illumina 转录组测序简明实验流程(PE-oligodT NEB)
转录组测序 步骤 流程

转录组测序步骤流程Transcriptome sequencing, also known as RNA-Seq, is a powerful technique used to study the entire set of RNA molecules transcribed by an organism. It provides valuable insights into gene expression levels, alternative splicing, and novel transcript discovery. The process oftranscriptome sequencing involves several steps, which Iwill explain in detail.Firstly, the RNA molecules need to be isolated from the biological sample of interest. This can be done using various methods, such as using a commercial RNA extraction kit or employing a phenol-chloroform extraction protocol. The quality and integrity of the RNA are crucial at this stage to ensure accurate downstream analysis.Once the RNA is extracted, it needs to be convertedinto complementary DNA (cDNA) through reverse transcription. This step involves the use of reverse transcriptase enzyme and random primers or oligo(dT) primers. The cDNA synthesisallows for the amplification of RNA molecules and reduces the presence of ribosomal RNA, which is highly abundant and can hinder sequencing efficiency.Next, the cDNA library preparation is carried out. This involves fragmentation of the cDNA and the addition of sequencing adapters, which contain barcode sequences for sample identification. The fragmented cDNA is then subjected to size selection to obtain the desired insert size range for sequencing. This step ensures that only fragments within the desired range are sequenced, improving the quality of the data obtained.After library preparation, the sequencing step takes place. This can be done using various sequencing platforms, such as Illumina, Ion Torrent, or PacBio. The choice of platform depends on factors like sequencing depth, read length, and cost considerations. During sequencing, the cDNA fragments are amplified on a solid support and undergo cycles of nucleotide incorporation, resulting in the generation of millions of short reads.Once the sequencing is complete, the generated reads need to be processed and aligned to a reference genome or transcriptome. This step involves bioinformatics analysis, where the reads are mapped to the reference using alignment algorithms. This allows for the identification of known transcripts and the discovery of novel transcripts.Following alignment, differential gene expression analysis can be performed to compare gene expression levels between different samples or conditions. This analysis involves statistical methods to identify genes that are significantly upregulated or downregulated. It provides valuable insights into the biological processes and pathways that are affected under different conditions.In conclusion, transcriptome sequencing is a multi-step process that involves RNA extraction, cDNA synthesis, library preparation, sequencing, alignment, and analysis.It provides a comprehensive view of gene expression and transcriptome dynamics. By understanding the steps involved in transcriptome sequencing, researchers can gain valuable insights into gene regulation and biological processes.转录组测序,也被称为RNA-Seq,是一种用于研究生物体转录的所有RNA分子的强大技术。
illumina转录组测序简明实验流程(PE-oligodTNEB)
illumina转录组测序简明实验流程(PE-oligodTNEB)illumina 转录组测序简明实验流程一、实验基本流程图mRNA Library Construction二、mRNA建库流程1.材料准备1.2.1.3.2.样品准备和QC选择质量合格的Total RNA作为mRNA测序的建库起始样品,其质量要求通过Agilent 2100 BioAnalyzer检测结果RIN≥7,28S和18S的RNA 的比值大于或等于1.5:1,起始量的要求范围是0.1∽1ug。
用QUBIT RNA ASSAY KIT对起始的Total RNA进行准确定量。
3.建库实验步骤3.1.mRNA纯化和片段化3.1.1.mRNA纯化纯化原理是用带有Oligod(T)的Beads对Total RNA中mRNA进行纯化。
3.1.2.mRNA片段化3.2.1st Strand cDNA 合成3.3.2nd Strand cDNA 合成根据下表制备反应体系,然后在PCR仪上运行Program3,然后将第2链cDNA合成产物用144uL AMPure XP Beads进行纯化,最后用60μL的Nuclease free water进行重悬,取出55.5μL以备下一步使用;3.4.Perform End Repair/dA-tail3.5.Adaptor Ligation根据下表制备反应体系,然后在PCR仪上运行Program5、Program6,然后100uL AMPure XP Beads进行纯化后用52.5μL的Resuspension Buffer进行重悬,再用50uL AMPure XP Beads3.6.PCR扩增根据下表制备反应体系,然后在PCR仪上运行Program7,然后再45μL用AMPure XP Beads 进行纯化,最后用23μL的Resuspension Buffer进行重悬,取出20μL以备下一步使用;3.7. PCR 产物质控用QUBIT DNA HS ASSAY KIT 对PCR 产物进行准确定量。
安捷伦二代测序操作方法

安捷伦二代测序操作方法安捷伦二代测序(Illumina sequencing)是一种高通量测序技术,也称为第二代测序技术。
它是目前最常用的测序技术之一,具有高效、高精度和经济实惠的特点。
下面将详细介绍安捷伦二代测序的操作方法。
安捷伦二代测序的操作主要包括样品准备、文库构建、测序芯片负载和测序运行、数据分析等步骤。
首先是样品准备。
样品可以是DNA、RNA或其它核酸。
如果是DNA样品,首先需要进行DNA提取和纯化。
对于RNA样品,一般需要进行RNA提取和转录成cDNA。
提取过程需要注意样品的完整性和纯度,以保证后续步骤的准确性和可靠性。
接下来是文库构建。
文库是指将样品DNA或RNA片段连接到测序适配体上的过程。
适配体是一种DNA片段,其中包含引物序列和测序平台特异性序列。
文库构建可以为无法测序的DNA或RNA片段增加引物序列,并通过PCR扩增获得足够的文库量。
文库构建的关键是要控制适配体和样品DNA的比例,以避免过度扩增或不足。
完成文库构建后,可以将样品的文库片段装载到测序芯片上。
测序芯片是一种固定了上千万个DNA克隆簇的玻璃或硅片。
每个克隆簇都含有相同的文库片段,在测序过程中可以通过化学或光学方法进行扩增和测序。
装载完成后,测序芯片被放入测序仪中进行测序运行。
安捷伦二代测序采用的是桥式扩增法(bridge amplification),即在每个克隆簇上通过引物的控制使得DNA扩增成桥状结构,然后进行测序。
测序过程中使用碱基特异性荧光探针,通过测量碱基的荧光强度来确定碱基序列。
测序结束后,得到的原始测序图像数据需要进行图像处理和碱基识别。
图像处理包括去除背景噪音和探针交叉干扰等步骤,以提高测序数据的质量。
碱基识别则是根据荧光信号的强度和位置信息,将测序图像转化为碱基序列。
得到碱基序列后,可以进行测序数据的分析。
常见的分析包括比对参考基因组、寻找变异位点、计算基因表达水平等。
这些分析需要结合相关的生物信息学软件和数据库进行。
转录组测序 步骤 流程

转录组测序步骤流程英文回答:Transcriptome sequencing, also known as RNA sequencing (RNA-seq), is a powerful technique used to study the transcriptome of an organism. The process involves several steps that are essential for obtaining accurate andreliable results.Firstly, the RNA molecules are extracted from the cells or tissues of interest. This step is crucial as it ensures that the RNA represents the gene expression profile of the sample. Various methods can be used for RNA extraction, such as phenol-chloroform extraction or commercial kits.Once the RNA is extracted, it needs to be purified to remove any contaminants, such as genomic DNA or proteins. This purification step is important to ensure that the sequencing reads obtained are specific to the RNA molecules and not from other sources.Next, the purified RNA is converted into complementary DNA (cDNA) through a process called reverse transcription. This step involves the use of reverse transcriptase enzyme to synthesize cDNA from the RNA template. The cDNA represents a copy of the RNA molecules and can be used for sequencing.After obtaining the cDNA, it is then fragmented into smaller pieces to facilitate sequencing. This fragmentation can be achieved through physical methods, such as sonication or enzymatic methods, such as restriction enzyme digestion. The fragmented cDNA is then ready for sequencing library preparation.Library preparation involves adding specific adapters to the fragmented cDNA molecules. These adapters contain sequences that are recognized by the sequencing platform and allow for the attachment of the cDNA fragments to the sequencing flow cell. This step is crucial for the subsequent sequencing process.Once the library is prepared, it is loaded onto the sequencing platform, such as Illumina or PacBio. The sequencing process generates millions of short reads or long reads, depending on the platform used. These reads represent fragments of the cDNA molecules and are used to reconstruct the original RNA sequences.After sequencing, the reads are aligned to a reference genome or transcriptome to determine their origin and quantify gene expression levels. This step involves bioinformatics analysis, where specialized software tools are used to process the sequencing data and generate meaningful results.Finally, the results of the transcriptome sequencing experiment can be interpreted to gain insights into gene expression patterns, alternative splicing events, and other transcriptomic features. This information can be used to study gene function, identify biomarkers, or understand disease mechanisms.中文回答:转录组测序,也被称为RNA测序(RNA-seq),是一种用于研究生物体转录组的强大技术。
简述illumina测序原理和流程

简述illumina测序原理和流程下载温馨提示:该文档是我店铺精心编制而成,希望大家下载以后,能够帮助大家解决实际的问题。
文档下载后可定制随意修改,请根据实际需要进行相应的调整和使用,谢谢!并且,本店铺为大家提供各种各样类型的实用资料,如教育随笔、日记赏析、句子摘抄、古诗大全、经典美文、话题作文、工作总结、词语解析、文案摘录、其他资料等等,如想了解不同资料格式和写法,敬请关注!Download tips: This document is carefully compiled by theeditor. I hope that after you download them,they can help yousolve practical problems. The document can be customized andmodified after downloading,please adjust and use it according toactual needs, thank you! In addition, our shop provides you with various types ofpractical materials,such as educational essays, diaryappreciation,sentence excerpts,ancient poems,classic articles,topic composition,work summary,word parsing,copy excerpts,other materials and so on,want to know different data formats andwriting methods,please pay attention! Illumina测序是一种广泛应用的高通量测序技术,其原理和流程可以概括如下:原理:Illumina测序基于“边合成边测序”(Sequencing by Synthesis, SBS)的原理,利用荧光标记的可逆终止脱氧核苷三磷酸(dNTPs)在DNA合成过程中实现序列的测定。
illumina 转录组测序简明实验流程(PE-oligodT NEB)
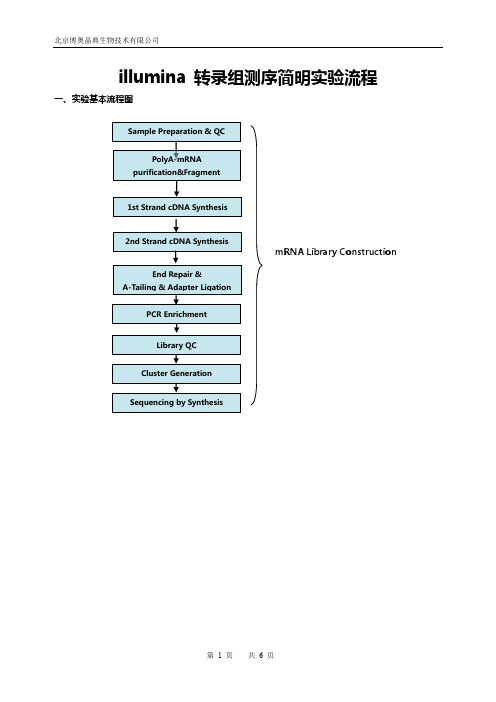
illumina 转录组测序简明实验流程一、实验基本流程图mRNA Library Construction二、mRNA建库流程1.材料准备1.2.1.3.2.样品准备和QC选择质量合格的Total RNA作为mRNA测序的建库起始样品,其质量要求通过Agilent 2100 BioAnalyzer检测结果RIN≥7,28S和18S的RNA 的比值大于或等于1.5:1,起始量的要求范围是0.1∽1ug。
用QUBIT RNA ASSAY KIT对起始的Total RNA进行准确定量。
3.建库实验步骤3.1.mRNA纯化和片段化3.1.1.mRNA纯化纯化原理是用带有Oligod(T)的Beads对Total RNA中mRNA进行纯化。
3.1.2.mRNA片段化3.2.1st Strand cDNA 合成3.3.2nd Strand cDNA 合成根据下表制备反应体系,然后在PCR仪上运行Program3,然后将第2链cDNA合成产物用144uL AMPure XP Beads进行纯化,最后用60µL的Nuclease free water进行重悬,取出55.5µL以备下一步使用;3.4.Perform End Repair/dA-tail3.5.Adaptor Ligation根据下表制备反应体系,然后在PCR仪上运行Program5、Program6,然后100uL AMPure XP Beads进行纯化后用52.5µL的Resuspension Buffer进行重悬,再用50uL AMPure XP Beads3.6.PCR扩增根据下表制备反应体系,然后在PCR仪上运行Program7,然后再45µL用AMPure XP Beads 进行纯化,最后用23µL的Resuspension Buffer进行重悬,取出20µL以备下一步使用;3.7. PCR 产物质控用QUBIT DNA HS ASSAY KIT 对PCR 产物进行准确定量。
转录组测序的流程

转录组测序的流程转录组测序是一种用于研究RNA转录本的高通量测序技术,它可以帮助科研人员了解生物体内部的基因表达情况,从而揭示基因调控、代谢途径等重要生物学过程。
本文将介绍转录组测序的流程,包括样本准备、RNA提取、建库、测序和数据分析等步骤。
1. 样本准备。
转录组测序的第一步是样本准备,样本的选择和处理对后续的实验结果至关重要。
首先需要确定研究的对象,是细胞、组织还是整个生物体,然后采集样本并进行保存。
在采集样本的过程中,需要注意避免RNA的降解和污染,可以使用RNAlater等试剂来稳定RNA。
此外,还需要记录样本的相关信息,如采集时间、处理方法等。
2. RNA提取。
RNA提取是转录组测序的关键步骤,它可以从样本中纯化出RNA,并去除DNA、蛋白质和其他杂质。
常用的RNA提取方法包括酚/氯仿法、硅胶柱法和磁珠法等。
在进行RNA提取时,需要注意保持样本的完整性和纯度,避免外源性RNA的污染。
此外,还需要对提取得到的RNA进行定量和质量检测,确保其可以用于后续的实验。
3. 建库。
建库是将提取得到的RNA转录本转化为可以进行测序的DNA文库的过程。
建库的关键步骤包括RNA的反转录、cDNA合成、末端修复、连接接头、文库扩增和纯化等。
在建库的过程中,需要注意避免外源DNA的污染,确保文库的纯度和完整性。
此外,还需要对建库得到的DNA文库进行定量和质量检测,以确保其可以用于高通量测序。
4. 测序。
建库完成后,就可以进行高通量测序了。
目前常用的转录组测序技术包括RNA-seq和全长转录组测序。
RNA-seq可以对RNA转录本进行定量和差异表达分析,全长转录组测序可以获取RNA的全长序列信息。
在进行测序时,需要选择合适的测序平台和测序深度,确保可以获得足够的数据量用于后续的数据分析。
5. 数据分析。
测序数据的分析是转录组测序的最后一步,它包括数据的质控、比对、表达定量和差异分析等。
在进行数据分析时,需要选择合适的分析软件和算法,对数据进行准确的处理和解释。
转录组测序与单细胞测序的实验流程

转录组测序与单细胞测序的实验流程转录组测序实验流程包括样品提取、RNA提取和纯化、RNA测序文库构建、测序、数据分析和生物信息学分析等步骤。
The experimental process of transcriptome sequencing includes sample extraction, RNA extraction and purification, RNA sequencing library construction, sequencing, data analysis, and bioinformatics analysis.首先,样品提取需要选择合适的组织或细胞,并使用合适的方法来提取RNA。
First, sample extraction requires selecting appropriate tissues or cells and using suitable methods to extract RNA.其次,RNA提取和纯化需要使用RNA提取试剂盒来提取总RNA,然后通过反转录酶将RNA转录成cDNA,最后利用PCR扩增和纯化得到RNA测序文库。
Second, RNA extraction and purification require using RNA extraction kits to extract total RNA, then transcribing RNA into cDNA using reverse transcriptase, and finally obtaining RNA sequencing libraries through PCR amplification and purification.接下来,测序是将RNA测序文库进行高通量测序,通常使用Illumina测序技术。
Next, sequencing is the high-throughput sequencing of RNA sequencing libraries, usually using Illumina sequencing technology.然后,得到的测序数据需要进行质量控制、比对、基因表达量分析、差异表达基因分析等生物信息学分析。
- 1、下载文档前请自行甄别文档内容的完整性,平台不提供额外的编辑、内容补充、找答案等附加服务。
- 2、"仅部分预览"的文档,不可在线预览部分如存在完整性等问题,可反馈申请退款(可完整预览的文档不适用该条件!)。
- 3、如文档侵犯您的权益,请联系客服反馈,我们会尽快为您处理(人工客服工作时间:9:00-18:30)。
illumina 转录组测序简明实验流程一、实验基本流程图
mRNA Library Construction
二、mRNA建库流程
1.材料准备
1.2.
1.3.
2.样品准备和QC
选择质量合格的Total RNA作为mRNA测序的建库起始样品,其质量要求通过Agilent 2100 BioAnalyzer检测结果RIN≥7,28S和18S的RNA 的比值大于或等于1.5:1,起始量的要求范围是0.1∽1ug。
用QUBIT RNA ASSAY KIT对起始的Total RNA进行准确定量。
3.建库实验步骤
3.1.mRNA纯化和片段化
3.1.1.mRNA纯化
纯化原理是用带有Oligod(T)的Beads对Total RNA中mRNA进行纯化。
3.1.2.mRNA片段化
3.2.1st Strand cDNA 合成
3.3.2nd Strand cDNA 合成
根据下表制备反应体系,然后在PCR仪上运行Program3,然后将第2链cDNA合成产物用144uL AMPure XP Beads进行纯化,最后用60µL的Nuclease free water进行重悬,取出
55.5µL以备下一步使用;
3.4.Perform End Repair/dA-tail
3.5.Adaptor Ligation
根据下表制备反应体系,然后在PCR仪上运行Program5、Program6,然后100uL AMPure XP Beads进行纯化后用52.5µL的Resuspension Buffer进行重悬,再用50uL AMPure XP Beads
3.6.PCR扩增
根据下表制备反应体系,然后在PCR仪上运行Program7,然后再45µL用AMPure XP Beads 进行纯化,最后用23µL的Resuspension Buffer进行重悬,取出20µL以备下一步使用;
3.7. PCR 产物质控
用QUBIT DNA HS ASSAY KIT 对PCR 产物进行准确定量。
(1) 2100质控
用2100 Bioanalyzer chip 判断PCR 切胶产物片段大小是否符合后续测序的要求。
(2) 文库摩尔浓度准确定量
通过q-PCR 标准品的摩尔浓度对构建的文库进行摩尔浓度的绝对定量,以保证文库上机用量的准确性,采用Illumina 推荐的KAPA 定量试剂盒(Cat no.KK4602)。
3.8. 混合文库 (Optional)
根据文库的定量及定性结果将每个library 的摩尔浓度统一调整到2nM ,然后根据实验设计选择性的将需要混合的library 进行等量等浓度混合,保证混合后的library 的终浓度也是2nM
,体积大于等于10µL ,然后将文库进行-80℃保存。
三、上机测序 1. 材料准备 1.1.
1.2. 2. 文库上机样品准备
2.1. 准备新鲜配置的NaOH ,将其浓度调整到0.1N ;取0.1N 的NaOH (10µL)与2nM 的文库(10µL)
的进行混合vortex ,离心,室温放置5min ,然后冰上放置;
2.2. 然后将20µL 已经变性成单链DNA 的Library 加入到980µL 预冷的HT1(Hybridization buffer)
中,使文库的终浓度为20pM ,冰上放置;
2.3.
3.Phix control样品准备
3.1.Phix control 2 nM(10µL)的体系,即
3.2.取0.1N的NaOH(10µL)与2nM的Phix control(10µL)的进行混合vortex,离心,室温
放置5min,然后冰上放置;
3.3.然后将20µL已经变性成单链DNA的Phix control加入到980µL预冷的HT1(Hybridization
buffer)中,使Phix control的终浓度为20pM,冰上放置;然后再将Phix control浓度稀释到5pM,即20pM的Phix control(600µL)+HT1(400µL);
4.簇生成
使用TruSeq Rapid PE Cluster Kit将Flowcell和准备好的文库在cBot上进行簇生成,即文库中的分子与Flowcell上固定的引物结合进行桥式PCR扩增,然后才能再HiSeq2500上进行测序。
5.边合成边测序
将完成做好簇生成的Flowcell转移到HiSeq2500仪器上准备测序,根据不同的测序类型以及测序长度,选择正确的Recipe,平均完成1个碱基的测序大概需要9分钟,所以测序周期与测序的长度直接相关。
测序程序正式运行前,首先根据First Base Report判断每条Lane上A,T,C,G碱基信号是否正常,从而判断测序引物结合是否有问题;其次根据First Base Report中cluster数量估测生成的数据量是否满足测序实验要求。
