Transient Expression of Exogenous Gene into Plant Cell Mediated by PEI Nanovector
参考文献及对应bioworld抗体-8月2号
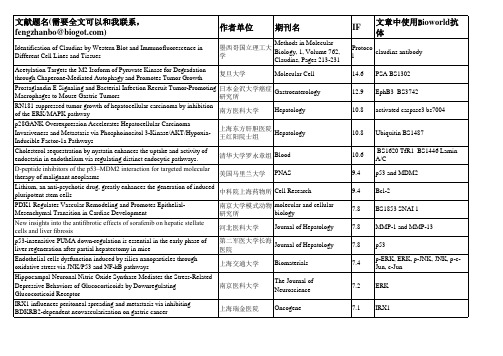
中科院上海药物所 Cell Research 南京大学模式动物 molecular and cellular biology 研究所 河北医科大学 Journal of Hepatology
第二军医大学长海 Journal of Hepatology 医院 上海交通大学 南京医科大学 上海瑞金医院 Biomaterials The Journal of Neuroscience Oncogene
德国Greifswald大 Free Radical Biology & Medicine 学 Journal of Controlled 复旦大学药学院 Release 中科院苏州纳米所 Nanotoxicology 南京大学模式动物 The Journal of Biological Chemistry 研究所 The Journal of Biological 台湾医学科学院 Chemistry 台湾大学 南京医科大学 The Journal of Biological Chemistry The Journal of Biological Chemistry
文献题名(需要全文可以和我联系, 文献题名 需要全文可以和我联系, 需要全文可以和我联系 fengzhanbo@)
Identification of Claudins by Western Blot and Immunofluorescence in Different Cell Lines and Tissues Acetylation Targets the M2 Isoform of Pyruvate Kinase for Degradation through Chaperone-Mediated Autophagy and Promotes Tumor Growth Prostaglandin E Signaling and Bacterial Infection Recruit Tumor-Promoting Macrophages to Mouse Gastric Tumors RN181 suppressed tumor growth of hepatocellular carcinoma by inhibition of the ERK/MAPK pathway p28GANK Overexpression Accelerates Hepatocellular Carcinoma Invasiveness and Metastasis via Phosphoinositol 3-Kinase/AKT/HypoxiaInducible Factor-1a Pathways Cholesterol sequestration by nystatin enhances the uptake and activity of endostatin in endothelium via regulating distinct endocytic pathways. D-peptide inhibitors of the p53–MDM2 interaction for targeted molecular therapy of malignant neoplasms Lithium, an anti-psychotic drug, greatly enhances the generation of induced pluripotent stem cells PDK1 Regulates Vascular Remodeling and Promotes EpithelialMesenchymal Transition in Cardiac Development New insights into the antifibrotic effects of sorafenib on hepatic stellate cells and liver fibrosis p53-insensitive PUMA down-regulation is essential in the early phase of liver regeneration after partial hepatectomy in mice Endothelial cells dysfunction induced by silica nanoparticles through oxidative stress via JNK/P53 and NF-kB pathways Hippocampal Neuronal Nitric Oxide Synthase Mediates the Stress-Related Depressive Behaviors of Glucocorticoids by Downregulating Glucocorticoid Receptor IRX1 influences peritoneal spreading and metastasis via inhibiting BDKRB2-dependent neovascularization on gastric cancer
一氧化氮NO胁迫诱导水稻转座子发生遗传和表观遗传变异

transposition.However,expression of at least one of these retrotransposons,i.e.,
Tosl 7,was up-regulated in the NO-derived plants,which Was accompanied by
1998年的诺贝尔生理学或医学奖授予Robert E Furchgott、Louis J.Ignarro和 Fetid Murad三位药理学家,表彰他们在“一氧化氮作为心血管系统的信号分子" 上的突破性研究。这引起科学家对NO及其生物效应和作用机制的广泛关注,近 几年来的研究进展更是层出不穷。
decrease in DNA demethylation at the LTR regions.This suggests that either the
transcript level did not reach to the threshold required for transpositional activation or
本研究发现,在NO胁迫条件下水稻品种松前子二代植株中检测到8个变异 单株。变异株中,转座子roping发生了转座激活:共检测到32个roPing变异位 点,其中15个跳出位点,17个插入位点。在这些植株中,roping的转座酶供体 一自主型转座子尸D略也发生了转座激活。变异植株roPing和Pong的激活都可 以稳定遗传给其后代。水稻中另一类活跃的转座子,反转座子Tosl7则发生了变 异,包括序列变异和DNA甲基化变异。变异株的Tosl7在转录水平的表达增强, 伴随着LTR区甲基化水平明显降低,说明Tosl7的表达受到LTR区甲基化的调 控:但非变异株中Tosl7表达水平与甲基化程度并不存在一一对应的关系,说明 Tosl7的表达也受到其它机制的调控。我们虽然在转录水平检测到Tosl7的表达 增强,但并没有检测到其转座激活,可能由于转录产物没有达到可导致转座激活 的域值或存在转录后水平的调控,如siRNA介导的转录本降解等。此外,Osr 类反转座子Osr23和Osr35也发生了变异。
植物学报 2020年 第55卷 总目次

植物学报2020年第55卷总目次第1期 (2020年1月)1 基因组学技术大发展助力园艺植物研究取得新进展唐嘉瓅, 邱杰, 黄学辉5 “绿色革命”新进展: 赤霉素与氮营养双重调控的表观修饰助力水稻高产高效育种韩美玲, 谭茹姣, 晁代印9 转昆虫抗冻蛋白基因增强甘薯抗冻能力赖先军, 张义正, 古英洪, 颜朗21 外源物质对茶树耐寒及蔗糖代谢关键基因表达的影响杨小青, 黄晓琴, 韩晓阳, 刘腾飞, 岳晓伟, 伊冉31 烟草叶片中呼吸电子传递途径在缓解叶绿体PSII光抑制中的作用罗蛟, 李玉婷, 张子山, 车兴凯, 梁英, 李月楠, 李滢, 赵世杰, 高辉远38 甘薯盐胁迫响应基因IbMYB3的表达特征及生物信息学分析李格, 孟小庆, 李宗芸, 朱明库49 水杨酸调控盐胁迫下羽衣甘蓝种子萌发的机理曹栋栋, 陈珊宇, 秦叶波, 吴华平, 阮关海, 黄玉韬62 植物中验证蛋白相互作用的Pull-down和Co-IP技术徐重益69 萤火素酶互补实验检测蛋白互作赵燕, 周俭民76 植物蛋白磷酸化的检测方法朱丹, 曹汉威, 李媛, 任东涛83 植物蛋白SUMO化修饰检测方法曲高平, 金京波90 怀牛膝细胞悬浮培养条件的优化李萍, 董亚辉, 李成龙, 何雨龙, 李明军96 植物凝集素类受体蛋白激酶研究进展王梦龙, 彭小群, 陈竹锋, 唐晓艳106 薄壳山核桃酚类代谢物研究进展贾晓东, 许梦洋, 莫正海, 宣继萍, 翟敏, 郭忠仁第2期 (2020年3月)123 小麦抗赤霉病利器——他山之石周俭民126 磷酸肌醇激酶FAB1调控拟南芥根毛伸长姚玉婷, 马家琦, 冯晓莉, 潘建伟, 王超137 小麦TaLCD基因的克隆及其对渗透胁迫的调节作用张扬, 刘华杰, 薛瑞丽, 李海霞, 李华147 拟南芥花药绒毡层细胞中具有基因簇特征的基因进化和功能分析左泽远, 刘琬琳, 许杰163 蕨类植物的鳞片特征及演化I: 凤尾蕨科顾钰峰, 金冬梅, 刘保东, 戴锡玲, 严岳鸿177 生物发光成像无损伤研究植物生物钟的方法于英俊, 徐航, 王雷182 双管基因枪介导的基因瞬时表达技术在拟南芥中的应用赵华, 邵广达, 高文鑫, 顾彪192 羊草成熟胚诱导愈伤组织及植株再生系统的优化肖燕, 王振兴, 李东明, 齐艳华, 恩和巴雅尔199 新疆地区发展大豆生产的可行性和初步建议冯锋, 战勇, 田志喜205 miR172-AP2模块调控植物生长发育及逆境响应的研究进展王劲东, 周豫, 余佳雯, 范晓磊, 张昌泉, 李钱峰,刘巧泉216 蓝色花形成分子机理研究进展张泰然, 张和臣, 武荣花228 C3和C4植物的氮素利用机制张璐, 何新华240 拟南芥NPH3/RPT2-Like (NRL)家族蛋白在向光素信号转导通路中的作用研究进展赵青平, 马世凡, 李芮茜, 王田雨, 赵翔第3期 (2020年5月)257 2019年中国植物科学若干领域重要研究进展左建儒, 漆小泉, 林荣呈, 钱前, 顾红雅, 陈凡, 杨淑华, 陈之端, 白永飞, 王雷, 王小菁, 姜里文, 萧浪涛, 种康, 王台270 小RNA, 大本领: 22 nt siRNAs在植物适应逆境中的重要作用武亮, 戚益军274 质疑、创新与合理性——纪念《植物学通报》创刊主编曹宗巽先生诞辰100周年白书农279 一个新的OsBRI1弱等位突变体的鉴定及其调控种子大小的功能研究管柳蓉, 刘祖培, 徐冉, 段朋根, 张国政, 于海跃,李静, 罗越华, 李云海287 蕨类植物rpoC1内含子缺失及其分子进化速率彭阳, 苏应娟, 王艇299 毛竹不同截短U3启动子的克隆及表达分析凡惠金, 金康鸣, 卓仁英, 乔桂荣308 森林生态系统细根周转规律及影响因素赵佳宁, 梁韵, 柳莹, 王玉珏, 杨倩茹, 肖春旺318 芳香堆心菊离体再生体系的建立罗虹, 温小蕙, 周圆圆, 戴思兰329 模式识别受体的胞内转运及其在植物免疫中的作用崔亚宁, 钱虹萍, 赵艳霞, 李晓娟340 转录因子调控植物萜类化合物生物合成研究进展董燕梅, 张文颖, 凌正一, 李靖锐, 白红彤, 李慧,石雷351 植物次生细胞壁生物合成的转录调控网络张雨, 赵明洁, 张蔚369 植物激素研究中的化学生物学思路与应用徐佳慧, 代宇佳, 罗晓峰, 舒凯, 谭伟明382 水稻根系遗传育种研究进展章怡兰, 林雪, 吴仪, 李梦佳, 张晟婕, 路梅, 饶玉春, 王跃星第4期 (2020年7月)397 独脚金内酯信号途径的新发现——抑制子也是转录因子姚瑞枫, 谢道昕403 360度群体遗传变异扫描——大豆泛基因组研究祝光涛, 黄三文407 拟南芥黏连蛋白RAD21对增强UV-B辐射后细胞分裂的响应贺芳芳, 陈慧泽, 冯金林, 高琳, 牛娇, 韩榕421 拟南芥AtR8 lncRNA对盐胁迫响应及其对种子萌发的调节作用张楠, 刘自广, 孙世臣, 刘圣怡, 林建辉, 彭疑芳,张晓旭, 杨贺, 岑曦, 吴娟430 不同抗性苹果品种应答轮纹病菌胁迫的差异蛋白质组分析张彩霞, 袁高鹏, 韩晓蕾, 李武兴, 丛佩华442 4种模式植物LRR VIII-2亚家族基因的鉴定和进化历史分析闫晨阳, 陈赢男457 基于多个叶绿体基因序列片段重建广义苋科系统发育关系黄久香, 陈文娜, 李玉玲, 姚纲468 植物转录因子与DNA互作研究技术杨立文, 刘双荣, 李玉红, 林荣呈475 染色质免疫共沉淀实验方法王泓力, 焦雨铃481 AP2/ERF转录因子调控植物非生物胁迫响应研究进展洪林, 杨蕾, 杨海健, 王武497 NLR及其在植物抗病中的调控作用杨程惠子, 唐先宇, 李威, 夏石头505 走出歌德的阴影: 迈向更加科学的植物系统学王鑫, 刘仲健, 刘文哲, 廖文波, 张鑫, 刘忠, 胡光万, 郭学民, 王亚玲513 纳米农药在植物中的吸收转运研究进展李晶, 郭亮, 崔海信, 崔博, 刘国强第5期 (2020年9月)533 踏破铁鞋无觅处——一类新型抗真菌剂的发现周俭民, 曹立冬537 WUSCHEL介导的固有免疫: 植物干细胞抵御病毒侵害的新机制杜斐, 焦雨铃541 植物类LORELEI糖基磷脂酰肌醇锚定蛋白研究进展李思佳, 张咏雪, 贾明生, 李莹, 戴绍军551 芒果胶孢炭疽病菌应答菌丝机械损伤产生无性孢子的分子机制王丽妍, 卢梦瑶, 童悦, 徐祥斌, 张正科, 孟兰环,史学群, 宋海超564 航天搭载对武夷名丛相关生理及生长特性的影响刘建福, 陈育才, 王文建, 王河川, 蔡金福, 王明元, 李丹丹, 张斌, 黄昆573 大麦抗叶锈病慢锈性鉴定技术及抗性评价方法车明哲, 王亚军, 马创新, 漆小泉577 水稻稻瘟病和纹枯病抗性鉴定方法贺闽, 尹俊杰, 冯志明, 朱孝波, 赵剑华, 左示敏,陈学伟588 早花百子莲叶片器官发生和胚胎发生再生体系的建立岳建华, 董艳, 王小画, 孙佩霞, 王思颖, 张新年,张琰596 根尖整体透明技术改良马龙, 李桂林, 李师鹏, 蒋苏605 长白落叶松体胚发生再生体系优化刘建飞, 刘炎, 刘克俭, 池阳, 霍志发, 霍永洪, 由香玲613 禾本科作物芒遗传研究进展亓斐, 邢丕一, 鲍印广, 王洪刚, 李兴锋623 一氧化氮对豆科植物结瘤及固氮的影响机制张卫勤, 邹杭, 张妮娜, 林雪媛, 陈娟634 植物嫁接愈合分子机制研究进展谢露露, 崔青青, 董春娟, 尚庆茂644 芳香植物精油的抗菌性及在动物生产中的应用郝渊鹏, 李静一, 杨瑞, 李慧, 白红彤, 石雷第6期 (2020年11月)661 豆科植物SHR-SCR模块——根瘤“奠基细胞”的命运推手刘承武, 赵忠666 再生水补给河道内芦苇的光谱特征及其对水体氮和磷含量的响应赵睿, 卜红梅, 宋献方, 高融瑾677 干旱胁迫下表观遗传机制对转C4型PEPC基因水稻种子萌发的影响宋凝曦, 谢寅峰, 李霞693 北京地区芦苇资源状态及其多样性张茜, 裘天航, 王安安, 周华健, 袁敏, 李利, 白素兰, 崔素霞705 亏缺灌溉对板蓝根叶片光合生理特性及产量的影响王泽义, 张恒嘉, 王玉才, 陈谢田, 巴玉春715 生物信息学分析方法I: 全基因组关联分析概述赵宇慧, 李秀秀, 陈倬, 鲁宏伟, 刘羽诚, 张志方,梁承志733 微区XRF技术分析无机元素在植物中的原位分布林梵宇, 尹希杰, 梁毓娜, 黄杰超740 P700氧化还原动力学的测量方法及原理张春艳749 万寿菊再生体系的建立及优化王亚琴, 韦陆丹, 王文静, 刘宝骏, 张春玲, 张俊卫, 何燕红760 香鳞毛蕨的组织培养和快速繁殖体系构建张冬瑞, 卜志刚, 陈玲玲, 常缨768 转座子来源的植物长链非编码RNA王益军, 王亚丽, 陈煜东777 木葡聚糖及其在植物抗逆过程中的功能研究进展肖银燕, 袁伟娜, 刘静, 孟建, 盛奇明, 谭烨欢, 徐春香788 植物根系分泌物主要生态功能研究进展李佳佳, 樊妙春, 上官周平CHINESE BULLETIN OF BOTANY Vol. 55 2020 CONTENTSNo. 1 (January, 2020)1 The Development of Genomics Technologies DrivesNew Progress in Horticultural Plant ResearchJiali Tang, Jie Qiu, Xuehui Huang5 A New Progress of Green Revolution: EpigeneticModification Dual-regulated by Gibberellin and Ni-trogen Supply Contributes to Breeding of High Yieldand Nitrogen Use Efficiency RiceMeiling Han, Rujiao Tan, Daiyin Chao9 Transformation of Insect Derived Antifreeze Geneinto Sweet Potato (Ipomoea batatas) and EnhancedIts Freeze-toleranceXianjun Lai, Yizheng Zhang, Yinghong Gu, Lang Yan21 Effect of Exogenous Substances on Cold Toleranceand Key Sucrose Metabolic Gene Expression in Ca-mellia sinensisXiaoqing Yang, Xiaoqin Huang, Xiaoyang Han, Tengfei Liu,Xiaowei Yue, Ran Yi31 Effects of the Respiratory Electron Transport Path-ways in Relieving Photoinhibition of Chloroplast PSIIin Tobacco LeavesJiao Luo, Yuting Li, Zishan Zhang, Xingkai Che, Ying Liang,Yuenan Li, Ying Li, Shijie Zhao, Huiyuan Gao38 Expression Patterns and Bioinformatic Analyses ofSalt Stress Responsive Gene IbMYB3 in IpomoeabatatasGe Li, Xiaoqing Meng, Zongyun Li, Mingku Zhu 49 Regulatory Mechanism of Salicylic Acid on SeedGermination Under Salt Stress in KaleDongdong Cao, Shanyu Chen, Yebo Qin, Huaping Wu,Guanhai Ruan, Yutao Huang62 Pull-down and Co-immunoprecipitation Assays ofInteracting Proteins in PlantsChongyi Xu69 Luciferase Complementation Assay for DetectingProtein InteractionsYan Zhao, Jianmin Zhou76 Protocols for Analyzing Plant Phospho-proteinsDan Zhu, Hanwei Cao, Yuan Li, Dongtao Ren83 Detection of SUMOylation in PlantsGaoping Qu, Jingbo Jin90 Optimization of Cell Suspension Culture Conditionsof Achyranthes bidentataPing Li, Yahui Dong, Chenglong Li, Yulong He, Mingjun Li 96 Research Advances on Lectin Receptor-like Kinasein PlantsMenglong Wang, Xiaoqun Peng, Zhufeng Chen, XiaoyanTang106 Recent Advances in Phenolic Metabolites in Pecan Xiaodong Jia, Mengyang Xu, Zhenghai Mo, Jiping Xuan,Min Zhai, Zhongren GuoNo. 2 (March, 2020)123 FightingFusarium Head Blight in Wheat—a Reme-dy from AfarJian-MinZhou126 A Role of Arabidopsis Phosphoinositide Kinase, FAB1, in Root Hair GrowthYuting Yao, Jiaqi Ma, Xiaoli Feng, Jianwei Pan, Chao Wang 137 Cloning of Wheat TaLCD Gene and Its Regulation on Osmotic StressYang Zhang, Huajie Liu, Ruili Xue, Haixia Li, Hua Li147 Evolution and Functional Analysis of Gene Clusters in Anther Tapetum Cells of Arabidopsis thalianaZeyuan Zuo, Wanlin Liu, Jie Xu163 Morphology Characters and Evolution of Ferns Scale Ι: PtaridaceaeYufeng Gu, Dongmei Jin, Baodong Liu, Xiling Dai, Yue-hong Yan177 A Non-invasive Method for Measuring and Analy-zing Circadian Phenotype in Living PlantsYingjun Yu, Hang Xu, Lei Wang182 The Application of Double-barreled Particle Bom-bardment for Transient Gene Expression in Arabi-dopsisHua Zhao, Guangda Shao, Wenxin Gao, Biao Gu192 Optimization of Tissue Culture and Plant Regene-ration System of Mature Embryo of Leymus chinen-sisYan Xiao, Zhenxing Wang, Dongming Li, Yanhua Qi,Enhebayaer199 The Feasibility and Recommendation for Improving Soybean Production in XinjiangFeng Feng, Yong Zhan, Zhixi Tian205 Advances in the Regulation of Plant Growth and Development and Stress Response by miR172-AP2 ModuleJindong Wang, Yu Zhou, Jiawen Yu, Xiaolei Fan, Chang-quan Zhang, Qianfeng Li, Qiaoquan Liu216 Recent Advances on Blue Flower FormationTairan Zhang, Hechen Zhang, Ronghua Wu228 Nitrogen Utilization Mechanism in C3 and C4 Plants Lu Zhang, Xinhua He240 Advances of NPH3/RPT2-Like (NRL) Family Pro-teins in Phototropin-mediated Signaling in Arabidop-sis thalianaQingping Zhao, Shifan Ma, Ruixi Li, Tianyu Wang, XiangZhaoNo. 3 (May, 2020)257 Achievements and Advance in Chinese Plant S-ciences in 2019Jianru Zuo, Xiaoquan Qi, Rongcheng Lin, Qian Qian,Hongya Gu, Fan Chen, Shuhua Yang, Zhiduan Chen,Yongfei Bai, Lei Wang, Xiaojing Wang, Liwen Jiang,Langtao Xiao, Kang Chong, Tai Wang270 Small RNA, No Small Feat: Plants Deploy 22 nt siRNAs to Cope with Environmental StressLiang Wu, Yijun Qi274 Critical Thinking, Alternative Interpretation, and Lo-gic Consistency—To Commemorate the 100 Birth-day of then Professor Tsunghsing Tsao (Zong-XunCao), the Founding Editor-in-Chief of the ChineseBulletin of BotanyShunong Bai279 Identification of a New OsBRI1 Weak Allele and Analysis of its Function in Grain Size ControlLiurong Guan, Zupei Liu, Ran Xu, Penggen Duan, GuozhengZhang, Haiyue Yu, Jing Li, Yuehua Luo, Yunhai Li287 Intron Loss and Molecular Evolution Rate of rpoC1 in FernsYang Peng, Yingjuan Su, Ting Wang299 Cloning and Expression Analysis of Different Trun-cated U3 Promoters in Phyllostachys edulisHuijin Fan, Kangming Jin, Renying Zhuo, Guirong Qiao 308 Patterns and Influence Factors of Fine Root Turn-over in Forest EcosystemsJianing Zhao, Yun Liang, Ying Liu, Yujue Wang, QianruYang, Chunwang Xiao318 EstablishmentofIn Vitro Regeneration System of Helenium aromaticumHong Luo, Xiaohui Wen, Yuanyuan Zhou, Silan Dai329 Intracellular Trafficking in Pattern Recognition Re-ceptor-triggered Plant ImmunityYaning Cui, Hongping Qian, Yanxia Zhao, Xiaojuan Li 340 Advances in Transcription Factors Regulating Plant Terpenoids BiosynthesisYanmei Dong, Wenying Zhang, Zhengyi Ling, Jingrui Li,Hongtong Bai, Hui Li, Lei Shi351 Transcriptional Regulatory Network of Secondary Cell Wall Biosynthesis in PlantsYu Zhang, Mingjie Zhao, Wei Zhang369 Thoughts and Applications of Chemical Biology in Phytohormonal ResearchJiahui Xu, Yujia Dai, Xiaofeng Luo, Kai Shu, Weiming Tan 382 Research Progress on Genetics and Breeding of Rice RootsYilan Zhang, Xue Lin, Yi Wu, Mengjia Li, Shengjie Zhang,Mei Lu, Yuchun Rao, Yuexing WangNo. 4 (July, 2020)397 New Insight into Strigolactone SignalingRuifeng Yao, Daoxin Xie403 A 360-degree Scanning of Population Genetic Va-riations—a Pan-genome Study of SoybeanGuangtao Zhu, Sanwen Huang407 ResponseofArabidopsis Cohesin RAD21 to Cell Division after Enhanced UV-B RadiationFangfang He, Huize Chen, Jinlin Feng, Lin Gao, Jiao Niu,Rong Han421 ResponseofAtR8 lncRNA to Salt Stress and Its Regulation on Seed Germination in ArabidopsisNan Zhang, Ziguang Liu, Shichen Sun, Shengyi Liu,Jianhui Lin, Yifang Peng, Xiaoxu Zhang, He Yang, Xi Cen,Juan Wu430 Proteome Analysis of Different Resistant Apple Cultivars in Response to the Stress of Ring RotDiseaseCaixia Zhang, Gaopeng Yuan, Xiaolei Han, Wuxing Li,Peihua Cong442 Identification and Evolution of LRR VIII-2 Subfamily Genes in Four Model Plant SpeciesChenyang Yan, Yingnan Chen457 Phylogenetic Study of Amaranthaceae sensu lato Based on Multiple Plastid DNA FragmentsJiuxiang Huang, Wenna Chen, Yuling Li, Gang Yao468 Methods for Examining Transcription Factor-DNA Interaction in PlantsLiwen Yang, Shuangrong Liu, Yuhong Li, Rongcheng Lin 475 Protocols for Chromatin ImmunoprecipitationHongli Wang, Yuling Jiao481 Research Advances in AP2/ERF Transcription Fac-tors in Regulating Plant Responses to Abiotic St-ressLin Hong, Lei Yang, Haijian Yang, Wu Wang497 NLR and Its Regulation on Plant Disease Resis-tanceChenghuizi Yang, Xianyu Tang, Wei Li, Shitou Xia505 Stepping out of the Shadow of Goethe: for a More Scientific Plant SystematicsXin Wang, Zhongjian Liu, Wenzhe Liu, Wenbo Liao, XinZhang, Zhong Liu, Guangwan Hu, Xuemin Guo, YalingWang513Research Progress on Uptake and Transport of Nano-pesticides in PlantsJing Li, Liang Guo, Haixin Cui, Bo Cui, Guoqiang LiuNo. 5 (September, 2020)533 Antifungal Compounds Come in HandyJian-Min Zhou, Lidong Cao537WUSCHEL-mediated Innate Immunity in Plant S-tem Cells Provides a Novel Antiviral StrategyFei Du, Yuling Jiao541 Advances of LORELEI-like Glycosylphosphatidy-linositol-anchor (LLG) Proteins in PlantsSijia Li, Yongxue Zhang, Mingsheng Jia, Ying Li, ShaojunDai551 Molecular Mechanism of the Generation of Asexual Spores of the Mango Fungal Pathogen (Colletotri-chum gloeosporioides) Induced by Mechanical In-juriesLiyan Wang, Mengyao Lu, Yue Tong, Xiangbin Xu,Zhengke Zhang, Lanhuan Meng, Xuequn Shi, HaichaoSong564 Effects of Space Treatment on Biological and Grow-th Characteristics of Camellia sinensisJianfu Liu, Yucai Chen, Wenjian Wang, Hechuan Wang,Jinfu Cai, Mingyuan Wang, Dandan Li, Bin Zhang, KunHuang573 Methods for Identification and Resistance Evalua-tion of Barley Slow Rusting to Leaf RustMingzhe Che, Yajun Wang, Chuangxin Ma, Xiaoquan Qi 577 Methods for Evaluation of Rice Resistance to Blast and Sheath Blight DiseasesMin He, Junjie Yin, Zhiming Feng, Xiaobo Zhu, JianhuaZhao, Shimin Zuo, Xuewei Chen588 A Regeneration System for Organogenesis and Somatic Embryogenesis Using Leaves of Agapan-thus praecox as ExplantsJianhua Yue, Yan Dong, Xiaohua Wang, Peixia Sun,Siying Wang, Xinnian Zhang, Yan Zhang596 An Improved Protocol for Whole Mount Clearing of Plant Root TipLong Ma, Guilin Li, Shipeng Li, Su Jiang605Optimization of the Regeneration System from So-matic Embryogenesis in Larix olgensisJianfei Liu, Yan Liu, Kejian Liu, Yang Chi, Zhifa Huo,Yonghong Huo, Xiangling You613 Advances in Genetic Studies of the Awn in Cereal CropsFei Qi, Piyi Xing, Yinguang Bao, Honggang Wang, Xing-feng Li623 Influence Mechanisms of Nitric Oxide on Nodula-tion and Nitrogen Fixation in LegumesWeiqin Zhang, Hang Zou, Nina Zhang, Xueyuan Lin, JuanChen634 Recent Advances in Molecular Mechanisms of Plant Graft Healing ProcessLulu Xie, Qingqing Cui, Chunjuan Dong, Qingmao Shang 644 Antimicrobial Activity of Aromatic Plant Essential Oils and Their Application in Animal ProductionYuanpeng Hao, Jingyi Li, Rui Yang, Hui Li, Hongtong Bai,Lei ShiNo. 6 (November, 2020)661 The Legume SHR-SCR Module Predetermines No-dule Founder Cell IdentityChengwu Liu, Zhong Zhao666 SpectralCharacteristicsofPhragmites australis and Its Response to Riverine Nitrogen and Phos-phorus Contents in River Reaches Restored byReclaimed WaterRui Zhao, Hongmei Bu, Xianfang Song, Rongjin Gao677 Effects of Epigenetic Mechanisms on C4Phos-phoenolpyruvate Carboxylase Transgenic Rice (Ory-za sativa) Seed Germination Under Drought StressNingxi Song, Yingfeng Xie, Xia Li693 Morphology and Genetic Diversity of Phragmites australis in BeijingXi Zhang, Tianhang Qiu, Anan Wang, Huajian Zhou, MinYuan, Li Li, Sulan Bai, Suxia Cui705Effects of Deficit Irrigation on the Photosynthetic and Physiological Characteristics of Leaves andYield of Isatis tinctoriaZeyi Wang, Hengjia Zhang, Yucai Wang, Xietian Chen,Yuchun Ba715 An Overview of Genome-wide Association Studies in PlantsYuhui Zhao, Xiuxiu Li, Zhuo Chen, Hongwei Lu, YuchengLiu, Zhifang Zhang, Chengzhi Liang733 AnalysisofIn Situ Distribution of Inorganic Ele-ments in Plants by Micro-XRFFanyu Lin, XijieYin, Yuna Liang, Jiechao Huang 740 The Measurement Methods and Principles of P700 Redox KineticsChunyanZhang749The Establishment and Optimization of a Rege-neration System for Marigold (Tagetes erecta)Yaqin Wang, Ludan Wei, Wenjing Wang, Baojun Liu,Chunling Zhang, Junwei Zhang, Yanhong He760Establishment of a Tissue Culture and Rapid Pro-pagation System of Dryopteris fragransDongrui Zhang, Zhigang Bu, Lingling Chen, Ying Chang 768 Transposon-derived Long Noncoding RNA in PlantsYijun Wang, Yali Wang, Yudong Chen777 Xyloglucan and the Advances in Its Roles in Plant Tolerance to StressesYingyan Xiao, Weina Yuan, Jing Liu, Jian Meng, QimingSheng, Yehuan Tan, Chunxiang Xu788 Research Advances in the Main Ecological Func-tions of Root ExudatesJiajia Li, Miaochun Fan, Zhouping Shangguan致谢审者本刊编辑部衷心感谢以下审稿专家对我刊工作的支持和帮助!(统计时间2019年11月1日至2020年11月1日。
Activation Tagging Identifies a Conserved MYB Regulator of

The Plant Cell, Vol. 12, 2383–2393, December 2000, © 2000 American Society of Plant Physiologists Activation Tagging Identifies a Conserved MYB Regulator of Phenylpropanoid BiosynthesisJustin O. Borevitz,a,1 Yiji Xia,a,b,1 Jack Blount,b Richard A. Dixon,b and Chris Lamb a,2a Plant Biology Laboratory, Salk Institute for Biological Studies, La Jolla, California 92037b Plant Biology Division, Samuel Roberts Noble Foundation, Ardmore, Oklahoma 73401Plants produce a wide array of natural products, many of which are likely to be useful bioactive structures. Unfortu-nately, these complex natu ral produ cts u su ally occu r at very low abu ndance and with restricted tissu e distribu tion,thereby hindering their evaluation. Here, we report a novel approach for enhancing the accumulation of natural prod-ucts based on activation tagging by Agrobacterium-mediated transformation with a T-DNA that carries cauliflower mo-saic virus 35S enhancer sequences at its right border. Among ف5000 Arabidopsis activation-tagged lines, we found a plant that exhibited intense purple pigmentation in many vegetative organs throughout development. This upregulationof pigmentation reflected a dominant mu tation that resu lted in massive activation of phenylpropanoid biosyntheticgenes and enhanced accumulation of lignin, hydroxycinnamic acid esters, and flavonoids, including various anthocya-nins that were responsible for the purple color. These phenotypes, caused by insertion of the viral enhancer sequencesadjacent to an MYB transcription factor gene, indicate that activation tagging can overcome the stringent genetic con-trols regulating the accumulation of specific natural products during plant development. Our findings suggest a func-tional genomics approach to the biotechnological evaluation of phytochemical biodiversity through the generation ofmassively enriched tissue sources for drug screening and for isolating underlying regulatory and biosynthetic genes. INTRODUCTIONEthnobotany and limited screens of medicinal plants indi-cate that the huge repertoire of chemical diversity in plants contains many potentially useful bioactive structural princi-ples for developing novel drug s, flavors, frag rances, and other specialty chemicals. Unfortunately, these complex natural products usually occur in very low abundance and with a restricted tissue distribution. Moreover, almost all of this phytochemical biodiversity resides in exotic, unculti-vated species. Whereas drug s such as taxol, vinblastine, and vincristine illustrate the potential of plants as sources of new drugs, the development of rational approaches for the g eneration of useful amounts of complex natural products for industrial evaluation remains an unsolved problem. In particular, an intense 30-year effort using cell and tissue cul-tures from medicinal plants has failed to g enerate useful quantities of complex products for the commercial produc-tion of established drugs in vitro or for high-throughput, mul-tiplex screening of phytochemicals (Facchini and Deluca, 1995; McCaskill and Croteau, 1998). This failure probably reflects the string ent spatial and temporal transcriptional controls governing the biosynthesis of specific natural prod-ucts during plant development (Fowler, 1983; Robins, 1994; Facchini and Deluca, 1995). Transg enic manipulation to override these genetic controls thus may provide the key to enhancing natural product biosynthesis for industrial evalu-ation and exploitation.Activation tagging with the enhancer from the cauliflower mosaic virus 35S transcript promoter (35Se) is an emerging technology in plant functional genomics (Weigel et al., 2000). This approach, based on Agrobacterium-mediated transfor-mation, can create transgenic plants in which the T-DNA car-rying 35Se at its right border is spliced into the plant genome at random sites. In each independent transgenic line, 35Se strongly activates the plant gene to which, by chance, it lies adjacent. Activation of a gene in this fashion may lead to ob-servable effects on the modified plant, providing important clues about the function of the activated g ene. Screening larg e collections of independent, activation-tag g ed lines thus represents a powerful way of surveying the g enome and isolating genes that affect traits of interest.Using activation tagging, we have isolated a bright-purple mutant, pro ductio n o f antho cyanin pigment 1-Dominant (pap1-D), in which genes encoding enzymes involved in the biosynthesis of phenylpropanoid natural products exhibit massive and widespread activation throughout plant de-velopment. The pap1-D phenotype, which is caused by1These authors contributed equally to this work.2To whom correspondence should be addressed. E-mail mb @; fax 44-1603-456844.2384The Plant CellFigure 1.pap1-D Phenotypes.(A)pap1-D (left) and Col-0 (right) flowers.(B) Roots of pap1-D (left) and Col-0 (right) plants.(C) Six-week-old adult pap1-D (front) and Col-0 (back) plants.Activation Tagging of PAP12385 overexpression of a g ene encoding an MYB transcriptionfactor, indicates that activation tagging can be used to over-come the stringent genetic controls regulating the develop-mental accumulation of specific natural products. Thesefindings suggest a new approach for the systematic biotech-nological evaluation of phytochemical biodiversity through theg eneration of massively enriched tissue sources for drugscreening and for isolation of the underlying regulatory andbiosynthetic genes.RESULTSMutant CharacterizationApproximately 5000 activation-tagged primary lines of Ara-bidopsis ecotype Columbia (Col-0) were generated by usingpSKI015, which contains four copies of 35Se at the rig htborder of the T-DNA, pBluescript KSϩ for plasmid rescue,and the BAR g ene for Basta resistance as a selectablemarker (Kardailsky et al., 1999; Weigel et al., 2000). A singlebright-purple plant was observed in this collection, and itsseed was collected for prog eny analysis. T2 plants seg re-g ated for the purple coloration characteristic of anthocya-nins in a 3:1 ratio, which is consistent with this trait being determined by a single dominant allele, an allele we named pap1-D (see above). The whole plant, including the roots, stems, leaves, primary and secondary branches, and cauline leaves as well as sepals, anthers, and carpels, ex-hibited purple pigmentation (Figures 1B and 1C). The purple coloration was more pronounced when plants were grown under high-intensity light or other stress conditions, such as droug ht and pathog en infection (data not shown). Under these conditions, leaves and stems of wild-type plants also show a slig ht pig mentation, sug g esting that the pap1-D phenotype might in part reflect enhancement of an endoge-nous stress response. However, we never observed pig-mentation in the roots of wild-type plants—in marked contrast to the strong pigmentation at the base of pap1-D roots (Fig ure 1B). Except for very weak pig mentation in flower petals (Figure 1A), enhanced pigmentation in pap1-D was observed throughout development. No other morpho-logical phenotypes were observed.Because anthocyanins are a subclass of flavonoid natural products derived from the phenylpropanoid skeleton, we examined the expression of phenylpropanoid biosynthetic g enes and the accumulation of natural products. RNA g el blot analysis showed massive enhancement of the expres-sion of phenylpropanoid biosynthetic g enes in pap1-D plants (Figure 2). The amounts of transcripts encoding chal-cone synthase (CHS), the entry point enzyme into the fla-vonoid branch pathway, and dihydroflavonol reductase, an enzyme of flavonoid biosynthesis specific for anthocyanins, were greater in pap1-D plants than in wild-type Col-0 plants.Transcripts encoding g lutathione S-transferase, which has been implicated in the transport of anthocyanins into the vacuole (Alfenito et al., 1998), also were expressed in in-creased amounts. Moreover, the accumulation of tran-scripts that encode phenylalanine ammonia-lyase (PAL), the first enzyme of the overall phenylpropanoid biosynthetic pathway, also was markedly enhanced, indicating that tran-scriptional activation was not confined to the flavonoid branch.To determine the extent of changes in anthocyanins and other phenylpropanoid-derived compounds in pap1-D, we extracted and analyzed soluble and cell wall–bound phe-nolic compounds by HPLC. Analysis of the soluble fraction, which was obtained by extraction in acetone, revealed sev-eral quantitative differences between pap1-D and wild-type Col-0 plants—in particular, increased concentrations of cer-tain flavonol g lycosides, including Glc-rhamnose (Rha)-quercetin, Glc-Rha-kaempferol, and unidentified conjugates of kaempferol and quercetin (Figures 3A and 3B). After alka-line hydrolysis of the residue that was obtained after ace-tone extraction, one portion was freeze-dried for analysis of anthocyanidins (anthocyanin aglycones); the remainder was partitioned into ethyl acetate for determination of cell wall–bound hydroxycinnamic acids. Anthocyanidins were present in g reater concentrations in pap1-D than in Col-0 (Fig ures 3C and 3D), as were coumaric and sinapic acids measured in alkaline hydrolysates of the wall-bound phenolic fraction (Fig ures 3E and 3F). Thus, pap1-D is characterized by strongly increased concentrations of glycosylated anthocya-nins, flavonols, and cell wall–esterified hydroxycinnamic acidsin comparison with wild-type Col-0.Figu re 2.Enhanced Expression of Phenylpropanoid Biosynthetic Genes in pap1-D.RNA g el blot hybridization was conducted with total RNA isolated from 6-week-old pap1-D and Col-0 wild-type plants. DFR, dihydrofla-vonol reductase; GST, glutathione S-transferase; UBQ, ubiquitin.2386The Plant CellThe observation of increased wall-bound hydroxycin-namic acids in pap1-D prompted us to measure the content and composition of lignin, which is derived from hydroxycin-namic acid precursors. Lig nin was analyzed by derivati-zation followed by reductive cleavag e, which helps to determine the absolute amounts of guaiacyl (G, monomethyl-ated) and syringyl (S, dimethylated) lignin monomers (Lu and Ralph, 1997). The results in Table 1 indicate increases in both total G and total S residues in the cell wall fraction of pap1-D compared with those in Col-0, but the S/G ratio var-ied little. The change in lignin monomers could reflect an in-crease in lignin content or a change in composition that led to more efficient monomer extractability.Changes in lignin content and composition have been en-g ineered in transg enic plants by downreg ulation of PAL,caffeic acid O -methyltransferase, and caffeoyl-CoA O -meth-yltransferase, enzymes of the lig nin branch of phenylpro-panoid biosynthesis (Atanassova et al., 1995; Zhong et al.,1998). PAL activity in stems of pap1-D plants was approxi-mately twice that found in stems of wild-type plants,whereas the activities of the two O -methyltransferases dif-fered little between the two (Table 1). Thus, the changes in lig nin composition and increased concentrations of wall-bound sinapic acid in pap1-D reflect the change in PAL ac-tivity but do not appear to be associated with increases in lignin O -methyltransferase activities.The Arabidopsis transparent testa glabra1-1 ( ttg1-1 ) mu-tation blocks anthocyanin accumulation and trichome for-mation (Koornneef, 1981). TTG1 encodes a WD40 repeat protein homolog ous with an AN11-encoded protein from petunia, which also controls anthocyanin production (deVetten et al., 1997; Walker et al., 1999). To test the geneticFigure 3.Effect of pap1-D Mutation on Accumulation of Phenylpropanoid Products.(A) to (F) HPLC profiles of phenylpropanoid metabolites in extracts from wild-type Col-0 ([A], [C], and [E]) and pap1-D ([B], [D], [F]) plants.(A) and (B) Soluble phenolics: peak 1, rhamnose (Rha)-Glc-Rha-quercetin; peak 2, quercetin conjugate; peak 3, Rha-Glc-Rha-kaempferol; peak 4, Glu-Rha-quercetin; peak 5, Rha-Rha-quercetin; peak 6, Glc-Rha-kaempferol; peaks 7 and 8, kaempferol conjugates; peak 9, sinapic acid;peak 10, Rha-Rha-kaempferol.(C) and (D) Anthocyanidins; the inset in (D) shows the UV light absorption spectrum of the major anthocyanidin eluting at 21.5 min.(E) and (F) Wall-bound phenolics: peak 1, trans -4-coumaric acid; peak 2, sinapic acid; peak 3, cis -4-coumaric acid.Activation Tagging of PAP12387relationship between TTG1 and PAP1, we crossed ttg1-1 with pap1-D. The pap1-D allele was tracked by Basta resis-tance, and the ttg1-1 mutation was scored visibly. The dou-ble mutant F2 plants failed to accumulate anthocyanins, indicating that TTG1 is required for the production of antho-cyanins mediated by PAP1 overexpression and acts either downstream from or at the same step as PAP1.Cloning of PAP1In a population of Ͼ100 seg reg ating T2 plants, each plant that had the pap1-D phenotype showed resistance to Basta, and all plants with a wild-type phenotype (i.e., lacking purple pig mentation) were sensitive to Basta, indicating that the mutation was tightly linked to the T-DNA insert. Moreover, hybridization of DNA gel blots of pap1-D genomic DNA that had been digested with EcoRI or KpnI showed that the 35Se sequences were localized to single fragments of 10 and 12 kb, respectively (data not shown), indicating that the mutant contained a single, simple insertion. The 12-kb KpnI and 10-kb EcoRI fragments were cloned by plasmid rescue (Weigel et al., 2000), g enerating pPAP1-K1 and pPAP1-E1, respec-tively. Nucleotide sequencing and restriction analysis showed that the 12-kb KpnI frag ment fully overlapped the smaller EcoRI frag ment. Probing the Arabidopsis CD4-7 cDNA li-brary at high stringency with pPAP1-K1 resulted in isolation of a single 956-bp cDNA, which defined three exons in the genomic DNA of pPAP1-K1 encoding an MYB transcription factor (Figures 4A and 4B). In the pap1-D line, the 35Se se-quences had inserted 5.1 kb 3Ј to the start of this gene, des-ignated PAP1, and RNA gel blot hybridization with the PAP1 cDNA revealed a sing le 1-kb transcript, which was mas-sively overexpressed in the pap1-D mutant (Figure 4C).To confirm that overexpression of PAP1 caused the pap1-D phenotype, we cloned a 3-kb g enomic frag ment spanning the three PAP1 exons and flanking sequences into pMN20-2, which contains two copies of 35Se (Weig el et al., 2000), thereby creating pMN-PAP1. Transformation of wild-type Col-0 with this construct, which mimics the g enomic con-text of the pap1-D allele, generated multiple transgenic lines with the characteristic purple phenotype (Fig ure 5B). As would be expected from position effects, these transgenic lines represented an allelic series ranging from the wild type to an even more intense phenotype than pap1-D, in some cases having strong purple pigmentation in the petals (Fig-ure 5E). In contrast, transformation with pMN20-2 as an empty vector control gave no enhanced pigmentation phe-notype (Figures 5A and 5D).Sequence alig nments with the Arabidopsis databases showed that PAP1 is a member of the R2, R3 MYB family, which is estimated to have Ͼ100 members in Arabidopsis (Kranz et al., 1998; Romero et al., 1998) and Ͼ80 membersin maize (Rabinowicz et al., 1999). PAP1 is identical to ATMYB75 (Kranz et al., 1998) except that ATMYB75 con-tains a sequencing error (1-bp deletion at position 695), cre-ating an early stop codon. The PAP1 protein shares hig h homology with other MYB-like transcription factors that reg-ulate anthocyanin production (Figures 4B and 4D). PAP1 is closely related to the product of the petunia AN2 g ene, showing 82% identity throug h the MYB reg ion and 50% identity overall. The products of the maize anthocyanin MYB genes C1 and pl are 64% identical through the MYB region, with 38% identity overall. The MYB transcription factor GLABAROUS1 from Arabidopsis and MIXTA from snap-dragon both control trichome development (Oppenheimer et al., 1991; Glover et al., 1998) and are each 58% identical to PAP1 in the MYB domain and 33% identical overall. A phy-log enetic tree constructed with these full-leng th MYB pro-teins shows that PAP1 belong s to a branch involved in anthocyanin biosynthesis (Figure 4D).The PAP1 gene was mapped to 0.2 centimorgan (cM) up from mi303 at 83.7 cM on chromosome 1 by using an Xba1 restriction frag ment leng th polymorphism and the Col/Ler recombinant inbred lines (Nottingham University Stock Cen-tre, Nottingham, UK). The sequencing project recently came to PAP1 on bacterial artificial chromosome F25P12 just be-low mi303 at 85 cM.PAP2Also discovered in the Arabidopsis database was 193M15, an expressed sequence tag with very hig h homolog y withTable 1.Enhanced PAL Activity and Lignin Levels in pap1-D PlantsPlant PAL Activity(mkat/g FW)aStemCOMT b Activity(kat/g FW)StemCCOMT c Activity(pkat/g FW)StemLignin CompostionTotal G(mol/g)Total S(mol/g)Total G and S(mol/g)S/GWild type30.544.250.421.6 2.524.10.12 pap1-D70.344.460.630.6 5.035.60.16a FW, fresh weight; kat, katal.b COMT, caffeic acid O-methyltransferase.c CCOMT, caffeoyl-CoA O-methyltransferase.2388The Plant CellFigure 4.Molecular Characterization of PAP1.Activation Tagging of PAP12389PAP1. 193M15 encodes a protein with 93% identity to PAP1 in the MYB domain and 77% identity overall. To test whether overexpression of 193M15 also could cause the production of anthocyanin pig ments, we made a 35S pro-moter::cDNA fusion construct, pCHF3-PAP2. Transgenic plants containing pCHF3-PAP2 had purple leaves and stems (Fig-ure 5C), althoug h the pig mentation was somewhat weaker than that of the pap1-D mutant or some pMN-PAP1 lines. In view of the sequence similarity between the PAP1 protein and that encoded by 193M15 (Figures 4B and 4D) and similar overexpression phenotypes (Fig ure 5C), we named the 193M15 cDNA PAP2. PAP2 is on bacterial artificial chromo-some T27F4 near mi424 on chromosome 1, ف9 cM below PAP1. PAP2 is also a member of the Arabidopsis MYB family reported as ATMYB90 (Kranz et al., 1998).Overexpression of PAP1 and PAP2 in TobaccoTo test whether PAP1 and PAP2 could enhance pigmenta-tion in other plants, we transformed tobacco cv xanthi with pCHF3-PAP1 and pCHFS-PAP2. Both constructs g ener-ated purple tobacco plants, indicating that the Arabidopsis PAP1 and PAP2 genes could activate production of antho-cyanin pig ments in another species (Fig ures 5G and 5H). These transgenic tobacco plants also produced flowers with much more pig mentation than did the pCHF3 transg enic control plants (Figures 5I and 5J). Tobacco transformed with pCHF3 as an empty vector control did not have an in-creased pigmentation phenotype (Figures 5F and 5I). DISCUSSIONAccumulation of phenylpropanoid products during develop-ment is under tight transcriptional regulation, and the pap1-D phenotype represents a striking override of these g enetic controls. Thus, specific tranches of the overall pathway ap-pear to be controlled by separate sets of transcription fac-tors. For example, the maize myb g enes C1 and pl are involved in the reg ulation of anthocyanin synthesis from CHS onward but do not regulate PAL and other genes of the upstream central pathway (Cone et al., 1993a, 1993b; Mol et al., 1996), whereas P independently controls the 3-deoxy flavonoid branch pathway (Grotewold et al., 1994). In con-trast, the pap1-D phenotype, which results from overex-pression of the PAP1 MYB transcription factor, reflects massively enhanced expression of PAL as well as CHS, the g ene encoding dihydroflavonol reductase, and the g lu-tathione S-transferase gene. This broad transcriptional acti-vation of the overall pathway is accompanied by a corresponding ly broad pattern of enhanced product accu-mulation with increases in lig nin, wall-bound hydroxycin-namic acid esters, flavonols, and anthocyanins. Moreover, pathway activation in pap1-D was observed in all vegetative organs and maintained throughout development, in marked contrast to activation in wild-type plants of individual branch pathways at defined developmental stages and with charac-teristic cell-type, tissue-type, and organ specificities (Graham, 1991; Grotewold et al., 1994). The relatively modest in-crease in lig nin content probably reflects a major control point at the polymerization stag e (Bate et al., 1994) with consequent spillover of lig nin monomers and their precur-sors, which contributes to the marked accumulation of wall-bound hydroxycinnamic acid esters in pap1-D.MYB genes contribute to the control of flavonoid biosyn-thesis in a wide range of plant species, often in combination with other regulatory genes. The extensive sequence simi-larity with AN2, c1, and pl, together with the overexpression phenotypes, suggests that PAP1 and PAP2 may be the Ara-bidopsis orthologs of these petunia and maize myb genes, with genetically defined functions in phenylpropanoid regu-lation. In maize, c1 and pl but not P require R and B, encod-ing basic helix-loop-helix proteins, to activate transcription of maize flavonoid biosynthetic genes (Mol et al., 1996). Ba-sic helix-loop-helix proteins and MYB proteins also function together in the control of flower pigmentation in snapdragon (Goodrich et al., 1992) and petunia (Quattrocchio et al., 1998). Moreover, the WD40 proteins TTG1 and AN11 are re-quired for MYB control of flavonoid biosynthesis in bothFigure 4.(continued).(A) Genomic context of T-DNA insertion in pap1-D and structure of pMN-PAP1. Basta r, Basta resistance; Kan r, Kanamycin resistance; LB, left border; pBS, pBluescript KSϩ plasmid; RB, right border; 4 ϫ 35S denotes four copies of 35Se.(B) Sequence homology of R2R3 MYB. Proteins were aligned using the ClustalW software program. Red shading denotes 100% conserved res-idues, and yellow shading denotes matching residues with PAP1. R2 and R3 MYB domains are shown. GenBank accession numbers PAP1 (AF325123), PAP2 (AF325124), AN2 (AAF66727), C1 (AAA33482), P1 (AAB67720), P (AAC49394), GL1 (P27900), Mixta (CAA55725), and c-myb (AAA52031).(C) Gel blot hybridization of total RNA from 4-week-old pap1-D and Col-0 plants.1.0 denotes transcript size in kilobases.(D) Neighbor-joining phylogenetic tree built by using full-length proteins, showing the anthocyanin MYB family branch. The human gene c-myb is used as an outgroup.2390The Plant CellArabidopsis and petunia (de Vetten et al., 1997; Larkin et al.,1999; Walker et al., 1999), and the pap1-D phenotypes re-quire the WD40 gene TTG1. Despite the stringent and often complex g enetic control of phenylpropanoid biosynthesis,strong overexpression of PAP1 in the pap1-D line was suffi-cient to hyperactivate the pathway, which is reminiscent of the enhancement of flavonoid biosynthesis by deliberate ectopic expression of P in suspension cultures of maize cells (Grotewold et al., 1999). The pap1-D phenotypes may reflect involvement of PAP1 as the limiting factor in a novel reg ulatory circuit with atypically broad control functions in phenylpropanoid biosynthesis or may indicate functional spillover from one regulatory circuit to related circuits when PAP1 is massively overexpressed.A recent report describes the use of activation tagging in Catharanthus cell cultures to isolate ORCA3, a jasmonate-responsive transcriptional regulator of primary and second-ary metabolism, the upregulation of which promotes biosyn-thesis of indole alkaloids (Van der Fits and Memelink, 2000).These data, along with the present finding s, indicate that activation tag g ing can be used to g enerate novel g ain-of-function mutations that affect complex biosynthetic path-ways under polygenic control; as such, this presents a po-tentially powerful new approach for isolating the genes that reg ulate biosynthesis of plant natural products. Loss-of-function screens for transparent testa have been saturated,and no mutations map to the PAP1 or PAP2 loci (Shirley et al., 1995). Moreover, examination of Ͼ100 PAP1 antisense lines showed no visible phenotype (data not shown). The similar overexpression phenotypes of PAP1 and PAP2 sug-gest that these genes may be functionally redundant, such that only activation tagging or some other gain-of-function screen could have readily revealed their key attributes.Activation tag g ing as a g ene discovery tool based on g ain-of-function is intrinsically oriented toward biotechno-logical utility, and the ability to activate a biosynthetic path-way that will lead to the enhanced accumulation of several distinct subclasses of natural products has several impor-tant potential applications. Thus, hyperactivated tissues or org ans provide massively enriched sources for passag e through multiplex drug screens with the potential for discov-ery of novel activities based on what combinatorial effectsFigure 5.Overexpression of PAP1 or PAP2 Enhances Pigmentation in Arabidopsis and Tobacco.(A) to (E) Arabidopsis plants transformed with pMN20-2 ([A] and [D]), pMN-PAP1 ([B] and [E]), and pCHF3:PAP2 (C). (A) to (C) show six-week-old plants. (D) and (E) show flowers on 12-week-old plants.(F) to (J) Tobacco plants transformed with pCHF3 ([F] and [I]), pCHF3:PAP1 (G), and pCHF3:PAP2 ([H] and [J]). Plantlets in (F) to (H) were pho-tographed at age 4 weeks, and flowers in (I) and (J) at 10 weeks after transfer to soil. pCHF3-PAP1 plants had brilliant flower pigmentation, iden-tical to that of pCHF3-PAP2 (data not shown).Activation Tagging of PAP12391might arise from complex mixtures as well as allowing con-venient isolation and characterization of individual bioactive components. This approach in principle could be aug-mented by feeding studies using pathway intermediates or synthetic derivatives. Moreover, activation of phenylpro-panoid biosynthesis in pap1-D reflects massively enhanced expression of g enes encoding pathway enzymes; hence, these tissues provide a correspondingly enriched source for isolating the cDNAs that encode key biosynthetic enzymes not readily identified by biochemical approaches.Although the plant kingdom has a remarkable diversity of natural products, the underlying pathway regulatory mecha-nisms appear to be at least partially conserved between species (Mol et al., 1996; Quattrocchio et al., 1998). For ex-ample, the maize anthocyanin reg ulatory g ene R functions appropriately when expressed in Arabidopsis or tobacco (Lloyd et al., 1992). Likewise, ectopic expression of PAP1 or PAP2 in transg enic tobacco caused phenotypes similar to those observed in Arabidopsis. Therefore, convenient, readily transformed genetic model species, such as Arabi-dopsis, can be used to isolate candidate regulatory genes for direct evaluation in medicinal plants and other exotic species or as a platform for the identification of ortholog s and potentially useful, related genes in target species.The serendipitous discovery of pap1-D among a larg e collection of activation-tag g ed lines was possible because activation of PAP1 enhanced the accumulation of anthocya-nin pigments, which was easily scored. Several other plant natural products, such as the isoprenoids lycopene and car-otene and the alkaloid sang uinarine, also are colored. Hence, genetic activation of these tranches of plant metab-olism also could be scored by visual inspection, but this is not a generally applicable approach. In principle, activation-tag g ed lines with enhanced accumulation of natural prod-ucts of interest could be identified by high-throughput meta-bolic profiling. However, a more promising general strategy may be to make transg enic plants that express easily screened marker genes under the control of promoters from g enes encoding enzymes involved in the biosynthesis of natural products of interest.METHODSPlant Growth and TransformationArabidopsis thaliana ecotype Columbia (Col-0) plants were grown in growth rooms at 22ЊC in long days (16 hr of light) or short days (9 hr of light) and received 250 E from three 35-W cool white bulbs and one 35-W Sylvania GrowLux bulb (Osram Sylvania, Danvers, MA). Nico tiana tabacum cv xanthi plants were reg enerated under 24-hr-light conditions at 25ЊC and then transferred to the greenhouse. Tobacco and Arabidopsis transformation was performed as previ-ously described (Neff et al., 1999; Weig el et al., 2000) except that 0.02% Silwet-L77 (Lehle Seeds, Round Rock, TX) was used for the latter. Basta was obtained from AgrEvo (Montvale, NJ).Gel Blot HybridizationDNA and RNA gel blot hybridizations were performed according to standard procedures (Sambrook et al., 1989). RNA samples used in the gel blot analysis shown in Figure 2 were from vegetative leaves of pap1-D and Col-0 plants g rown under short-day conditions for 4 weeks and long-day conditions for 2 weeks. Probes were full-length cDNA fragments of PAP1, the glutathione S-transferase gene, and the g ene encoding ubiquitin. The chalcone synthase (CHS) probe was a polymerase chain reaction product amplified by using the primers 5Ј-TGGTCTCCGTCCTTCCGTCAA and 5Ј-CCCTCAAATGTCCGT-CTATGGAA. The phenylalanine ammonia-lyase (PAL) probe was am-plified by using the primers 5Ј-CTATACGCTTACCTACCAACAAAC and 5Ј-TCTCCGATGAGAAGTAGCACCAA, and the dihydroflavonol reductase probe was amplified with primers 5Ј-AAAAAGATGACA-GGATGGGT-3Ј and 5Ј-CCCCTGTTTCTGTCTTGTTA-3Ј.Enzyme AssaysPAL activity was measured by using a microcuvette spectrophoto-metric assay (Blount et al., 2000). Caffeic acid O-methyltransferase and caffeoyl-CoA O-methyltransferase activities were assayed by standard methods (Inoue et al., 1998). Protein concentrations were determined by the procedure of Bradford (1976).Phenylpropanoid AnalysisSoluble and wall-bound phenolics in whole-plant extracts as well as extracts of individual tissues were analyzed by HPLC (Blount et al., 2000). The aqueous phase, which remained after ethyl acetate ex-traction of the wall-bound phenolics, was lyophilized and resus-pended in 70% methanol for analysis. The HPLC eluates were monitored by absorbance at 270, 310, and 550 nm, and the peaks were identified by comparing their retention times and UV light spec-tra with those of known standards. Lignin was assayed by derivatiza-tion followed by reductive cleavage (Lu and Ralph, 1997).pMN-PAP1, pCHFS-PAP1, and pCHFS-PAP2 ConstructsA PAP1 genomic fragment was amplified by using 5Ј-AACTACTGC-AGCTAGAGCGTAGAGG-3Ј and 5Ј-TCAAACTGCAGAAACTAAGCC-CA-3Ј to construct 5Ј and 3Ј Pst sites. This fragment was cloned into pMN20-2 (Weigel et al., 2000), which contains two copies of 35Se to create pMN-PAP1. PCHF3-PAP1 was created by amplifying the PAP1 cDNA with primers 5Ј-ACTGGTACCTTTTACAATTTGTTTA-3Јand 3Ј-AAGGGATCCTATACACAAACGCA-5Ј and cloning it into the KpnI and BamHI sites of pCHF3, a pPZP211-based plant expression vector carrying the cauliflower mosaic virus 35S promoter and a pea ribulose 1,5-bisphosphate carboxylase/oxyg enase terminator (C. Fankhauser, K. Hanson, and J. Chory, unpublished data). The PAP2 cDNA was excised from expressed sequence tag clone 193M15 with KpnI and BamHI and was cloned into pCHF3 to create pCHF3-PAP2.ACKNOWLEDGMENTSWe thank the following (all at Salk Institute unless otherwise noted): Tseg aye Dabi for help with transg enic tobacco; Mary Anderson at。
用于植物基因表达载体构建的质粒改造及其应用

用于植物基因表达载体构建的质粒改造及其应用安韶雅;虎娟;张虹;孙放;马霞;王晨;陈任【摘要】为克服目前常用于植物基因表达载体构建的质粒所具有的酶切位点有限,目的基因片段难于插入和连接,缺少植物基因表达所必须的启动子、终止子、筛选标记等功能元件的缺点,本研究构建了一个用于植物基因表达载体构建的质粒栽体pNULPGE200.该质粒载体引入了植物基因表达最常用的CaMV 35S启动子(cauliflowermosaic virus 35S promoter)和NOS终止子(nopaline synthase terminator),以及之间的多克隆酶切位点MCS(multiple cloning site).利用pNULPGE200构建植物基因表达栽体,经PCR等方法克隆得到的目的基因可以直接连接到35S启动子与NOS终止子之间,使目的基因能够在植物体内稳定表达;同时该质粒载体还具有独立表达的卡那霉素NPT Ⅱ (neomycinphosphotransferase Ⅱ)耐性基因和sGFP (synthetic green-fluorescent protein with S65T mutation)绿色荧光蛋白报告基因,可用于基因转化时的筛选.本研究以假单胞菌(Pseudomonas putida)携带质粒的二甲苯单加氧酶(xylene monooxygenase)编码基因为材料,分别利用本文质粒载体和常规的质粒栽体pBI121构建了植物表达栽体,验证了文中质粒栽体的实用性.%In order to overcome the defects that the commonly used plasmids have a limited number of restriction sites for target gene cloning, and lack expressing elements such as promoter, terminator and selection marker genes, a plasmid named pNULPGE200 for construction of plant gene expression vector was modified. The plasmid contains cauliflower mosaic virus (CaMV) 35S promoter, nopaline synthase (NOS) ter-minator, and a multiple cloning site between them. The target gene amplified by PCR can beinserted directly between the 35S promoter and the NOS terminator, and can be expressed in plants stably. pNULPGE200 also contains two independent expression marker genes, encoding neomycin phosphotransferase Ⅱ(NPT Ⅱ) and synthetic green-fluorescent protein with S65T mutation (sGFP), which can be used for mutual selection in plant transformation. The practicability of the plasmid was confirmed by comparing with a conventional plas-mid pBI121 in the construction of the gene encoding xylene monooxygenase from Pseudomonas putida to create a plant gene expression vector.【期刊名称】《生命科学研究》【年(卷),期】2018(022)002【总页数】8页(P114-121)【关键词】质粒改造;植物基因表达载体;载体构建;二甲苯单加氧酶基因【作者】安韶雅;虎娟;张虹;孙放;马霞;王晨;陈任【作者单位】宁夏大学宁夏优势特色作物现代分子育种重点实验室,中国宁夏银川750021;宁夏大学西部特色生物资源保护与利用教育部重点实验室,中国宁夏银川750021;宁夏大学生命科学学院,中国宁夏银川 750021;宁夏大学宁夏优势特色作物现代分子育种重点实验室,中国宁夏银川 750021;宁夏大学西部特色生物资源保护与利用教育部重点实验室,中国宁夏银川 750021;宁夏大学生命科学学院,中国宁夏银川 750021;宁夏大学宁夏优势特色作物现代分子育种重点实验室,中国宁夏银川 750021;宁夏大学西部特色生物资源保护与利用教育部重点实验室,中国宁夏银川 750021;宁夏大学生命科学学院,中国宁夏银川 750021;宁夏大学宁夏优势特色作物现代分子育种重点实验室,中国宁夏银川 750021;宁夏大学西部特色生物资源保护与利用教育部重点实验室,中国宁夏银川 750021;宁夏大学生命科学学院,中国宁夏银川 750021;宁夏大学宁夏优势特色作物现代分子育种重点实验室,中国宁夏银川 750021;宁夏大学西部特色生物资源保护与利用教育部重点实验室,中国宁夏银川 750021;宁夏大学生命科学学院,中国宁夏银川 750021;宁夏大学宁夏优势特色作物现代分子育种重点实验室,中国宁夏银川 750021;宁夏大学西部特色生物资源保护与利用教育部重点实验室,中国宁夏银川 750021;宁夏大学生命科学学院,中国宁夏银川 750021;宁夏大学宁夏优势特色作物现代分子育种重点实验室,中国宁夏银川 750021;宁夏大学西部特色生物资源保护与利用教育部重点实验室,中国宁夏银川 750021;宁夏大学生命科学学院,中国宁夏银川 750021【正文语种】中文【中图分类】Q782自1984年首次获得转基因烟草[1]以来,以转基因技术为代表的植物基因工程技术的广泛应用为植物的遗传改良开拓了广阔的前景。
杆状病毒

Trypsin cleavage of the baculovirus occlusion-derived virus attachment protein P74is prerequisite in per os infectionJeffrey M.Slack,1Susan wrence,2Peter J.Krell3and Basil M.Arif1Correspondence Jeffrey M.Slack jslack@nrcan.gc.ca 1Great Lakes Forestry Centre,Sault Ste Marie,ON P6A2E5,Canada2US Department of Agriculture,Beltsville,MD20705-2350,USA3Department of Molecular and Cellular Biology,University of Guelph,ON N1G2W1,CanadaReceived28March2008 Accepted25June2008Baculovirus occlusion-derived virions(ODVs)contain a number of infectivity factors essential for the initiation of infection in larval midgut cells.Deletion of any of these factors neutralizes infectivity by the per os route.We have observed that P74of the group I alphabaculovirus Autographa californica multiple nucleopolyhedrovirus(AcMNPV)is N-terminally cleaved when a soluble form of the protein was incubated with insect midgut tissues under alkaline conditions and that cleavage was prevented by soybean trypsin inhibitor(SBTI).Presently,biological assays were carried out that suggest SBTI inhibits and trypsin enhances baculovirus per os infectivity.We developed a method to rescue per os infectivity of a P74null virus involving co-transfection of viral DNA with a plasmid that transiently expresses p74.We used this plasmid rescue method to functionally characterize P74.A series of site-directed mutants were generated at the N terminus to evaluate if trypsin cleavage sites were necessary for function.Mutagenesis of R195,R196and R199compromised per os infectivity and rendered P74resistant to midgut trypsin.INTRODUCTIONBaculoviruses are a group of arthropod-specific viruses (Zanotto et al.,1993)that have been applied as insecticides and as vectors for the expression of exogenous genes.They have a biphasic replication strategy producing two distinct viral phenotypes:the budded virion(BV)and the occlusion-derived virion(ODV).Both virion phenotypes have bilayer lipid envelopes surrounding bacillus-shaped nucleocapsids and contain double-stranded,circular DNA genomes.However,the integral protein composition of their envelopes and their roles in infection are distinct. BVs spread viral infection throughout host tissues by attaching to and entering host cells via receptor-mediated endocytosis(Volkman&Goldsmith,1985).Endosomal acidification triggers envelope fusion with the endosomal membrane to release the viral nucleocapsid into the host cell(Leikina et al.,1992).In the case of group I alphabaculoviruses(Jehle et al.,2006),budding,attach-ment and envelope fusion are mediated by the viral protein GP64(Blissard&Wenz,1992)and for other baculovirus types these processes are mediated by an F protein (Pearson et al.,2000;Westenberg et al.,2002).ODVs are required in the horizontal transmission of baculoviruses between insect hosts(Kozlov et al.,1986). The ODVs of all baculoviruses are occluded into proteinaceous occlusion bodies(OBs)prior to release from the host(Rohrmann,1986).The distinctive protein structure of OBs has a dual function.It serves to protect virions from deleterious environmental factors,and also acts as a delivery mechanism to transport the ODVs to the alkaline midgut where the cells are susceptible to infection. ODV envelopes are derived from the inner nuclear membrane(Braunagel&Summers,1994)and contain envelope proteins that can survive the protease-rich environment of the insect midgut.ODVs attach to midgut columnar epithelial cells and fuse their envelopes directly with the cell membrane(Kawanishi et al.,1972;Tanada et al.,1975;Granados,1978;Horton&Burand,1993).A number of ODV envelope proteins are essential per os infectivity factors(PIFs),including P74(Kuzio et al., 1989),PIF1(Kikhno et al.,2002),PIF2(Pijlman et al., 2003)and PIF3(Ohkawa et al.,2005).PIF1,PIF2and P74 have been implicated to be involved in ODV attachment to midgut cells(Haas-Stapleton et al.,2004;Yao et al.,2004; Ohkawa et al.,2005).P74,the first identified PIF(Kuzio et al.,1989),is N-terminally exposed on the ODV surface and C-terminally anchored in the ODV envelope(Faulkner et al.,1997; Rashidan et al.,2005).These investigations are thePublished online ahead of print on23July2008as DOI10.1099/vir.0.2008.002543-0.Supplementary tables are available with the online version of this paper.Journal of General Virology(2008),89,2388–2397DOI10.1099/vir.0.2008/002543-0 23882008/002543Printed in Great Britaincontinuation of previous ones that showed the N terminus of P74to be specifically cleaved by insect midgut trypsins (Slack&Lawrence,2005).In the present study we found that Trichoplusia ni larvae were more susceptible to per os infection by Autographa californica multiple nucleopolyhe-drovirus(AcMNPV)when trypsin was added to the diet and they were less susceptible in the presence of soybean trypsin inhibitor(SBTI).We have developed a transient plasmid-based assay to evaluate the ability of P74mutants to rescue per os infectivity of a P74null virus.It has been established that a P74fused with the green fluorescent protein can function in place of native P74(Yao et al.,2004).In this assay,insect cells were co-transfected with P74null virus DNA and a plasmid expressing the p74gene fused in-frame with the enhanced green fluorescent protein(egfp)gene. Earlier,we proposed that P74is cleaved at arginine residue R156(Slack&Lawrence,2005)and this study examines the functional effects on P74of mutating R156and nearby trypsin cleavage site residues.We co-transfected insect cells with mutant p74–EGFP-expressing plasmids and P74D virus DNA.OBs produced in these cells were harvested and fed to insects.It was clearly shown that R156is not required for P74function and that instead a nearby cluster of trypsin cleavage sites at residues R195,R196and R199 were important for function.Experiments with insect brush border membrane vesicles(BBMV)confirmed that absence of these trypsin cleavage sites eliminated the specific cleavage of P74.METHODSCell lines and viruses.Spodoptera frugiperda Sf9cell lines were propagated in10%v/v fetal bovine serum,2.5%w/v tryptose broth-supplemented Grace’s(FBS/Grace’s)media(Sigma-Aldrich).The parental AcMNPV virus used in this study was isolate HR3(Brown et al.,1979).The P74null(P74D)AcMNPV virus used in this study was AcLP4(Faulkner et al.,1997).This AcLP4virus has the b-galactosidase reporter gene inserted into the middle of the native p74 ORF of AcMNPV and it produces the N-terminal194aa of P74fused in-frame with b-galactosidase.Plasmid constructs.The plasmids,pBAC-5-EGFP and pBAC-5-p74-EGFP,were previously described as pBAC-5-GFP and pBAC-5-p74-GFP(Slack et al.,2001).pBAC-5-p74-EGFP contains the AcMNPV p74ORF fused at the39end in-frame with the jellyfish EGFP ORF(Zhang et al.,1996).pBAC-5-p74-EGFP is based on the pBAC-5baculovirus transfer plasmid(Novagen).Site-directed mutagenesis.All mutants were made using muta-genic PCR primers and the Deep Vent polymerase(New England Biolabs).PCR amplifications were done with35cycles of95u C 1min,45u C1min30s and72u C1min45s.The seven P74mutants,R114Q,R119Q,R132Q,K138Q,R156Q, RK184/186QQ and RRR195/196/199QQQ,were generated by amp-lifying two PCR products that,when ligated together,yield an823bp fragment of the59end of the p74ORF.This823bp fragment contained Nco I and Bst XI sites that permitted cloning in place of the corresponding p74ORF region in pBAC-5-p74-EGFP.The first or‘left’PCR products were made with a common Nco I site (underlined)-containing primer,p74M1NcoI-59(59-AAGCACCA-TGGCGGTTTTAACAGCCGT-39)and phosphorylated mutagenic primers.The second or‘right’PCR products were made with a common Bst XI site(underlined)-containing primer,p74D274BstXI-39(59-AAGCACCATGGCGGTTTTAACAGCCGT-39)and primers that corresponded to the positions immediately on the59end of the mutagenic primers(see Supplementary Table S1,available with the online version of this paper).Left and right PCR products were fractionated by agarose gel electrophoresis and purified using Qiaex II glass milk(Qiagen).The corresponding left and right PCR products were ligated using T4 DNA ligase and the ligation was amplified by PCR using the primers p74M1NcoI-5and p74D274BstXI.The pBAC-5-p74-EGFP plasmid contained two Bst XI sites and was partially digested with Bst XI prior to Nco I digestion.The resulting vector minus the original p74Nco I/ Bst XI fragment was fractionated by agarose gel electrophoresis and purified by glass milk.The PCR product was cut with Nco I and Bst XI and ligated into the corresponding sites in the p74ORF.The P74mutants R195Q,R196Q,R199Q,R195Q/R196Q,R196Q/ R199Q and R195Q/R199Q were made within a395bp Eco RI/Sac II fragment of the p74ORF.All of these mutagenic PCR products were amplified with the Sac II-containing primer p74R319SacII-39(59-CGTTACCGCGGTAATTGTATGCGATC-39)and an Eco RI site containing mutagenic primer(see Supplementary Table S1).PCR products were fractionated by agarose gel electrophoresis and purified using glass milk.The pBAC-5-p74-EGFP plasmid and PCR products were cut with Eco RI and Sac II,fractionated by agarose gel electrophoresis,glass milk-purified and ligated together.All p74mutants were sequenced(University of Guelph Molecular Supercentre)in both directions using primers p74Sequencing1LP36 (59-AAACCCAAGTTCAGTCTGC-39)and p74Sequencing2RP37(59-AAGTGACAAAGATCGTGTC-39).OB preparation.OBs were obtained from infected Sf9insect cells. Infected cells were suspended in media and then centrifuged for 5min at1000g.Pellets were suspended in PBS(120mM NaCl, 10mM Na2HPO4,2.5mM KCl,pH6.2)and the OBs were released with SDS(0.3%w/v)and centrifuged for10min at2000g.OB pellets were washed several times in PBS(pH6.2),counted with a haemocytometer and diluted to desired concentrations.Insect bioassays.Bioassays were all done in128-well assay trays(C-D International).Artificial insect diet and T.ni larvae were obtained from the Great Lakes Forestry Centre,Insect Production Service, Ontario,Canada.Assay tray wells contained1ml volumes of diet. OBs were suspended in PBS(pH6.2)and20m l were applied to the diet surface(2.3cm2)and allowed to dry onto the diet surface for1h under shaded rvae were place onto diet,incubated at28u C and mortality data were collected at7days.DNA purification and transfections.Transfections were done with P74D virus DNA and plasmid DNA.Plasmid DNA was purified by CsCl density-gradient centrifugation(Slack&Lawrence,2002).P74D virus DNA was purified from ODVs by methods adapted from O’Reilly et al.(1992).Six150cm2T-flasks,each containing26107 Sf9cells,were infected with AcLP4at a m.o.i.of1p.f.u.per cell and OBs were purified at5days post-infection,cells were harvested and OBs were purified.Suspensions of OBs were pelleted for45min at 1200g and suspended in20ml H2O and were released by the addition of0.5ml of500mM EDTA and2.5ml freshly made2M Na2CO3. After10min,2ml Tris-OH(1M,pH6.8)was added and the insoluble debris was removed at1200g centrifugation for5min.The supernatant was centrifuged at112500g for1h at15u C.The ODV Trypsin cleavage of baculovirus envelope protein P742389pellet was suspended in400m l TE buffer(1mM EDTA,10mM Tris-OH pH8.0)and disrupted by addition of20m l EDTA(500mM), 10m l SDS(10%w/v)and12.5m l proteinase K(10mg ml21).After 2h at37u C,viral DNA was extracted by phenol and chloroform/ isoamyl alcohol(24:1).The DNA was dialysed against TE at4u C in 10K molecular weight cut-off Slide-A-Lyzer cassettes(Pierce). Transfections were done with the transfection reagent,ExGen500 (Fermentas)using methods adapted from Ogay et al.(2006).Sf9cells were seeded onto6-well plates at a density of1.256106cells per well in2ml FBS/Grace’s media.Medium was supplemented with antibiotics/antimycotics(100U penicillin G ml21,100m g streptomy-cin sulfate ml21and250ng amphotericin B ml21)(Sigma-Aldrich). The transfection mixtures included1600ng plasmid DNA,600ng viral DNA,8m l NaCl(1M),1077m l NaCl(150mM)and55m l ExGen500transfection reagent.A transfection mixture volume of 200m l was added to each well and then plates were centrifuged for 5min at500g.At5days post-transfection,cells and media were harvested and processed for either OB purification or SDS-PAGE. Brush border membrane vesicles preparation.BBMV were prepared from80fourth instar T.ni larvae using methods described previously(Slack&Lawrence,2005).Final BBMV preparations were suspended in buffer B(8.5mM MgCl2,150mM D-sorbitol,5mM EGTA,17mM Tris-OH,pH7.4),diluted to1mg ml21total protein and stored at280u C.BBMV total protein was determined with a Bradford reagent assay(Bio-Rad Laboratories).ODV purification and interactions with BBMV.ODVs were produced in Sf9cells that had been co-transfected with P74D virus DNA and P74–EGFP plasmid DNA.Five150cm2T-flasks(1.36108 Sf9cells)were transfected with160m g plasmid DNA and3.6m g P74D virus DNA.DNA was suspended in11ml NaCl(150mM)and550m l ExGen500was added and incubated for15min.The solution was added to110ml FBS/Grace’s media-suspended Sf9cells(1.26106 cells ml21).Cells were then distributed into five150cm2T-flasks (22ml per flask).After5days,OBs were purified as described,but with three additional PBS washes to ensure complete removal of SDS. The final yield was561011OBs per transfection group.ODVs were released from OBs by alkali treatment prior to experiments and were prepared on a small scale in2ml poly-propylene tubes.To a total of1.361010OBs in165m l PBS(pH6.2), 300m l freshly prepared carbonate-buffered potassium was added (CBK)(500mM KCl,100mM Na2CO3,pH10.5).The final solution was incubated at room temperature for5min.Tubes were placed on ice and then centrifuged in an Eppendorf5417R microcentrifuge for 45min,21000g at4u C.The supernatant was aspirated off and ODV pellets remained on ice no more than30min until their suspension in BBMV solutions.Frozen BBMV were thawed on ice and then diluted50%in CBK buffer.The resulting BBMV incubation(BI)buffer contained 250mM KCl,50mM Na2CO3,75mM D-sorbitol,1.25mM EGTA, 4mM Tris-OH and4mM MgCl2.The pH of BI buffer was10.5.In some experimental groups,BBMV were either preheated to95u C for 5min or type II SBTI(Sigma-Aldrich)was added to a final concentration of500m g ml21.BBMV were further diluted in BI buffer to desired concentrations of BBMV proteins and55m l volumes of the vesicles were added to the ODV pellets.Control groups just received BI buffer.ODV pellets were suspended by brief vortexing and incubated for20min at28u C and then placed on ice.This was followed by the addition of55m l26Laemmli SDS-PAGE disruption buffer and heating at95u C for10min.Western blots.Proteins were fractionated under reducing condi-tions in8.75%w/v acrylamide:bis(37:1)SDS-PAGE gels and transferred to nitrocellulose.Western blotting was done using an enhanced chemifluorescence(ECF)Western blotting kit(AmershamBiosciences).Primary antibodies were either the GP64-specific mono-clonal antibody,AcV5(Hohmann&Faulkner,1983)or a EGFP-specificmonoclonal antibody JL-8(Clontech).Blots were blocked overnight at 4u C with10%w/v powdered milk in PBS(pH7.4)and were probedfor2h at room temperature with either AcV5or JL-8diluted1:500inPBS(pH7.4),0.05%v/v tween-20(PBS-T).Blots were probed for2hwith secondary anti-mouse,fluorescein conjugated antibody at1:500 dilution in PBS-T and then probed for2h with tertiary anti-fluorescein,alkaline phosphatase(AP)conjugated antibody at1:2000dilution in PBS-T.After tertiary antibody probing,the blots were washed briefly with1mM MgCl2,10mM Tris-OH(pH9.5)and thenreacted with1.25m g ml21dimethylacridinone phosphate(MolecularProbes)fluorescent substrate.Blots were scanned using a TyphoonTrio+laser bed scanner(GE Healthcare Life Sciences)(633nm excitation,670nm BP30nm emission).Protein sequence analysis and sequence sources.Alignment ofthe amino acid sequences of the P74homologues was done using MEGALIGN5.08(DNASTAR).The CLUSTAL W method(Thompson et al., 1994)and Gonnet250protein weight matrix were used in alignments(gap penalty10,gap length penalty0.2).Hydrophobicity profile(15aa window)was determined using theprogram Protean5.08(DNASTAR).For transmembrane(TM)domainprediction,the site http://www.cbs.dtu.dk/services from the Centerfor Biological Sequence Analysis at the Technical University of Denmark was accessed and the program TMHMM2.0(Krogh et al., 2001)was used.RESULTSRole of trypsin in the infectivity of lepidopteran baculovirusesP74is essential for per os infection and is cleaved at a specific location by lepidopteran midgut trypsins.As a model system for other baculoviruses,we wanted to ascertain the role of indigenous midgut trypsin in per os infectivity.The experiments involved feeding T.ni larvae with parental AcMNPV OBs along with either SBTI or trypsin.Varying amounts of OBs were fed to groups of insects such that lethal concentration for50%(LC50) values could be estimated.It was observed that,relative to control groups of insects that were fed only OBs,the SBTI increased the LC50threefold and the presence of trypsin decreased the LC50fivefold(Fig.1).Differences in the LC50 in the presence of either additives clearly demonstrate that trypsin plays an important role in virus infectivity.Development of a transient expression-based assay for characterization of P74A simple method to evaluate P74mutants without constructing new recombinant viruses was developed.It involved transfecting cells with plasmids that transiently express P74in the presence of a co-infecting P74D virus. This was similar to a successful approach involving p74-expressing insect cell lines complementing a P74D virus (Wilson,1997).We used pBAC-5-based plasmids to transiently express the p74ORF fused in-frame with the 59end of the EGFP ORF(Fig.2a).These plasmids wereJ.M.Slack and others2390Journal of General Virology89previously used to study P74localization in infected cells (Slack et al.,2001).Though designed as baculovirus shuttle vectors,pBAC-5-based plasmids are excellent for transient gene expression in insect cell culture (Ogay et al.,2006),due to the presence of an AcMNPV gp64early/latepromoter (Whitford et al.,1989;Blissard &Rohrmann,1991).Homologous recombination of plasmids such as pBAC-5with circularized viral DNA is an infrequent event (less than 1%)(Kitts et al.,1990)and thus would not be significant for these transient experiments.Also,pBAC-5plasmids are designed to eliminate the polh gene after recombination and any incidental recombinant viruses would not produce the polyhedra that were used to perform experiments.The C-terminal tagging of P74with EGFP enabled monitoring of transfections and also provided an epitope tag for immunodetection (Fig.2b).The OBs produced from P74D virus/P74–EGFP plasmid co-transfections appeared by microscopic examination to be similar to parental virus OBs.Western blotting confirmed that P74–EGFP proteins co-purified with ODVs (Fig.2b,ODV lane).More importantly,the P74D virus was rescued for per os infectivity by P74–EGFP,but not by EGFP (Table 1).Trypsin cleavage site mutantsThe previous hypothesis of midgut-specific P74cleavage at R156(Slack &Lawrence,2005)was based on the estimated size of tryptic digest fragments in Western blots and on conservation of the R156trypsin cleavage site among P74homologues.It was not possible to obtain enough material to do N-terminal sequencing of P74trypticdigestFig. 1.Effects of trypsin and SBTI on susceptibility of T.ni larvae to AcMNPV infection.Second instar T.ni larvae were fed dosages of AcMNPV OBs in the presence of trypsin or SBTI and compared to OBs alone (control).Test animals received 1mg/diet well porcine parvovirus tested (1:250)trypsin or 1mg/diet well type II SBTI.(a)Cumulative %mortality is plotted against OB amounts that were added to diet wells.(b)The LC 50of each group is graphed and was determined by probit analysis using the program PoloPlus1.0(LeOra Software).Numerical bioassay data are provided in Supplementary TableS2.Fig. 2.Control co-transfection experiments.Initial experiments involved co-transfection of the P74D virus DNA with plasmid DNA expressing either EGFP or p74–EGFP .(a)The EGFP and P74–EGFP ORFs are schematically illustrated along with the predicted molecular masses of their products.(b)We confirmed the synthesis of EGFP and P74–EGFP by Western blotting with an anti-EGFP ne M is mock-infected Sf9cells,lane 1is cells transfected with P74D virus DNA only,lane 2is cells transfected with pBAC-5-EGFP and P74D virus DNA,lane 3is cells transfected with pBAC-5-p74-EGFP and P74D virus DNA.The lane labelled ‘ODV’is purified ODVs from cells that had been co-transfected with pBAC-5-p74-EGFP and P74D virus DNA.The positions and molecular masses in kDa of markers are indicated on the sides.Table 1.Mortality of 2nd instar T.ni larvae 7days post feeding OBs (100000OBs per well)from P74D virus,P74D virus/plasmid co-transfections or wt AcMNPVVirus Plasmid Mortality (dead/total)P74D AcMNPV 20/16P74D AcMNPV EGFP 0/16P74D AcMNPV p74–EGFP16/16AcMNPV 216/16Mock20/16Trypsin cleavage of baculovirus envelope protein P742391fragments and site-directed mutagenesis was chosen to verify if R156or nearby trypsin cleavage sites are required for P74function.In the vicinity of R156,there are11 consensus trypsin cleavage site motifs(R^X,K^X except R^P,K^P)(Fig.3)and all are present in AcMNPV P74at R114,R119,R132,K138,K147,R156,R184,K186,R195, R196and R199.A series of sited-directed mutants were constructed by substituting R or K residues with glutamine(Q)residues. Glutamine was chosen because its side chain size is similar to R or K residues.Glutamine is a polar amino acid residue and R and K are positively charged residues.However,in the highly alkaline conditions of the insect gut where P74is to function,K and R residues would also become polar residues.Also,this Q substitution occurs naturally in some P74homologues at K138and R186(Fig.3).In the first group of mutants,each of the residues R114,R119,R132, K138,K147and R156were changed to Q residues.It was initially opted to make multiple mutations where trypsin cleavage sites were clustered close to each other(R184/ K186and R195/R196/R199).Site-directed mutants were made in P74–EGFP plasmids that were then co-transfected with P74D virus DNA into insect cells.It was confirmed by Western blot with EGFP-specific monoclonal antibody that similar amounts of P74–EGFP were being translated from each mutant P74–EGFP plasmid(Fig.4a).As an internal control to virus replication,Westerns were also probed with anti-GP64 monoclonal antibody,AcV5(Hohmann&Faulkner,1983; Monsma&Blissard,1995).OBs from each co-transfection group were fed to2nd instar T.ni larvae at several dosages.The bioassay results of the first group of P74mutants are plotted as cumulative mortality(Fig.4c).It was evident from this experiment and others that most of the P74–EGFP mutants rescued per os infection of the P74D virus as effectively as the native P74–EGFP protein.R156was not required for P74’s function in oral infection.One clear and interesting observation is that cells producing the multiple site P74mutant R195Q/ R196Q/R199Q resulted in OBs that were1000-fold less infectious than OBs derived from the other groups and warranted furtherinvestigation.Fig.3.Alignment of predicted trypsin cleavage sites.A portion of the N terminus of AcMNPV P74was aligned with other P74 homologues.The consensus sequence(Cons)is shown at the top.The R and K residues of predicted trypsin cleavage sites are highlighted.P74homologues are arranged into group I alphabaculoviruses(I),group II alphabaculoviruses(II), betabaculoviruses(B),hymenopteran-specific gammabaculoviruses(G),the dipteran-specific deltabaculovirus(D),Cuni and Hz-1.The group is indicated on the right side.The numbers along the top correspond to amino acid positions of AcMNPV P74.For this alignment,short form virus source names are indicated on the left side.Ac/Px,Cf/Op,Hear/Hz,and Co/Cf GV are baculovirus species with P74homologues that are identical in the region shown and are represented by single sequences.Also Splt(2)and Sf(2)indicates there are two virus isolates with P74homologues that are identical in the region shown.P74 homologue source baculovirus full names and GenBank protein accession numbers are listed in Supplementary Table S5.J.M.Slack and others2392Journal of General Virology89P74site-directed mutants in region R195/R196/R199The multiple mutations in this region of the protein could have caused conformational changes that precluded the ability of the mutant to rescue infectivity.Also,the P74D virus used in these experiments contained a p74gene region corresponding to M1to C194.It is possible homologous recombination of our p74–EGFP plasmids with P74D virus DNA rescued P74mutations between R114and K186and that mutations beyond C194were not.We,therefore,generated a second series of site-directedmutants that covered all combinations of mutations in the R195/R196/R199region.Bioassays were carried out as previously described and the results are summarized in Fig.4(d).The single mutants R195Q,R196Q and R199Q were as functional as native P74–EGFP and could rescue per os infectivity of the P74D virus.The double mutants R195Q/R196Q and R195Q/R199Q were also able to rescue per os infectivity.However,the double mutant R196Q/R199Q produced OBs that were at least 100-fold less infectious than the native P74–EGFP group.These data suggestthatFig.4.Bioassay of P74site-directed mutants.Sf9cells were co-transfected with P74D virus DNA and the p74–EGFP plasmid or p74–EGFP plasmids with mutations in the P74ORF.Numbers refer to the amino acid position where R or K residues were changed to Q residues.The control group was just P74D virus DNA.(a and b)Western blotting was done with two different antibodies to confirm the presence of P74–EGFP proteins or to detect the virus-specific protein,GP64.The blots were probed with either anti-GFP monoclonal antibody (a -GFP)or with the AcV5anti-GP64monoclonal antibody (a -AcV5).Antibody reacting portions of blots are shown in upper two panels (102.6kDa P74–EGFP and 64kDa GP64).Also in the lower panel of (a)and (b),SDS-PAGE gels were stained with Coomassie brilliant blue and the dominant staining 29kDa protein which is likely to be polyhedrin is shown (Polh).(c and d)In two separate bioassays,purified OBs from mutant groups were fed to 2nd instar T.ni larvae at varying dosages.The cumulative mortality curve is plotted.Most P74–EGFP mutants rescued per os infectivity of the P74D virus like wt P74–EGFP except for mutants P74(R195Q/R196Q/R199Q)-EGFP (c and d)and P74(R196Q/R199Q)–EGFP (d).Numerical bioassay data are provided in Supplementary Tables S3and S4.Trypsin cleavage of baculovirus envelope protein P742393R196and R199may be functionally redundant for the per os infectivity.None of the mutants compromised P74function as much as mutant R195Q/R196Q/R199Q,indicating that the three R residues may work in concert.BBMV-specific cleavage of P74BBMV from T.ni larvae were used to evaluate the effects of insect trypsins on P74.By using azocasein assays (Slack &Lawrence,2005)and different proteinase inhibitors,it was determined that BBMV from T.ni contained almost entirely trypsin protease activity (data not shown).In experiments,P74–EGFP or mutant P74(R195Q/R196Q/R199Q)–EGFP-containing ODVs were purified and incu-bated with BBMV,followed by fractionating the proteins on SDS-PAGE.P74–EGFP cleavage products were detected by Western blot using an anti-EGFP antibody (Fig.5).BBMV-specific trypsin cleavage of native P74–EGFP was compared with that of the mutant P74(R195Q/R196Q/R199Q)–EGFP.After incubation with BBMV,the 102.6kDa native P74–EGFP protein produced a cleavage product of 80kDa (Fig.5,lanes 9and 11)whereas the mutant P74–EGFP protein did not produce this product (Fig.5,lanes 10and 12).The presence of SBTI inhibitedthe cleavage of P74–EGFP by the BBMV (Fig.5,lanes 7and 8).Although our ODV preparations were partially purified,these experiments suggest that P74–EGFP cleavage occurs in the context of the ODV and that the cleavage takes place at residues R195,R196and R199.DISCUSSIONThe data presented here clearly suggest that trypsin activates the per os infectivity of baculovirus ODVs by cleaving P74at the R195/R196/R199vicinity.Tryptic activation has been documented for other viruses including coronaviruses (Frana et al.,1985),rotaviruses (Vonderfecht et al.,1988),Sendai virus (Muramatsu &Homma,1980)and poxviruses (Ichihashi &Oie,1982).The lepidopteran midgut is rich in trypsins (Johnston et al.,1991;Terra &Ferreira,1994;Oliveira et al.,2005;Pereira et al.,2005)and it is probable that baculoviruses have evolved to exploit this environment.Trypsin is a serine protease and,in past studies,serine proteases have been found associated with insect-derived baculovirus OBs (Eppstein &Thoma,1975;Eppstein et al.,1975;Langridge &Balter,1981;Maeda et al.,1983).Site-directed mutagenesis data and BBMV cleavage data suggest that P74is a target of midgut trypsins and point to R196and R199being alternate primary trypsin cleavage sites on P74.So long as R196or R199are present,the full P74function is retained.Residues R196and R199are present among P74homologues of alphabaculoviruses,betabaculoviruses and gammabaculoviruses.The only exception is one isolate of Spodoptera frugiperda multiple nucleopolyhedrovirus which has a P74homologue lacking R196(Sf-CV,Fig.3).There was insufficient material for N-terminal sequencing of BBMV-specific P74–EGFP cleavage products,but the molecular masses of the products correspond well with cleavage in the R195/R196/R199region.This is supported by lack of a specific cleavage product in the R195Q/R196Q/R199Q mutant.Two studies (Haas-Stapleton et al.,2004;Yao et al.,2004)have concluded that P74is a viral attachment protein.Haas-Stapleton et al.(2004)showed that the P74D virus could not compete with wt virus ODVs for binding to the midgut and Yao et al.(2004)identified a 30kDa midgut receptor protein for P74.P74cleavage by midgut proteases may expose a midgut receptor binding domain on P74.However P74binds to BBMV in the presence of serine protease inhibitors (Yao et al.,2004)and thus trypsin cleavage of P74is not prerequisite to virus attachment.P74has characteristics of proteolytically activated virus envelope proteins such as influenza virus haemagglutinin (HA)protein and paramyxovirus F protein.Like P74,these proteins are C-terminally anchored in the virion envelope and have a large surface-exposed N-terminal domains.When influenza virus HA proteins and paramyxovirus F proteins are cleaved by host trypsins,a hydrophobic membrane insertion N-terminal domain isexposedFig. 5.Western blot of ODV-associated native P74or P74mutant R195Q/R196Q/R199Q after incubation with BBMV.An experiment was done to examine midgut BBMV-specific proteo-lytic cleavage of wt P74–EGFP and P74mutant (R195Q/R196Q/R199Q)–EGFP in context of the ODV.OBs were purified from Sf9cells that had been co-transfected with P74D virus DNA and either P74–EGFP (wt)or P74(R195Q/R196Q/R199Q)–EGFP (MT)plasmids.The ODVs were released from OBs and incubated with varying amounts of BBMV prepared from T.ni larvae midguts.Proteins were then fractionated by SDS-PAGE and Western blotted with a -EGFP monoclonal antibody.The result is shown here.The positions of molecular mass standards are indicated on the ne 1is the BBMV alone and lane 2is the P74D virus ODVs alone.The 3rd and 4th lanes are wt and MT P74–EGFP-containing ODVs incubated alone.We incubated ODVs in BBMV that had been previously boiled (5th and 6th lanes)or treated in SBTI (7th and 8th lanes).Lanes 9through 12are ODVs incubated with two different amounts of BBMV.In lanes we indicate the amount of BBMV used in incubations as the number of ng of BBMV-specific protein.The positions of full-length P74–EGFP (102.6kDa)and a cleavage product of interest (80.0kDa)are indicated on the right side.J.M.Slack and others2394Journal of General Virology 89。
碧云天生物技术一氧化氮检测试剂盒说明书
碧云天生物技术/Beyotime Biotechnology 订货热线:400-1683301或800-8283301 订货e-mail :******************技术咨询:*****************网址:碧云天网站 微信公众号一氧化氮检测试剂盒产品编号 产品名称包装 S0021S 一氧化氮检测试剂盒 500次 S0021M一氧化氮检测试剂盒2500次产品简介:碧云天生产的一氧化氮检测试剂盒采用了经典的Griess Reagent ,并对其测定的溶液体系进行了优化,使检测下限达到1µM ,在1-100µM 范围内有非常完美的线性关系。
检测速度极快,完成一条标准曲线或5-10个样品的测定只需3分钟。
样品范围广,可以检测细胞或组织及其培养液中的一氧化氮的含量,酚红和10%血清均对测定无明显干扰,也可以检测血清、血浆和尿液中一氧化氮的含量。
包装清单:产品编号 产品名称 包装 S0021S-1 1M NaNO 2 1ml S0021S-2 Griess Reagent I 25ml S0021S-3 Griess Reagent II25ml —说明书1份产品编号 产品名称 包装 S0021M-1 1M NaNO 2 1ml S0021M-2 Griess Reagent I 125ml S0021M-3Griess Reagent II125ml —说明书1份保存条件:-20ºC 避光保存,一年有效。
4ºC 避光保存,半年有效。
注意事项:本产品对人体有害,操作时请小心,并注意有效防护以避免直接接触人体或吸入体内。
如保存不当导致溶液变色或沉淀,则说明该溶液已经失效,请购买新的试剂盒。
不建议使用RIPA 裂解液对细胞或者组织进行裂解,使用RIPA 裂解液可能在后续反应中产生沉淀,影响测试。
推荐使用碧云天的细胞与组织裂解液(一氧化氮检测用)(S3090)或Western 及IP 细胞裂解液(P0013)。
pull down buffer经典配方
Arabidopsis EPSIN1Plays an Important Role in VacuolarTrafficking of Soluble Cargo Proteins in Plant Cells via Interactions with Clathrin,AP-1,VTI11,and VSR1WJinhee Song,Myoung Hui Lee,Gil-Je Lee,Cheol Min Yoo,and Inhwan Hwang1Division of Molecular and Life Sciences and Center for Plant Intracellular Trafficking,Pohang University of Scienceand Technology,Pohang790-784,KoreaEpsin and related proteins play important roles in various steps of protein trafficking in animal and yeast cells.Many epsin homologs have been identified in plant cells from analysis of genome sequences.However,their roles have not been elucidated.Here,we investigate the expression,localization,and biological role in protein trafficking of an epsin homolog, Arabidopsis thaliana EPSIN1,which is expressed in most tissues we examined.In the cell,one pool of EPSIN1is associated with actinfilaments,producing a network pattern,and a second pool localizes primarily to the Golgi complex with a minor portion to the prevacuolar compartment,producing a punctate staining pattern.Protein pull-down and coimmunoprecipitation experiments reveal that Arabidopsis EPSIN1interacts with clathrin,VTI11,g-adaptin-related protein(g-ADR),and vacuolar sorting receptor1(VSR1).In addition,EPSIN1colocalizes with clathrin and VTI11.The epsin1mutant,which has a T-DNA insertion in EPSIN1,displays a defect in the vacuolar trafficking of sporamin:greenfluorescent protein(GFP),but not in the secretion of invertase:GFP into the medium.Stably expressed HA:EPSIN1complements this trafficking defect.Based on these data,we propose that EPSIN1plays an important role in the vacuolar trafficking of soluble proteins at the trans-Golgi network via its interaction with g-ADR,VTI11,VSR1,and clathrin.INTRODUCTIONAfter translation in eukaryotic cells,a large number of proteins are transported to subcellular compartments by a variety of different mechanisms.Newly synthesized vacuolar proteins that are delivered to the endoplasmic reticulum(ER)by the cotrans-lational translocation mechanism are transported to the vacuole from the ER by a process called intracellular trafficking.Traffick-ing of a protein to the vacuole from the ER occurs through two organelles,the Golgi complex and the prevacuolar compartment (PVC)(Rothman,1994;Hawes et al.,1999;Bassham and Raikhel, 2000;Griffiths,2000).Transport of a protein from the ER to the Golgi complex is performed by coat protein complex II vesicles. Transport from the trans-Golgi network(TGN)to the PVC occurs via clathrin-coated vesicles(CCVs)(Robinson et al.,1998;Tang et al.,2005;Yang et al.,2005).Transport of a protein from the ER to the vacuole/lysosome requires a large number of proteins,including components of vesicles,factors involved in vesicle generation and fusion,reg-ulators of intracellular trafficking,adaptors for the cargo proteins, and other accessory proteins(Robinson and Kreis,1992;Bennett, 1995;Schekman and Orci,1996;da Silva Conceic¸a˜o et al.,1997;Kirchhausen,1999;Sever et al.,1999;Bassham and Raikhel, 2000;Griffiths,2000;Jin et al.,2001;Robinson and Bonifacino, 2001).Most of these proteins are found in all eukaryotic cells from yeast,animals,and plants,suggesting that protein traffick-ing mechanisms from the ER to the vacuole/lysosome may be highly conserved in all eukaryotic cells.Of the large number of proteins involved in intracellular traf-ficking,a group of proteins that have the highly conserved epsin N-terminal homology(ENTH)domain have been identified as playing a critical role at various trafficking steps in animal and yeast cells(Chen et al.,1998;De Camilli et al.,2002;Wendland, 2002;Overstreet et al.,2003;Legendre-Guillemin et al.,2004). The ENTH domain binds to phosphatidylinositols(PtdIns), although the lipid binding specificity differs with individual members of the epsin family.For example,epsin1binds to PtdIns(4,5)P2,whereas EpsinR and Ent3p bind to PtdIns(4)P and PdtIns(3,5)P2,respectively(Itoh et al.,2001).The ENTH domain is thought to be responsible for targeting these proteins to specific compartments and also for introducing curvature to the bound membranes to assist in the generation of CCVs(Legendre-Guillemin et al.,2004).However,the exact steps of intracellular trafficking in which ENTH-containing proteins play a role are complex.Epsin homologs can be divided into two groups based on the pathway in which they play a role.One group,which includes epsin1in animal cells and Ent1p and Ent2p in yeast cells,is involved in endocytosis from the plasma membrane (Chen et al.,1998;De Camilli et al.,2002;Wendland,2002).The other group,which includes EpsinR/clint/enthoprotin in animal cells and Ent3p and Ent4p in yeast cells,is involved in protein trafficking from the TGN to the lysosome/vacuole as well as1To whom correspondence should be addressed.E-mail ihhwang@postech.ac.kr;fax82-54-279-8159.The author responsible for distribution of materials integral to thefindings presented in this article in accordance with the policy describedin the Instructions for Authors()is:Inhwan Hwang(ihhwang@postech.ac.kr).W Online version contains Web-only data./cgi/doi/10.1105/tpc.105.039123The Plant Cell,Vol.18,2258–2274,September2006,ª2006American Society of Plant Biologistsretrograde trafficking from the early endosomes to the TGN (Kalthoff et al.,2002;Wasiak et al.,2002;Hirst et al.,2003; Chidambaram et al.,2004;Eugster et al.,2004;Saint-Pol et al., 2004).Another common feature of epsin-related proteins is that they play a role in CCV-mediated protein trafficking at both the TGN and the plasma membrane.These proteins can bind directly to clathrin through their multiple clathrin binding motifs;thus,they may recruit clathrin to the plasma membrane or the TGN to generate CCVs(Rosenthal et al.,1999;Wendland et al.,1999; Drake et al.,2000).In addition,these proteins interact with many other proteins,such as heterotetrameric clathrin adaptor complexes(APs),monomeric adaptor Golgi-localized,g-ear–containing Arf binding proteins(GGAs),and soluble NSF attach-ment protein receptors(SNAREs).Epsin1interacts with AP-2, Epsin15,and intersectin(Chen et al.,1998;Legendre-Guillemin et al.,2004),whereas EpsinR/enthoprotin/clint and Ent3p interact with SNAREs such as vti1b and vti1p,respectively (Chidambaram et al.,2004)and with adaptor proteins such as GGAs and AP-1(Duncan et al.,2003;Mills et al.,2003).In addition,epsin homologs have ubiquitin-interacting motifs and are ubiquitinated(Oldham et al.,2002;Shih et al.,2002).Protein ubiquitination acts as a signal for endocytosis from the plasma membrane and trafficking from the TGN through the endosome/ PVC to the lysosome/vacuole(Polo et al.,2002;Horak,2003; Raiborg et al.,2003;Scott et al.,2004).The binding of epsin homologs to ubiquitin raises the possibility that epsin homologs may bind directly to cargo proteins that are destined for the vacuole/lysosome from either the plasma membrane or the TGN (Chen and De Camilli,2005;Sigismund et al.,2005).In plant cells,sequence analysis of the entire Arabidopsis thaliana genome reveals several proteins with the highly con-served ENTH domains(Holstein and Oliviusson,2005).However, their biological roles have not been addressed.In this study,we investigate the functional role of EPSIN1,an Arabidopsis epsin homolog,at the molecular level.In particular,we focus on its possible role in protein trafficking in plant cells.We demonstrate that EPSIN1interacts with clathrin,AP-1,VSR1,and VTI11and plays an important role in the vacuolar trafficking of a soluble protein from the Golgi complex to the central vacuole.RESULTSEPSIN1,a Member of the Epsin Family,Is Ubiquitously Expressed in ArabidopsisThe Arabidopsis genome encodes three highly similar epsin-related proteins,EPSIN1,EPSIN2,and EPSIN3(Holstein and Oliviusson,2005).In this study,we investigated the biological role of EPSIN1.EPSIN1has the highly conserved ENTH domain at the N terminus.However,the rest of the molecule is less similar to other epsin-related proteins,although it has motifs,such as LIDL and DPF,that may function as clathrin and AP-1binding motifs,respectively.To understand the biological role of EPSIN1,its expression in various plant tissues was examined.An antibody was raised against the middle domain of EPSIN1(amino acid residues153to 337).The antibody recognized a protein band at90kD,which was much larger than the expected size,60kD,of EPSIN1 (Figure1A).It was shown previously that epsin-related proteins migrate slower than expected in SDS-PAGE(Chen et al.,1998). The control serum did not recognize any protein bands.This re-sult suggested that the antibody specifically recognized EPSIN1. To confirm this,protoplasts were transformed with EPSIN1 tagged with HA at the N terminus(HA:EPSIN1)and protein extracts from the transformed protoplasts were analyzed by protein gel blotting using anti-HA and anti-EPSIN1antibodies. The anti-HA antibody specifically recognized a protein band from the transformed protoplasts,but not from the untransformed protoplasts,at90kD(Figure1B).In addition,the90-kD protein species was recognized by the anti-EPSIN1antibody,confirming that the90-kD band was EPSIN1.The expression of EPSIN1in various tissues was examined using the anti-EPSIN1antibody. Protein extracts were prepared from various tissues at different stages of plant growth and used for protein gel blot analysis. EPSIN1was expressed in all of the tissues examined,with the highest expression in cotyledons andflowers(Figure1C). EPSIN1Produces Both Network and PunctateStaining PatternsTo examine the subcellular distribution of EPSIN1,total protein extracts from leaf tissues were separated into soluble and membrane fractions and analyzed by protein gel blotting using anti-EPSIN1antibody.EPSIN1was detected in both membrane (pellet)and soluble fractions(Figure2A).As controls for the fractionation,Arabidopsis aleurain-like protease(AALP)and Arabi-dopsis vacuolar sorting receptor(VSR)were detected with anti-AALP and anti-VSR antibodies,respectively(Sohn et al.,2003). AALP is a soluble protein present in the vacuolar lumen,and VSR is a membrane protein that is localized primarily to the PVC with a minor portion to the Golgi complex(da Silva Conceic¸a˜o et al., 1997;Ahmed et al.,2000).As expected,AALP and VSR were detected in the supernatant and pellet fractions,respectively. These results indicated that EPSIN1localized to multiple loca-tions,consistent with the behavior of other epsin-related proteins (Legendre-Guillemin et al.,2004).Next,we defined the subcellular localization of EPSIN1.Our initial attempts to localize the endogenous EPSIN1with the anti-EPSIN1antibody failed.Thus,we determined the localization of EPSIN1protein transiently expressed in protoplasts.EPSIN1 was tagged with the HA epitope,greenfluorescent protein(GFP), or redfluorescent protein(RFP).The amount of total EPSIN1 protein was determined using various amounts of HA:EPSIN1 plasmid DNA by protein gel blot analysis with anti-EPSIN1an-tibody and was found to be proportional to the amount of plasmid used(Figure2B).For the localization,we used a minimal amount(5to10m g)of EPSIN1plasmid DNAs.Protoplasts were transformed with HA:EPSIN1,and localization of EPSIN1 was determined by immunostaining with anti-HA antibody.HA: EPSIN1produced primarily a punctate staining pattern(Figure 2Ca).In addition to punctate stains,we occasionally observed weakly stained strings that connected punctate stains(Figure 2Cc,arrowheads).By contrast,the nontransformed controls did not produce any patterns(Figure2Ce).In protoplasts trans-formed with EPSIN1:GFP and EPSIN1:RFP,both EPSIN1fusionEPSIN1in Vacuolar Trafficking2259proteins produced a network pattern with punctate stains (Fig-ures 2Cg and 2Ch),whereas GFP and RFP alone produced diffuse patterns (Figures 2Dh and 2Di),indicating that EPSIN1produces the network pattern with punctate stains.These results were further confirmed by cotransforming the protoplasts with either EPSIN1:GFP and HA:EPSIN1or EPSIN1:GFP and EPSIN1:RFP .The punctate staining pattern of EPSIN1:GFP closely over-lapped that of HA:EPSIN1(Figures 2Da to 2Dc).In addition,the network and punctate staining patterns of EPSIN1:GFP closely overlapped those of EPSIN1:RFP (Figures 2De to 2Dg).However,the fine networks revealed by EPSIN1:GFP in the live protoplasts were nearly absent in the fixed protoplasts.Thus,the differences in the staining patterns between fixed and live protoplasts may be attributable to the fact that the network pattern of live protoplasts are not well preserved under the fixing conditions used.In addi-tion,the strings occasionally observed in the fixed protoplasts may represent the remnants of the network pattern revealed by HA:EPSIN1.These results strongly suggest that EPSIN1is re-sponsible for the network pattern as well as the punctate stains.The network pattern was reminiscent of the ER or actin pattern in plant cells (Boevink et al.,1998;Jin et al.,2001;Kim et al.,2005),whereas the punctate staining pattern suggested that EPSIN1may localize to the Golgi complex or endosomes,as observed previously with epsin homologs in animal and yeast cells (Wasiak et al.,2002;Chidambaram et al.,2004;Saint-Pol et al.,2004).Therefore,protoplasts were cotransformed with EPSIN1:RFP and GFP:talin ,a marker for actin filaments consist-ing of GFP and the actin binding domain of mouse talin (Kost et al.,1998;Kim et al.,2005).As expected,GFP:talin produced the network pattern (Figure 3A)(Kost et al.,1998;Kim et al.,2005).Furthermore,the red fluorescent network pattern of EPSIN1:RFP closely overlapped the green fluorescent network pattern of GFP:talin (Figure 3A),raising the possibility that EPSIN1:GFP bound to the actin filaments rather than to the ER.To confirm this,the EPSIN1:RFP pattern was examined after treatment with latrunculin B (Lat B),a chemical agent known to disrupt actin filaments (Spector et al.,1983).Lat B–treated protoplasts produced the diffuse green fluorescent pattern of GFP:talin (Figure 3A),an indication of solubilized actin filaments,as observed previously (Kim et al.,2005).In addition,the Lat B–treated protoplasts displayed a diffuse red fluorescent pattern of EPSIN1:RFP (Figure 3A),indicating that EPSIN1is associated with actin filaments but not with the ER.Furthermore,the punc-tate staining pattern of EPSIN1:RFP also was not observed in the presence of Lat B,indicating that actin filaments played a role in yielding the punctate staining pattern of EPSIN1.In the same conditions,BiP:GFP,an ER marker (Lee et al.,2002),produced a network pattern,indicating that Lat B does not disrupt the ER network patterns (Figure 3Ai).To identify the organelle responsible for the punctate staining pattern of EPSIN1,its localization was compared with that of ST:GFP and PEP12p/SYP21.ST:GFP,a chimericproteinFigure 1.EPSIN1Is Expressed in Various Arabidopsis Tissues.(A)Generation of anti-EPSIN1antibody.The middle domain,corresponding to amino acid residues 153to 337,was expressed as the Hisx6-tagged form in E.coli and used to raise antibody in a rabbit.Control serum was obtained from the rabbit before immunization.Total protein extracts were obtained from leaf tissues and used to test the anti-EPSIN1antibody.(B)Specificity of the anti-EPSIN1antibody.Protein extracts were obtained from protoplasts expressing EPSIN1tagged with the HA epitope at the N terminus and used for protein gel blot analysis using anti-HA and anti-EPSIN1antibodies.(C)Expression of EPSIN1in various tissues.Total protein extracts from the indicated tissues were analyzed by protein gel blotting using anti-EPSIN1antibody.Leaf tissues were harvested 11and 20d after germination.Cotyledons were obtained from 5-d-old plants.The membranes were stained with Coomassie blue to control for protein loading.RbcL,large subunit of the ribulose-1,5-bis-phosphate carboxylase/oxygenase (Rubisco)complex.2260The Plant CellFigure 2.EPSIN1Produces Both Network and Punctate Staining Patterns.(A)Subcellular fractionation of EPSIN1.Total (T)protein extracts of leaf tissues were separated into soluble (S)and pellet (P)fractions and analyzed by protein gel blotting using anti-EPSIN1,anti-AALP,and anti-VSR antibodies.(B)Expression level of EPSIN1in transformed protoplasts.Protoplasts were transformed with various amounts of HA:EPSIN1DNA,and the level of EPSIN1was determined by protein gel blotting with anti-EPSIN1antibody.Protein extracts from untransformed protoplasts were used as a control.The membrane was also stained with Coomassie blue to control for loading.(C)Localization of EPSIN1.Protoplasts were transformed with the indicated constructs (5to 10m g),and the localization of EPSIN1was examined either by immunostaining with anti-HA antibody or by direct detection of the GFP or RFP signal.Untransformed protoplasts were immunostained with anti-HA antibody as a control.Bars ¼20m m.(D)Colocalization of EPSIN1proteins.The localization of EPSIN1protein was examined in protoplasts transformed with HA:EPSIN1and EPSIN1:GFP or with EPSIN1:GFP and EPSIN1:RFP .As controls,GFP and RFP alone were transformed into protoplasts.Bars ¼20m m.EPSIN1in Vacuolar Trafficking 2261亚细胞定位可以荧光观察也可以做western 检测Figure 3.Localization of EPSIN1in Protoplasts.2262The Plant Cellbetween rat sialyltransferase and GFP,localizes to the Golgi complex,and PEP12p,a t-SNARE,localizes to the PVC(da Silva Conceic¸a˜o et al.,1997;Boevink et al.,1998;Jin et al.,2001). Protoplasts were cotransformed with HA:EPSIN1and ST:GFP. The localization of these proteins was examined after staining with anti-HA antibody.ST:GFP was observed directly with the greenfluorescent signals.A major portion of the HA:EPSIN1-positive punctate stains closely overlapped with those of ST:GFP (Figures3Ba to3Bc).To further confirm the Golgi localization of HA:EPSIN1,protoplasts transformed with HA:EPSIN1were treated with brefeldin A(BFA),a chemical known to disrupt the Golgi complex(Driouich et al.,1993),and the localization of HA:EPSIN1was examined.In the presence of BFA,HA:EPSIN1 yielded a largely diffuse pattern with aggregates,but not the punctate staining pattern,indicating that BFA affects EPSIN1 localization(Figure3Be).In the same conditions,ST:GFP pro-duced a network pattern with large aggregates(Figure3Bg), confirming that the Golgi complex was disrupted.These results support the notion that EPSIN1localizes to the Golgi complex. Next,we examined the possibility of EPSIN1localizing to the PVC.Protoplasts were cotransformed with EPSIN1:GFP and PEP12p:HA.The localization of PEP12p:HA was examined after staining with anti-HA antibody.EPSIN1:GFP was observed di-rectly with the greenfluorescent signals.Only a minor portion of the EPSIN1:GFP-positive punctate stains overlapped with the PEP12p:HA-positive punctate stains(Figures3Bi to3Bk,ar-rows).These results indicated that EPSIN1localized primarily to the Golgi complex with a minor portion to the PVC.To obtain independent evidence for the localization,we ex-amined the colocalization of EPSIN1with VTI11,a v-SNARE that is distributed equally to both the TGN and the PVC(Zheng et al., 1999;Bassham et al.,2000;Kim et al.,2005).Protoplasts were cotransformed with EPSIN1:GFP and VTI11:HA,and the local-ization of these proteins was examined by immunostaining with anti-HA antibody.EPSIN1-positive punctate stains largely colo-calized with those of VTI11:HA(Figures3Bm to3Bo),confirming that EPSIN1localizes to both the Golgi complex and the PVC. EPSIN1Binds to and Colocalizes with ClathrinThe members of the epsin family have two clathrin binding motifs (Rosenthal et al.,1999;Wendland et al.,1999;Drake et al.,2000). Sequence analysis indicated that EPSIN1has a potential clathrin binding motif.To explore the possibility that EPSIN1binds to clathrin,glutathione S-transferase–fused EPSIN1(GST:EPSIN1) was constructed for a protein pull-down assay(Figure4A).GST: EPSIN1was expressed in Escherichia coli and purified from E. coli extracts(Figure4B).The purified GST:EPSIN1was mixed with protein extracts obtained from leaf tissues.Proteins pelleted with glutathione–agarose were analyzed by protein gel blotting using anti-clathrin antibody.GST:EPSIN1,but not GST alone, precipitated from the plant extracts a180-kD protein species that was recognized by anti-clathrin antibody(Figure4C),indi-cating that EPSIN1bound to clathrin.To further examine its binding to clathrin,EPSIN1was divided into two regions,the ENTH and the remainder of the molecule (EPSIN1D N)(Figure4A).These regions were expressed in E.coli as GST fusion proteins,GST:ENTH and GST:EPSIN1D N,re-spectively(Figure4B).Protein pull-down experiments using leaf cell extracts were performed with purified GST:ENTH and GST: EPSIN1D N.GST:EPSIN1D N,but not GST:ENTH,precipitated clathrin from the plant extracts(Figure4C).To identify the clathrin binding motif,the C-terminal region containing the putative clathrin binding motif,LIDL(Lafer,2002),as well as GST:RIDL, which contained an Arg substitution of thefirst Leu residue in the motif,were expressed as GST fusion proteins in E.coli(Figures 4A and4B).GST:LIDL,but not GST:RIDL,precipitated clathrin from protein extracts(Figure4C),indicating that the LIDL motif functioned as a clathrin binding motif.The in vitro binding of EPSIN1with clathrin strongly suggested that EPSIN1was likely to colocalize with clathrin.Therefore, immunohistochemistry for the localization of EPSIN1and clathrin was performed.Protoplasts were transformed with HA:EPSIN1, and the localization of HA:EPSIN1and clathrin was examined by staining with anti-HA and anti-clathrin antibodies,respectively. The anti-clathrin antibody produced a punctate staining pattern (Figure4D).A majority(60to70%)of the HA:EPSIN1-positive punctate stains closely overlapped with a pool(40to50%)of clathrin-positive punctate stains(Figure4D),consistent with an interaction between EPSIN1and clathrin.There was also a pool of clathrin-positive punctate stains that lacked the HA:EPSIN1 signal,suggesting that clathrin also was involved in an EPSIN1-independent process.To further characterize the interaction between EPSIN1and clathrin,we examined whether or not EPSIN1is permanently associated with CCVs.Protein extracts from leaf tissues were first separated into soluble and pellet fractions by ultracentrifu-gation.The pellet fraction was treated with Triton X-100and further fractionated by gelfiltration,and the fractions were ana-lyzed by protein gel blotting using anti-clathrin,anti-EPSIN,and anti-VSR antibodies.Clathrin was detected in a peak between 443and669kD(see Supplemental Figure1online).Interestingly, VSR,the vacuolar cargo receptor,was eluted at the same posi-tion with clathrin.By contrast,EPSIN1was eluted at90kD. These results suggest that EPSIN1is not permanently associ-ated with CCVs.Figure3.(continued).(A)Colocalization of EPSIN1with actinfilaments.Protoplasts were transformed with the indicated constructs,and the localization of these proteins was examined in the presence(þLat B)and absence(ÿLat B)of Lat B(10m M).Bars¼20m m.(B)Localization of EPSIN1to the Golgi complex and the PVC.Protoplasts were transformed with the indicated constructs,and localization of the proteins was examined after immunostaining with anti-HA.The GFP signals were observed directly in thefixed protoplasts.For BFA treatment,BFA(30 m g/mL)was added to the transformed protoplasts at24h after transformation and incubated for3h.Arrows indicate the overlap between EPSIN1:GFP and PEP12p:HA.Bars¼20m m.EPSIN1in Vacuolar Trafficking2263Figure 4.EPSIN1Binds to and Colocalizes with Clathrin.(A)Constructs.GST was fused to the N terminus.ENTH,the epsin N-terminal homology domain.DLF and DPF motifs are similar to AP-1and AP-3binding motifs,respectively.Q11indicates a stretch of 11Glu residues.The clathrin binding motif (LIDL)and the Leu-to-Arg substitution in the clathrin binding motif (RIDL)are shown in the C-terminal region.The numbers indicate amino acid positions.(B)Expression of GST-fused EPSIN1proteins.Constructs were introduced into E.coli ,and their expression was induced by isopropylthio-b -galactoside.GST fusion proteins were purified from E.coli extracts with glutathione–agarose beads.Purified proteins were stained with Coomassie blue.(C)Interaction of EPSIN1with clathrin.GST-fused EPSIN1proteins were mixed with protein extracts from leaf tissues.EPSIN1binding proteins were precipitated using glutathione–agarose beads and analyzed by protein gel blotting using anti-clathrin antibody.Supernatants also were included in the protein gel blot analysis.Subsequently,the membranes were stained with Coomassie blue.Bead,glutathione–agarose beads alone;P,pellet;S,supernatant (10%of total).(D)Colocalization of EPSIN1with clathrin.Protoplasts transformed with HA:EPSIN1were fixed with paraglutaraldehyde,and the localization of HA:EPSIN1and clathrin was examined by immunostaining with anti-HA and anti-clathrin antibodies,respectively.Bar ¼20m m.2264The Plant CellEPSIN1Interacts with VTI11Epsin-related proteins in animal and yeast cells are involved in either endocytosis or vacuolar/lysosomal protein trafficking(Chen et al.,1998;De Camilli et al.,2002;Wendland,2002;Overstreet et al.,2003;Legendre-Guillemin et al.,2004).To elucidate the pathway of EPSIN1involvement,binding partners of EPSIN1 were examined.In animal and yeast cells,epsin-like proteins have been shown to interact with SNAREs(Chen et al.,1998; Chidambaram et al.,2004).Because EPSIN1localized to the Golgi complex and the PVC,EPSIN1interactions with Arabidop-sis VTI11and VTI12(formerly At VTI1a and At VTI1b,respectively) were examined.VTI11is a v-SNARE that localizes to the TGN and travels to the PVC(Zheng et al.,1999;Bassham et al.,2000). VTI11and VTI12were tagged with HA at the C terminus and introduced into protoplasts.The expression of VTI11:HA and VTI12:HA in protoplasts was confirmed by protein gel blot analysis using anti-HA antibody.The anti-HA antibody detected protein bands at33and35kD(Figure5A),the expected positions of VTI11:HA and VTI12:HA,respectively.Purified GST:EPSIN1 from E.coli extracts was mixed with plant extracts from the VTI11:HA-or VTI12:HA-transformed protoplasts,and GST: EPSIN1-bound proteins were precipitated from the mixture using glutathione–agarose beads.The pellet fraction was analyzed by protein gel blotting using anti-HA antibody.VTI11:HA,but not VTI12:HA,was detected from the pellet(Figure5A).GST alone did not precipitate VTI11:HA from the plant extracts.These results indicated that although VTI11and VTI12are highly similar to each other,EPSIN1specifically binds to VTI11:HA.To further confirm this interaction,we performed a reciprocal protein pull-down experiment(i.e.,pull-down of EPSIN1with VTI11)using protein extracts obtained from protoplasts transformed with VTI11:HA and EPSIN1:GFP.VTI11:HA-bound proteins were immunoprecipitated with anti-HA antibody,and the immunopre-cipitates were analyzed by protein gel blotting using anti-HA, anti-GFP,and anti-calreticulin antibodies.Anti-calreticulin anti-body was used as a negative control.In addition to VTI11:HA, EPSIN1:GFP was detected in the immunoprecipitates(Figure 5B).However,calreticulin was not detected in the pellet.These results further confirm the interaction between VTI11and EPSIN1. To determine the VTI11binding domain of EPSIN1,proteinpull-down experiments were performed using GST:ENTH and GST:EPSIN1D N.GST:ENTH,but not GST:EPSIN1D N,precipi-tated VTI11:HA from the plant extracts(Figure5C),indicating that the ENTH domain contained the VTI11binding motif.Similarly,in animal and yeast cells,EpsinR and Ent3p have been shown to bind to vti1b and vti1p,respectively(Chidambaram et al.,2004). EPSIN1Binds to the Arabidopsis Homolog of g-Adaptinof AP-1Epsin homologs bind to adaptor proteins(APs)(Duncan et al., 2003;Mills et al.,2003).In animal cells,EPSIN1binds to the a-adaptin of AP-2via the D F F/W(where F indicates a hydro-phobic amino acid)and FXDXF motifs(Figure4A)(Brett et al., 2002).Arabidopsis EPSIN1has three DPF motifs to which a-adaptin of AP-2could bind.In addition,EPSIN1has two regions with motifs similar to the acidic Phe motif for binding AP-1and GGAs(Duncan et al.,2003).Therefore,the interactions of EPSIN1with AP complexes were examined.We isolated the Arabidopsis proteins g-adaptin related protein(g-ADR),a-ADR, and d-ADR,which were most closely related to g-adaptin, a-adaptin,and d-adaptin of AP-1,AP-2,and AP-3,respectively. These Arabidopsis proteins were tagged with GFP and ex-pressed transiently in protoplasts.Protein extracts from the transformed protoplasts were mixed with purified GST:EPSIN1, and the GST:EPSIN1-bound proteins were precipitated.The pellet was analyzed by protein gel blotting using anti-GFP antibody.GFP:g-ADR,but not a-ADR:GFP or d-ADR:GFP,was detected in the pellet(Figure6A).The control for the protein pull-down assay,GST alone,did not precipitate any of these proteins. These results strongly suggested that EPSIN1interacts with g-ADR specifically.To further confirm the interaction between EPSIN1and g-ADR,we performed a reciprocal protein pull-down experiment(i.e.,pull down of EPSIN1proteins with Figure5.EPSIN1Binds to VTI11.(A)Protein extracts were prepared from VTI11:HA-and VTI12:HA-transformed protoplasts and mixed with GST alone or GST:EPSIN1. EPSIN1-bound proteins were precipitated from the mixture with gluta-thione–agarose beads and analyzed by protein gel blotting using anti-HA antibody.(B)Coimmunoprecipitation of EPSIN1:GFP with VTI11:HA.Protein ex-tracts from protoplasts cotransformed with VTI11:HA and EPSIN1:GFP were used for immunoprecipitation with anti-HA antibody.The immuno-precipitates were analyzed by protein gel blotting with anti-HA,anti-GFP, and anti-calreticulin antibodies.P,immunoprecipitate;S,supernatant;T, total protein extracts(5%of the input).(C)For binding experiments,protein extracts from protoplasts trans-formed with VTI11:HA were mixed with GST alone,GST:ENTH,and GST:EPSIN1D N.Proteins were precipitated with glutathione-agarose beads and analyzed by protein gel blotting using anti-HA antibody.The amount of the input proteins is indicated.EPSIN1in Vacuolar Trafficking2265。
植物干细胞调控研究新进展
植物干细胞调控研究新进展中国细胞生物学学报Chinese Journal of Cell Biology 2015, 37(7): 1021–1028DOI: 10.11844/cjcb.2015.07.0024收稿日期: 2015-01-15 接受日期: 2015-04-07973计划前期研究专项(批准号: 2014CB160306)、重庆市教委创新团队建设基金(批准号: KJTD201307)和重庆师范大学引进人才启动基金项目(批准号: 12XLR36)资助的课题*通讯作者。
Tel: 023-********, E-mail: hanmazhang@/doc/f017810486.html, Received: January 15, 2015 Accepted: April 7, 2015This work was supported by the National Grand Fundamental Research Pre-973 Program of China (Grant No.2014CB160306), the Innovation T eam Fund of the Education Department of Chongqing Municipality (Grant No.KJTD201307) and a Start-Up Fund from Chongqing Normal University (Grant No.12XLR36)*Corresponding author. Tel: +86-23-65912976, E-mail: hanmazhang@/doc/f017810486.html, 网络出版时间: 2015-07-01 16:53 URL: /doc/f017810486.html,/kcms/detail/31.2035.Q. 20150701.1653.001.html植物干细胞调控研究新进展赵中华南文斌梁永书张汉马*(植物环境适应分子生物学重庆市重点实验室, 重庆师范大学生命科学学院, 重庆 401331)摘要植物干细胞是植物胚后发育形成各种组织和器官的细胞来源和信号调控中心, 其调控机理是植物学研究的重要内容。
分子生物学名词解释(Molecularbiologicalterms)
分子生物学名词解释(Molecular biological terms)The beanmail polar tropical fish is set to exitDouban-douban-douban-douban-douban-douban-douban-douban-dou ban-douban-douban-douban stationDouban searchThe home page of my douban my group of the city browse discoveryExplanation of molecular biology terms.Confused の detectiveThe 2010-05-21 20:43:54 from: confused の detective (if you want to let me live please give me happy pain)Title: molecular biology noun explanation (personal arrangement, only a lot of ~ exams are risky, review should be cautious ~ true love life, far away from the point of birth)Meristematic name solutionProbe: molecular hybridization and the tagged nucleotide chain with specific sequences of nucleotide nucleotide nucleotides can be used to detect specific genes in nucleic acid samples.Molecular hybridization: a technique for qualitative or quantitative analysis of DNA or RNA using the basic properties of DNA degeneration and renature.Gene chip: the support of a specific piece of DNA that is closely aligned in a unit area.Gene library: a clone group that contains the entire DNA sequence of an object in a lifetime.CDNA library: it is contain a tissue cells under certain conditions all mRNA expression by the reverse transcription and synthesis of cDNA sequence of clone population, it is stored in the form of cDNA fragments with the tissue cell gene expression information.Genomic DNA library: a clone population stored in the form of fragments of DNA (including all coding and non-coding regions) of the genome of a living organism.Transgenic technology: gene transfer technology is used to integrate the target genes into the fertilized egg cells or embryonic stem cells, then the cells are imported into the animal's uterus to develop into individual technology.Transgenic: the gene that is being imported in transgenic technologyTransgenic animals: the receptor animals that are genetically engineered to be genetically engineeredNuclear transfer technique: an individual cell nucleus of an animal is introduced into the activated egg cell of another individual to develop into an individual, namely, clone.Gene elimination: a technique for removing certain genes in animals based on homologous recombination.Functional cloning: cloning the pathogenic gene by understanding the function of a pathogenic gene.Location cloning: gradually narrowing the range from the chromosomal localization of a pathogenic gene and finally cloning the gene.Gene diagnosis: direct detection of gene structure and its expression level is normal, so as to diagnose the disease.Gene therapy: an exogenous gene that functions as a defective cell can be used to correct or compensate for its genetic defects to achieve therapeutic purposes.Viral oncogene: a type of gene that is present in tumor viruses (mostly retroviruses) that can cause malignant transformation of the target cell.Proto-oncogene: is the oncogene in normal cells, and its expression products regulate the normal growth and differentiation of cells. When activated, can cause cell growth differentiation abnormality, form tumour. Also called cell carcinoma genes.Oncogene: a normal gene in a living organism or in a cell, which controls the growth and differentiation of cells. Cell carcinogenesis can only be caused when its structure changes or expresses an abnormality.Tumor suppressor gene: inhibits the proliferation and proliferation of cells and thus inhibits the genes of tumor formation.Transformation action: by automatically obtaining or artificially supplying exogenous DNA, the cells obtain a new genetic phenotype, which is called transformation.Conjugation: when a cell or bacterium interacts with the bacteria, the plasmid DNA can be transferred from one cell (bacteria) to another (bacteria). This DNA transfer is called the conjugation.Transduction function: when the release of the virus from infected cells, infected again another cell, occurred in donor cells and DNA transfer and recombination between receptor cells is called transduction.Plasmid: small ring double stranded DNA molecule outside the bacterial chromosomeHomologous recombination: a recombination between the homologous sequences, which is also called fundamental recombinationSite specific recombination: the integration of integrase catalysis between the specific sites of two DNA sequences.Transposition: the translocation or rearrangement of genes mediated by insertion sequence and transposon is referred toas transpositionTransposon: a discrete sequence of repeated sequences that can be transferred from one chromosome site to another.Clone: a collection of identical copies or copies from the same ancestor.Cloning: the process of obtaining the same copy, i.e. asexual reproduction.DNA cloning: the method of application enzymology to reorganize the target gene and carrier DNA in vitro, transform or transfect host cells, and gain a large number of genes. It is also called gene cloning and recombinant DNAGenetic engineering: the methods and related work for gene cloning are described as genetic engineering.Compatibility terminal: some of the restriction enzyme recognition sequences are not exactly the same, but after cutting DNA, they produce the same sticky end, which is called the compatibility terminal.Restriction endonuclease: a specific sequence of DNA that identifies a DNA, and an enzyme that cuts double strand DNA around the identification site or its surroundings.CDNA: transcriptional synthesis of single stranded DNA that complements mRNA. Double stranded cDNA can be synthesized with single stranded cDNA as template and polymerized.Genomic DNA: a complete set of genetic information (chromosomes and mitochondria) of a cell or organism.Gene carrier: some of the DNA molecules used to reproduce or express a meaningful protein for the purpose of carrying a target gene.Gene: a genetic base unit located on a chromosome that carries a DNA fragment of a specific genetic information that can encode a single biological product, including RNA and polypeptide chains.Genome: a complete set of genetic information from a living organism, which is the entire genetic information or whole gene that a cell or virus carries.Gene expression: the process of transcription and translation of genesTime specificity: the expression of a specific gene takes place in a certain chronological order according to the function, which is called the time specificity of gene expressionSpace specificity: in the whole process of individual growth, a certain gene product appears in the order of individual tissue space, which is called the spatial specificity of gene expressionHousekeeping genes: some genes continue to be expressed in almost all cells of an individual, often referred to ashousekeeping genesConstitutive gene expression: usually a gene expression similar to the expression of a butler gene, also known as basic expression.Coordinate expression: under certain mechanism control,A set of genes related to function, no matter how they are expressed, should be coordinated and expressed in a coordinated manner.Trans action: the protein factor expressed by one gene interacts with the specific cis-acting component of another gene to regulate its expression. This conditioning is known as the trans action.Cis: protein factor can recognize, regulate the sequence of its own genes, regulate the expression of its own genes, and call it cis.Self - control: regulation of protein is generally used in automrna, inhibiting the synthesis of itself, and self-controlMonocistron: a coding gene transcribed to generate an mRNA molecule that translates into a polypeptide chain.Protein biosynthesis is the process of synthesizing proteins in the sequence of nucleotides in mRNA molecules.S - D sequence: in prokaryotes initiation codon upstream 8 to13 AUG nucleotide site there are 4-9 consensus sequence, high in purine bases, is with small ribosome binding sites of mRNA, known as the S - D sequence.Ribosomal circulation: the peptide chain is extended continuously in the nucleoprotein body, also known as prolongation. This includes carry, peptide and peptide.Polyribosome: the polymer that mRNA forms with multiple nucleosomes is called polyribosomeMolecular partner: the molecular partner is a nonnatural conformation that identifies a type of protein in the cell that can recognize the right folding of the functional domain and the whole protein.Cistron: a genetic unit that codes for a polypeptide.Signal sequence: all sorting signals exist in the targeted delivery of protein structure, mainly was the specific N terminal amino acid sequence, may guide protein metastasize to the appropriate target cells, the sequences are called signal sequence.Open reading framework: the sequence of nucleotide sequences from the mRNA initiation codon AUG to the termination codon.The degeneracy of the genetic code: an amino acid can have two or more codons coded for it, a feature known as the degeneracy of the genetic code.Enhancers: a sequence of DNA that binds specific gene regulation proteins to promote the expression of specific genes near or far away. The distance of the enhancement subscriptional start point varies greatly, but it always ACTS on the most recent promoter.Transcription is the process by which an organism USES DNA as a template to synthesize RNAStructural genes: segments of RNA that are transcribed from DNA molecules called structural genesAsymmetric transcription: in the genome, the genes are transcribed only by the genes of different developmental timing, conditions and physiological needs of the cell. In the DNA molecule double strand, a strand is used as a template for transcription, and the other strand is not transcribed. The template chain is not always on the same chain.Manipulation: transcription is not continuous. Each transcriptional block can be considered as a transcriptional unit, called the operator. The manipulators include several structural genes and their upstream regulatory sequences.Cis-acting elements: the sequence of DNA that is involved in transcriptional regulation at the beginning of the transcription starting point, consisting of promoters, enhancers, and silences.Anti-type action factor: the ability to recognize and combine the homeopathic components, and reverse the transcriptionaleukaryotic proteins that are transcribed by other genes.Transcription factor: in the trans action factor,The direct or indirect combination of RNA polymerase is called transcription factor.Exon: a sequence of nucleic acid sequences of mature rnas in eukaryotic organisms that appear in the fault gene and its primary transcription products.Intron: linear expression of the partition gene in eukaryotes and the sequence of nucleic acids removed during the splicing process.Nuclease: RNA that has enzymatic activity is called a nucleaseReproduction: refers to the generation of genetic material, the process of synthesizing subchain DNA by the mother chain DNA.Semi - reserved replication: when DNA biosynthesis, the mother chain DNA is unwound into two single strands, each acting as a template by the base pairing rule, and the subchain complemented by the template. The DNA of the daughter cell, a single strand is fully accepted from the parent, and the other single strand is completely resynthesized. The DNA of the two subcells is identical to the parental DNA base sequence. This replication method is called semi-retained replication.Bidirectional replication: when the prokaryote replicates, the DNA dissolves the chain from the starting point to the twodirections, forming the opposite of the two directions of the replication fork, called bidirectional replication.Initiator: a compound structure formed at the beginning of DNA replication, containing helicases, DnaC proteins, primers, and DNA replication initiation regions.Replicator: the unit that completes the replication independently, from the replication point to the replication endpoint.Okazaki fragment: a discontinuous fragment formed in the following chain replication during DNA replication.The double strand of DNA is divided into two segments, each acting as a template, and the subchain lengthens the formed y-font structure along the template to be called a replication fork.Half-discontinuous replication: the lead chain replicates continuously and the attendant chain discontinuous replication is called semi-discontinuous replication.Reverse transcription: the process of synthesizing DNA with RNA as a template for reverse transcriptase.Telomere: the structure of the end of the linear DNA molecule of the eukaryotic biosomatic chromosome, which makes the ends of chromosomes become granule.Transcriptional initiation complex: a compound formed by thebinding of the prokaryotic RNA polymerase, the transcriptional pppGpN - product and template DNA.Boundary sequence: the eukaryotic introns start with GU at 5 'end and AG is 3' end. 5 '- GU... Ag-oh3 is called a boundary sequence, also known as a splice interface.Splicing: removing introns from RNA molecules so that exons can be connected together.Promoter: a sequence of DNA in the upstream of the transcription starting point of RNA polymerase. If the RNA polymerase is combined with it, it can initiate transcription.Central rule: the law of transmission of genetic information from DNA to RNA to protein.Transcriptase: transcriptional complex, which is composed of the nuclease of RNA polymerase and the product of its template DNA, transcribed.Silencing: the negative regulating element in the eukaryotic element, which ACTS as a repressor when it binds the specific protein factor.Gene: the structure of eukaryotic gene, by a number of coding and non-coding area interval with each other, but embedded in a row, after connected to again unless the coding regions, can translate the continuous complete protein amino acid composition. These genes are called broken genes.[figure]DavidDavid (I'm looking for the book direction)It is also the common touch of biochemistryYour response? Who? Who? Who? Who? Who? , ah ha... Good thing ~ plus go> medical student's home groupLatest topic:Molecular biology noun explanation (individual, only a lot more to test a risk... (confused の detective)N reasons you attended medical school (breba)I was depressed. (hh)How microbes learn?Everybody work first or take the exam first. (MaoMaoQ)My man is studying medicine (summer)The division room chooses the direction, especially lost... (ssu xiaawn)Are there any graduate students in Concorde? Seek counsel (summercool)> to report bad information2005-2010 , all rights reserved about , contact us · disclaimer · help center · douban service (API) · mobile phone douban · brand club。
- 1、下载文档前请自行甄别文档内容的完整性,平台不提供额外的编辑、内容补充、找答案等附加服务。
- 2、"仅部分预览"的文档,不可在线预览部分如存在完整性等问题,可反馈申请退款(可完整预览的文档不适用该条件!)。
- 3、如文档侵犯您的权益,请联系客服反馈,我们会尽快为您处理(人工客服工作时间:9:00-18:30)。
Agricultural Sciences in China 201 1,10(6):820—826
Available online at、^ vw.sciencedirect.com 撇一 秽 喾 秽ScienceDirect June 2011
Transient Expression of Exogenous Gene into Plant Cell Mediated by PEI Nanovector
LI Ying,cuiHai—xin,SONGYu,LIYaoandHUANGJin—li Institute ofAgricultural Environment and Sustainable Development,Chinese Academy ofAgricultural Sciences,Beijing 100081,P.R.China
Abstract This study was carried out to investigate the transfection effect of exogenous gene into plant protoplast cell mediated by polyethylenimine(PEI)nanovector,based on PEI gene delivery system in the field of medical science.PEI/DNA complexes were prepared by using PEI polymer to bind the plant expression plasmid,pCM1205-GFPn.The ability of PEI combining and protecting DNA was investigated by agarose gel electrophoresis retardation assay.The surface characteristics of PEI/DNA complexes were observed with transmission electron microscope.The transfection efficiency of Arabidopsis thaliana protoplasts mediated by PEI/DNA complexes at different N/P ratios was analyzed based on observation of transient expression of green fluorescent protein with confocal laser scanning microscope.PEI could bind and condense DNA.and form stable 100.200 nm PEI/DNA complexes when the proportion ofPEl and DNA is in the range of 5:1.1:4. Transfection efhciency of PEI/DNA complexes increased with N/P ratios in range of N/P<5 and reached the highest at N/P=5.and began to decrease beyond N/P>5 as higher toxicity to cells.The transfection efhciency of PEI/DNA complexes at N/P 5 was higher than PEG.This study confirmed that PEI nanovector could effectively mediate foreign gene entering into A.thaliana protoplast cell to obtain transient expression,which may be developed as a hopeful and novel transgenic method combined with plant protoplast regeneration.
Key words:polyethylenimine,gene nanovector,plant cell,transient expression,Arabidopsis thaliana protoplast
lNTRODUCTION Since transgenic tobacco come out in I 983(Herrera— Estrella et a1.1 984),plant transgenic technology has developed extremely rapidly and become the core tools of plant biotechnology and genetic engineering(Bretell 1 995).It provides effective means for breeding new varieties of crops with the traits of stress resistance, high yield and excellent quality.and breaking the limits of gene exchange between species.Developing novel gene vectors and transgenic methods is an important
researching area of modem plant genetic engineering, which also has great significance for the development ofmolecular biology,molecular genetics,and other fun— damental biology. So far,over 1 0 kinds ofplant transgenic method were established,which can be divided into two categories: (i)vector—mediated method:such as Agrobacterium mediated transformation(Shaw et a1.1 983),virus—me— diated transformation(Wu et a1.1 988),liposome—me— diated transformation(Hussain et a1.1985;Antonelli and Stadle 1996),etc.;(ii)direct DNA uptake method: such as polyethylene glycol(PEG)transformation
This paper is translated from its Chinese version in Scientia Agricultura Sinica. LI Ying,Ph D,Tel:+86-10-82105997,E—mail:liyinglucky2001@yahoo.com.cn;correspondence CUI Hai-xin,Professor,Tel:+86—10-82106013,E—mail zhongzhengc@yahoo.eom.cn
@2011.CAAs All rights reserved PublishedbyElsevierLtd doi:10 1016/¥1671-2927(11 0067-9 Transient Expression of Exogenous Gene into Plant Cell Mediated by PEI Nanovector 821 (Lazzeri et a1.1 99 1),e1ectroporation(Fromm et a1. 1 985),biolistic system(Sanford et a1.1 987),pollen— tube pathway(Wang and Fang 1 998),microinjection (Lu and Gong 1 998),etc.But,only two methods, Agrobacterium.mediated method and biolistic system are mainly used for plant breeding.As a vector, Agrobacterium method is a relative mature technology, but mainly confined to dicotyrledon due to the materials and host restriction。Biolistic system can introduce for— eign genes directly into plant cells by bombarding DNA micro—shells,which has no plant species restriction, but with relatively low transformation efficiency and high cost.Therefore.the development of high—e筒一 ciency gene delivery vectors and transformation method is of great significance to the innovation of plant ge— netic engineering.Since 1995,Boussif and many re— searchers fLungwitz et a1.2005)have proved that PEI can be used as non—virus gene vector for DNA delivery as it may s ̄ongly combine and condense DNA to form nanoscale PEI/DNA complexes by its high—density of positive charges(Dunlap et a1.1997;Tiyaboonchai et a1. 2003).Besides,PEI has‘'proton sponge’’effect(Kichler Pf a1.200 1 1.which is conducive to introduce the for— eign gene into cell by endocytosis,and improve the transfection e ciency by 1 0—30 times(Oh et a1.2002; Jiang et a1.2006).However,up to now,the research reports about PEI as gene vector were only focused on gene therapy and animal cell transfection in the field of medical study.There are still no similar research re— ports on PEI as gene vector in mediating plant genetic transformation.In view of plant cell protoplasts own many characteristics similar to animal cells and are able to regenerate plantlet,this study aims to explore a new method for plant transgenic by using PEI as gene vec— tor for foreign gene delivery into plant protoplast cells.
