 37°C); 1.5 μM of oligomycin, 2 μM of FCCP, and 0.5 μM of rotenone/antimycin A were loaded in injection ports A, B, and C, respectively. For the detection of extracellular acidification rate, 3 × 105 CD45.2+ SoNar-high and -low FL hematopoietic cells were incubated in the 37°C carbon dioxide–free incuba tor in 175 μL of assay medium (XF Base Medium with 1 mM of glutamine [pH, 7.4]; 37°C); 10 mM of glucose, 1.5 μM of oligomycin, and 100 mM of 2-DG were loaded into injection ports A, B, and C, respectively. ATP level was analyzed using the ATP Bioluminescence Assay Kit HS II (Roche) according to the manufacturer’s protocol, and data were normalized to cell count. T o analyze the mitochondrial DNA (mtDNA) copy numbers, total genomic DNA was extracted from the indicated cells for comparing the copies of the mitochondrial-specific mt-ND4 gene with those of the nuclear B2m gene. The primer sequences used are shown in supplemental Table 1.For intracellular and extracellular pyruvate and lactate assays, the extracts were prepared from 5 × 106 CD45.2+ FL hematopoiet ic cells using 300 μL of ice-cold 0.5-M perchloric acid for each sample. Extracts were centrifuged at 10 000 g for 5 minutes at 4°C, and the supernatant was neutralized with 5 M ofKOH and centrifuged at 10 000 g for 5 minutes at 4°C. The supernatant was removed for assay. For the pyruvate assay, 180 μL of assay buffer (100 mM of potassium chloride [pH, 6.7], 1 mM of EDTA, 0.1% bovine serum albumin, 10 μM of flavin adenine dinucleotide, 0.2 mM of thiamine pyrophosphate, 0.5 U of pyruvate oxidase, 0.2 U of h orseradish peroxidase, and 50 μM of AmplexRed) was added to a 96-well plate containing 20 μL of the cell extract or medium containing extracellular pyruvate. Changes in fluorescence were measured every 30 seconds for 15 minutes at 37°C by a Synergy 2 Multi-Mode Microplate Reader with an excitation filter of 530 BP 40 nm and emission filter of 590 BP 35 nm at 37°C. Calibration experiments were performed with 20 μL of pyruvate standards (0, 10, 20, 40, 60, 100, and 200 μM per well). For the lactate assay, 180μL of assay buffer (PBS [pH, 7.4], 0.1% bovine serum albumin, 500 μM of NAD+, 0.5 U of lactate dehydrogenase (LDH), 0.2 U of diaphorase, and 10 μM of resazurin) was added to a 96-well plate containing 20 μL of the cell extract or medium containing extracellular lactate. Changes in fluorescence were measured every 30 seconds for 15 minutes at 37°C by a Synergy 2 Multi-Mode Microplate Reader with an excitation filter of 540 BP 25 nm and emission filter of 590 BP 35 nm at 37°C. Calibration experiments were performed with 20 μL of lactate standards (0, 10, 20, 40, 60, 100, and 200 μM per well). All samples were diluted to fit within the range of the standard curve and run in triplicate. For the evaluation of NADH/NAD+ ratios by a biochemical assay, 1 million SoNar-high and -low FL hematopoietic cells were sorted from E14.5 FLs and subjected to measurement of NADH/NAD+ level using a commercially available NADH/NAD+ assay kit (Sigma #MAK037) according to the manufacturer’s instructions.Immunoblot analysisSingle cell colony formingThe single CD45.2+ SoNar-high or -low FL hematopoietic cell was freshly isolated and plated onto a 35-mm poly-D-lysine hydrobromide-coated glass-bottom dish (Cellvis) and cultured in Stemspan serum-free medium (Stemcell Technologies) containing 10 ng/mL of murine SCF and 10 ng/mL of murine TPO (Peprotech) for 96 hours. The ratios of daughter cells derived from a single parent cell were recorded with excitation at 405 and 488 nm using Nikon A1 confocal microscopy and analyzed with Image J software.Luciferase reporter assaysThe luciferase reporter vector pGL4.27 containing the Mdh1 promoter was constructed to identify transcriptional activation of Mdh1 by STAT3. Indicated doses of pLVX-Stat3 (or negative control vector) plasmid along with pGL4.27-mdh1 promoter vector were cotransfected into 293T cells. Luciferase activities were measured according to the manufacturer’s instructions (Promega #E1910) by using a luciferase reporter system (GloMax Multi Instrument) 24 hours after transfection. ChIP assaysChromatin immunoprecipitation (ChIP) assays were performed using the ChIP Assay Kit (Beyotime #P2078). Briefly, 293T cells were overexpressed with pGL4.27-mdh1 promoter vector and STAT3 (with Strep II tag) crosslinked with 1% formaldehyde (S igma) at 37°C for 10 minutes, and precleared DNA was then used for immunoprecipitation with 4 mL of anti–Strep II antibody (Genescript) or rabbit control immunoglobulin G (CST) at 4°C overnight. For the sample input, 1% of the sonicated pre-cleared DNA was purified at the same time withthe precipitated immune complex. The ChIP samples were purified by the Gel and PCR Clean Up Kit (Necleospin). The STAT3-binding sequence was amplified by semiquantitative PCR using primers specific for the Mdh1 promoter region as listed in supplemental Table 1.Methylation-specific PCR assayGenomic DNA was extracted from 500 000 CD45.2+ SoNar-high and -low FL hematopoietic cells using a DNA extraction kit (Generay Biotech #GK0122). The promoter methylation status of MDH1 was determined by sodium bisulfate to convert unmethylated (but not methylated) cytosine to uracil, followed by analysis by methylation-specific PCR to amplify specifically either methylated or unmethylated DNA using the Zymoresearch kit (EZ DNA Methylation-Direct Kit D5020) according to the manufacturer’s instruction. The methylation-specific PCR primers are listed in supplemental Table 1.Statistical analysisStatistical analysis was performed using GraphPad and SPSS software (version 19.0). Data are represe nted as mean ± standard error of the mean. n represents the number of independent experiments or the number of cells or mice per group from independent experiments. All experiments were performed independently 3 to 5 times. Data were analyzed with a Student t test (2 tailed), 1-way analysis of variance with Tukey’s multiple comparison test, or 2-way analysis of variance with Sidak’s multiple comparison test according to the experimental design, and statistical significance was set at P < .05.Establishment of pan-tissue SoNar transgenic mice.Figure 2.SoNar indicates metabolically distinct populations of FL hematopoietic cells.Figure 3.SoNar-low FL hematopoietic cells exhibit similar glycolytic but enhanced mitochondrial activity compared with SoNar-high cells.Figure 4.Functional HSCs are enriched in SoNar-low FLhematopoietic cells.Figure 5.FL-HSCs respond differently to AOA stimulation in the BM niche compared with adult HSCs.Figure 6.MDH1 enhances the malate-aspartate NADH shuttle and decreases NADH/NAD+ level in SoNar-low FL-HSCs.Figure 7.STAT3 transactivates Mdh1 expression to maintain FL-HSC activities.。
苹果酸-天冬氨酸穿梭名词解释

苹果酸-天冬氨酸穿梭名词解释
苹果酸-天冬氨酸穿梭(Malate-Aspartate Shuttle,也称为苹果酸穿梭)是真核细胞中一个转运胞质NADH的还原性氢进入线粒体,参与氧化磷酸化的穿梭代谢途径。
苹果酸-天冬氨酸穿梭是真核细胞中一个重要的生物化学体系,它能够将胞质NADH的电子通过线粒体内膜上的苹果酸-天冬氨酸穿梭运送至线粒体,参与氧化磷酸化。
这个过程需要苹果酸接受胞质NADH脱氢,转化为苹果酸进入线粒体,在线粒体中重新氧化成草酰乙酸,生成的NADH进入呼吸链,草酰乙酸通过转氨反应以天冬氨酸的形式回到胞液,完成穿梭。
生化习题(名解、间答题)答案

第1章绪论习题答案三、名词解释1.生物化学:即生命的化学,它是分子水平上研究生物体的化学组成和生命过程中化学变化规律的一门科学。
2.分子生物学:它是生物化学的重要组成部分,它主要是研究生物大分子的结构与功能,代谢及其调控的一门学科。
3.生物大分子:是由基本结构单位按一定顺序和方式连接而成的,相对分子质量在104以上具有特定构象与特异功能的生物分子(多聚体)。
四、简答题1简述近代生物化学的发展简史答:近代生物化学的发展经历了三个发展阶段(时期)。
即(1)初期(生化萌芽时期):从18世纪中叶至20世纪初。
这一时期主要是研究生物体的化学组成,客观描述组成生物体的物质含量、分布、结构、性质与功能,故又称为叙述生化。
(2)蓬勃发展期:从20世纪次至20世纪的50年代的几十年间,生物化学发展迅猛。
这一时期,除了在营养、内分泌及酶学等方面有许多重大发现与进展外,更主要的进展是物质的代谢与调节,许多代谢途径基本弄清楚,故此阶段又称为动态生化阶段。
(3)分子生物学时期:20世纪50年代以来,生物化学的发展进入一个新的高潮。
这一时期生化领域的重大事件层出不穷。
2.简述当代生物化学研究的主要内容答:(1)生物分子的结构与功能生物体是由一些生物分子所组成,生物分子复杂多样,有无机物和有机物,有机物中又有小分子的有机物和生物大分子。
(2)物质代谢及其调节生物体内的物质代谢非常活跃,错综复杂,但又有条不紊。
(3)基因(遗传)信息传递及其调控。
3.简述生物化学与医学的关系答:生物化学称为医学学科的基础,是一门重要的医学必修课程所有医学(基础医学、临床医学、预防医学、药学、口腔医学、护理学)都必须修该门课程。
因为生物化学与分子生物学的理论与技术已渗入到所有医学(当代医学、大医学)的各个领域,促进了现代医学突飞猛进的发展,反过来,现代医学各学科又不断地向生物化学与分子生物学提出问题和挑战,从而推动生物化学与分子生物学不断深入研究与发展。
生化名词解释

20. PFK2 06
磷酸果糖激酶2:磷酸果糖激酶可作用于果糖-6-磷酸,使ATP的磷酸基转移到果糖-6-磷酸上。磷酸果糖激酶有两种,分为1和2,其中磷酸果糖激酶1作用可得果糖-1,6-二磷酸。 由磷酸果糖激酶2作用可得果糖-2,6-二磷酸
30. one-carbon group 05
氨基酸分解代谢中产生含有一个碳原子的基团的集合。一碳单位是嘌呤和嘧啶的合成原料,是氨基酸和核苷酸联系的纽带。它具有两个特点:1.不能在生物体内以游离形式存在;2.必须以四氢叶酸为载体。
31. melting temperature of DNA 05
15. glucokinase(in liver) 07&04 见生化笔记
葡萄糖激酶:催化葡萄糖磷酸化形成葡萄糖-6-磷酸的没有两个:一个是己糖激酶,另一个是葡萄糖激酶。其中己糖激酶专一性不强,被产物G-6-P别构抑制;葡萄糖激酶存在于肝脏中,对葡萄糖有专一活性,且不被G-6-P抑制。它是一种诱导酶,由胰岛素促使合成。
4.PRPP 2007&04 p392下册
全称5-磷酸核糖-1-焦磷酸, 是组氨酸和色氨酸生物合成中的关键性中间产物,是核苷酸中核糖磷酸部份的供体. 它也参与多种生物合成.
5. γ-carboxyglutamate 2007 p440上册γ羧基谷氨酸
谷氨酸经过依赖维生素K的谷氨酰羧化酶催化可变为γ羧基谷氨酸,它是血液凝固中所必需的,因为只有谷氨酸经羧化才能结合Ca2+,进而与膜中的磷脂结合,凝血酶原才能被蛋白酶水解为凝血酶,才能发挥凝血功能。经常流鼻血就有可能是维生素K缺乏导致。
苹果树一天冬氨酸穿梭作用名词解释

苹果树一天冬氨酸穿梭作用名词解
释
苹果树是我们常见的果树之一,它能够在不同的季节提供给我们不同口感的水果。
在苹果树成长的过程中,它需要各种营养物质的支持才能快速、健康地生长。
而冬氨酸就是其中一个重要的营养物质。
冬氨酸(aspartic acid)是一种天然的α-氨基酸,它是身体内蛋白质的合成和生长的关键所在,也是重要的人体代谢产物。
冬氨酸在苹果树中担任很多重要的角色,其中最重要的是它在大量存在于苹果树中的酸性质的结构中扮演着重要的角色。
苹果汁中的pH基本为3.5,非常酸性,但是苹果树却可以健康地生长,其中一个原因就是冬氨酸可以在其生长中发挥重要的作用。
冬氨酸是在众多蛋白质中的一种氨基酸,能够汇集在一起,形成苹果树中多肽,起到维持苹果树生命的任务。
这种氨基酸也可以很好地与其他氨基酸结合,形成更复杂的蛋白质结构。
不仅如此,冬氨酸还能够帮助苹果树在寒冷的冬季中存活下来。
在寒冷的冬季,冬氨酸会发挥“穿梭作用”,把温度更低的地方导向暖和的地方,这样就能够维持苹果
树内部的温度稳定,保证苹果树能够在严寒的冬天中继续生长。
但如果过少摄取冬氨酸,苹果树就可能面临一些问题,比如酸性结构的维护不足、树木无法在寒冷天气中良好生长等。
因此在苹果树栽培中,及时补充冬氨酸非常重要。
总的来说,冬氨酸在苹果树的生长过程中有很重要的作用。
它可以帮助苹果树维持酸性结构、维持苹果树内部的温度稳定、形成复杂的蛋白质结构、帮助苹果树在严寒的冬天中继续生长等。
因此,苹果树栽培中,正确的冬氨酸摄取方式是非常重要的。
2022生化思考题详细答案解析(医学本科生适用)
生物化学思考题1、叙述L-α氨基酸结构特征,比较各种结构异同并分析结构与性质的关系。
结构特点:氨基酸是较酸分子的a-氢原子被氨基取代直接形成的有机化合物,即当氨基酸的氨基与殁基连载同一个碳原子上,就成为a-氨基酸。
氨基酸中与竣基直接相连的碳原子上有个氨基,这个碳原子上连的集团或原子都不一样,称手性碳原子,当一束偏振光通过它们时,光的偏振方向将被旋转,根据旋转的方向分为左旋和右旋即D系和L系,L-a-氨基酸再被骗争光照射时,光的偏正方向为左旋。
R为侧链,连接-COOH的碳为a-碳原子为不对称碳原子(除了甘氨酸)不同的氨基酸其R基团结构各异。
根据测链结构可分为:①含煌链的为非极性脂肪族氨基酸,如丙氨酸;②含极性不带电荷的为极性中性氨基酸,如半胱氨酸;③含芳香基的为芳香族氨基酸,如酪氨酸;④含负性解离基团的为酸性氨基酸,如谷氨酸;⑤含正性解离基团的为碱性氨基酸,如精氨酸。
2、简述蛋白质一级结构、二级结构、三级结构、四级结构基本概念及各结构层次间的内在关系。
蛋白质的一级结构就是蛋白质多肽链中氨基酸残基的排列顺序,也是蛋白质最基本的结构。
主要化学键是肽键,二硫键也是一级结构的范畴。
蛋白质的二级结构是指多肽链中主链原子的局部空间排布即构象,不涉及侧链部分的构象。
主要化学键为氢犍。
蛋白质的多肽链在各种二级结构的基础上再进一步盘曲或折迭形成具有一定规律的三维空间结构,称为蛋白质的三级结构,蛋白质三级结构的稳定主要靠次级键,包括氢键、疏水键、盐键以及范德华力等。
具有二条或二条以上独立三级结构的多肽链组成的蛋臼质,其多肽链间通过次级键相互组合而形成的空间结构称为蛋白质的四级结构,其中,每个具有独立三级结构的多肽链单位称为亚基。
层次之间的关系:一级结构是空间构象的基础,决定高级结构;氨基酸的残基影响二级结构的形成,二级结构以一级结构为基础;在二级结构的基础上,肽链还按照一定的空间结构进一步形成更复杂的三级结构;具有三级结构的多肽链按一定空间排列方式结合在一起形成的聚集体结构称为蛋白质的四级结构。
生物化学问答题(含答案)
蛋白质化学1.蛋白质:是一类生物大分子,有一条或多条肽链构成,每条肽链都有一定数量的氨基酸按一定的序列以肽键连接形成。
蛋白质是生命的物质基础,是一切细胞和组织的重要组成成分。
2.标准氨基酸:是可以用于合成蛋白质的20种氨基酸。
7.氨基酸的等电点:氨基酸在溶液中的解离程度受PH值的影响,在某一PH值条件下,氨基酸解离成阳离子和阴离子的程度相等,在溶液中的氨基酸以间性离子形式存在,且净电荷为0,此时溶液的PH值成为该氨基酸的等电点9.缀合蛋白质:含有非氨基酸成分的蛋白质10.蛋白质的辅基:缀合蛋白所含有的非氨基酸成分12.肽键:存在与蛋白质和肽分子中,是有一个氨基酸的ɑ-羧基与另外一个氨基酸的ɑ-氨基缩合时形成的化学键14.肽:是指由2个或多个氨基酸通过肽键连接而成的分子15.氨基酸残基:肽和蛋白质中的氨基酸是不完整的,氨基失去了氢,羧基失去了羟基,因而称为氨基酸残基16.多肽:由10个以上氨基酸通过肽键连接而成的肽18.生物活性肽:是指具有特殊生理功能的肽类物质,它们多为蛋白质多肽链的一个片段,当被降解释放之后就会表现出活性,例如参与代谢调节、神经传导。
食物蛋白质的消化产物也有生物活性肽,它们可以被直接吸收。
20.蛋白质的一级结构:通常叙述为蛋白质多肽链种氨基酸的链接顺序,简称为氨基酸序列,蛋白质的一级结构反应蛋白质分子的共价键结构21.蛋白质的二级结构:是指蛋白质多肽链局部片段的构象,该片段的氨基酸序列式连续的,主链构象通常是规则的23.蛋白质的超二级结构:又称模体基序,是指几个二级结构单元进一步聚合和结合形成的特定构象单元,如ɑɑ、βɑβ、ββ、螺旋-转角-螺旋、亮氨酸拉链等24.蛋白质的三级结构:是指蛋白质分子整条肽链的空间结构,描述其所有原子的空间分布,蛋白质三级结构的形成是肽链在二级结构的基础上进一步折叠的结果。
26.蛋白质的亚基:许多蛋白质分子可以用物理方法分离成不止一个结构单位,每个结构单位可以有不止一条肽链构成,但都有特定且相对独立的三级结构,且是由一个共价键连接的整体,该结构单位称为该蛋白质的一个亚基27.蛋白质的四级结构:多亚基蛋白的亚基与亚基通过非共价键结合,形成特定的空间结构,这一结构层次称为该蛋白质的四级结构35.变构蛋白:具有下列特性蛋白质的统称:它们有两种或多种构象,有两个或多个配体结合位点,配体与其中一个结合位点结合导致蛋白质变构,及从一种构象转换成另一种构象,这种变构影响到其他配体结合位点与配体的结合36.变构剂:导致变构蛋白变构的物质,多为小分子42.蛋白质的等电点:蛋白质是两性的电解质其解离状态受溶液的PH值影响,在某一PH值条件下,蛋白质的净电荷为0,该PH值称为该蛋白质的等电点44.蛋白质变性:由于稳定蛋白质构象的化学键被破坏,造成其四级结构三级结构甚至二级结构被破坏,结果其天然构象部分或全部改变,变性导致蛋白质理化性质改变,生物活性丧失。
包含苹果酸-天冬氨酸穿梭抑制剂及抗癌剂作为有效成分的癌症预防
专利名称:包含苹果酸-天冬氨酸穿梭抑制剂及抗癌剂作为有效成分的癌症预防及治疗用药学组合物
专利类型:发明专利
发明人:金铢烈
申请号:CN201880024463.8
申请日:20180413
公开号:CN110636841A
公开日:
20191231
专利内容由知识产权出版社提供
摘要:本发明涉及将苹果酸‑天冬氨酸穿梭(MAS,malate‑aspartate sh uttle)抑制剂用作癌症治疗剂的技术,更具体地,提供包含作为苹果酸‑天冬氨酸穿梭抑制剂的苯基琥珀酸(Phenyl Succinic Acid)、甲基丙二酸(Methyl Malonic Acid)、N‑(1‑芘基)马来酰亚胺(N‑(1‑pyrenyl)maleimide)及邻羧基苯乙酮酸(Phthalonic Acid)或上述苹果酸‑天冬氨酸穿梭及抗癌剂的混合物作为有效成分的癌症预防及治疗用药学组合物。
申请人:国立癌症研究中心
地址:韩国京畿道
国籍:KR
代理机构:北京聿宏知识产权代理有限公司
更多信息请下载全文后查看。
中医大16年6月考试《生物化学(本科)》复习题
中医大16年6月考试《生物化学(本科)》复习题一.单选题1.下列哪一种氨基酸是亚氨基酸A.脯氨酸B.焦谷氨酸C.亮氨酸D.丝氨酸E.酪氨酸2.关于蛋白质分子三级结构的描述,其中错误的是A.天然蛋白质分子均有这种结构B.有三级结构的多肽链都具有生物学活性C.三级结构的稳定性主要是次级键维系D.亲水基团聚集在三级结构的表面E.决定盘曲折叠的因素是氨基酸残基3.含有Ala,Asp,Lys,Cys的混合液,其pI依次分别为6.0,2.77,9.74,5.07,在pH 为9环境中电泳分离这四种氨基酸,自正极开始,电泳区带的顺序是A.Ala,Cys,Lys,AspB.Asp,Cys,Ala,LysC.Lys,Ala,Cys,AspD.Cys,Lys,Ala,AspE.Asp,Ala,Lys,Cys4.亚基与亚基之间呈特定的三维空间排布,并以非共价键相连接的结构称之为A.模体B.二级结构C.三级结构D.四级结构E.结构域5.形成稳定的肽链空间结构,非常重要的一点是肽键中的四个原子以及和它相邻的两个α-碳原子处于A.不断绕动状态B.可以相对自由旋转C.同一平面D.随不同外界环境而变化的状态E.不同平面6.下列有关谷胱甘肽叙述正确的是A.谷胱甘肽N端的羧基是主要的功能基团B.谷胱甘肽中谷氨酸的α-羧基是游离的C.谷胱甘肽是体内重要的氧化剂D.谷胱甘肽的C端羧基是主要的功能基团E.谷胱甘肽所含的肽键均为α肽键7.蛋白质的等电点是A.蛋白质带正电荷或负电荷时的溶液pH值B.蛋白质溶液的pH值等于7时溶液的pH值C.蛋白质分子呈兼性离子,净电荷为零时溶液的pH值D.蛋白质分子呈正离子状态时溶液的pH值E.蛋白质分子呈负离子状态时溶液的pH值8.关于蛋白质亚基的描述,哪项正确A.整条多肽链呈螺旋结构B.两条以上多肽链形成三级结构C.两条以上多肽链与辅基结合成蛋白质D.每个亚基都有各自的三级结构E.每个亚基都具有生物学功能9.下列关于肽单元的叙述,正确的是A.是组成多肽链的三级结构的基本单位B.C=O、N-H四个原子在同一个平面C.肽键中的氢和氧总是顺式结构D.在肽单元中与α-碳相连的单键不能自由旋转E.相邻两个肽单元的相互位置与α-碳两侧单键旋转无关10.DNA分子的腺嘌呤含量为20%,则胞嘧啶的含量应为A.20%B.30%C.40%D.60%E.80%11.酶的活性中心是指酶分子A.其中的必需基因B.其中的辅基C.与底物结合部位D.催化底物变成产物的部位E.结合底物并发挥催化作用的关键性三维结构区12.关于酶的化学修饰的叙述,错误的是A.有活性与无活性两种形式B.有放大效应C.两种形式的转变有酶催化D.两种形式的转变有共价变化E.化学修饰不是快速调节13.某一酶促反应的速率为最大速率的80%时,Km等于A.[S]B.1/2[S]C.1/4[S]D.0.4[S]E.0.8[S]14.反竞争性抑制剂的动力学效应是使A.K m值升高,V max不变B.K m值降低,V max不变C.K m值不变,V max升高D.K m值不变,V max降低E.K m值和V max均降低15.全酶是指A.酶蛋白—底物复合物B.酶蛋白—抑制剂复合物C.酶蛋白—别构剂复合物D.酶蛋白的无活性前体E.酶蛋白—辅助因子复合物16.同工酶的特点是A.催化作用,分子组成和理化性质相同,但组织分布不同的酶B.催化作用分子组成相同,辅酶不同C.多酶体系中酶组分的统称D.一类催化作用相同,分子组成和理化性质不同的酶E.催化同一底物起不同反应的酶的总称17.有机砷化合物对酶的抑制作用,可用下列哪种方法解毒A.加入适量的半胱氨酸B.加入过量的GSHC.加入适量的甲硫氨酸D.加入适量的二巯基丙醇E.超滤18.1分子葡萄糖有氧氧化时,经底物水平磷酸化方式可产生的ATP是A.2B.3C.4D.6E.819.与丙酮酸在线粒体内氧化无关的酶促反应是A.苹果酸酶反应B.琥珀酸脱氢酶反应C.异柠檬酸脱氢酶反应D.丙酮酸脱氢酶反应E.α-酮戊二酸脱氢酶反应20.磷酸戊糖途径的限速酶是A.6-磷酸葡萄糖酸脱氢酶B.内酯酶C.6-磷酸葡萄糖脱氢酶D.己糖激酶E.转酮醇酶21.存在于肌肉、脂肪组织中的葡萄糖转运体是A.GLUT1B.GLUT2C.GLUT3D.GLUT4E.GLUT522.与脂酸β-氧化无关的酶是A.脂酰辅酶A脱氢酶B.β-羟脂酰辅酶A脱氢酶C.Δ2-烯酰辅酶A水化酶D.β-酮脂酰辅酶A硫解酶E.β-酮脂酰还原酶23.关于酮体的叙述,正确的是哪一项A.不能为机体所利用B.是甘油在肝脏代谢的特有中间产物C.主要为肝脏本身利用D.在肝细胞的线粒体中生成E.在血中与清蛋白结合运输24.合成酮体的限速酶是A.HMGCoA合酶B.HMGCoA裂解酶C.HMGCoA还原酶D.脂酰CoA合成酶E.乙酰CoA羧化酶25.催化甘油磷脂水解变为溶血磷脂的酶是A.磷脂酶AB.磷脂酶B1C.磷脂酶B2D.磷脂酶CE.磷脂酶D26.胆固醇合成原料乙酰CoA从线粒体运至胞液的途径是A.蛋氨酸循环B.葡萄糖-丙氨酸循环C.鸟氨酸酸循环D.柠檬酸-丙酮酸循环E.嘌呤核苷酸循环27.蛋白质含量最高的血浆脂蛋白是A.HDLB.IDLC.LDLD.VLDLE.CM28.Apo C II可激活A.LPLB.LCATC.HLD.ACATE.HSL29.有关甘油磷脂的叙述正确的是A.除脑组织外,几乎全身各组织均可合成B.其结构特点是2位通常结合不饱和脂酸C.心磷脂是细胞膜中最丰富的磷脂之一D.在PLB的作用下产生溶血磷脂E.其主要生理功能是氧化供能30.苹果酸-天冬氨酸穿梭涉及下列哪种氨基酸A.AsnB.GluC.AlaD.LysE.Val31.不是琥珀酸氧化呼吸链的成分为A.铁硫蛋白B.FMNC.CoQD.CytcE.Cytc132.呼吸链中不具有质子泵功能的是A.复合体ⅠB.复合体ⅡC.复合体ⅢD.复合体ⅣE.复合体Ⅰ和复合体Ⅳ33.1分子NAD+在电子传递链中作为递氢体,可接受A.2个氢原子B.2个电子C.2个氢原子和1个电子D.2个氢原子和2个电子E.1个氢原子和1个电子34.S-腺苷甲硫氨酸的主要作用是A.生成腺嘌呤核苷B.合成四氢叶酸C.补充甲硫氨酸D.合成同型半胱氨酸E.提供甲基35.体内转运一碳单位的载体是A.维生素B12B.叶酸C.四氢叶酸D.生物素E.S-腺苷甲硫氨酸(S-腺苷蛋氨酸)36.决定蛋白质营养价值高低的是A.氨基酸的种类B.氨基酸的数量C.必需氨基酸的数量D.必需氨基酸的种类E.必需氨基酸的数量、种类及比例37.下列不属于一碳单位的是A.=CH-B.-CH2-C.―CH3D.―CHOE.CO238.可与谷丙转氨酶共同催化丙氨酸和α-酮戊二酸反应产生游离氨的酶是A.谷氨酸脱氢酶B.谷草转氨酶C.谷氨酰胺酶D.谷氨酰胺合成酶E.α-酮戊二酸脱氢酶39.精氨酸分解的产物除了尿素外还有1分子A.鸟氨酸B.瓜氨酸C.天冬氨酸D.谷氨酸E.延胡索酸40.ALT活性最高的组织为A.心肌B.脑C.肝D.骨骼肌E.肾41.以整个分子掺入嘌呤环的氨基酸是A.丝氨酸B.天冬氨酸C.甘氨酸D.丙氨酸E.谷氨酸42.HGPRT(次黄嘌呤-鸟嘌磷酸核糖转移酶)参与下列哪种反应A.嘌呤核苷酸从头合成B.嘌呤核苷酸补救合成C.嘧啶核苷酸从头合成D.嘧啶核苷酸补救合成E.嘌呤核苷酸分解代谢43.别嘌呤醇治疗痛风的机制是该药抑制A.黄嘌呤氧化酶B.腺苷脱氨酸C.尿酸氧化酶D.鸟嘌呤脱氢酶E.黄嘌呤脱氢酶44.嘧啶环中第二位C原子来自B.天门冬氨酸C.氨甲酰磷酸D.乙酸E.CO245.在体内能分解为β-氨基异丁酸的核苷酸是A.CMPB.AMPC.TMPD.UMPE.IMP46.生物体内物质代谢调节最基础的水平是A.细胞水平B.激素水平C.神经调节D.整体水平E.器官水平47.关于糖、脂、氨基酸代谢的叙述,错误的是A.乙酰CoA是糖、脂、氨基酸分解代谢共同的中间代谢物B.三羧酸循环是糖、脂、氨基酸分解代谢的最终途径C.当摄入糖量超过体内消耗时,多余的糖可转变为脂肪D.当摄入大量脂类物质时,脂类可大量异生为糖E.糖、脂不能替代蛋白质48.糖类、脂类、氨基酸氧化分解时,进入三羧酸循环的主要物质是A.异柠檬酸B.α-酮酸C.苹果酸D.乙酰CoAE.草酰乙酸49.底物对酶含量的影响,通常是A.阻遏酶蛋白的合成B.诱导酶蛋白的合成C.促进酶蛋白的降解D.抑制酶蛋白的降解E.使酶蛋白的含量不变50.通过细胞膜受体起作用的激素是A.雌激素B.肾上腺素C.孕激素D.糖皮质激素E.甲状腺素51.DNA复制中,不需要下列哪种酶A.DNA指导的DNA聚合酶B.DNA指导的RNA聚合酶D.拓扑异构酶E.限制性核酸内切酶52.DNA连接酶的作用是A.解决复制解链过程中的打结、缠绕现象B.合成RNA引物C.使DNA形成负超螺旋结构D.连接DNA双链中的单链缺口E.去除引物,填补空缺53.冈崎片段的生成是由于A.真核生物具有多个复制起始点B.DNA复制速度过快C.RNA引物合成不足D.随从链的复制方向与解链方向相反E.DNA连接酶缺失54.真核生物复制延长过程中,主要起催化作用的聚合酶是A.DNA-polαB.DNA-polβC.DNA-polγD.DNA-polδE.DNA-polε55.DNA复制的方向性是指A.新链只能从5'端向3'端延长B.DNA双螺旋上两股单链走向相反,一股从5'→3'延长,一股从3'→5'延长C.两股单链走向相同D.两股均为连续复制E.多个复制起始点,有多种复制方向56.转录是A.以前导链为模板B.以DNA的两条链为模板C.以编码链为模板D.以DNA的一条链为模板E.以RNA为模板57.原核生物转录作用生成的mRNA是A.多顺反子B.内含子C.插入子D.单顺反子E.间隔区序列58.AATAAA是A.启动子的辨认序列B.真核生物的顺式作用元件C.真核生物的反式作用因子D.真核生物转录加尾修饰点E.线粒体的起始密码序列59.体内核糖核苷酸链合成的方向是A.3'→5'B.C→NC.N→CD.5'→3'E.既可自3'→5,亦可自5'→3'60.能特异性抑制原核细胞mRNA聚合酶的是A.假尿嘧啶B.鹅膏蕈碱C.亚硝酸盐D.氯霉素E.利福平61.真核细胞的TATA BOX是A.DNA合成的起始位点B.RNA聚合酶与DNA模板稳定结合处C.RNA聚合酶的活性中心D.翻译起始点E.转录起始点62.真核生物mRNA的聚腺苷酸尾巴A.由模板DNA上的聚T序列转录生成B.是输送到胞质之后才加工接上的C.可直接在初级转录产物的3'-OH末端加上去D.维持DNA作为转录模板的活性E.先切除部分3'端的核苷酸,然后加上去63.遗传密码的阅读方向A.没有方向性B.3'→5'C.5'→3'D.N末端→C末端E.C末端→N末端64.嘌呤霉素抑制蛋白质生物合成的机制是A.抑制氨基酰-tRNA合成酶的活性,阻止氨基酰-tRNA的合成B.其结构与酪氨酰-tRNA相似,可和酪氨酸竞争与mRNA结合C.抑制转肽酶活性D.可与核糖体大亚基受位上的氨基酰-tRNA形成肽酰嘌呤霉素E.可与给位上的肽酰-tRNA形成肽酰嘌呤霉素65.真核生物蛋白质合成时,正确的叙述是A.70S核蛋白体首先进入核蛋白体循环B.70S核蛋白体不能进入核蛋白体循环C.50S核蛋白体亚基首先进入核蛋白体循环D.60S核蛋白体亚基首先进入核蛋白体循环E.80S核蛋白体解离后进入核蛋白体循环66.管家基因的表达A.不受环境因素影响B.较少受环境因素影响C.极少受环境因素影响D.有时受,也有时不受环境因素影响E.特别受环境因素影响/doc/431850385.html,c阻遏蛋白由A.Z基因编码B.Y基因编码C.A基因编码D.I基因编码E.以上都不是68.基因表达的基本调控点是A.基因活化B.转录后加工C.转录起始D.翻译后加工E.翻译起始69.下列哪一种不是操纵子的组成部分A.结构基因B.启动子C.操纵基因D.阻遏物E.pribnow盒70.基因表达过程中仅在原核生物中出现而真核生物没有的是A.tRNA的稀有碱基B.AUG用作起始密码子C.冈崎片段D.DNA连接酶E.σ因子71.转录因子A.是原核生物RNA聚合酶的组分B.是真核生物RNA聚合酶的组分C.有α、β、γ等各亚基D.是转录调控中的反式作用因子E.是真核生物的启动子72.在已知序列信息的情况下,获取目的基因的最方便方法是A.化学合成法B.差异显示法C.基因组文库法D.cDNA文库法E.聚合酶链反应(PCR)73.直接针对目的DNA进行筛选的方法是A.青霉素抗药性B.分子筛C.电源D.分子杂交E.氨苄青霉素抗药性74.某限制性核酸内切酶切割5'-GGGGGG↓AATTCC-3'序列后产生A.5'突出末端B.3'突出末端C.平末端D.5'及3'突出末端E.5'或3'突出末端75.下面有关肾上腺素受体的描述,正确的是A.具有催化cAMP生成的功能B.与激素结合后,释出催化亚基C.与催化cAMP生成的蛋白质是各自独立的D.特异性不高,可与一些激素结合E.与激素共价结合76.关于第二信使Ca2+的叙述不正确的是A.Ca2+能激活Ca2+-CaM激酶B.Ca2+能激活PKCC.细胞外Ca2+浓度远大于细胞内D.IP3可促使内质网的Ca2+释入胞质E.内分泌等变化持续数小时,细胞内Ca2+也升高数小时77.下列哪种因素不参与血红素合成代谢的调节A.促红细胞生成素(EPO)B.ALA合酶C.ALA脱水酶D.亚铁鳌合酶E.原卟啉原Ⅸ氧化酶78.血红素的合成部位在A.线粒体与胞液B.胞液与内质网C.微粒体与核糖体D.线粒体E.内质网与线粒体79.下列哪一种胆汁酸是次级胆汁酸A.甘氨鹅脱氧胆酸B.甘氨胆酸C.牛磺鹅脱氧胆酸D.牛磺胆酸E.脱氧胆酸80.能编码具有GTP酶活性的癌基因是A.myc基因B.ras基因C.sis基因D.src基因E.myb基因二.名词解释1.蛋白质的三级结构2.肽单元3.T m值4.DNA变性5.核酸酶6.多酶体系7.必需基团8.酶的变构调节9.糖酵解10.糖异生11.Cori循环12.HSL13.呼吸链14.ATP合酶15.氮平衡16.必需氨基酸17.物质代谢调节18.应激19.基因20.复制叉21.领头链22.随从链23.中心法则24.点突变25.cDNA26.结构基因27.内含子28.剪接29.不对称转录30.操纵子31.增强子32.结构基因33.基因组文库34.转导作用35.PCR36.克隆37.DNA重组38.受体39.第二信使40.小G蛋白三.填空题1.按照分子形状分类,蛋白质分子形状的长短轴之比小于10的称为(),蛋白质分子形状的长短轴之比大于10的称为()。
