ABA 信号 6 (1)
【国家自然科学基金】_脱落酸(aba)_基金支持热词逐年推荐_【万方软件创新助手】_20140802

科研热词 脱落酸 植物激素 内源激素 水稻 棉花 鹰嘴豆 锌指蛋白 表达分析 萌发 茉莉酸 膜脂过氧化 胁迫 突变体 水分胁迫 根系 干旱 大豆幼苗 基因克隆 休眠 uv-b辐射 la(ⅲ) aba 高温胁迫 高效液相色谱法 高效液相色谱 马尾松毛虫 马尾松 雨生红球藻 部分根区灌溉 遗传转化 过氧化氢 赤霉素 诱导 补偿生长 虾青素 葡萄糖 营养物质 荧光 花铃期 花芽分化 脱落 脯氨酸 细胞分裂素 纤维素酶 紫球藻 粒位 米质 种子包衣 种子 离子阱串联质谱 盐胁迫 番茄
53 54 55 56 57 58 59 60 61 62 63 64 65 66 67 68 69 70 71 72 73 74 75 76 77 78 79 80 81 82 83 84 85 86 87 88 89 90 91 92 93 94 95 96 97 98 99 100 101 102 103 104 105 106
科研热词 推荐指数 脱落酸 8 激素 3 aba 3 非生物胁迫 2 镉 2 生物胁迫 2 玉米 2 水稻 2 植物激素 2 拟南芥 2 内源激素 2 乙烯 2 高效液相色谱 1 顶端优势 1 青花菜 1 长春花 1 钙调素(cam) 1 钙调素 1 钙 1 酶活性 1 赤霉素 1 计算机网络 1 覆草旱种 1 覆膜旱种 1 表达模式 1 蚕豆根尖微核 1 蔗糖酸性转化酶 1 蒸腾速率 1 茉莉酸 1 茉莉素 1 脱落酸(abscisic acid,aba) 1 脱落酸(aba) 1 胼胝质沉积 1 胡杨 1 胚乳细胞 1 细胞信号转导 1 粒位 1 稻米品质 1 种子萌发 1 瞬时表达分析 1 瞬时表达 1 盐胁迫 1 生态毒性 1 烟草悬浮细胞 1 烟草叶片 1 淹水胁迫 1 淀粉积累 1 水稻条纹病毒 1 水杨酸 1 氮素营养 1 氮素 1 气孔运动 1
ABA生理功能与信号转导相关综述

科技资讯科技资讯S I N &T NOLOGY I NFORM TI ON2008N O .10SCI ENC E &TECH NOLOG Y I N FOR M A TI ON工程技术植物作为多细胞的复杂有机体,在维持其正常的新陈代谢,生长发育,以及对外界多种环境刺激与胁迫的适应过程中,各个细胞,组织,器官之间进行着错综复杂的信息交流和多种因子的调节植物激素担任着信号感受和信号传递的重要任务。
脱落酸(absi csi c aci d,A B A)作为植物体内一种重要的植物激素参与多种信号转导途径,尤其在植物抵御外界不良环境影响,如干旱,低温等逆境下起着尤为重要的作用,素有“逆境激素”之称。
1A B A 的发现ABA 最早是在成熟的干棉壳中分离纯化得到的,W .C.l i u 等研究发现它能使棉花幼龄脱落,认为它是一种“脱落素”。
同时C.F.E agl es 等从桦树中提取出了一种抑制生长诱导休眠的物质,命名为“休眠素”。
后经鉴定,二者为同一化学物质,最终被定名为脱落酸。
2A B A 在植物体内的分布及生理作用AB A 在植物体内广泛存在,在植物体的不同部位分配存在着差异,正常植株中,根系比叶片往往含有更多的ABA 。
在细胞水平上,水分充足时细胞内ABA 呈均匀分布[1]。
放射免疫分析表明细胞溶质、核、叶绿体和细胞壁中都存在标记A B A ,并且标记量没有差异[2]。
干旱导致ABA 重新分布:法国薰衣草受胁迫后(-2.6M P a 水势)叶片总AB A 从900pm ol /g 鲜重增加到3600pm ol /g 鲜重,其中细胞壁A B A 水平增加T 4倍,增值最多。
W i l ki nson 也发现干旱使质外体AB A 水平增加[3]。
最初有人认为是由于叶绿体膜破裂导致ABA 外泄,但后来研究发现叶绿体和核ABA 都有增加(分别为2倍和3倍)。
Re n s bu r g (1996)则报告胁迫下植物叶绿体A BA 含量没有改变,因此质外体A BA 浓度增加可能使根的释放量或合成增加[4]。
信号完整性仿真流程

信号完整性仿真流程
信号完整性仿真是一种通过计算机辅助工程(CAE)软件模拟电子系统中信号质量的过程,主要关注的是高速数字信号在传输过程中受到的各种干扰对信号质量的影响。
简要流程如下:
1. 模型建立:根据设计需求,创建电路板、连接器、电缆等模型,并定义元器件参数及互连结构。
2. 设置边界条件:设定电源网络、信号激励(如上升沿、下降沿、数据眼图等)、负载条件等边界条件。
3. 选择仿真类型:进行瞬态仿真分析信号时域行为,如延时、振铃、过冲等;进行频域仿真分析信号频谱特性,如插入损耗、串扰、反射系数等。
4. 执行仿真:运行仿真软件,计算并输出仿真结果,如眼图、时序图、S参数等。
5. 结果分析:解读仿真结果,评估信号完整性是否满足设计要求,如是否满足建立保持时间、是否存在严重的噪声干扰或信号衰减等。
6. 优化设计:根据仿真结果对设计方案进行优化调整,如调整布线拓扑、添加端接电阻、优化电源/地平面布局等,然后再进行仿真验证,直至满足信号完整性要求。
内源ABA信号水平动态调控的分子机制

表达 的协 同控制,多 因素 共 同参 与 内源 A A信 号水平 的动 态调控 。 B 关 键 词 : A A信 号水 平;合成调 节;代谢调 节; 馈调 节;转运调节; iN B 反 m R A途径 ; 同控制 协
M o e ul r m e ha im o y m i e ul to f e d g no s ABA l c a c n s f r d na c r g a i n o n o e u
sg a e e in l v l l
W EIKa — CHEN ua iFa , J n ,CHEN n. n W U n . u n , I W l . uo Ya Fe g , Li g J a J A e S n
1De at n ilgc l cecsa d oeh ooy Z a gh uN r l nvri , h nz o 6 0 0 C ia . pr me tfBoo i ine n Bit n lg , h nz o oma iest Z ag h u3 3 0 , hn ; o aS c U y
g n sNC D, RGS n A2 C l b ci ae n r s o s o s e s s Ho v r t e e p e so fg n se c d n e e e E At 1 a d AB , a e a t t d i e p n e t t s e . we e , x r si n o e e n o i g d — l v r h g a a i e e z me ,i cu i g 7 r d t n y s n l d n ,8 v ,9 - y r x ls n l c s l a se a e e a i ey r g lt s ABA c u lt n ' d o y a e a d g u o y t n fr s ,n g t l e u ae h r v a c mu a i . o M e n i , h x r s i n ft e s n h s s s c sZE n a wh l t e e p e so so y t a e , u h a P a d NCE e h D3 a ei d c d b n r a i g e d g n u , r n u e y ic e s n o e o sABA o - n c n tn s e t.Ad i o al ,t e a a y e f g n x r s i n a d s u c -ik d n mi s i d c t s h t s san d u p y fo dt n l i y h n l s s o e e e p e so n o r e sn y a c n iae t a u t i e s p l m r ro—o re o ts u c d ABA e u r d f rt ema n e a c fla i r q i i t n n eo f s e o h e ABA y a i o 1 I i n tb et a i NAss o l ei v l e d n m cp o . t s o a l t R h m h u d b ov d n
植物 PP2C 蛋白磷酸酶 ABA 信号转导及逆境胁迫调控机制研究进展

植物 PP2C 蛋白磷酸酶 ABA 信号转导及逆境胁迫调控机制研究进展张继红;陶能国【摘要】蛋白磷酸酶(protein phosphatase,PP)是蛋白质可逆磷酸化调节机制中的关键酶,而 PP2C 磷酸酶是一类丝氨酸/苏氨酸残基蛋白磷酸酶,是高等植物中最大的蛋白磷酸酶家族,包含76个家族成员,广泛存在于生物体中。
迄今为止,在植物体内已经发现了4种 PP2C 蛋白磷酸酶。
蛋白激酶和蛋白磷酸酶协同催化蛋白质可逆磷酸化,在植物体内信号转导和生理代谢中起着重要的调节作用,蛋白质的磷酸化几乎存在于所有的信号转导途径中。
大量研究表明,PP2Cs 参与多条信号转导途径,包括PP 2C 参与 ABA 调控,对干旱、低温、高盐等逆境胁迫的响应,参与植物创伤和种子休眠或萌发等信号途径,其调控机制不同,但酶催化活性都依赖于 Mg2+或Mn2+的浓度。
植物 PP2C 蛋白的 C 端催化结构域高度保守,而 N 端功能各异。
文中还综述了高等植物PP 2C 的分类、结构、ABA 受体与 PP2Cs 蛋白互作、PP2C 基因参与 ABA 信号途径以及其他逆境信号转导途径的研究进展。
%Protein phosphatase is the most important and pivotal enzymes in reversible protein phosphorylation regula-ting mechanisms.While the PP2C phosphatase is a kind of serine/threonine residues of protein phosphatase,is the largest protein phosphatase family in higherplants,there are 76 family members,widely exists in living organisms. So far,four kinds of PP2C protein phosphatases have been found in plants.Protein kinase and protein phosphatase catalyzed reversible protein phosphorylation,play an important role in plant signal transduction and physiological me-tabolism,protein phosphorylation exist in almost thesignal transduction pathway.Numerous academic studies have shown that plant PP2Cs are involved in multiple signal transduction pathways including PP 2C involved in ABA sig-naling pathway,the response to drought,low temperature,salt stress,participated in the plant wound and seed dor-mancy or germination signal pathway,and exist the different regulation mechanism and the enzyme catalytic activity were dependent on the concentrations of Mg2+ or Mn2+ .In plant PP2Cs protein C-terminal,there are a highly con-served catalytic domains,as well as in their N-terminal,their function are different.The review would provide a brief overview of classification,structure of PP 2Cs ,the interaction between ABA receptor and PP2Cs protein,the recent progresses about their roles in ABA and other stress signal transduction pathway in higher plant.【期刊名称】《广西植物》【年(卷),期】2015(000)006【总页数】7页(P935-941)【关键词】蛋白磷酸酶;ABA;信号转导;逆境胁迫;研究进展【作者】张继红;陶能国【作者单位】湘潭大学化工学院生物工程系,湖南湘潭 411105;湘潭大学化工学院生物工程系,湖南湘潭 411105【正文语种】中文【中图分类】Q945.78环境胁迫是限制植物生长和作物产量的重要因素,蛋白质激酶在植物逆境胁迫信号转导中的作用已有较多报道,而关于催化其可逆反应的蛋白磷酸酶与干旱等逆境胁迫的报道却不多,而且不一致(Schweighofer et al.,2004)。
植物激素——脱落酸(ABA)的细胞信号转导机制

图 1 植 物 激 素 脱 落 酸 的 分 子 结 构
AB A作 为一 种 植 物 激 素 在 植 物 生长 发 育 过
程 中起 重要 作用 , 如促 进果 实 与叶 片脱 落 、 官 衰 器 老 和气 孔关 闭 , 响植 物开 花 、 影 调节 种子 和胚 的发 育 等生 理 功 能 ] 。但 是 , A 通 过 何 种 机 制 发 AB 挥其 生 理作 用 , 目前还 不得 而 知 。近年来 , 用模 利 式植 物 拟 南 芥 ( a io s h l n ) 其 它 相 Ar bd p i ta i a 和 s a 关 植物 , AB 细胞 信 号 转导 进 行 了广泛 研 究 , 对 A
16 9 3年 F T. d ic nt 从棉 铃 ( os p - . a dn o t 等 G sy i u is tm) m h ruu 中提纯 了一种 物质 能显 著促 进 棉苗 外植 体 叶 柄 脱 落 , 为 脱 落 素 I。 1 6 称 I 9 5年 证 实 , 该 脱落 素 I 和休 眠 素 为同 一 种 物质 , 一 命 名 为 I 统 脱 落酸 (b cs cd AB , 一 种 具 有 倍 半 萜 a si ca i, A) 是 i 结 构 的植 物激素 。天然植 物激 素脱 落 酸是顺 式 结 构, 系统名 是 2} ,一 式--1羟基一一 代一 , - 顶式 4反 5( 一 4氧 2 6 6三 甲基一一 己烯一~ )3甲基一 ,一 二烯 酸 ,- 2环 1基 一一 2 4戊
黑龙 江 农 业 科 学 2 1 ( ) 1 ~ 1 0 1 7 :4 7 He o gi g A r u t rl c n e i n j n gi l a S i cs l a c u e
植 物 激 素
应用行为分析报告法ABA
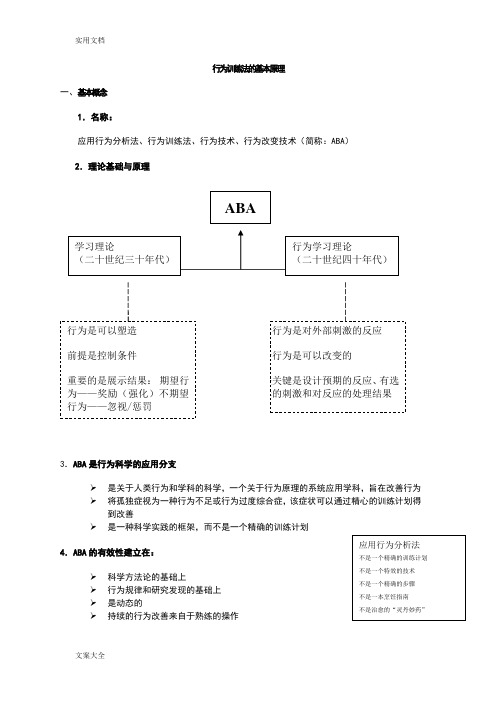
行为训练法的基本原理一、基本概念1.名称:应用行为分析法、行为训练法、行为技术、行为改变技术(简称:ABA ) 2.理论基础与原理3.ABA 是行为科学的应用分支是关于人类行为和学科的科学,一个关于行为原理的系统应用学科,旨在改善行为 将孤独症视为一种行为不足或行为过度综合症,该症状可以通过精心的训练计划得到改善是一种科学实践的框架,而不是一个精确的训练计划4.ABA 的有效性建立在:科学方法论的基础上行为规律和研究发现的基础上 是动态的持续的行为改善来自于熟练的操作5.ABA 干预的基本原则提高学习能力,即“学习如何学习”不仅在于教新的行为。
还涉及到用更合适的行为替代问题行为强调的是如何获得适应性行为。
因为当孩子的适应性行为能力越强时,出现问题行为的几率就越低通过数据的收集了解学生的进步 附:关于“行为”的几个重要表述ABA 不仅仅是针对孤独症的一种干预方法,有着悠久的历史,并且在许多不同的领域都被采用:儿童管理、发育障碍、教育和特殊教育、康复、临床心理、自我管理、预防、运动心理、健康行为、老人医学。
二、行为改变技术1、行为改变的公式:——将ABA 的原理放进实践操作之中应用(ABA)在实践种操作行为的改变主要包含一下几个步骤:控制结果(行为发生之后的结果) 改变或调整三个元素种的一项或两项2、回合操作教学法(DTT)DTT五元素:指令、辅助、反应、结果(强化)、停顿DTT回合公式:(辅助)指令→ 反应→ 结果→ 停顿↑∣下一回合刺激三要素(指令、刺激物、环境)反应过程中注意:1反应标准一致 2注意反应中的不良行为三、ABA与孤独症1.为什么ABA适合孤独症孤独症儿童的障碍最终体现在行为上他们有严重的信息输入障碍医学上因病因不明而无法“对症”,行为改变则为矫治孤独症儿童提供了切入点2.ABA的四大设计的特点孤独症的人际关系障碍是因直觉障碍导致行为训练从障碍结果入手非专业人员也可操作训练的效果可以预测和量化3.ABA的四个操作特点对行为进行分解,在DTT中操作操作中伴随指令和辅助,对孩子的反应有预期性的行为标准(目标行为)反复教,注重巩固和泛化从“一对一”开始,逐步进展到“小组”和“集体课”ABA训练的主要技巧(回合式教学法DTT)应用行为分析法(ABA)是根据行为理论发展演变出的一套完整的行为训练方法。
植物ABA信号转导与植物抗逆性研究

植物ABA信号转导与植物抗逆性研究概述植物生长发育和响应环境胁迫的各种信号都需要通过信号转导通路来实现。
ABA是一种在水分胁迫、高温、低温、重金属等环境胁迫下显著积累的类激素分子,对植物的抗逆性具有关键作用。
本文将就植物ABA信号转导及参与植物抗逆的机制进行讨论。
ABA合成和信号传递ABA是由MEP途径合成的,最初的化合物是萜类化合物,然后到达ABA的核酸类化合物。
ABA需要在植物体内快速传导,以便在环境压力下迅速响应。
ABA信号传递与ABA感受受体和共同的信号分子有关,其中包括Ca2+、ROS以及与ABA共同响应的信号分子,例如亚硫酸和腺苷酸。
ABA在植物细胞内高浓度激活胞浆型和细胞核型的钙离子通道,并且促使ROSCa2+信号的范围扩大。
钙信号进一步促进了一系列ABA响应基因的表达,例如抗氧化基因和受体激酶基因。
此外,ABS-RESISTANT6(ABI6)和ABA INSENSITIVE4(ABI4),是ABRE家族转录因子的主要成员,它们通过ABA介导的途径表达,并且参与抗逆性响应。
ABA信号转导及抗逆性的机制植物ABA信号的转导途径涉及了复杂数字级联级反应,并与ABA受体和一系列ABA响应因子有关。
另外,ABA在调节细胞水位方面的重要性贡献众所周知。
ABA能够介导离子/水研究中重要基因的表达,例如钙离子通道CNGC2、ABI2和黄酮基苷酸酯酶(FLAVIN-CONTAINING MONOOXYGENASE 1, FMO1)等。
ABA通过激活Ca2+信号的通路调节离子渗透调节因子(IonOsmosisRegulatoryFactors,IORFs)和活性氧抗氧化酶(ActiveOxygenantioxidative enzyme, AOE),以及由ABA受体介导的细胞中心体(Centrin)和黄素类胡萝卜素,从而提高植物生长发育和抗逆力。
此外,ABA信号通路中一直具有关键作用的信号分子为MPK3/MAPK6和SnRK2蛋白激酶。
- 1、下载文档前请自行甄别文档内容的完整性,平台不提供额外的编辑、内容补充、找答案等附加服务。
- 2、"仅部分预览"的文档,不可在线预览部分如存在完整性等问题,可反馈申请退款(可完整预览的文档不适用该条件!)。
- 3、如文档侵犯您的权益,请联系客服反馈,我们会尽快为您处理(人工客服工作时间:9:00-18:30)。
ABA-dependent and ABA-independent signaling in response to osmotic stress inplantsTakuya Yoshida,Junro Mogami and Kazuko Yamaguchi-ShinozakiPlants have adaptive robustness to osmotic stresses such as drought and high salinity.Numerous genes functioning in stress response and tolerance are induced under osmotic conditions in diverse plants.Various signaling proteins,such astranscription factors,protein kinases and phosphatases,play signal transduction roles during plant adaptation to osmotic stress,with involvement ranging from stress signal perception to stress-responsive gene expression.Recent progress has been made in analyzing the complex cascades of geneexpression during osmotic stress response,and especially in identifying specificity and crosstalk in abscisic acid (ABA)-dependent and ABA-independent signaling pathways.In this review,we highlight transcriptional regulation of geneexpression governed by two key transcription factors:AREB/ABFs and DREB2A operating respectively in ABA-dependent and ABA-independent signaling pathways.AddressesLaboratory of Plant Molecular Physiology,Graduate School ofAgricultural and Life Sciences,The University of Tokyo,Tokyo 113-8657,JapanCorresponding author:Yamaguchi-Shinozaki,Kazuko (akys@mail.ecc.u-tokyo.ac.jp )Current Opinion in Plant Biology 2014,21:133–139This review comes from a themed issue on Cell signalling and gene regulation 2014Edited by Xiangdong Fu and Junko Kyozuka/10.1016/j.pbi.2014.07.0091369-5266/#2014Published by Elsevier Ltd.All right reserved.IntroductionAs sessile organisms facing various environmental chal-lenges,higher plants have adaptive robustness at mol-ecular,cellular and physiological levels to environmental stress.Water deficit is a major factor restricting plant growth,survival,yield and distribution.Under osmotic stress conditions such as drought and high salinity,numerous genes functioning in stress response and tol-erance are induced,and abscisic acid (ABA),a key plant stress-signaling hormone,is accumulated [1–3].Osmotic stress-responsive gene expression is regulated byABA-dependent and ABA-independent pathways [2].The cis -acting element,ABA-responsive element (ABRE),and a group of transcription factors,ABRE-binding protein/ABRE-binding factors (AREB/ABFs),have pivotal functions in ABA-dependent gene expres-sion.Similarly,a cis -element,dehydration-responsive element/C-repeat (DRE/CRT),and DRE-/CRT-binding protein 2(DREB2)transcription factors play key roles in ABA-independent gene expression in response to osmotic stress.In this review,we focus on recent progress in the study of transcriptional regulatory networks governed by AREB/ABFs and DREB2A transcription factors in osmo-tic stress signaling,with particular emphasis on the func-tional interaction of ABA-dependent and ABA-independent pathways.Various signaling proteins,such as transcription factors,kinases and phosphatases,are involved in expression of osmotic stress-responsive genes;their roles have been extensively reviewed elsewhere [4,5].The pivotal role of AREB/ABF transcription factors in ABA-dependent gene expressionAmong group-A bZIP transcription factors in Arabidopsis thaliana (Arabidopsis ),nine AREB/ABF transcription fac-tors have a bZIP domain and four conserved domains containing Ser/Thr kinase phosphorylation sites [6 ].Consistent with increased AREB1/ABF2,AREB2/ABF4and ABF3expression induced by drought,high salinity and ABA in vegetative tissues [7],overexpression studies have shown that these three AREB/ABFs are positive regulators of ABA signaling under drought stress con-ditions [7–10].These studies and investigation of the areb1areb2abf3triple mutant [11]have jointly demon-strated that the three AREB/ABFs function as master transcription factors cooperatively regulating ABRE-de-pendent gene expression in ABA signaling during response to drought stress.AREB/ABF transcription factors are activated in an ABA-dependent manner through multiple-site phosphoryla-tion of their conserved domains by SNF1-related kinase 2s (SnRK2s)[10,12,13].Nine of 10Arabidopsis SnRK2s are activated by osmotic stress,with 3subclass III SnRK2s,SRK2D/SnRK2.2,SRK2E/SnRK2.6/OST1and SRK2I/SnRK2.3,also strongly activated by ABA [14].The three SnRK2s co-localize and interact with AREB/ABFs in plant cell nuclei [11,15].Furthermore,expres-sion of most downstream genes of AREB1/ABF2,Available online at ScienceDirectAREB2/ABF4and ABF3is substantially impaired in the srk2d/e/i triple mutant [15],and ABA-dependent phos-phorylation of AREB/ABFs is completely eliminated [16,17].These findings support the view that the three subclass III SnRK2s regulate ABA-responsive gene expression through phosphorylation of AREB/ABFs under osmotic stress conditions (Figure 1).The srk2d/e/i triple mutant displays an extreme ABA-insensitive phenotype in regard to seed germination,seedling growth,stomatal regulation and osmotic stress-responsive gene expression [15,16,18].AREB1/ABF2,AREB2/ABF4and ABF3regulate expression of one-third of SnRK2downstream genes [15];other than these three AREB/ABFs,however,little is known about the transcription factors functioning downstream of SnRK2s.To gain a comprehensive understanding of signaling networks governed by the three subclass III SnRK2s,two phosphoproteome studies using the srk2d/e/itriple mutant have been performed [19 ,20 ].A com-parative phosphoproteome analysis of the srk2d/e/i mutant and wild-type plants after dehydration or ABA treatment identified 32phosphopeptides as candidates for SnRK2substrates,including two phosphopeptides corresponding to AREB1/ABF2,AREB2/ABF4,ABF3or ABF1[19 ].Although phosphopeptides corresponding to other tran-scription factors were undetected,SNRK2-SUB-STRATE 1(SNS1)was identified as a novel SnRK2substrate (Figure 1).The sns1mutant shows increased sensitivity to ABA in post-germination stages,with the expression of ABA-responsive genes such as RD29B and RAB18being slightly increased.Given that SNS1is of unknown function,further analyses are required to reveal its precise roles in ABA signaling.Another phosphoproteome study using the srk2d/e/i mutant identified 84phosphopeptides as possible SnRK2sub-strates,including eight proteins classified as transcription134Cell signalling and gene regulation 2014Figure 1ABAABA-independentpathway ABA-dependentpathwayAREB/ABFsInteraction???PYR/PYL/RCARPP2CSubclass III SnRK2Other substratesAREB1DREB2AInduction ofstress-responsive genes Inhibition of stomatal openingModulation of ABA sensitivityOther responsesCE ABRE Target geneAREB2FBH3/AKS1KAT1mRNASNS1KAT1AREB3EELTAF5ABF3ABF1DRE Current Opinion in Plant BiologyABA-dependent signaling pathway and crosstalk with ABA-independent signaling in response to osmotic stress.The four AREB/ABF transcription factors,AREB1,AREB2,ABF3,and ABF1,play key roles in ABA-dependent gene expression.As pivotal positive regulators downstream of PP2C-PYR/PYL/RCAR ABA receptor complexes in ABA signaling,subclass III SnRK2s phosphorylate a variety of substrates,including AREB/ABFs,FBH3/AKS1,and SNS1,and modulate their activities.Recent studies suggest that crosstalk occurs between the AREB/ABF-SnRK2pathway in ABA signaling and the ABA-independent pathway.Transcription factors and DNA-binding proteins are shown in colored ellipses.Dashed lines indicate possible although unconfirmed routes.PYR/PYL/RCAR,pyrabactin resistance1/PYR1-like/regulatory components of ABA receptor;PP2C,protein phosphatase 2C;CE,coupling element.factors and DNA-binding proteins[20 ].Results of previous studies imply that physiological roles of the remaining proteins,including AREB3,ENHANCED EM LELVEL(EEL),FLOWERING BHLH3(FBH3) and TATA BINDING PROTEIN-ASSOCIATED FAC-TOR5(TAF5),but not AREB1/ABF2,are associated with developmental processes rather than osmotic stress responses[6 ,21,22](Figure1).Although10RNA-binding proteins were also identified as candidate SnRK2sub-strates,their roles in ABA signaling are still undetermined. Collectively,these phosphoproteome analyses suggest that AREB/ABFs are predominant transcription factors func-tioning downstream of the three subclass III SnRK2s during ABA signaling in response to osmotic stress.The view derived from the phosphoproteome studies has been further supported by a reverse genetic approach showing that ABF1is a transcription factor that functions down-stream of SnRK2s[23 ].Despite lower expression levels of ABF1compared with AREB1/ABF2,AREB2/ABF4and ABF3,even under stress conditions,the areb1areb2abf3 abf1quadruple mutant displays increased sensitivity to drought,decreased sensitivity to ABA in primary root growth,and impaired expression of ABA-responsive genes and osmotic stress-responsive genes compared with the areb1areb2abf3triple mutant.Transcriptome analyses have revealed that70%of downstream genes of the three subclass III SnRK2s are down-regulated in the areb1areb2 abf3abf1quadruple mutant.These results favor the view that these SnRK2s regulate ABA-responsive gene expres-sion under osmotic stress primarily through the four AREB/ ABF transcription factors(Figure1). Phosphoproteome and transcriptome approaches have pro-vided further new insights into the role of the three subclass III SnRK2s in ABA signaling.FBH3,identified as a possible SnRK2substrate,and its homologs are bHLH transcription factors activating CONSTANS(CO)expres-sion duringfloral regulation[21].Interestingly,FBH3and CO expression is impaired in both late-flowering areb1areb2 abf3abf1and early-flowering srk2d/e/i mutants[20 ,23 ], although their phenotypes are contradictory.Given that otherflowering-associated proteins,such as MOS3and XRN3,are also possible SnRK2substrates[20 ],SnRK2s may intricately andfinely regulate thefloral transition in ABA signaling via a variety of substrates,including AREB/ ABFs and FBH3.Surprisingly,FBH3and CO are involved in stomatal opening[24,25].FBH3is identical to ABA-responsive kinase substrate1(AKS1),which is negatively regulated in guard cells by SRK2E/SnRK2.6/OST1-de-pendent phosphorylation[24](Figure1).FBH3/AKS1is a transcriptional activator of KAT1,which encodes the major inward-rectifying K+channel involved in stomatal opening.Because KAT1is also negatively regulated by SnRK2E/SnRK2.6/OST1-dependent phosphorylation [26],SnRK2-mediated ABA signaling might inhibit sto-matal opening through transcriptional and posttranscrip-tional mechanisms.Studies exploiting these recently identified putative SnRK2substrates are needed to under-stand the global regulatory roles of subclass III SnRK2s in ABA signaling.Negative regulatory mechanisms of DREB2A activity in ABA-independent gene expression DREB2proteins are members of the AP2/ERF family of plant-specific transcription factors.Among eight DREB2s in Arabidopsis,DREB2A and DREB2B are highly induced by drought,high salinity and heat stress[27,28].Despite the instability of DREB2A proteins in planta,studies of Arabidopsis overexpressing a constitutively active form of DREB2A,which harbors an internal deletion of the domain involved in protein stability,have revealed that DREB2A plays pivotal roles in ABA-independent gene expression under osmotic and heat stress conditions [28,29].Because DREB2A,which induces many genes encoding proteins involved in stress response and tolerance,has adverse effects on plant growth,its transcript and protein levels are tightly regulated.An attempt to elucidate tran-scriptional regulatory mechanisms of DREB2A has ident-ified GROWTH-REGULATING FACTOR7(GRF7)as a transcriptional repressor of osmotic-stress responsive genes including DREB2A[30 ](Figure2).GRF proteins are putative transcription factors harboring two conserved domains.Among nine GRFs in Arabidopsis,GRF7interacts with a short DREB2A promoter region that suppresses its expression under unstressed growth conditions.GRF7 knockout and knockdown mutants,showing increased DREB2A expression,display enhanced salinity tolerance as well as growth retardation.In addition,transcriptome analysis has revealed that hundreds of osmotic stress-responsive genes are up-regulated in the grf7knockout mutant compared with wild-type plants grown under nor-mal conditions,suggesting that GRF7is a repressor reg-ulating a wide range of osmotic-stress responsive genes during unstressed growth conditions.In addition to transcriptional repression under normal growth conditions,a ubiquitin–proteasome pathway is pro-posed to degrade leaky expression of the DREB2A protein. DREB2A-INTERACTING PROTEIN1(DRIP1),a ubi-quitin E3ligase harboring a C3HC4-type RING domain, has been identified as an interacting protein of DREB2A by yeast two-hybrid screening[31].Whereas DRIP1over-expression delays the expression of DREB2A downstream genes in response to dehydration,their expression is increased in double knockout mutants of DRIP1and its homolog DRIP2.Furthermore,greenfluorescent protein (GFP)-DREB2A fusion proteins are stably expressed in the drip1drip2mutant,which displays growth retardation under normal growth conditions and enhanced drought stress tolerance.These results suggest that DRIP1and DRIP2negatively regulate drought stress-responsive gene expression by targeting DREB2A to26S proteasomeCrosstalk in ABA signaling Yoshida,Mogami and Yamaguchi-Shinozaki135proteolysis (Figure 2).Despite the crucial role of DRIPs in DREB2A degradation,a recent study has shown that DREB2A protein levels,which are rapidly increased in response to heat stress,are reduced by prolonged heat stress even in the drip1drip2mutant [32].Given that stabilization of DREB2A is required,but not sufficient for substantial induction of its downstream genes [32],DREB2A degra-dation and activation might be regulated by many yet-to-be-identified proteins in addition to DRIPs.DREB2A and DRIPs are a well-characterized transcrip-tion factor/ubiquitin E3ligase combination that regulates osmotic stress-responsive genes.A similar regulatoryinteraction has recently been identified between the transcription factor AtERF53and two homologous C3HC4-type RING E3ligases,RING domain ligase 2(RGLG2)and RGLG1[33].The expression of AtERF53,an AP2/ERF transcription factor belonging to a non-DREB2subfamily [34],is induced significantly by drought and high salinity but only slightly by ABA treat-ment [33,35].AtERF53interacts and co-localizes with RGLG2and RGLG1,both of which mediate AtERF53ubiquitination in vitro .By contrast to wild-type plants,AtERF53-GFP fusion proteins are stably expressed in the rglg1rglg2mutant that displays enhanced drought stress tolerance.Because AtERF53is involved in induction of136Cell signalling and gene regulation 2014Figure 2Current Opinion in Plant BiologyABA-independentpathway ABA-dependentpathwayABA PYR/PYL/RCARPP2C Subclass III SnRK2StabilizationGRF7?GTE GTEHSE ABREDREB2A DREB2A (inactive)DREB2A (active)DREB2A??Ub OtherE3ligases?DRIP 1/2Proteolysis26S proteasomeDRE Target geneTranscriptionUbUbUbUb UbOsmotic stress-responsive geneTranscriptionAREB/ABFsActivationABA-independent signaling pathway and crosstalk with ABA-dependent signaling in response to osmotic stress.DREB2A is the key transcription factor in ABA-independent gene expression in response to osmotic stress.DREB2A protein levels are tightly regulated by ubiquitin E3ligases such as DRIP1and DRIP2.In addition,DREB2A expression is repressed by GRF7under unstressed growth conditions.Crosstalk between ABA-independent and ABA-dependent pathways is suggested on the basis of recent findings that GRF7is involved in repression of ABA-inducible genes and osmotic stress-inducible genes and that DREB2A is induced by AREB/ABFs.Transcription factors and DNA-binding proteins are shown in colored ellipses.Dashed lines indicate possible although unconfirmed routes.PYR/PYL/RCAR,pyrabactin resistance1/PYR1-like/regulatory components of ABA receptor;PP2C,protein phosphatase 2C;GTE,GRF7-targeting cis -element;HSE,heat shock element;Ub,ubiquitin.stress-responsive genes such as COR15A and COR15B[35],AtERF53and RGLGs may function as positiveand negative regulators,respectively,of osmotic stress-responsive gene expression.As each group of AP2/ERFtranscription factors and RING-type ubiquitin ligasescomprises many genes[34,36],comprehensive analysesof these genes may provide further insights into transcrip-tional networks in response to osmotic stress. Crosstalk between ABA-dependent and ABA-independent pathwaysAs evidenced by the fact that multiple dehydration-inducible genes are induced by exogenous ABA treat-ment[2],the global transcriptional network activated inresponse to osmotic stress is cooperatively but not exclu-sively regulated by ABA-dependent and ABA-indepen-dent pathways.Despite the importance of crosstalkbetween ABA-dependent and ABA-independent path-ways,knowledge of how the two signaling pathwaysregulate each other has been limited until recently.The three subclass III SnRK2s likely participate in theconvergence of ABA-dependent and ABA-independentsignaling[6 ](Figure1).Activated by osmotic stress aswell as by ABA,these SnRK2s substantially regulateABA-dependent and ABA-independent gene expression[15].Analysis of multiple knockout mutants of all10SnRK2s in Arabidopsis has further implied that the threeSnRK2s are essential to both osmotic stress response andABA signaling[37].In-gel kinase assay results,however,indicate that osmotic stress activates SnRK2s in an ABA-independent manner[38,39].Consistent with theseresults,a recent phosphoproteome study has revealedthat phosphopeptides corresponding to the three subclassIII SnRK2s are phosphorylated after5min of treatmentwith ABA but not by osmotic stresses such as NaCl andmannitol[40 ].On the other hand,short-term osmoticstress has been found to phosphorylate a phosphopeptidecorresponding to SRK2A/SnRK2.4,SRK2B/SnRK2.10,SRK2G/SnRK2.1or SRK2H/SnRK2.5,which are subclassI SnRK2s not activated by ABA,and phosphopeptidescorresponding to MAP3K and MAP4K.Although subclassIII SnRK2s appear to be activated by autophosphoryla-tion during ABA signaling,novel proteins not yet charac-terized may be responsible for SnRK2activation inresponse to osmotic stress.Analyses of DREB2A promoters have provided newinsights into crosstalk between ABA-dependent andABA-independent signaling(Figure2).Given that anABRE motif-containing short promoter region is requiredfor dehydration-responsive DREB2A expression,transientexpression and chromatin immunoprecipitation(ChIP)analyses have demonstrated that DREB2A is regulatedby AREB1/ABF2,AREB2/ABF4and ABF3during ABAsignaling under osmotic stress[41].Interestingly,severalAP2/ERF transcription factors including DREB2A interactwith AREB1/ABF2,AREB2/ABF4and ABF3[42](Figure1),but it is unclear whether these interactions have functional significance in gene expression.It is note-worthy that expression of ABA-inducible genes(including AREB2/ABF4)and osmotic stress-inducible genes is increased in the grf7mutant[30 ].Thisfinding implies that GRF7has roles in repressing both ABA-dependent and ABA-independent gene expression(Figure2).By contrast to DREB2A,transcriptional regulatory mechan-isms of AREB/ABF s are poorly understood.Further studies of AREB/ABF promoters may help to elucidate whole transcriptional networks governed by ABA-dependent and ABA-independent signaling.Transcriptional networks under osmotic stress have been recently investigated using large-scale data analyses.A differential network analysis based on machine learning, an intelligent data mining technique,was used to reana-lyze an abiotic stress-responsive gene expression data set. The analysis identified two novel genes whose mutants are sensitive to salt stress[43].Integrated transcriptome and metabolome analyses have revealed the roles of ABA-dependent and ABA-independent signaling in metabolic alterations under osmotic stress conditions in Arabidopsis and rice[44,45].Cytokinin signaling is also reportedly decreased by dehydration stress in these two species[45]. Thisfinding supports the idea that crosstalk between ABA and cytokinin plays key roles in osmotic stress signaling[46].Furthermore,transcriptome and metabo-lome profiling of Arabidopsis grown under mild osmotic stress conditions has shown that cell proliferation and expansion are mainly regulated by ethylene and gibber-ellin signaling in developing leaves[47].Importantly,a large-scale phenotyping study,which measured the rosette size of Arabidopsis grown in soil using an auto-mated platform,has suggested that improved drought stress tolerance under lethal conditions is not well corre-lated with superior growth under moderate drought con-ditions[48].Thus,more attention must be focused on the roles of other plant hormones besides ABA to understand the molecular mechanisms used by plants to rigorously control their tolerance and growth under osmotic stress conditions.ConclusionsIn this review,we demonstrated that four AREB/ABF transcription factors,AREB1/ABF2,AREB2/ABF4, ABF3and ABF1,regulate most downstream genes of three subclass III SnRK2s in ABA-dependent gene expression. Under unstressed conditions,transcriptional and protein levels of DREB2A,a key transcription factor in the ABA-independent pathway,are properly regulated by GRF7 and DRIPs,respectively.Given that AREB/ABF and DREB2A expression is regulated by the three subclass III SnRK2s,these kinases might serve as a convergence point in the crosstalk between ABA-dependent and ABA-independent gene expression.Because AREB/ABF and DREB2A transcription factors are conserved in land plantsCrosstalk in ABA signaling Yoshida,Mogami and Yamaguchi-Shinozaki137[6 ,34],these transcription factors are available for applied research.Their orthologous genes have indeed been shown to participate in stress-responsive gene expression in crops such as rice and soybean as well as Arabidopsis [6 ,49].For future application of these transcription factors to various crop,timber and other plants,research involving large-scale analyses,such as transcriptome and phospho-proteome analyses,should provide further insights into transcriptional networks under osmotic stress conditions. AcknowledgmentsWe thank E.Toma for skillful editorial assistance.This work was supported by a Grant-in-Aid for Scientific Research on Innovative Areas(no. 22119004)from the Ministry of Education,Culture,Sports,Science and Technology of Japan(MEXT),the Program for the Promotion of Basic Research Activities for Innovative Biosciences(BRAIN)of Japan,and the Science and Technology Research Partnership for Sustainable Development(SATREPS)of the Japan Science and Technology Agency (JST)/Japan International Cooperation Agency(JICA). References and recommended readingPapers of particular interest,published within the period of review, have been highlighted as:of special interestof outstanding interest1.Bartels D,Sunkar R:Drought and salt tolerance in plants.CritRev Plant Sci2005,24:23-58.2.Yamaguchi-Shinozaki K,Shinozaki K:Transcriptional regulatorynetworks in cellular responses and tolerance to dehydration and cold stresses.Annu Rev Plant Biol2006,57:781-803.3.Finkelstein R:Abscisic acid synthesis and response.Arabidopsis Book2013,11:e0166/10.1199/tab.0166.4.Fujita Y,Fujita M,Shinozaki K,Yamaguchi-Shinozaki K:ABA-mediated transcriptional regulation in response to osmoticstress in plants.J Plant Res2011,124:509-525.5.Qin F,Shinozaki K,Yamaguchi-Shinozaki K:Achievements andchallenges in understanding plant abiotic stress responsesand tolerance.Plant Cell Physiol2011,52:1569-1582.6. Fujita Y,Yoshida T,Yamaguchi-Shinozaki K:Pivotal role of the AREB/ABF-SnRK2pathway in ABRE-mediated transcription in response to osmotic stress in plants.Physiol Plant2013, 147:15-27.This current review focuses on the roles of AREB/ABF transcriptionfactors and SnRK2kinases in ABA signaling.This paper also summarizes the nomenclature and phylogenetic relationships of the AREB/ABFs andSnRK2s to help promote the link between basic and applied studies.7.Fujita Y,Fujita M,Satoh R,Maruyama K,Parvez MM,Seki M,Hiratsu K,Ohme-Takagi M,Shinozaki K,Yamaguchi-Shinozaki K:AREB1is a transcription activator of novel ABRE-dependent ABA signaling that enhances drought stress tolerance inArabidopsis.Plant Cell2005,17:3470-3488.8.Kang JY,Choi HI,Im MY,Kim SY:Arabidopsis basic leucinezipper proteins that mediate stress-responsive abscisic acid signaling.Plant Cell2002,14:343-357.9.Kim S,Kang JY,Cho DI,Park JH,Kim SY:ABF2,an ABRE-binding bZIP factor,is an essential component of glucosesignaling and its overexpression affects multiple stresstolerance.Plant J2004,40:75-87.10.Furihata T,Maruyama K,Fujita Y,Umezawa T,Yoshida R,Shinozaki K,Yamaguchi-Shinozaki K:Abscisic acid-dependent multisite phosphorylation regulates the activity of atranscription activator AREB1.Proc Natl Acad Sci U S A2006, 103:1988-1993.11.Yoshida T,Fujita Y,Sayama H,Kidokoro S,Maruyama K,Mizoi J,Shinozaki K,Yamaguchi-Shinozaki K:AREB1,AREB2,and ABF3are master transcription factors that cooperatively regulateABRE-dependent ABA signaling involved in drought stresstolerance and require ABA for full activation.Plant J2010,61:672-685.12.Uno Y,Furihata T,Abe H,Yoshida R,Shinozaki K,Yamaguchi-Shinozaki K:Arabidopsis basic leucine zipper transcriptionfactors involved in an abscisic acid-dependent signaltransduction pathway under drought and high-salinityconditions.Proc Natl Acad Sci U S A2000,97:11632-11637. 13.Fujii H,Verslues PE,Zhu JK:Identification of two proteinkinases required for abscisic acid regulation of seedgermination,root growth,and gene expression inArabidopsis.Plant Cell2007,19:485-494.14.Boudsocq M,Barbier-Brygoo H,Laurie`re C:Identification of ninesucrose nonfermenting1-related protein kinases2activated by hyperosmotic and saline stresses in Arabidopsis thaliana.J Biol Chem2004,279:41758-41766.15.Fujita Y,Nakashima K,Yoshida T,Katagiri T,Kidokoro S,Kanamori N,Umezawa T,Fujita M,Maruyama K,Ishiyama K et al.: Three SnRK2protein kinases are the main positive regulators of abscisic acid signaling in response to water stress inArabidopsis.Plant Cell Physiol2009,50:2123-2132.16.Fujii H,Zhu JK:Arabidopsis mutant deficient in3abscisic acid-activated protein kinases reveals critical roles in growth,reproduction,and stress.Proc Natl Acad Sci U S A2009,106:8380-8385.17.Fujii H,Chinnusamy V,Rodrigues A,Rubio S,Antoni R,Park SY,Cutler SR,Sheen J,Rodriguez PL,Zhu JK:In vitro reconstitution of an abscisic acid signalling pathway.Nature2009,462:660-664.18.Nakashima K,Fujita Y,Kanamori N,Katagiri T,Umezawa T,Kidokoro S,Maruyama K,Yoshida T,Ishiyama K,Kobayashi Met al.:Three Arabidopsis SnRK2protein kinases,SRK2D/SnRK2.2,SRK2E/SnRK2.6/OST1and SRK2I/SnRK2.3,involved in ABA signaling are essential for the control of seed development and dormancy.Plant Cell Physiol2009,50:1345-1363.19.>Umezawa T,Sugiyama N,Takahashi F,Anderson JC,Ishihama Y, Peck SC,Shinozaki K:Genetics and phosphoproteomics reveala protein phosphorylation network in the abscisic acidsignaling pathway in Arabidopsis thaliana.Sci Signal2013,6:rs8. This is thefirst study integrating genetics with phosphoproteomics to investigate protein phosphorylation networks in Arabidopsis.By applying a phosphoproteomic approach to the triple knockout mutant of the three subclass III SnRK2kinases,the authors identifies multiple components of the ABA-responsive protein phosphorylation network governed by the SnRK2s,including AREB/ABFs,AtMPK1,AtMPK2and SNS1.20.Wang P,Xue L,Batelli G,Lee S,Hou YJ,Van Oosten MJ,Zhang H, Tao WA,Zhu JK:Quantitative phosphoproteomics identifiesSnRK2protein kinase substrates and reveals the effectors of abscisic acid action.Proc Natl Acad Sci U S A2013,110:11205-11210.This phosphoproteomic study using the triple knockout mutant of the three subclass III SnRK2s also identifies candidate substrates of the SnRK2s in ABA signaling,including proteins involved inflowering time regulation,RNA and DNA binding,miRNA and epigenetic regulation, signal transduction,chloroplast function and many other cellular pro-cesses.Given that the srk2d/e/i triple mutant shows the earlyflowering phenotype,the authors suggest that the SnRK2s are important for flowering time regulation.21.Ito S,Song YH,Josephson-Day AR,Miller RJ,Breton G,Olmstead RG,Imaizumi T:FLOWERING BHLH transcriptionalactivators control expression of the photoperiodicflowering regulator CONSTANS in Arabidopsis.Proc Natl Acad Sci U S A 2012,109:3582-3587.22.Mougiou N,Poulios S,Kaldis A,Vlachonasios K:Arabidopsisthaliana TBP-associated factor5is essential for plant growth and development.Mol Breed2012,30:355-366.23.Yoshida T,Fujita Y,Maruyama K,Mogami J,Todaka D,Shinozaki K,Yamaguchi-Shinozaki K:Four Arabidopsis AREB/ ABF transcription factors function predominantly in geneexpression downstream of SnRK2kinases in abscisic-acidsignaling in response to osmotic stress.Plant Cell Environ2014 /10.1111/pce.12351.138Cell signalling and gene regulation2014。
