A Large Maize (Zea mays L.) SNP Genotyping Array
Axiom Chicken Genotyping Array说明书

Axiom Chicken Genotyping ArrayHigh-density array for genotyping layers and broilers across multiple breedsThe Applied Biosystems ™ Axiom ™ Chicken Genotyping Array is the first commercially available high-density chicken genotyping array. This array includes hundreds of thousands of polymorphic markers that are present in commercial layer and broiler chickens as well as outbred noncommercial populations.Highlights• Only chicken genotyping array openly available as a catalog product, allowing effective data sharing • Enables variation detection both within and between poultry breeds in broilers, white egg layers, brown egg layers, and outbred noncommercial breeds• 580,961 highly polymorphic genetic variants chosen from a screening of 1.8 million markers• Designed in collaboration with leading academic institutions and commercial poultry companies that include The Roslin Institute, Aviagen Ltd, Hy-Line International, and the German Synbreed projectWide range of applicationsThe Axiom Chicken Genotyping Array can be used for predicting breeding values in both layers and broilers, for genome-wide association studies, high-resolution genetic mapping, Mendelian trait mapping, and selection signature analysis. The parental high-density information from the array can be used to make decisions on breeding poultry to gain incremental improvements in feed conversion ratio, growth rate, saleable eggs, white meat yield, and understanding inheritance traits across and within multiple breeds. The Axiom Chicken Genotyping Array alsocontains markers associated with wild outbred lines that can be useful in conducting functional research studies, and observing phenotypic effects across outbred andcrossbred lines for better management of indigenous chickens as well as for studying genetic diversity among populations.Maximum coverage of genetic diversitySNP discovery was carried out by resequencing 243chickens from 24 lines representing both elite commercial and experimental lines (Table 1). Sequencing data was aligned to the reference genome Gallus_gallus-4.0.Array designThe SNPs discovered through resequencing were genotype-tested on the Applied Biosystems ™ Axiom ™genotyping platform by screening 1.8 million SNPs across 300 samples that included 186 commercial lines, wild out-group samples, and 32 parent-offspring trios for Mendelian inheritance.DATA SHEETAxiom Chicken Genotyping ArrayTable 1. Description of sequenced individuals.Ordering informationCat. No.Axiom Genome-Wide Chicken Array Contains one 96-array plate; reagents and GeneTitan Multi-Channel Instrument consumables sold separately902148Axiom GeneTitan Consumables Kit Contains all GeneTitan Multi-Channel Instrument consumables required to process one Axiom array plate901606Axiom 2.0 Reagent KitIncludes all reagents (except isopropanol) for processing 96 DNA samples901758For Research Use Only. Not for use in diagnostic procedures. © 2017 Thermo Fisher Scientific Inc. All rights reserved. All trademarks are the property of Thermo Fisher Scientific and its subsidiaries unless otherwise specified. FTA is a trademark of Whatman. COL31709 0617Find out more at/microarraysTable 2. Genotyping performance on commercial samples.Figure 1. PCA analysis of markers showing closely related individuals grouped together.Markers were selected for inclusion on the Axiom Chicken Genotyping Array based on the following criteria:• Markers exhibited a range of minor allele frequencies to ensure representation of both rare and common variants • Markers were uniformly distributed across the genome in layers and broilers• Greater than 98% call rate across tested samples • Markers exhibited Mendelian inheritance; highly polymorphic markers were prioritized• Annotation information was available for defining the effect of the selected variantsPrincipal component analysis (PCA) shows that markers for layers, broilers, and out-group lines can group the closely related individuals, demonstrating the success of SNP selection criteria (Figure 1).Superior performanceThe Axiom Chicken Genotyping Array is part of theApplied Biosystems ™ Axiom ™ Genotyping Solution, which generates robust and reliable genotypes with minimal user intervention while helping to reduce costs and processing complexity. Three hundred customer samples representing commercial and experimental lines were genotyped on the array. The samples were prepared manually and processed on the Applied Biosystems ™ GeneTitan ™ Multi-Channel (MC) Instrument. Array performance was measuredacross 559,249 SNPs. The SNPs were filtered as per the Best Practice Supplement to Axiom Genotyping Solution Data Analysis (P/N 703083). The results are summarized in Table 2.Sample types supported • Blood on FTA ™ cards • BloodGenomics presentationGheyas AA et al. (2012) Development and characterization of a high-density SNP genotyping assay for the chicken. The Roslin Institute, University of Edinburgh. Posterpresentation at the Plant and Animal Genome Conference, San Diego, CA, USA.。
NodetrimentaleffectofBtmaizepollencontainingCry1Ab

Journal of Integrative Agriculture2019,18(4):893-899Available on line at ScienceDirectELSEVIERRESEARCH ARTICLENo detrimental effect of Bt maize pollen containing Cry1Ab/2Aj or CrylAc on adult green lacewings Chrysoperla sinica TjederLIU Yan-min1,LI Yun-he1,CHEN Xiu-ping1,SONG Xin-yuan2,SHEN Ping3,PENG Yu-fa11State Key Laboratory for Biology of Plant Diseases and Insect Pests,Institute of Plant Protection,Chinese Academy of Agricultural Sciences,Beijing100193,P.R.China2Jilin Academy of Agricultural Sciences,Changchun130124,P.R.China3Science and Technology Development Center,Ministry of Agriculture,Beijing100122,P.R.ChinaAbstractAdult Chrysoperla sinica Tjeder is a common pollen feeder in maize fields.They are thus directly exposed to insecticidal proteins by consumption of genetically engineered maize pollen containing Bacillus thuringiensis(Bt)proteins.Here we assessed the potential effects of Cry1Ab/2Aj-or Cry1Ac-containing Bt maize pollen on the fitness of adult C.sinica via a dietary-exposure assay under laboratory conditions.Survival,pre-oviposition,fecundity and adult dry weight did not differ between adult C.sinica consuming Bt or the corresponding non-Bt maize pollen.The stability of the Cry protein in the food sources and uptake of the Cry protein by adult C.sinica duri n g the feedi n g experime n t were con f irmed by ELISA.These results dem o n s trate that adult C.sinica are not affected by the consumpti o n of Cry1Ab/2Aj-or Cry1Ac-c o ntaining maize pollen, suggest!ng that production of Bt maize expressing cry1Ab/2Aj or cry1A c genes will pose a negligible risk to adult C.sinica.Keywords:non-target effect,environmental risk assessment,ELISA,dietary exposure assay1.IntroductionMaize(Zea mays L.)is one of the most important cereal crops,serving as food,feed,and in d ustrial material in China(Pechanova and Pechan2015).In maize production, insect pests pose a serious con s training factor,in particular lepidopteran pests,such as Ostrinia furnacalis(GuenQe)Received14September.2017Accepted18December,2017 LIU Yan-min,E-mail:lym09b1@;Corrrespondence LI Yun-he,Tel:+86-10-62815947,Fax:+86-10-62896114,E-mail: liyun h e@caas.c n©2019CAAS.Published by Elsevier Ltd.This is an open access article under the CC BY-NC-ND license(http:// creativecomm o n /lice n ses/by-nc-nd/4.0/).doi:10.1016/S2095-3119(17)61887-2and Helicoverpa armigera(Hubner),cause large yield loses annually.O.furnacalis is the most destructive pest in China, causing yield losses of about10to20%in an ordinary year and resulting in no harvest in an outbreak year(He et al. 2003).Therefore,con t rolling O.furnacalis is an essential part of maize production.The traditional measure to control the pest largely relies on the application of chemical insecticides,which has brought multiple problems such as insect resista n ee evolution and en v ironme n tal polluti o n (Murray et al.2002).Genetic engineering(GE)technology has been regarded as a promising new tool for pest control and improving crop productivity.GE maize lines producing crystal endotoxins (Cry)derived from Bacillus thuringiensis(Bt)have been widely planted around the world(James2016).For efficient control of lepidopteran maize pest and reducing chemical insecticide application,China has devoted a great effort to894LIU Yan~min et al./Journal of Integrative Agriculture2019,18(4):893-899develop Bt maize lines.To date,multiple Bt maize lines have been developed(Liu Q S et al.2016),most of which,such as Shuangkang12-5(expressing a fusion cry1Ab/2Ajgene), BT-799(expressing a modified cry1Acger\e)and IE09S034 (expressing a crylie gene),have proven to be effective in control of the target lepidopteran pest(Chang et al.2013; Zhang et al.2013;Wang et al.2014,2016).Prior to the commercialization of any GE line,it must be subjected to rigorous risk assessment to guarantee safety to human health and the environment(Garcia-Alonso et al. 2006;Romeis et al.2008;Li et al.2014c).A part of the risk assessment concerns potential effects of the insecticidal proteins produced by insect-resistant GE crops on nontarget arthropods that provide important ecosystem services in e luding biological control,polli nation,and decompositi o n (Romeis et al.2011;Devos et al.2015;Li et al.2015, 2017a,b;Wang et al.2015,2017;Yang et al.2015).The green lacewing,Chrysoperla sinica Tjeder(Neuroptera: Chrysopidae)is a prevale n t predator in many non-crop and crop habitats,including maize,rice,cotton and vegetables distributed throughout China(Bai et al.2005;Jiang and Xiao2010).The larvae are generalist predators of many important agricultural pests(including aphids,mites, leafhoppers,and eggs and young larvae of lepidopterans). In contrast to larvae,C.sinica adults are not predators and feed mainly on pollen,honeydew and plant nectar(Wang et al.2012).Therefore,adult C.sinica is directly exposed to plant-produced insecticidal proteins by feeding on Bt maize pollen.In the current study,the potentiml impacts of feeding Bt maize pollen containing Cry1Ab/2Aj or CrylAc protein on adult C.sinica were assessed using dietary exposure assay in the laboratory with the expectation of providing information valuable for regulatory decisi o n making reg a r d i n g possible commercial use of these Bt maize lines.2.Materials and methods2.1.InsectChrysoperla sinica was derived from a colony maintained in the laboratory at the conditions of(26±1)°C,(75±5)%RH with a16h L:8h D photoperiod at the experimental field station of the Institute of Plant Protect!on,Chinese Academy of Agricultural Scien c es(CAAS),near Lmngfmng City,Hebei Province,China(39.5°N,116.6°E).Larvae of C.sinica were fed on soybean seedlings in t ested with Aphis glycines Matsumura(Hemiptera:Aphididae).Adults were reared on an artificial diet containing sucrose and brewers'yeast at a ratio of1:1.The aphids were replaced daily,guaranteeing an ad libitum food supply for the larvae.Newly emerged C.sinica adult(<12h after emergence)were used for the following experiments.2.2.Maize plants and pollen collectionTwo transgenic Bt maize lines,SK12-5(Shuangkang12-5) and BT-799,and their corresponding non-transformed near isolines,ZD958(Zhengdan958)and Z58(Zheng58), respectively,were used in the current study.SK12-5plants express a cry1Ab/2Aj^s\on gene driven by the constitutive pZmUbi-1promoter,and BT-799plants express a modified mcrylAc gene driven by the constitutive CaMV35s promoter.Cry proteins produced by the Bt maize lines target lepidopteran maize pests including O.furnacalis.The seeds of ZD958and SK12-5(Cry1Ab/2Aj)were offered by Prof. Shen Zhicheng(Zhejiang University,Hangzhou,China),and the seeds of Z58and BT-799(CrylAc)were offered by Prof. Lai Jin she n g(China Agricultural University,Beiji ng,China).The maize lines were planted at the experimental field station of the Jilin Academy of Agricultural Sciences in Gongzhuling City,Jilin Province,China(43°19'N,124O29*E) in2014.The maize seeds were sown in12adjace n t plots (20m><20m)in a completely randomized design(three plots per cultivar)on25May2014.The plants were man a ged according to common local agricultural practices without applicati o n of in s ecticides for the whole growing seas o n.During the maize anthesis period in late July2014,maize pollen was collected using the procedure as described in our previous study(Li et al.2017a).2.3.Pollen feeding bioassayOur previous experiments showed that feeding on rice pollen together with2mol L1sucrose solution is highly nutritious for adult C.sinica and can sustain their normal survival and development(Wang et al.2012).In the current dietary exposure assay,we used maize polle n in s tead of rice polle n, and our preliminary experime n t con f irmed the validati o n of the maize pollen diet used in the feeding assay.Four dietary treatments were set up,namely2mol L_1sucrose solution together with(i)SK12-5maize pollen containing Cry1Ab/2Aj;(ii)ZD958maize pollen(the non-Bt control pollen for SK12-5);(iii)BT-799maize pollen containing CrylAc;or(iv)Z58maize pollen(the non-Bt control pollen for BT-799).The sexes of freshly emerged C.sinica adults were identified and then single randomly selected pairs of males and females were confined in transparent plastic containers(8.0cm in diameter,8.0cm in height). Maize pollen mnd2mol sucrose solution were supplied separately in small Petri dishes(3.0cm in diameter,1.0cm in height),which were placed on the bottom of each container. In addition,a water-saturated cotton ball was supplied as a water source on the bottom of each container.Pollen wasLIU Yan-min et al./Journal of Integrative Agriculture2019,18(4):893-899895replaced every day,and the sucrose solution and cotton ball were replaced every2days.Each container was covered with a layer of cotton gauze which served as an oviposition substrate.Forty pairs of C.sinica adults were tested in each of the pollen treatments.Survival,pre-oviposition period, and daily and total fecundity were recorded or calculated based on daily observations.The experiment terminated after25days,when all of the alive adults were lyophilized, and the dry weight was determined using an electronic balance(CPA224S;Sartorius AG;readability=0.1mg, repeatability<±0.1mg).Subsequently,the Cry1Ab/2Aj and CrylAc protein concentrations contained in the in s ects were detected by ELISA.The experime n t was con d ucted in an environmental chamber at(26±1)°C,(75±5)%RH and a 16h L:8h D photoperiod.To identify the temporal stability of the Cry proteins in the maize pollen during the feeding bioassay,five pollen subsamples were taken from each of the Bt maize pollen before and after being exposed to C.sinica for1day.The Cry protei n con t ents in polle n samples were measured by ELISA.2.4.ELISA measurementsThe contents of the fusion protein Cry1Ab/2Aj and the protein CrylAc in maize pollen and lacewings were measured using ELISA Kit from EnviroLogix(Portiand,ME, USA;catalog number AP003).The ELISA Kit was originally used for quantitative detection of CrylAb and CrylAc,but they were confirmed to be appropriate for quantification of Cry1Ab/2Aj in our previous study(Liu Y M et al.2016). The same CrylAb calibrators were used for developing the stmndard curve,and the concentrmtions were adjusted to the equivalent Cry1Ab/2Aj concentrations of0, 1.0, 5.0,and 10.0pg L~1because the molecular weight of Cry1Ab/2Aj is two times that of CrylAb.The kit could be directly used for detection of CrylAc with the same CrylAb calibrators,but the con c entrati o ns were adjusted to the equivale n t CrylAc concentrations of0, 1.5,10.0,and25.0mg L_1according to the manufacturer's instructions.Prior to the analyses,all in s ect samples were washed in phosphate-buffered sali ne Tween(PBST)to remove any Bt protein from their outer surface.For extraction of Cry proteins,samples of insects or maize pollen were weighed and then mixed in1mL PBST with a micro-mortar and pestle on ice.After centrifugation and appropriate dilution of the supernatants,ELISA was conducted following the manufacturer's instructions.A microplate spectrophotometer(PowerWave XS2,BioTek, Winooski,VT,USA)was used for reading the optical density (OD)values.The contents of Cry proteins in the samples were calculated by com pari ng the OD values to the standard curves.2.5.Statistical analysesStatistical comparisons were made between each Bt maize pollen treatment and the corresponding non-Bt maize pollen treatment.The data on adult survival and pre-oviposition period were analyzed using Chi-square test and Mann-Whitney L/-test,respectively,since the data did not satisfy the assumptions for parametric analyses.Repeated-measures an a lysis of varia n ee(AN O VA)was con d ucted for analyzing the daily fecundity data.Student's t-test was used to compare the total fecundity and adult dry weight and to compare the Cry protein contents in maize pollen before and after1day feeding exposure.All statistical analyses were carried out with the software package SPSS(version 13;SPSS.Inc.,Chicago,IL,USA).3.Results3.1.Effects on life table parametersOver83%of the adult C.sinica survived the25-day pollen feeding period,and the survival rates did not differ between the Bt and the corresponding non-Bt pollen treatments(/2-test;both P>0.05)(Table1).Similarly,the pre-oviposition period and total fecundity were not significantly affected by ingestion of Bt maize pollen(pre-oviposition period, Cry1Ab/2Aj:1/=545,P二0.163,CrylAc:1/=572.5,P=0.141; total fecundity,Cry1Ab/2Aj:f=1.39,df=71,P=0.17; CrylAc:f=0.33,df=73,P=0.74).The daily fecundity was not significantly affected by feeding on Bt maize pollen (Cry1Ab/2Aj:F—产1.92,P=0.17;CrylAc:F173=0.11, P二0.74)(Fig.1).The student's f-tests revealed no statistical differences between Bt and non-Bt pollen treatments for adult dry weight(Cry1Ab/2Aj:f=0.15,c//=19,P二0.88for females and f二0.30,df=23,P=0.77for males;CrylAc:匸0.49,df=22,P=0.63for females and f=0.76,df=26,P二0.47 for males)(Table1).3.2.Uptake of Cry proteins by C.sinicaWhen the experiment was terminated after25days,ELISA measurements showed that all adults fed upon Bt maize pollen containing Cry protein.The mean concentrations (±SE)of Cry1Ab/2Aj in C.sinica females and males were (2.06±0.37)pg g1dry weight(DW)and(0.15±0.03)pg g_1 DW,respectively.The mean concentrations(±SE)of CrylAc in C.sinica females and males were(0.0028±0.0008)and (0.0005±0.0001)pg g_1DW,respectively.896LIU Yan-min et al. / Journal of Integrative Agriculture 2019, 18(4): 893-899Table 1 Effect of consumption of pollen from Bt maize (SK12-5 containing Cry1Ab/2Aj and BT-799 containing CrylAc) or from the correspond!ng non-transformed varieties (ZD958 and Z58) on life-table parameters of adult Chrysoperla sinica^1) Survival rate was analysed by Chi-square test, pre-oviposition period was analysed by Mann-Whitney L/-test, total fecundity and adultdry weight were analysed by Student's f-test.Data are mean±SE. Number of replicates is given in parenthesis.Maize cultivar Survival rate (%)Pre-oviposition periodToal fecundityAdult dry weight (mg)FemaleMale SK12-5 (Cry1Ab/2Aj)85.1 (74) 6.21±0.25 (37)307.4±27.66 (37) 4.45±0.34 (15) 2.34±0.15 (14)ZD95883.3 (72) 5.83±0.26 (36)361.4±27.45 (36) 4.63±0.26 (12) 2.40±0.10(11)BT-799 (CrylAc)85.1 (74) 5.71±0.20 (37)318.3127.24 (37) 4.2410.32 (13) 2.40±0.11 (15)Z5886.5 (74)6.35±0.31 (37)330.5±25.58 (37)4.28±0.22 (15) 2.57±0.20(13)Fig. 1 Daily fecundity of Chrysoperla sinica fed pollen from Bt maize (SK12-5 containing Cry1Ab/2Aj and BT-799 containing CrylAc) or its corresponding non-transformed maize plants (ZD985 and Z58). In s ects were provided 2 mol L _1 sucrose solution together with pollen. Values are mean±SE,门=36, 37.3.3. Stability of Cry proteins in maize pollen The ELISA results showed that the original concentrations (mean±SE) of Cry1Ab/2Aj in SK12-5 pollen and CrylAc in BT-799 pollen were (22.111.3) and (0.034±0.0017) pgg _1 DW, respectively. After 1-day feeding exposure, the Cry1Ab/2Aj content decreased to (18.6±0.9) pg g _1 DW,and the differenee was marginally significant (=2.24, df=8, P=0.055), while the CrylAc content decreased to(0.028±0.002) pg g 1 DW, with a non-significant difference (f=2.19, ¢//=8, P=0.06). No Bt protein was detected in pollen from the control maize cultivars (ZD958 and Z58).4. DiscussionOur pollen feeding bioassay showed that the fitness of adult C. sinica was not altered by ingestion of Bt maize pollen expressing Cry1Ab/2Aj or CrylAc compared to thatof lacewings feeding on the corresponding non-Bt maize pollen. Since the Bt proteins are orally active compounds,they have to be ingested by test organisms to assess toxicity (Li et al. 2014c). To confirm that adult of C. sinica indeed ingested Cry proteins during the feeding bioassay, ELISAassay was con d ucted to detect Cry protei n concentrations inthe insects. Considerable contents of Cry1 Ab/2Aj or Cry1 Ac were detected in C. sinica females after 25 days of feeding upon Bt maize pollen. The concentrations of Cry1 Ab/2Ajand CrylAc in C. sinica females were more than 13 and 5 times, respectively, higher than those in males. The resultswere consistent with those of a previous study in which theconcentration of Cry2Aa detected in C. sinica females was over 7 times higher than in males after 26 days of pollenfeeding (Wang et al. 2012). The concentrations of CrylAb or Cry3Bb1 detected in C. carnea females were over 25times higher than in males after feeding upon Bt maizepollen for 28 days (Li et al. 2008). This observation is notsurprising due to the fact that females need carbohydrates as an energy source and also pollen as a protein source forreproduction, while males likely survive well on carbohydrate alone; thus, males ingested much fewer pollen grains thanfemales (Li et al. 2008, 2010).In addition, to clarify whether adult C. sinica was exposedto a con sistent level of Cry protei n s, we further measuredthe contents of Cry proteins in maize pollen before and after exposure to the in s ects. The results indicated that there wasno significant degradation of Cry proteins in maize pollen duri ng the durati o n of feedi n g. However, the con t ent of Cryproteins in maize pollen may rapidly decrease by exposure to rainfall and sun l ight un d er field conditi o ns (Pusztai et al.1991; Sims and Ream 1997). For example, the CrylAbconcentration in Bt maize pollen rapidly declined to 38% of the initial concentration after the pollen was shed andaccumulated in the leaf axil (Xing et al. 2008). In addition,C. sinica adults were consistently fed on pollen for more than 3 weeks, which was 1.5-3 times longer than the pollen-shedding period in maize fields that usually lasts1-2 weeks (Sears et al. 2001). Hence, C. sinica adultswere exposed to much higher levels of Bt proteins in thelaboratory experiment than those foraging pollen in maize fields. However, no detrimental effect was detected in thecurrent study, suggesting that adults of C. sinica are not sensitive to Cry1Ab/2Aj and CrylAc, and consumption of Btmaize pollen containing such Cry proteins do not adversely affect theirfitness.LIU Yan-min et al./Journal of Integrative Agriculture2019,18(4):893-899897The curre nt results are con sistent with those of previous studies regarding effects of Bt crop pollen on different green lacewing species.For example, C.sinica adults were not adversely affected by consumption of Cry1C-containing pollen from Bt rice line T1C-19,Cry2Aa-containing pollen from Bt rice lines T2A-1,or Cry1Ab-containing pollen from Bt rice lines KMD1and KMD2(Bai et al.2005;Wang et al. 2012;Li et al.2014a).Other green lacewing species,such as Chrysoperla carnea(Stephens)and C.plorabunda(Fitch) (both Neuroptera:Chrysopidae),have also been tested in risk assessments of GE crops.Consumption of Bt maize pollen from Event176containing CrylAb or MON88017 containing Cry3Bb1does not harm adult C.carnea(Li et al.2008).Although significantly reduced daily fecundity and Iongevity of C.plorabunda(Fitch)was reported when they were fed MON810pollen containing CrylAb(Mason et al.2008),the fecundity of C.carnea was not affected by consumption of Event176maize pollen(Sears et al.2001) even though the CrylAb content in Event176was over10 times greater tha n in MON810.Again s t this background, Mason et al.(2008)suggested that the decreased fecundity and Iongevity were unlikely due to the CrylAb protein toxicity,but rather to some unknown differences in the composition of MON810maize pollen.Due to their ecological importanee as predators in agricultural systems, C.sinica larvae have received considerable attention in risk assessment of Bt crops,even though they are only indirectly exposed to Cry proteins by con s umption of prey in Bt crop fields,especially after adverse effects of CrylAb on C.carnea larvae were first reported in 1998(Hilbeck et al.1998a,b).However,subsequent studies con f irmed that CrylAb has no directly toxicity to gree n lacewing larvae,and the reported n e gative effects of CrylAb protein on lacewing larvae were attributed to the reduction of the nutritional quality of Cry protein-toxicated prey items (Romeis et al.2014).Such an effect was also found on C.sinica larvae(Li et al.2013). C.sinica larvae were negatively affected by feeding on Chilo suppressalis larvae that had been reared on an artificial diet containing Cry2Aa protein at100pg g~1,but were not affected by directly feeding on sucrose solution containing Cry2Aa proteins even at500pg mL_1(Li et al.2013).These results demonstrate that the observed detrimental effects were caused by the decreased prey quality due to the sensitivity of C.suppressalis larvae to Cry2Aa(Li et al.2013).A sound dietary exposure assay was developed in which C.sinica larvae can be directly exposed to high concentrations of Cry proteins,and using this testing system,it was further confirmed that the currently used Cry proteins Cry1Ab,Cry1Ac,and Cry2Aa are not toxic to green lacewing larvae(Li et al.2014b).5.ConclusionTo our knowledge,the current study is the first to assess the potential effects of Cry1Ab/2Aj-or CrylAc-containing pollen on C.sinica.The results show that the consumption of Bt maize pollen containing Cry1Ab/2Aj or CrylAc does not negatively affect the fitness of adult C.sinica.Hence, product!on of Cry1Ab/2Aj or Cry1Ac-transgenic maize may pose a negligible risk to the green lacewings.AcknowledgementsWe thank Prof.Eric M.Hallerman(Virginia Tech University, USA)for his comments and revision on an earlier draft of the manuscript.The study was supported by the National GMO New Variety Breeding Program of China(2015ZX08013-003).ReferencesBai Y Y,Jiang M X,Cheng J A.2005.Effects of transgenic crylAb rice pollen on the oviposition and adult Iongevity of Chrysoperla sinica Tjeder.Acta Phytophylacica Sinica,32, 225-230.(in Chinese)Chang X,Liu G G,He K L,Shen Z C,Peng Y F,Ye G Y.2013.Efficacy evaluation of two transgenic maize events expressing fused proteins to Cry1Ab-susceptible and・resista nt Ostrinia furnacalis(Lepidoptera:Crambidae).Journal of Economic Entomology,106,2548-2556. Devos Y,Romeis J,Luttik R,Maggiore A,Perry J N,Schoonjans R,Streissl F,Tarazona J V,Brock T C M.2015.Optimising environmental risk assessme nts.EM B O Reports,16, 1060-1063.Garcia-Alonso M,Jacobs E,Raybould A,Nickson T E,Sowig P,Willekens H,Van Der Kouwe P,Layton R,Amijee F, Fuentes A M.2006.A tiered system for assessing the risk of genetically modified plants to non-target organisms.Environmental Biosafety Research,5,57-65.He K L,Wang Z Y,Wen L,Bai S X,Zhou D R,Zhu Q H.2003.Field evaluation of the Asian maize borer control inhybrid of transgenic maize event MON810.Agricultural Sciences in China,2,1363-1368.Hilbeck A,Baumgartner M,Fried P M,Bigler F.1998a.Effects of transgenic Bacillus thuringiensis maize-fed prey on mortality and development time of immature Chrysoperla maizeea (Neuroptera:Chrysopidae).Environmental Entomology, 27,480-487.Hilbeck A,Moar W J,Pusztai-Carey M,Filippini A,Bigler F.1998b.Toxicity of Bacillus thuringiensis CrylAb toxin to the predator Chrysoperla carnea(Neuroptera:Chrysopidae).Environmental Entomology,27,1255-1263.James C.2016.Global status of commercialized biotech/GM crops:2015.ISAAA Briefs.ISAAA,Ithaca,NY,USA. Jiang X B,Xiao G Y.2010.Diversity of arthropod community898LIU Yan-min et al.I Journal of Integrative Agriculture2019.18(4):893-899in the canopy of genetically modified herbicide-tolerant rice (Oryza sativa L.).Chinese Journal of Eco-Agriculture,18, 1277-1283.(in Chinese)Li Y H,Chen X P,Hu L,Romeis J,Peng Y F.2014a.Bt rice producing Cry1C protein does not have direct detrimental effects on the green lacewing Chrysoperla sinica(Tjeder).Environmental Toxicology and Chemistry,33,1391-1397. Li Y H,Hu L,Romeis J,Wang Y,Han L,Chen X P,Peng Y F.e of an artificial diet system to study the toxicity of gut-active insecticidal compounds on larvae of the green lacewing Chrysoperla sinica.Biological Control,69,45-51. Li Y H,Liu Y M,Yin X M,Romeis J,Song X Y,Chen X P, Gen L L,Peng Y F,Li Y H.2017a.Consumption of Bt maize pollen containing Cry1le does not negatively affect Propylea j aponica(Thunberg)(Coleoptera:Coccinellidae).Toxins,9,108.Li Y H,Meissle M,Romeis J.2008.Consumption of Bt maize pollen expressing CrylAb or Cry3Bb1does not harm adult green lacewings,Chrysoperla carnea(Neuroptera: Chrysopidae).PLoS ONE,3,e2909.Li Y H Meissle M,Romeis e of maize pollen by adult Chrysoperla carnea(Neuroptera:Chrysopidae)and fate of Cry proteins in Bt-tra nsgenic varieties.Journal of Insect Physiology,56,157-164.Li Y H.Romeis J,Wu K M,Peng Y F.2014c.Tier-1assays for assessing the toxicity of insecticidal proteins produced by genetically engineered plants to non-target arthropods.Insect Science,21,125-134.Li Y H,Wang Y Y,Romeis J,Liu Q S,Un K J,Chen X P,Peng Y F.2013.Bt rice expressing Cry2Aa does not cause direct detrimental effects on larvae of Chrysoperla sinica.Ecotoxicology,22,1413-1421.Li Y H.Zhang Q L,Liu Q S,Meissle M,Yang Y,Wang Y N, Hua H X,Chen X P,Peng Y F,Romeis J.2017b.Bt rice in China-focusi n g the non t arget risk assessment.Plant Biotechnology Journal,15,1340-1345.Li Y H,Zhang X J,Chen X P,Romeis J,Yin X M,Peng Y F.2015.Con s umption of Bt rice polle n containing Cry1C or Cry2A does not pose a risk to Propylea j aponica(Thunberg) (Coleoptera:Coccinellidae).Scientific Reports,5,7679. Liu Q S,Halierman E,Peng Y F,Li Y H.2016.Development of Bt rice and Bt maize in China and their efficacy in target pest control.International Journal of Molecular Sciences, 17,1561.Liu Y M,Liu Q S,Wang Y N,Chen X P,Song X Y,Romeis J,Li Y H,Peng Y F.2016.Ingestion of Bt maize pollen containing Cry1Ab/2Aj or CrylAc does not harm Propylea japonica larvae.Scientific Reports,6,23507.Mason C E,Sheldon J K,Pesek J,Bacon H,Gallusser R, Radke G,Slabaugh B.2008.Assessment of Chrysoperla plorabunda Iongevity,feeundity,and egg viability when adults are fed transgenic Bt maize pollen.Journal of Agricultural and Urban Entomology,25,265-278.Murray D,Wesseling C,Keifer M,Corriols M,Henao S.2002.Surveillanee of pesticide-related illness in the developing world:putting the data to work.International Journal ofOccupational and Environmental Health,8.243-248. Pecha nova O,Pechan T.2015.Maize-pathogen in t eracti o ns: An on going combat from a proteomics perspective.International Journal of Molecular Sciences,16,28429-28448.Pusztai M,Fast P,Gringorten L,Kaplan H,Lessard T,Carey P.1991.The mechanism of sunlight-mediated inactivation of Bacillus thuringiensis crystals.Biochemical Journal, 273,43-47.Romeis J,Bartsch D,Bigler F,Candolfi M P,Gielkens M M, Hartley S E,Hellmich R L,Huesing J E,Jepson P C,Layton R.2008.Assessment of risk of in s ect-resistant transge n ic crops to non t arget arthropods.Nature Biotechnology,26, 203-208.Romeis J,Hellmich R L,Candolfi M P,Carstens K,De Schrijver A,Gatehouse A M,Herman R A,Huesing J E,McLean M A,Raybould A.2011.Recommendations for the design of laboratory studies on non-target arthropods for risk assessment of genetically engineered plants.Transgenic Research,20.1-22.Romeis J,Meissle M,Naranjo S E,Li Y H,Bigler F.2014.The end of a myth-Bf(CrylAb)maize does not harm green lacewings.Frontiers in Plant Science,5,391.Sears M K,Hellmich R L,Stanley-Horn D E,Oberhauser K S, Pleasants J M,Mattila H R,Siegfried B D,Dively G P.2001.Impact of Bt corn polle n on mon a rch butterfly populati o ns:a risk assessment.Proceedings of the National Academy of Sciences of t he United States of A merica,98,11937-11942. Sims S R,Ream J E.1997.Soil inactivation of the Bacillus thuringiensis subsp.kurstaki CryllA insecticidal protein within transgenic cotton tissue:Laboratory microcosm and field studies.Journal of Agricultural and Food Chemistry, 45,1502-1505.Wang J,Wu FC,Liu X Y,Feng S D,Song X Y.2016.Evaluation of transgenic maize*Shuangkang12-5'with complex traits of insect-resistanee and glyphosate-resistanee for the resista nee to Ostrinia furnacalis and tolerance to glyphosate.Plant Protection,42,45-50.(in Chinese) Wang Y Q,He K L,Jiang F,Wang Y D,Zhang T T,Wang Z Y, Bai S X.2014.Resista n ee of tran s genic Bt maize variety BT-799to the Asian maize borer.Chinese Journal of A pplied Entomology,3,636-642.(in Chinese)Wang Y Y,Dai P L,Chen X P,Romeis J,Shi J,Peng Y F,Li Y H.2017.Ingestion of Bt rice pollen does not reduce the survival or hypopharyngeal gland development of Apis mellifera adults.Environmental Toxicology and Chemistry, 36,1243-1248.Wang Y Y,Li Y H,Huang Z Y,Chen X P,Romeis J,Dai P L,Peng Y F.2015.Toxicological,biochemical,and histopathological analyses dem o n s trating that Cry1C and Cry2A are not toxic to larvae of the honeybee,Apis mellifera.Journal of A gricultural and Food Chemistry,63,6126-6132. Wang Y Y,Li Y H,Romeis J,Chen X P,Zhang J,Chen H,Peng Y F.2012.Consumption of Bt rice pollen expressing Cry2Aa does not cause adverse effects on adult Chrysoperla sinica Tjeder(Neuroptera:Chrysopidae).Biological Control,61,。
BRAVILOR RENEGITE产品说明书

SAFETY DATA SHEET IN ACCORDANCE WITH 91/155/EEC Page 1 of 4DATASHEET RENEGITE1. Identification of the substance1.1 Trademark : - RENEGITE1.2 Description : - Sulphamic Acid (solid)1.3 Distributed by : - Bravilor Bonamat B.V. Pascalstraat 20NL-1704 RD Heerhugowaard (The Netherlands)- Tel. +31 (0)72 5751751- Fax. +31 (0)72 57517582. Composition / information on ingredients2.1 Description : - Sulphamic Acid (~100%) (Syn.: Aminosulfinic Acid)2.2 Formula : - H2NSO3H2.3 MM : - 97.09 g/mol2.4 CAS. No. : - 5329-14-62.5 EEC. No. : - 016-026-00-02.6 Einecs No. : - 226-218-83. Hazards identification3.1 Health hazards : - Irritating to the eyes and skin3.2 Ecology : - Harmful to aquatic organisms- May cause long-term adverse effects in the aquatic environment3.3 Fire hazard : - Non combustible4. First aid measures4.1 After skin contact : - Wash off with plenty of water / shower- Remove contaminated clothing- Do not remove clothing in case this sticks to the skin- Cover wounds in a sterile way4.2 After contact with the eyes : - Rinse out with plenty of water with the eyelids held wide open- Summon the help of a ophthalmologist- Do not use a chemical neutralising agent4.3 After inhalation : - Let the victim breathe fresh air4.4 After swallowing : - Rinse the mouth with plenty of water- Make the victim drink plenty of water- Try to prevent vomiting of the victim- Do not give a chemical antidote- Summon medical help immediately- Ingestion of large quantities: transport to hospital immediately- Show the package / vomit to the doctor / in the hospital5. Fire-fighting measures5.1 Suitable extinguishing media : -In adaptation to materials stored in the immediate neighbourhood5.2 Special risks: -Development of hazardous combustion gases or vapours is possiblein the event of fire5.3 Special protecting clothing : -Do not stay in the danger zone without suitable chemical protectionclothing and self contained breathing apparatus5.4 Other information : - Contain escaping vapours with water- Prevent fire-fighting water from entering surface water or groundwaterSAFETY DATA SHEET IN ACCORDANCE WITH 91/155/EEC Page 2 of 4DATASHEET RENEGITE6. Accidental release measures6.1 Person related precautions : -Avoid generation of dust-Do not inhale dust- Avoid substance contact6.2 Procedures for cleaning /absorption : -Take up dry-Forward for disposal-Clean up affected area with plenty of water-Clean the clothing and material used afterward6.3 Environment : - Prevent the substance from entering in the environment7. Handling and storage7.1 Handling : -No additional requirements7.2 Storage : -Store in a dry place- Store in tightly closed container-Storage temperature at +15° C to 25° C- The data on storage temperature applies to the entire pack7.3 Keep away from : -Heat sources- oxidising agents- (strong) bases- easy combustible materials- metals- halogens8. Personal Protection / exposure protection8.1 Personal protective equipment:Respiratory protection : -Required when dusts are generatedEye protection : -RequiredHand protection : -RequiredProtective clothing should be selected specifically for the working place, depending on concentration and quantity of the hazardous substance handled. The resistance of the protective clothing to chemicals should be ascertained with the respective supplier.8.2 Industrial hygiene : -Change contaminated clothing-Application of skin-protective barrier cream is recommended-Wash hands after working with the substance9. Physical and chemical propertiesForm : - SolidColour : - WhiteOdour : - OdourlessMelting temperature : - 205°C (decomposition)Boiling temperature : - Not availableDensity : - 2130 kg/m3Bulk density : - ~600 kg/m3Solubility in water : - 213 g/l (at 20° C)- 470 g/l (at 80° C)pH : - 1.18 (1 g/100 ml water at 25° C)Thermal decomposition : - 209 ° CSAFETY DATA SHEET IN ACCORDANCE WITH 91/155/EEC Page 3 of 4DATASHEET RENEGITE10 Stability and reactivity10.1 Conditions to be avoided : - Strong heating10.2 Substances to be avoided : - Halogens- Alkalis- Oxidising agents (i.e. Nitrates, Nitrites, Nitric acids)- Metals with water10.3 Hazardous decompositionproducts: - In the event of fire: Ammonia, Nitrous gases, Sulphur oxides11. Toxicological information11.1 Acute Toxicity : - LD50 (oral rat): 3160 mg/kg11.2 Specific symptoms withanimal tests : - Eye irritation test: strong irritant effect - Skin irritation test: strong irritant effect11.3 Further toxicologicalinformation : - After inhalation of dust: irritation in the respiratory tract, coughing, dyspnoea - After skin contact: irritant effect- After eye contact: strong irritant effect- After swallowing: irritation of mucous membranes n the mouth, pharynx, oesophagus and gastrointestinal tract11.4 Over sensitiveness : - No sensitising effect11.5 Further information : - The product should be handled with the care usual when dealing withchemicals12. Ecological information12.1 Ecotoxic effects : - Quantitative data on the ecological effects of this product are not available 12.2 Biological effects : - Harmful effect on aquatic organisms- Harmful effect due to pH-shift12.3 Further ecological data : - May cause long-term adverse effects in aquatic environments- Do not allow to enter waters, waste water or soil- When handled and used competently no ecological problems are tobe expected13. Disposal considerations13.1 Product : - There are no uniform EC-regulations for the disposal of chemicals orresidues. Chemical residues in general count as special waste, thedisposal of which is regulated in the EC member states throughharmonised laws and regulations. We recommend that the authoritiesin charge or an approved disposal company is contacted to advice onhow to dispose of this special waste.13.2 Packaging : - Handle contaminated packaging in the same way as the substance itself- Disposal in compliance with the official regulations- If not specified differently, non contaminated packaging may betreated like household waste or re-cycledSAFETY DATA SHEET IN ACCORDANCE WITH 91/155/EEC Page 4 of 4DATASHEET RENEGITE14. Transport information14.1 Transport over land ADR/RID and GGVS/GGVE (Germany)VS/GGVE class : - 8 Number and letter : - 16cGG ADR/RID class : - 8 Number and letter : - 16cMarking : - 2967 Sulfaminsäure14.2 Transport by inland vessel ADN/ADNRNo data available14.3 Transport over sea IMDG/GGVS (sea)IMDG/GGVS - class : - 8 UN - number : - 2967EMS : - 8-08 MFAG : - 700Correct technical marking : - SULPHAMIC ACID Packaging group : - III14.4 Transport by air ICAO-TI en IATA-DGRICAO/IATA - class : - 8 UN- / ID- number : - 2967Correct technical name : - SULPHAMIC ACID Packaging group : - III14.5 The transport regulations are cited according to international regulations and in the form applicable inGermany (GGVS/GGVE). Possible national deviations in other countries are not considered.15. Regulatory information15.1 Hazard symbol : - Xi - Irritating15.2 R-phrases : - 36/38 - Irritating to eyes and skin- 52/53 - Harmful to aquatic organisms, may cause long-termadverse effects in the aquatic environment15.3 S-phrases : - (2) - (Keep out of reach of children)26 In case of contact with eyes, rinse immediately with plentyof water and seek medical advice- 28 - After contact with the skin, wash immediately with plenty ofwater- 61 - Avoid release to the environment. Refer to specialinstructions/ safety data sheets15.4 NOTE :15.5 EEC LIST : - EC no. 226-218-8 (EC label)15.6 German regulationsWater polution class : - 1 (slightly polluting substance) (own classification)16. Additional informationDate of issue : - 06 March 2002The information contained herein is based on the present state of our knowledge. It characterises the product with regard to the appropriate safety precautions. It does not represent a guarantee of theproperties of the product. The receiver and the users of the product are responsible for a proper use in conformity with the existing legislation and regulations.。
211126681_基于光谱法的特级初榨橄榄油快速鉴伪技术

唐聪,邵士俊,温玉洁,等. 基于光谱法的特级初榨橄榄油快速鉴伪技术[J]. 食品工业科技,2023,44(9):309−316. doi:10.13386/j.issn1002-0306.2022060131TANG Cong, SHAO Shijun, WEN Yujie, et al. Rapid Identification of Extra Virgin Olive Oil by Spectrometry[J]. Science and Technology of Food Industry, 2023, 44(9): 309−316. (in Chinese with English abstract). doi: 10.13386/j.issn1002-0306.2022060131· 分析检测 ·基于光谱法的特级初榨橄榄油快速鉴伪技术唐 聪1,2,邵士俊2, *,温玉洁1,2,梁 卿2,董树清2,*(1.中国科学院兰州化学物理研究所,中国科学院西北植物资源化学重点实验室,甘肃省天然药物重点实验室,甘肃兰州 730000;2.中国科学院大学,北京 100049)摘 要:本文研究了特级初榨橄榄油中掺入不同比例橄榄果榨油(精炼橄榄油)、菜籽油、玉米油和大豆油的光谱特征,采用荧光光谱和紫外光谱,对掺假样品及纯油样品进行了快速检测。
结果表明,特级初榨橄榄油的光谱特征与其他植物油之间差异较大,且掺假体积与吸光度之间存在良好的线性关系(R 2>0.89),实现了特级初榨橄榄油的定性鉴别与定量检测,建立了特级初榨橄榄油质量控制体系及其掺假检测分析技术,最低检出限为1%,线性范围为5%~100%(v/v )。
系统聚类分析将所有特级初榨橄榄油准确地分为一个亚类,也佐证了此方法的稳定性与可靠性。
这种简单快捷的检测技术,有助于特级初榨橄榄油实时、在线橄榄油检测分析技术的研发,为我国橄榄油品质鉴定及产业发展提供有利的技术保障。
SNP基因型分析
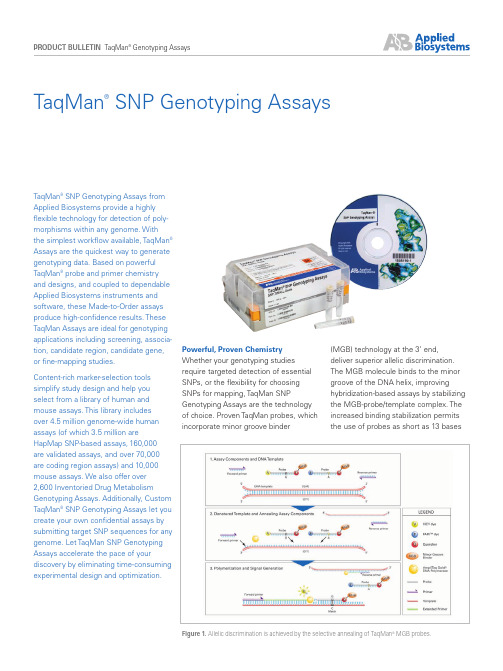
Powerful, Proven Chemistry Whether your genotyping studies require targeted detection of essentialSNPs, or the flexibility for choosingSNPs for mapping, TaqMan SNPGenotyping Assays are the technologyof choice. Proven TaqMan probes, whichincorporate minor groove binder(MGB) technology at the 3’ end,deliver superior allelic discrimination. The MGB molecule binds to the minor groove of the DNA helix, improvinghybridization-based assays by stabilizing the MGB-probe/template complex. The increased binding stabilization permits the use of probes as short as 13 basesTaqMan ®SNP Genotyping AssaysTaqMan ® SNP Genotyping Assays from Applied Biosystems provide a highly flexible technology for detection of poly-morphisms within any genome. With the simplest workflow available, TaqMan ® Assays are the quickest way to generate genotyping data. Based on powerful TaqMan ® probe and primer chemistry and designs, and coupled to dependable Applied Biosystems instruments and software, these Made-to-Order assays produce high-confidence results. These TaqMan Assays are ideal for genotyping applications including screening, associa-tion, candidate region, candidate gene,or fine-mapping studies.Content-rich marker-selection toolssimplify study design and help youselect from a library of human andmouse assays. This library includesover 4.5 million genome-wide humanassays (of which 3.5 million areHapMap SNP-based assays, 160,000 are validated assays, and over 70,000 are coding region assays) and 10,000 mouse assays. We also offer over 2,600 Inventoried Drug MetabolismGenotyping Assays. Additionally, Custom TaqMan ® SNP Genotyping Assays let you create your own confidential assays by submitting target SNP sequences for any genome. Let TaqMan SNP Genotyping Assays accelerate the pace of yourdiscovery by eliminating time-consuming experimental design and optimization.Figure 1. Allelic discrimination is achieved by the selective annealing of TaqMan MGB probes.Visit for more information.Over 10,000 mouse SnP Genotyping assaysThe Mouse TaqMan ® Pre-Designed SNP Genotyping Assays collection consists of over 10,000 assays, and can also be supplemented with assays designed using our Custom TaqMan ® SNP Genotyping Assays Service.T aqman ® Drug metabolism Genotyping assays Collection Over 2,600 assays that target high value polymorphisms in over 220 drug metabolism genes. These assays have proven performance in four different ethnic populations, comprised of 45 individuals each. To enable easyidentification, these assays have been mapped to the common public allele nomenclature Web sites where possible. Visit for more information.for improved mismatch discrimination and greater flexibility when designing assays for difficult or variable sequences. In addition to SNP detection, TaqMan probes can be designed to detect multiple nucleotide polymorphisms (MNPs) and insertion/deletions (InDels).Detection is achieved with proven 5’ nuc- lease chemistry by means of exonuclease cleavage of a 5’ allele-specific dye label, which generates the permanent assay signal (Figure 1). All MGB probes include a nonfluorescent quencher (NFQ) that virtually eliminates the backgroundfluorescence associated with traditional quenchers and provides a greater signal- to-noise ratio for superior assay sensitivity.T aqman ® SnP Genotyping assays CollectionTaqMan SNP Genotyping Assays are the world’s largest collection of single-tube, ready-to-use SNP assays available.The TaqMan SNP Genotyping Assays library consists of two human and one mouse assay collections, and can be supplemented with assays designed using our Custom TaqMan ® SNP Genotyping Assays Service (Table 1).Over 4.5 million Human SnP Genotyping assaysThis assay group contains over 4.5 million genome-wide SNPs providing unprecedented marker coverage.Included in this collection are 160,000 validated assays that have roughly 10-kb spacing across gene regions.These assays were subjected to a minor allele frequency test in 2–4 ethnic populations (45 individual samples per ethnic group) and offer the highest success rate because of this extensive testing. Over 70,000 assays are also included for the detection of nonsyn-onymous SNPs in coding regions, includ-ing many putative functional SNPs.Human assays Non-human Number of Reactions at Reactions at Assay mix Availabilitypart numbers assays part SNPs 5 μL volume 25 μL volume formulationnumbers (384-well plate) (96-well plate)TaqMan ® Pre-Designed SNP Genotyping Assays for Human and Mouse Small-scale 4351379 4351384* >4,500,000 1,500 300 40X Made-to-Order Medium-scale 4351376 4351382* >4,500,000 5,000 1,000 40X Made-to-Order Large-scale43513744351380*>4,500,000 12,0002,40080XMade-to-OrderTaqMan ® Drug Metabolism SNP Genotyping Assays Small-scale4362691N/A >2,600 750 150 20X InventoriedCustom TaqMan ® SNP Genotyping Assays Small-scale 4331349 4332077 ∞ 1,500 300 40X Made-to-Order Medium-scale 4332072 4332075 ∞ 5,000 1,000 40X Made-to-Order Large-scale43320734332076∞12,0002,40080XMade-to-OrderAll assays are quality control tested using a mass spectrophotometer to verify sequence and yield. All assays have one VIC ® and one FAM ™ dye-labeled probe and two target-specific primers. All assays, except Custom Assays, undergo bioinformatics evaluation of target SNP sequences.Functional testing against 20 unique genomic DNAs is performed on all human SNP Genotyping Assays. Validation testing against three populations with ~45 samples/population was performed on all Validated and Drug Metabolism Assays.All Validated and Coding Assays that were previously held in inventory are now available as Made-to-Order Pre-Designed Assays. Their assay IDs remain unchanged.*Over 10,000 mouse assays available.All TaqMan SNP Genotyping Assays were generated using next-generation algorithms from the Applied Biosystems bioinformatics pipeline. Bioinformatics evaluation of target SNP sequences includes the masking of adjacent SNPs and ambiguous bases so that assay design and subsequent performanceis not affected by underlying sequence quality. Lastly, the assay designs are aligned to the human genome using BLAST to ensure the assay is uniquely binding and targets only the intended polymorphism. As the Custom TaqMan SNP Genotyping Assay Service is a confidential and secure service, custom-ers perform their own bioinformatics analysis prior to submission of sequence for assay design.Free marker Selection T ools SNPbrowser™ Software for Human SNPs SNPbrowser™ Software for human SNPs simplifies study design by facilitating easy and intuitive selection of the optimal SNP assay set for each project. Y ou can download free SNPbrowser Software at /snpbrowser SNPbrowser Software provides a physical map view of the human genome.Two different genotype data sources are incorporated into the software togenerate unique linkage disequilibrium (LD) maps and haplotype block informa-tion. These two sources are the public HapMap Project and Applied Biosystems’ 20+ million genotypes generated during the development of the 160,000 TaqMan Validated SNP Genotyping Assays.This genotyping data also facilitates the use of tagging SNP methods, which allow more affordable study design by reducing the number of SNPs while conserving study statistical power (Figure 2). SNPbrowser Software allows you to optimize study design based on LD patterns, minor allele frequency require-ments, total number of cases and controls, and statistical power. Finally, the software includes a SNP density tool that prioritizes the selection of validated SNPs while supplementing with additional SNPs to fulfill the markerdensity requirements for your study.When you have identified your SNPsof interest, order TaqMan SNPGenotyping Assays by directly uploadingAssay IDs from the software to theApplied Biosystems Web site by goingto Mouse SNPbrowser™ SoftwareComplimentary Mouse SNPbrowser™Software enables efficient and easyselection of appropriate SNP setsto discriminate between strains ofinterest, with specific applications ingenotype/phenotype mapping and strainverification. It visualizes informativeSNPs that distinguish two selectedmouse strains at a user-definableresolution. There are ~10,000 mouseSNPs genotyped in 44 of the mostcommon strains.Mouse SNPbrowser selects fromPre-Designed SNP Genotyping assaysusing either TaqMan® Assay or SNPlex™Genotyping System Assay collectionsavailable from the Applied Biosystemsonline store. Y ou can download freeMouse SNPbrowser Software from/mousesnpbrowserMouse SNPbrowser Software supportsgenetic monitoring, genetic mapping,and speed congenics applications byoffering a variety of tools to search, view,export SNP information, and purchaseassays. The software displays the mousechromosome map so users can viewSNP location and obtain National Centerfor Biotechnology Information (NCBI)data for SNPs of interest. The softwareallows SNP selection for two applications:genetic monitoring and genetic mapping. Figure 2. Identifying Optimal Subsets of SNPs with the SNP Wizard. (A) Search and visualize your gene of interest, then activate the SNP Wizard. (B) Choose a tagging SNP method and set parameters to reducethe number of SNPs needed to represent common haplotypes. A summary panel appears that displays algorithm results and allows you to easily adjust your parameter selection. Selected tagging SNPs are shownin red on the graphical map.A.B.For genetic monitoring, the software selects optimal SNPs to distinguish between any number of strains. SNPs can be selected for each chromosome or across the genome. For genetic mapping, the software segments each chromosome into evenly sized blocks based on user-specified mapping resolu-tion, and searches for distinguishing SNP in each block to generate evenly-spaced SNP coverage throughout the genome. Mouse SNPbrowser Software also features a SNP wizard to assist defining search criteria. A help text file is provided within the tool to assist in familiarizing users of the extensive functionality offered.Custom assay Service for any Possible SnPCustom TaqMan SNP Genotyping Assays are available for any possible SNP in any organism. This service can generate assays for the detection of SNPs, MNPs, or insertions/deletions of up to six bases. Custom TaqMan SNP Genotyping Assays provide customers with a complete service that includes secure and confidential ordering, assay design and manufacturing, quality-control testingfor synthesis accuracy and formulation completeness. Additionally, custom human assays are subjected to a func-tional test on 20 unique DNA samples. Our complimentary File Builder Software makes it easy to submit your targetsfor design through our custom assay service. Download the latest versionof File Builder Software at/filebuilder Quality Design and manufacturing Probes and primers used in TaqMan SNP Genotyping Assays are designed byour rigorous bioinformatics pipeline. This proprietary group of algorithms has generated millions of TaqMan Assay designs by utilizing heuristic design rules deduced from both manufacturing and assay performance data. All assays are designed to perform under universal reaction conditions, as calculated probe and primer melting temperaturesare consistent and include contributionsfrom associated probe conjugates(i.e., dyes, MGB).After manufacturing, assay componentsundergo extensive laboratory testing atour state-of-the-art manufacturing facility(Figure 3). Quality-control testing includesmass spectrometry for sequenceFigure 4.A simple workflow and reliable instruments combine to generate fast, high-confidence results. Figure 3. Applied Biosystems high-throughput manufacturing facility.verification and formulation assessments of probe and primer concentrations. Additionally, we functionally test all human SNP genotyping assays with an allelic discrimination test.Simple Workflow for Quick Results TaqMan SNP Genotyping Assaysconstitute the simplest SNP genotyping technology available. Applied Biosystems delivers your ready-to-use SNP geno-typing assay in a convenient, single-tube format. The rest is easy. Just combine the assay with TaqMan ® Genotyping Master Mix or TaqMan ® Universal PCR Master Mix and your purified DNA sample. There is no need to optimize temperature or concentrations of probes, primers, or salts, because all assays run under universal reagent concentrations and thermal cycling conditions.After thermal cycling, no transfers, wash-es, or addition of reagents is required, and the plate remains sealed; just read the plate and analyze the genotypes (Figure 5). This reduces the chance of contamination, sample mix-up, and sample loss. Because of the simplicity in chemistry, you can easily automate the reaction for massively parallel genotyping studies, readily increasing the number of assays, number of samples, or both. Additionally, the analysis software allows you to auto-call genotypes, minimizing manual intervention.Reliable Real-Time PCR Platforms Applied Biosystems offers a suite of superior instrument platforms for processing and analyzing TaqMan SNP Genotyping Assays. These instruments, which meet all throughput needs and budgets, include the GeneAmp ® PCR System 9700 and Veriti ™ Thermal Cyclers and the Applied Biosystems 7300, 7500, 7500 Fast, 7900HT Fast, StepOne ™ or StepOnePlus ™ Real-Time PCR Systems. Following PCR amplification, an endpoint read can be performed on any Applied Biosystems Real-Time PCR System. All of these dependable instruments offer the advanced multicolor detection capabilities required for highly accurate and reproduc-ible allelic discrimination assays.Figure 5.The SNP auto-caller feature automatically determines sample genotypes and displays data.Figure 6. The flexible, high-throughput Applied Biosystems 7900HT Fast Real-Time PCR System.Table 2. InSTRumenT CaPaCITyInstrument PlatformsCapacityApplied Biosystems GeneAmp ® 60-, 96-, Dual 96-, or Dual 384-well PCR System 9700 Thermal Cycler blocksApplied Biosystems Veriti ™ Thermal Cycler 60-, 96- (Standard or Fast),or 384-well block Applied Biosystems 7300 Real-Time PCR System 96-well blockApplied Biosystems 7500/7500 Fast 96-well block (Standard and Fast) Real-Time PCR SystemApplied Biosystems 7900HT Fast 384-well block (Standard and Fast) Real-Time PCR System Applied Biosystems StepOne ™ 48-well block (Standard and Fast) Real-Time PCR SystemApplied Biosystems StepOnePlus ™ 96-well block (Standard and Fast)Real-Time PCR SystemHeadquarters850 Lincoln Centre Drive | Foster City, CA 94404 USA Phone 650.638.5800 | Toll Free 800.345.5224 International SalesFor our office locations please call the division headquarters or refer to our Web site at/about/offices.cfmFor Research Use Only. Not for use in diagnostic procedures. NOTICE TO PURCHASER: LIMITED LICENSEA license to perform the patented 5’ Nuclease Process for research is obtained by the purchase of (i) both Licensed Probe and Authorized 5’ Nuclease Core Kit, (ii) a Licensed 5’ Nuclease Kit, or (iii) license rights from Applied Biosystems.TaqMan SNP Genotyping Assays contain Licensed Probes. Use of these products is covered by one or more of the following US patents and corresponding patent claims outside the US: 5,538,848, 5,723,591, 5,876,930, 6,030,787, 6,258,569, and 5,804,375 (claims 1-12 only). The purchase of these products includes a limited, non-transferable immunity from suit under the foregoing patent claims for using only the amount of product specified for the purchaser’s own internal research. Separate purchase of an Authorized 5’ Nuclease Core Kit would convey rights under the applicable claims of US Patents Nos. 5,210,015 and 5,487,972, and corresponding patent claims outside the United States, which claim 5’ nuclease methods. No right under any other patent claim and no right to perform commercial services of any kind, including without limitation reporting the results of purchaser’s activities for a fee or other commercial consideration, is conveyed expressly, by implication, or by estoppel. These products are for research use only. Diagnostic uses under Roche patents require a separate license from Roche. Further information on purchasing licenses may be obtained from the Director of Licensing, Applied Biosystems, 850 Lincoln Centre Drive, Foster City, California 94404, USA.© 2008. Applied Biosystems. All Rights Reserved. Information subject to change without notice. Applera, Applied Biosystems, AB (Design), GeneAmp, and VIC are registered trademarks and FAM, StepOne, StepOnePlus, and Veriti are trademarks of Applera Corporation in the US and/or certain other countries.AmpliTaq Gold and TaqMan are registered trademarks of Roche Molecular Systems, Inc.Printed in the USA. 06/2008 Publication 135PB03-02Data analysis SoftwareThe sophisticated software package provided with all Applied Biosystems’ Real-Time PCR Systems facilitates experimental set-up, data collection, and assay performance analysis. The SDS software uses an advanced multi-component algorithm to calculate distinct allele/marker signal contributions from fluorescence measurements for each sample well during the assay plate read. The SNP auto-caller feature automatically determines samplegenotypes and generates a cluster plotFigure 7. The Applied Biosystems Web site ( ) facilitates convenient online ordering and multiple search options for all our genotyping assays, including keyword, batch, and location searches.that helps you visualize data across samples or populations (Figure 5).For large-scale studies, you can cycle samples on multiple Dual 384-Well GeneAmp ® 9700 or Veriti ™ Thermal Cyclers and then analyze them on a 7900HT System. The 7900HT System (Figure 6) facilitates high-throughput applications by allowing large sample batches (up to eighty-four 384-well plates) to be processed withoutmanual intervention using the optional Automation Accessory. Following data collection, you can accelerate andsimplify high volume data transfer and analysis with the optional SDS version 2.2 Enterprise Edition Software Suite.Ordering InformationSelecting and ordering TaqMan SNP Genotyping Assays is as simple as “point and click.” Use SNPbrowserSoftware to select the most informative SNPs for your genotyping studies. As you identify SNPs of interest, simply upload your selected TaqMan SNP Genotyping Assays to the Applied Biosystems Web site for ordering.The Applied Biosystems Web site (Figure 7) enables you to search for, select, and order from our catalog of over 4.5 million TaqMan SNP Genotyping Assays. Y ou can search for SNPs using several criteria options: National Center for Biotechnology Information (NCBI) gene ID, NCBI SNP reference ID (rs#) and gene symbol. Y ou can further refine your search by using SNP type (i.e., intragenic, 5’ or 3’ UTR, chromosome, etc.).Our Custom TaqMan SNP Genotyping Assays Service is provided to supply you with any SNP not available from our catalog, including any non-humanorganism. This service designs assays for all possible SNP , MNP , and InDeltargets. Our complementary File Builder Software conveniently formats your target sequence for submission to our manufacturing facilities. To order any custom assays, simply upload yoursubmission file to our Web site, or emailthe file to your local sales office.。
专题15-阅读之词义猜测题(原卷版)-备战2022年新高考英语一轮复习考点一遍过

考点15 阅读之词义猜测(原卷版)【命题趋势】词义猜测题型是高考中必考题型,每年考察1-2题。
所以词义猜测题型也是考生需要复习的考点。
词义猜测,即阅读中的完形填空。
可以通过前后句子中的关键词寻找相对应的词。
只要找到方法,拿下词义猜测轻而易举。
当然,着需要我们掌握足够的词汇量才能游刃有余!【重要考向】一、了解词义猜测题型的基本设题方式和基本提示词;二、掌握词义猜测解题技巧;考向一【2021年新高考全国卷Ⅰ阅读理解C篇】When the explorers first set foot upon the continent of North America, the skies and lands were alive with an astonishing variety of wildlife. Native Americans had taken care of these precious natural resources wisely. Unfortunately, it took the explorers and the settlers who followed only a few decades to decimate a large part of these resources. Millions of waterfowl ( 水禽) were killed at the hands of market hunters and a handful of overly ambitious sportsmen. Millions of acres of wetlands were dried to feed and house the ever-increasing populations, greatly reducing waterfowl habitat.29. What does the underlined word “decimate” mean in the first paragraph?A. Acquire.B. Export.C. Destroy.D. Distribute.了解词义猜测题型的基本设题方式(1) 代词类猜词题The underlined word "them" in Paragraph 2 refers to_______.The underlined word “it" in Para. 4 refers to _______.(2) 其他类猜测题What does the word X (in Paragraph X) mean?What does the underlined word X mean?What can we learn from the underlined sentence?The underlined word X probably means _______.考向二【2021年全国乙卷阅读理解C篇】Still, 55 percent of Australians have a landline phone at home and only just over a quarter (29%)rely only on their smartphones according to a survey (调查). Of those Australians who still have a landline, a third concede that it’s not really necessary and they’re keeping it as a security blanket — 19 percent say they never use it while a further 13 percent keep it in case of emergencies. I think my home falls into that category.25. What does the underlined word “concede” in paragraph 3 mean?A. Admit.B. Argue.C. Remember.D. Remark.1. 代词类猜词题做题方法第一步:优先看划线句第二步:前指向性第三步:代词单复数线索2. 其他类猜词题做题方法根据构词法:通过转化、合成、词根词缀等词根词缀常见前缀:前缀作用:通常前缀一般改变单词的意义,不改变词性常见后缀:后缀作用:通常会改变单词的词性,构成意义相近的其他词性(3) 因果法—— 根据因果关系常见表示因果的词汇:because, for, as, since, for the reason that, considering that (4) 并列法—— 根据并列顺承关系常见的并列词:and, as well as, not only…but also…, together/along with, when/while (5) 转折法——根据转折关系常见表示转折的词汇:while, not, but, however, yet, otherwise, though, despite, in spite of , in contrast, instead of, rather than1.【2021年浙江卷1月阅读理解C篇】“The vagueness of the gesture meanings suggests either that the chimps have little to communicate, or we are still missing a lot of the information contained in their gestures and actions,” she said. “Moreover, the meanings seem to not go beyond what other animals convey with non-verbal communication. So, it seems the gulf remains.”29. What does the underlined word “gulf” in the last paragraph mean?A. Difference.B. Conflict.C. Balance.D. Connection.2.【2021年全国甲卷阅读理解C篇】Southbank, at an eastern bend in the Thames, is the center of British skateboarding, where the continuous crashing of skateboards left your head ringing .I loved it. I soon made friends with the local skaters. We spoke our own language. And my favorite: Safe. Safe meant cool. It meant hello. It meant don't worry about it. Once, when trying a certain trick on the beam(横杆), I fell onto the stones, damaging a nerve in my hand, and Toby came over, helping me up: Safe, man. Safe. A few minutes later, when I landed the trick, my friends beat their boards loud, shouting: “Safe! Safe! Safe!” And that's what mattered—landing tricks, being a good skater.9. What do the underlined words “Safe! Safe! Safe!” probably mean?A. Be careful!B. Well done!C. No way!D. Don't worry!3.【2021年6月浙江卷阅读理解A篇】In the past few years, an increasing number of people and organisations have begun coming up with plans to counter this trend. A couple of years ago, film-maker David Bond realised that his children, then aged five and three, were attached to screens to the point where he was able to say "chocolate" into his three-year-old son's ear without getting a response. He realised that something needed to change, and, being a London media type, appointed himself"marketing director for Nature". He documented his journey as he set about treating nature as a brand to be marketed to young people. The result was Project Wild Thing, a film which charts the birth of the Wild Network a group of organisations with the common goal of getting children out into nature.26. Which of the following can replace the underlined word "charts" in paragraph 2A. recordsB. predictsC. delaysD. confirms4.【2021天津卷3月阅读理解B篇】The poignancy of Jordan retiring from his beloved basketball to play baseball and what had pushed him to make such a tough decision took me by surprise. As I watched him take off his basketball uniform and replace it with a baseball uniform, I saw him leaving behind the layer that no longer served him, just as our lizard had. Neither of them chose the moment that had transformed them. But they had to live with who they were after everything was different. Just like us. I realized that we have to learn to leave the past behind.43. The underlined part "leaving behind the layer" in Paragraph 8 can be understood as .A. letting go of the pastB. looking for a new jobC. getting rid of a bad habitD. giving up an opportunity5.【2020全国卷Ⅰ阅读理解B篇】While I tend to buy a lot of books, these three were given to me as gifts, which might add to the meaning I attach to them. But I imagine that, while money is indeed wonderful and necessary, rereading an author’s work is the highest currency a reader can pay them. The best books are the ones that open further as time passes. But remember, it’s you that has to grow and read and reread in order to better understand your friends.26. What does the underlined word "currency"in paragraph 4 refer to?A. Debt.B. Reward.C. Allowance.D. Face value.6. 【2020全国卷Ⅱ阅读理解C篇】Nutria were brought there from Argentina by fur farmers and let go into the wild. “The ecosystem down there can’t handle this non-native species(物种).It’s destroying the environment.It’s them or us." says Michael Massimi, an expert in this field.The fur trade kept nutria check for decades,but when the market for nutria collapsed in the late 1980s,the cat-sized animals multiplied like crazy.30. What does the underlined word “collapsed” in paragraph 5 probably mean?A. Boomed.B. Became mature.C. Remained stable.D. Crashed.7. 【2020全国卷Ⅱ阅读理解D篇】As I grew older and became a mother, the library took on a new place and an added meaning in my life. I had several children and books were our main source(来源) of entertainment. It was a big deal for us to load up and go to the local library, where my kids could pick out books to read or books they wanted me to read to them.33. What does the underlined phrase “an added meaning” in paragraph 3 refer to?A. Pleasure from working in the library.B. Joy of reading passed on in the family.C. Wonderment from acting out the stories.D. A closer bond developed with the readers.8. 【2020全国卷III阅读理解B篇】Yet "Apes" is more exception than the rule. In fact, Hollywood has been hot on live animals lately. One nonprofit organization, which monitors the treatment or animals in filmed entertainment, is keeping tabs on more than 2,000 productions this year. Already, a number of films, including "Water for Elephants," "The Hangover Part Ⅱ" and "Zookeeper," have drawn the anger of activists who say the creatures acting in them haven’t been treated properly.26. What does the underlined phrase "keeping tabs on" in paragraph 3 probably mean?A. Listing completely.B. Directing professionally.C. Promoting successfully.D. Watching carefully.9. 【2020全国新高考卷阅读理解C篇】His visit, however, ended up involving a lot more than that. Hence this book, Chasing the Sea: Lost Among the Ghosts of Empire in Central Asia, which talks about a road trip from Tashkent to Karakalpakstan, where millions of lives have been destroyed by the slow drying up of the sea. It is the story of an American travelling to a strange land, and of the people he meets on his way: Rustam, his translator, a lovely 24-year-old who picked up his colorful English in California, Oleg and Natasha, his hosts in Tashkent, and a string of foreign aid workers.9. What does the underlined word “that” in paragraph 2 refer to? ()A. Developing a serious mental disease.B. Taking a guided tour in Central Asia.C. Working as a volunteer in Uzbekistan.D. Writing an article about the Aral Sea.10. 【2020全国新高考卷阅读理解D篇】According to a recent study in the Journal of Consumer Research, both the size and consumption habits of our eating companions can influence our food intake. And contrary to existing research that says you should avoid eating with heavier people who order large portions(份), it's the beanpoles with big appetites you really need to avoid.13. What does the underlined word "beanpoles" in paragraph 1 refer to? ()A. Big eaters.B. Overweight persons.C. Picky eaters.D. Tall thin persons.11.【2019全国卷Ⅰ阅读理解B篇】But he’s nervous."I’m here to tell you today why you should … should…"Chris trips on the"-ld,"a pronunciation difficulty for many non-native English speakers. His teacher, Thomas Whaley, is next to him, whispering support."…Vo te for …me…"Except for some stumbles, Chris is doing amazingly well. When he brings his speech to a nice conclusion, Whaley invites the rest of the class to praise him.25. What does the underlined word "stumbles"in paragraph 2 refer to?A. Improper pauses.B. Bad manners.C. Spelling mistakes.D. Silly jokes.12.【2019全国卷Ⅱ阅读理解B篇】I guess that there's probably some demanding work schedule, or social anxiety around stepping up to help for an unknown sport. She may just need a little persuading. So I try again and tug at the heartstrings. I mention the single parent with four kids run ning the show and I talk about the dad coaching a team that his kids aren’t even on… At this point the unwilling parent speaks up, "Alright. Yes, I’ll do it."25. What does the underlined phrase "tug at the heartstrings" in paragraph 2 mean?A. Encourage team work.B. Appeal to feelings.C. Promote good deeds.D. Provide advice.13.【2019全国卷Ⅲ阅读理解B篇】"China is impossible to overlook," says Hill. "Chinese models are the faces of beauty and fashion campaigns that sell dreams to women all over the world, which means Chinese women are not just consumers of fashion —they are central to its movement. "Of course, only are today's top Western designers being influenced by China —some of the best designers of contemporary fashion are themselves Chinese." Vera Wang, Alexander Wang, Jason Wu are taking on Galiano, Albaz, Marc Jacobs-and beating them hands down in design and sales," adds Hill.26.What do the underlined words "taking on" in paragraph 4 mean?A. learning fromB. looking down onC. working withD. competing against14.【2019天津卷阅读理解D篇】The book turns out to be one that has appealed to the world for more than 350 years. That former prisoner was Cervantes, and the book was Don Quixote(《堂吉诃德》). And the story poses an interesting question: why do some people discover new vitality and creativity to the end of their days, while others go to seed long before?We’ve all known people who run out of steam before they reach life’s halfway mark. I’m not talking about those who fail to get to the top. We can’t all get there. I’m talking about people who have stopped learning on growing because they have adopted the fixed attitudes and opinions that all too often come with passing years.52. What does the underlined part in Paragraph 3 probably mean?A. End one’s struggle for liberty.B. Waste one’s energy taking risks.C. Miss the opportunity to succeed.D. Lose the interest to continue learning.1.【广东省广州市2021届高三一模】Nearly two hundred years later, Leeuwenhoek's discovery of microbes helped French chemist and biologist Louis Pasteur to develop his “theory of disease”. This concept suggested that disease originates from tiny organisms attacking and weakening the body. Pasteur’s theory later helped doctors to fight infectious diseases including anthrax, diphtheria, polio, smallpox, tetanus, and typhoid. All these breakthroughs were the result of Leeuwenhoek's original work. Leeuwenhoek did not foresee this legacy.5.The underlined phrase “this legacy” in paragraph 3 refers to_______.A.the discovery of microbes.B.Pasteur's theory of disease.C.Leeuwenhoek's contribution.D.the origin of the tiny organism.2.【广东省深圳市2021届高三调研】By the time I arrived, the sailing club had grown. At weekends, we would work on her all day, cleaning and painting her, and collapse into sleeping bags on the floor at night. Then came the big day when a school sailing trip was announced. Its destination was Saint-Malo. But it also meant that we had to refit the ship by removing every piece of ballast(压舱物)and replacing the bilge water(底舱污水)before we were allowed to sail.And I did learn the ropes on that school trip. I learned how to operate a ship in the high waves. I learned the meaning of teamwork and became a better team player. I also learned that the best fish is the one that you have caught and cooked yourself from a boat that you have rebuilt and sailed yourself.7.What does the underlined phrase “learn the ropes” in paragraph 4 mean?A.Learn to cook.B.Master useful skills.C.Ensure sailing safety.D.Use ropes properly.3.【广东湛江一中2021届高三模拟】On achieving success:“When you make a choice and say, ‘Come hell or high water, I am going to be this,’ then you should not be surprised when you are that. It should not be something that is intoxicating or out of character because you have seen this moment for so long that ... when that moment comes, of course it is here because it has been here the whole time, because it has been [in your mind] the whole time.”On failure:“I don’t mean to sound cavalier when I say that, but never. It’s basketball. I’ve practiced and practiced and played so many times. There’s nothing truly to be afraid of, when you think about it ... Because I’ve failed before, and I woke up the next morning, and I’m OK. People say bad things about you in the paper on Monday, and then on Wednesday, you're the greatest thing since sliced bread. I’ve seen that cycle, so why would I be nervous about it happening?”5.The word “cavalier” in the 3rd paragraph probably means __________.A.not serious or caring B.anxious and eagerC.worried and pessimistic D.not proud or arrogant4.【河北省石家庄市2021届高三质检】“This must be a simply vast wardrobe!” thought Lucy, going still further in. Then she noticed that there was something crunching under her feet. “Is that more mothball?” she thought, stooping down to feel it with her hand. But instead of feeling the hard, smooth wood, she felt something soft and powdery and extremely cold. “This is very queer,” she said, and went on a step or two further.7.What does the underlined word “queer” mean?A.Terrifying.B.Empty.C.Strange.D.Impressive.5.【湖南师范大学附中2021届高三模拟】In two-and-a-half years, the pair still have to replenish their incomes with some writing and editing, but the business is growing and within five years they hope to be helping fund half a dozen research projects. It is a big task. “Some days we think it would be great to turn it off,” MacCallum says, “but when we see the wonder on the face of a person who is experiencing somewhere like the Galapagos for the first time, we know we are living a wonderful life.”7.The underlined word “replenish" in Paragraph 4 means _________.A.consume B.supply C.reduce D.control6.【重庆市第八中学2021届高三二诊】Professor Sabbatucci received hundreds of calls from people hoping to trick him into handing them the cash. But there was one voice he recognized and he arranged to meet the man in a park. The robber, a 35-year-old unemployed father of two, gave back the suitcase and burst into tears. He could not believe what was happening. “Why didn’t you keep the money?” he asked. The professor replied, “I couldn’t because it’s not mine.” Then he walked off, spurning the thief’s offer of a reward.27.The underlined word “spurning” in the last sentence can be replaced by ________.A.accepting B.claimingC.rejecting D.cancelling7.【河北省张家口市2021届高三期末】In 1985. C'harlie Burrell inherited (继承)a piece of land in West Sussex, England, which is 3.500 acres. It had been in his family for more than 200 years. As it was a failing farm, he and his wife. Isabella Tree, were all set to run a traditional farm, but soon realized the costs far outweighed the profits. The land wasn't suitable for growing crops anymore.So. in 2001, the couple decided to try something else — an experiment in “rewilding”,or restoring environments to their natural state. They reintroduced native species, including Tamworth pigs, Exmoor ponies, longhorn cattle, and deer. Then they took out all the fences and let the animals wander freely.In the following years, the land went through an incredible change. The once neat fields were covered with bushes, creating habitats where new plant and animal species could grow.Now it's home to many different creatures, including two rare species of bats, nightingales, peregrine falcons, white storks, and even turtle doves, whose numbers have dropped by 98 percent in the U. K. over the last few decades!“We were living in a biological desert. Now, ecologists are blown away all the time by just the amount of life here," Isabella said. "It shows the potential that this kind of project has for changing trends of biodiversity decline.What's more, the land is now profitable! Charlie and Isabella offer multiple services there, including walking tours, safaris, photography classes, and rewilding workshops. They also sell meat to control their large animal populations, and even allow visitors to spend a night outside in the tents they set up.What a promising outcome! This is amazing news for conservationists and shows that our planet can be returned to its natural glory, if only we let it. Now, it's a beautiful reminder of how ecosystems can be restored by letting Mother Nature take the control.8.【河北省张家口市第一中学2021届高三期中】The end of the school year was in sight and spirits were high. I was back teaching after an absence of 15 years, dealing with the various kinds of "forbidden fruit" that come out of book bags. Now was the spring of the water pistol.I decided to think up a method of dealing with forbidden fruit."Please bring that pistol to me," I said. "I'm going to put it in my Grandma's Box.""What's that?" they asked."It's a large wooden chest full of toys for my grandchildren," I replied,"You don't have grandchildren," someone said."I don't now." I replied. "But someday I will. When I do, my box will be full of wonderful things for them."My imaginary Grandma's Box worked like magic that spring, and later. Sometimes. students would ask me to describe all the things I had in it. Then I would try to remember the different possessions I supposedly had taken away—since I seldom actually kept them. Usually the offender would appear at the end of the day, and I would return the belonging.25.What do the underlined words "the offender" in paragraph 8 refer to?A.The student's parent.B.The maker of the Grandma's Box.C.The author's grandchild.D.The owner of the forbidden fruit.9.【2020届8月贵州省贵阳市普通高中高三摸底】As it turned out, it not only shocked the US actress, but also brought criticism from those who believed Fanning was too young compared to other veterans in the industry. But Fanning’s acting experience was in fact a good argument for her jury membership. In fact, Fanning has been a Hollywood fixture for longer than you realize.4. What does the underlined word “veterans” probably mean?A. shocked actressesB. green hands C experienced people D. music producers10.【湖北省华中师范大学第一附属中学2021届高三期中】It’s been less than a month since Arnold, a Texas high school student, was forbidden from going to the prom and graduation unless he cut his locs. As his story spread on social media, celebrities and activists came to Arnold’s defence and encouraged him to stand up to his school’s hair discrimination. Now, in a significant turn of events, he’s proudly wearing his own hair-style at the 2020 Oscars.Arnold and his mother were invited to attend as the guests of director Matthew A.Cherry, who won the award for Best Animated Short Film for Hair Love tonight. Fittingly, the short film tells the story of a Black father learning to style his daughter’s natural hair.Arnold told us he couldn’t wait to walk the red carpet with the Hair Love team. “I’m so grateful. I never expected any of this,” Arnold says. “The message of that movie and my message go together so well. I think it’s really amazing how they reached out to me and how we can fight this together.”5.What does the underlined word “this” in Paragraph 3 probably mean?A.Violence.B.Discrimination.C.Panic.D.Starvation.11.【湖南省郴州市2021届高三第二次质量检测】While studying, I began exploring Mexico, crossing the country several times by bus. Now I work as a photojournalist in Mexico and along the border, documenting desperate situations that face everyday Mexicans. Though necessary and important, news stories don’t reflect the Mexico that I fell for and that is home to so many people I love. A task focused on the country’s natural beauty was a welcome reprieve for me.9.What can best replace the underlined word “reprieve” in Paragraph 2?A.Memory.B.Idea.C.Victory.D.Relief.12.【湖南省湖南师大附中2021届高三月考三】Clark, a rising fifth-grader who is Filipino-American, told HuffPost he's been dreaming about breaking Phelps' record ever since he started swimming competitively at age 7."I was so motivated,"Clark said about his win."I was so happy that I was able to beat that record.”Phelps competed in his first Olympics at age 15. He went on to become the most decorated Olympian in history, with 28 medals overall. “Everyone in the crowd was excited when they realized what a special swim they had just seen when we announced the long-standing record had been broken,"Cindy Rowland,Pacific Swimming's director, wrote in an email.5.Which of the following is closest in meaning to the underlined word“decorated”in Paragraph 4?A.Being popular.B.Being respected.C.Being talked about most.D.Being awarded medals.13.【辽宁省沈阳市郊联体2021届高三期末】The artists of Creative Growth will be on view throughout the month of August, coupled with musical performances and film screenings organized by L.A.Takedown. All proceeds from the month will benefit the artists and space. The money will be used to improve the condition of the studio and for the development of the artists. “L.A.doesn’t really have a place like Creative Growth,” Olson said, discussing his hopes for the show.14.What does the underlined word “proceeds” in the last paragraph mean?A.Activities.B.WorkC.Profits D.Arrangement14.【辽宁省沈阳市郊联体2021届高三期末】“I tested it at home and just couldn’t believe the result. I sprayed it in Sienna’ s room first and the floor was immediately silenced,” said Lanzarotti. “Eight months later, it was still silent. It felt like a huge relief. When Tracy saw the difference for the first time, her face just lit up. You can treat a couple of rooms in an hour.” he added. “It’ s a no-brainer for any parents with noisy floors, and it’ll be life-changing.”7.What do the underlined word “a no-brainer” in the last paragraph mean?A.A difficult decision.B.An unusual effort.C.A real headache.D.An easy job.15.【湖北省黄冈市2021届高三调研】I might be letting my young son watch too much television. I am certainly watching too many of his programs. They can really be perplexing as they force you to think carefully about the mysteries of life.For example, when Franklin (the turtle) wanted a pet, his parents didn’t want it to be a frog. They felt that frogs belong in a pond. Isn’t that where turtles belong?4.What does the underlined word “perplexing” in paragraph 1 probably mean?A.Boring.B.Humorous.C.Confusing.D.Curious.。
小学五年级上册第六次英语第1单元期末试卷(答案和解释)
小学五年级上册英语第1单元期末试卷(答案和解释)英语试题一、综合题(本题有50小题,每小题2分,共100分.每小题不选、错误,均不给分)1.Which of the following is a body part?A. ChairB. ArmC. TableD. Shoe2.What do we use to wash our hands?A. SpoonB. WaterC. PlateD. Cup3.Which of these is a mode of transport?A. BoatB. DogC. TreeD. Plate4.Which of the following is a color?A. TableB. GreenC. ChairD. Shoe5.It’s a sunny day, and we __________ (1) to the beach. My brother __________ (2) his surfboard, and I __________ (3) my swimming goggles. When we get there, we__________ (4) the water. I __________ (5) to swim for a long time, but my brother__________ (6) to play in the sand. After swimming, we __________ (7) some ice cream and __________ (8) a walk along the shore. We __________ (9) home in the evening, feeling very happy.6.Which of these is a body part?A. LegB. TableC. SpoonD. Plate7.Which of these is a type of transport?A. AirplaneB. SpoonC. PlateD. Table8.I live in a __________ house. It has a big __________ where we can play. There are three __________, a __________, and a __________ in my house. My bedroom is nextto the __________, and I have a __________ in my room where I keep my books. I love my house because it is __________ and cozy.9.You see a man on the street with a uniform, holding a big bag and delivering letters. What is his job?A. TeacherB. PostmanC. DoctorD. Driver10.I __________ (1) to play soccer on weekends. My friends __________ (2) with me, and we __________ (3) a game in the park. After the game, we __________ (4) some snacks and __________ (5) home.11.I __________ (be) very hungry when I __________ (wake) up this morning. So, I __________ (make) some pancakes for breakfast. After I __________ (eat), I__________ (decide) to go to the store because I __________ (need) to buy some fruit. I __________ (buy) apples, bananas, and oranges, and then I __________ (go) home to relax.12.What do we use to brush our hair?A. ToothbrushB. CombC. SpoonD. Fork13.What do we use to eat food from a plate?A. SpoonB. KnifeC. ForkD. Plate14.My sister __________ (have) a new bicycle. She __________ (get) it for her birthday. She __________ (love) riding it around the neighborhood. Last weekend, we __________ (ride) to the park together. We __________ (play) on the swings and__________ (have) a picnic there.15.Which one is a number?A. ThreeB. PenC. TableD. Chair16.Which one is the largest animal in the world?A. ElephantB. WhaleC. GiraffeD. Shark17.What is the opposite of "hot"?A. ColdB. WarmC. CoolD. Freezing18.We ________ breakfast at 7 oclock every day.A. haveB. havingC. hasD. is having19.David is at the zoo with his family. He is looking at a __________ that is eating some leaves from a tree. David’s little brother lik es the __________ because it can swingfrom tree to tree. They also see some __________ in the water, swimming around. David’s favorite animal is the __________.20.How many months are in a year?A. 12B. 10C. 11D. 921.This morning, I __________ (1) up at 7:00 AM. I __________ (2) my breakfast and __________ (3) my schoolbag. Then, I __________ (4) to school with my friends. We__________ (5) to talk and laugh on the way. I __________ (6) my math test today, and I __________ (7) it was easy.22.What is the opposite of "slow"?A. FastB. HeavyC. TallD. Light23.Which of these is a vegetable?A. TomatoB. AppleC. BananaD. Pear24.Which of these animals is known for having a long neck and eating leaves from tall trees?A. GiraffeB. ElephantC. TigerD. Zebra25.This morning, I _______ (wake) up early because I _______ (need) to finish my project. I _______ (work) on it for two hours before breakfast. After that, I _______ (take)a break and _______ (go) for a walk.26.I like to collect __. My collection includes different kinds of coins from various countries. I also have some __ that I found while traveling. My grandfather gave me a __ that belonged to him, and it is my most valuable item.27.David is excited because it is his first day at a new school. He wakes up early, eats some __________ for breakfast, and puts on his __________. His mom walks him to the school bus stop, and they wait together. When the bus arrives, David climbs on and finds a __________ to sit in. He says goodbye to his mom and looks out the window as the bus drives toward his __________.28.Which of these is a primary color?A. RedB. GreenC. OrangeD. Purple29.Which of these is a mammal?A. FishB. DogC. BirdD. Lizard30.Which is the smallest number?A. 5B. 0C. 1D. 331.Yesterday, we ______ (have) a test in math class. I ______ (study) hard for it, and I ______ (feel) confident. After the test, the teacher ______ (check) our answers and______ (say) we ______ (do) well. I ______ (be) very happy with my score.32.Which of these is used to measure time?A. RulerB. ClockC. SpoonD. Plate33.Which one is used for eating cereal?A. SpoonB. ForkC. KnifeD. Plate34.What do you call the big ball of fire in the sky?A. MoonB. SunC. EarthD. Star35.What do you use to clean the floor?A. KnifeB. BroomC. PlateD. Spoon36.What do you call the person who teaches you at school?A. DoctorB. TeacherC. CookD. Driver37.How do you say "谢谢" in English?A. HelloB. Thank youC. GoodbyeD. Sorry38.My sister __________ (study) at home today because she __________ (not/go) to school. She __________ (catch) a cold and __________ (need) to rest. She __________ (stay) in bed and __________ (watch) TV. I __________ (bring) her some tea, and she __________ (thank) me. I __________ (hope) she __________ (feel) better soon.39.Which of the following is the largest?A. DogB. CatC. ElephantD. Mouse40.My sister ______ (not like) vegetables when she was younger, but now she ______ (enjoy) eating them. Last week, she ______ (eat) spinach for the first time, and she______ (say) it was delicious.41.What do you wear on your head?A. ShirtB. ShoesC. HatD. Pants42.Today, Anna and her classmates are going on a __________ (1) to the farm. They will see many __________ (2) like cows, pigs, and __________ (3). Anna is excited tofeed the __________ (4) and collect __________ (5) from the chickens. After the visit, they will go to the __________ (6) to buy some fresh __________ (7).43.This weekend, we __________ (have) a party at home. My friends __________ (come) over, and we __________ (play) games and __________ (eat) snacks. I__________ (decorate) the house with balloons, and we __________ (sing) songs together. At the end of the party, we __________ (take) some photos to remember the fun time. It __________ (be) a wonderful day, and everyone __________ (enjoy) the party.44.Which of these is used to tell time?A. ClockB. PlateC. SpoonD. Shoe45.Which one is a holiday?A. MondayB. ChristmasC. RedD. Dog46.What do we use to drink water?A. PlateB. SpoonC. CupD. Knife47.I __________ (love) animals, especially dogs. I __________ (have) a dog named Max. Max __________ (be) very friendly and playful. Every morning, I __________ (take) Max for a walk in the park. He __________ (like) running and __________ (play) with other dogs. After the walk, we __________ (go) back home, and I __________ (feed) him. Max __________ (be) very happy when I __________ (play) with him.48.Which of these animals can live in water?A. DogB. FishC. TigerD. Monkey49.What color is an orange?A. redB. yellowC. orangeD. blue50.Which one is a mode of transportation?A. CarB. PlateC. SpoonD. Chair(答案及解释)。
IFU-068.1 REV 02 AccuMelt HRM SuperMix 说明书
AccuMelt™ HRM SuperMixCat No. 95103-250Size: 250 x 20-µL reactions (2 x 1.25 mL)Store at -25ºC to - 15°C 95103-0121250 x 20-µL reactions (10 x 1.25 mL)protected from light DescriptionAccuMelt HRM SuperMix is a 2X concentrated, ready-to-use reaction cocktail for detection of genetic variations using high resolution melting (HRM) analysis. It includes all required components except for primers and DNA template. HRM is a closed tube, rapid and cost effective procedure for characterization of sequence differences immediately following PCR amplification. It is based on the melting (dissociation) behavior of a PCR product as it transitions from double-stranded to single-stranded DNA in the presence of a fluorescent dsDNA-binding dye. The melting properties of a given PCR product are dependent upon the base composition, length, and strand base-pairing. HRM analysis tools exploit differences in melt curve shapes and DNA melting temperature (Tm) to discriminate sequence differences between samples.AccuMelt HRM SuperMix contains the green-fluorescent dye SYTO® 9 in a stabilized master mix that eliminates the need for time consuming optimization of critical PCR components. This dye provides strong fluorescent signal upon binding to dsDNA at saturating concentrations without inhibiting PCR. The unique chemical composition of this SuperMix further enhances and maximizes the impact of sequence variations on melt curve behavior. This facilitates discrimination of all sequence variations including difficult to resolve base-neutral transversions such as A>T class 4 SNPs. The dNTP mix in AccuMelt HRM SuperMix includes an optimized blend of dTTP and dUTP. This feature supports the optional use of uracil-DNA glycosylase (UNG) to prevent amplification of carry-over contamination, while providing high product yield and reliable PCR performance.Highly specific amplification with high product yield from complex genomic DNA template is critical for successful HRM studies. A key component of this SuperMix is AccuStart™ Taq DNA polymerase, which contains monoclonal antibodies that bind to the polymerase and keep it inactive prior to the initial PCR denaturation step. Upon heat activation at 95ºC, the antibodies denature irreversibly, releasing fully active, unmodified Taq DNA polymerase. This enables specific and efficient primer extension with the convenience of room temperature reaction assembly. AccuMelt HRM SuperMix can be used with all currently available HRM analysis systems. For HRM applications and more detailed product information, please visit our web site at .ComponentsAccuMelt HRM SuperMix (2X): 2X reaction buffer containing optimized concentrations of MgCl2, dNTPs(including dUTP), AccuStart Taq DNA Polymerase, SYTO 9 green-fluorescentdye, and stabilizers.[free Mg ++] = 0.8 mM at 1X final concentrationStorage and StabilityStore components in a constant temperature freezer at -25°C to -15°C protected from light upon receipt.For lot specific expiry date, refer to package label, Certificate of Analysis or Product Specification Form.Guidelines for PCR amplification and HRM analysis:The design of highly specific primers is the single most important parameter for successful PCR amplification and HRM analysis. The use of computer aided primer design programs is encouraged in order to minimize the potential for internal secondary structure and complementation at 3’-ends within each primer and the primer pair. Primer T m should be between 56 to 63ºC and the T m difference between the forward and reverse primers should be less than 2ºC.Amplicon size should be less than 250 bp. Smaller amplicons (60 to 100 bp) generally facilitate HRM discrimination of homozygote samples. We recommend designing and evaluating multiple primer set designs for any given application.Optimal primer concentration may vary between 100 and 500 nM. A final concentration of 300 nM for each primer is effective for most applications.Some primer set designs may require asymmetric primer concentrations.Always include a negative or no template control to evaluate the specificity of a given primer set and amplification protocol. Sequence specificity of the PCR should be confirmed by a method other than generation of a single melt peak. This includes confirmation of PCR product size, diagnostic restriction endonuclease fragment pattern, or sequencing.Preparation of a reaction cocktail is recommended to reduce pipetting errors and obtain reproducible HRM results. Assemble the reaction cocktail with all required components, except template DNA, and dispense equal aliquots into each reaction tube. Add DNA template as a final step. A minimum of 3 technical replicates for each DNA sample is recommended. Include appropriate positive controls for each sequence variant.Guidelines for PCR amplification and HRM analysis continued:Use approximately 10,000 copies of template DNA. A suggested input quantity for human genomic DNA is 10 to 30 ng.After sealing each reaction, vortex gently to mix contents. Centrifuge briefly to collect components at the bottom of the reaction tube.PCR amplification can be carried out in a conventional or real-time thermal cycler. Monitoring the reaction in real-time allows one to access the quality and PCR performance of a sample before HRM. All samples should produce comparable Cqs and fluorescence signal. Samples with delayed Cq (>30) or aberrant fluorescence signal should be excluded from the HRM analysis. Optimal cycling conditions will depend on the properties of your primers. Hold assembled reactions on ice, protected from light, if not proceeding immediately to PCR.Reaction AssemblyComponent Volume for 20-µL rxn. Final ConcentrationAccuMelt HRM SuperMix (2X) 10.0 µL 1xForward primer Variable 100 – 500 nMReverse primer Variable 100 – 500 nMNuclease-free water VariableDNA Template 2-5 µL ~10,000 copiesFinal Volume (µL) 20 µLPCR Cycling Protocol2-Step Cycling Protocol 3-Step Cycling Protocol Initial Denaturation 95ºC, 5 min*95ºC, 5 min*PCR cycling (40 to 45 cycles)Denaturation 95ºC, 5 to 10 s 95ºC, 5 to 10 sAnnealing60ºC, 30 s†15 s at 55 to 65ºCExtension 10 to 30s at 70ºC†HRM analysis§Consult instructions for your instrument* Full activation of AccuStart Taq DNA polymerase occurs within 30s at 95ºC; however, optimal initial denaturation time is template dependent and will affect PCR efficiency and sensitivity. Amplification of genomic DNA or supercoiled DNA targets may require 5 to 10 min at 95ºC to fully denature the template.† If monitoring PCR in real-time, collect and analyze kinetic PCR data at the end of the extension step. Extension time is dependent upon amplicon length and minimal data collection time requirement for your qPCR instrument. Some primer sets may require a 3-step cycling protocol for optimal performance. Optimal annealing temperature and time may need to be empirically determined for any given primer set.§ High Resolution Melting Analysis should be carried out immediately following PCR amplification. Please consult the instructions for your HRM instrument for procedural details. If not proceeding immediately to HRM, store plates at +4C protected from light. Mix and centrifuge samples immediately before HRM.Quality ControlKit components are free of contaminating DNase and RNase. AccuMelt HRM SuperMix is functionally tested to amplify a single copy gene in human genomic DNA and resolve homozygote samples for class 4 (A>T) and class 3 (C>G) SNPs in a model test system.Limited Label LicensesUse of this product signifies the agreement of any purchaser or user of the product to the following terms:1.The product may be used solely in accordance with the protocols provided with the product and this manual and for use with components contained in the kitonly. QIAGEN Beverly, Inc. grants no license under any of its intellectual property to use or incorporate the enclosed components of this kit with any components not included within this kit except as described in the protocols provided with the product, this manual, and additional protocols available at . Some of these additional protocols have been provided by Quantabio product users. These protocols have not been thoroughly tested or optimized by QIAGEN Beverly, Inc.. QIAGEN Beverly, Inc. neither guarantees them nor warrants that they do not infringe the rights of third-parties.2.Other than expressly stated licenses, QIAGEN Beverly, Inc. makes no warranty that this kit and/or its use(s) do not infringe the rights of third-parties.3.This kit and its components are licensed for one-time use and may not be reused, refurbished, or resold.4.QIAGEN Beverly, Inc. specifically disclaims any other licenses, expressed or implied other than those expressly stated.5.The purchaser and user of the kit agree not to take or permit anyone else to take any steps that could lead to or facilitate any acts prohibited above. QIAGEN Beverly,Inc. may enforce the prohibitions of this Limited License Agreement in any Court, and shall recover all its investigative and Court costs, including attorney fees, in any action to enforce this Limited License Agreement or any of its intellectual property rights relating to the kit and/or its components.The use of this product is covered by at least one claim of U.S. Patent No. 7,687,247 owned by Life Technologies Corporation. The purchase of this product conveys to the buyer the non-transferable right to use the purchased amount of the product and components of the product in research conducted by the buyer (whether the buyer is an academic or for-profit entity). The buyer cannot sell or otherwise transfer (a) this product, (b) its components, or (c) materials made by the employment of this product or its components to a third party or otherwise use this product or its components or materials made by the employment of this product or its components for Commercial Purposes. Commercial Purposes means any activity for which a party receives or is due to receive consideration and may include, but is not limited to: (1) use of the product or its components in manufacturing; (2) use of the product or its components to provide a service, information, or data; (3) use of the product or its components for therapeutic, diagnostic or prophylactic purposes; or (4) resale of the product or its components, whether or not such product or its components are resold for use in research. The buyer cannot use this product, or its components or materials made using this product or its components for therapeutic, diagnostic or prophylactic purposes. Further information on purchasing licenses under the above patents may be obtained by contacting the Licensing Department, Life Technologies Corporation, 5791 Van Allen Way, Carlsbad, CA 92008. Email: *************************©2018 QIAGEN Beverly Inc. 100 Cummings Center Suite 407J Beverly, MA 01915Quantabio brand products are manufactured by QIAGEN, Beverly Inc.Intended for molecular biology applications. This product is not intended for the diagnosis, prevention or treatment of a disease.AccuMelt and AccuStart are trademarks of QIAGEN Beverly, Inc. SYTO is a registered trademark of Life Technologies Corporation (Molecular Probes Labeling and Detection Technologies).。
American Journal of Medical Genetics 110234–242 (2002) Candidate Genes Involved in Cardiov
American Journal of Medical Genetics110:234–242(2002) Candidate Genes Involved in Cardiovascular Risk Factors by a Family-Based Association Study on the Island of Kosrae,Federated States of MicronesiaZhihua Han,1Simon C.Heath,2Dvora Shmulewitz,1Wentian Li,1Steve B.Auerbach,3Maude L.Blundell,1Thomas Lehner,1Jurg Ott,1Markus Stoffel,1Jeffrey M.Friedman,1and Jan L.Breslow1*1Starr Center Human Genetics,Rockefeller University,New York,New York2Lab Mathematical Genetics,MSKCC,New York,New York3HRSA,Department of Health and Human Services,New York,New YorkAltered plasma levels of lipids and lipopro-teins,obesity,hypertension,and diabetes are major risk factors for atherosclerotic cardiovascular disease.To identify genes that affect these traits and disorders,we looked for association between markers in candidate genes(apolipoprotein AII(apo AII),apolipoprotein AI-CIII-AIV gene clus-ter(apo AI-CIII-AIV),apolipoprotein E(apo E),cholesteryl ester transfer protein(CETP), cholesterol7a-hydroxylase(CYP7a),hepatic lipase(HL),and microsomal triglyceride transfer protein(MTP))and known risk factors(triglycerides(Tg),total cholesterol (TC),apolipoprotein AI(apo AI),apolipo-protein AII(apo AII),apolipoprotein B(apo B),body mass index(BMI),blood pressure (BP),leptin,and fasting blood sugar(FBS) levels.)A total of1,102individuals from the Pacific island of Kosrae were genotyped for the following markers:Apo AII/MspI,Apo CIII/SstI,Apo AI/XmnI,Apo E/HhaI,CETP/ TaqIB,CYP7a/BsaI,HL/DraI,and MTP/ HhpI.After testing for population stratifica-tion,family-based association analysis was carried out.Novel associations found were: 1)the apo AII/MspI with apo AI and BP levels,2)the CYP7a/BsaI with apo AI and BMI levels.We also confirmed the followingassociations:1)the apo AII/MspI with Tglevel;2)the apo CIII/SstI with Tg,TC,andapo B levels;3)the Apo E/HhaI E2,E3,and E4 alleles with TC,apo AI,and apo B levels;and4)the CETP/TaqIB with apo AI level.Wefurther confirmed the connection betweenthe apo AII gene and Tg level by a nonpara-metric linkage analysis.We therefore con-clude that many of these candidate genesmay play a significant role in susceptibilityto heart disease.ß2002Wiley-Liss,Inc.KEY WORDS:lipids and lipoproteins;can-didate genes;family-basedassociationINTRODUCTIONAltered plasma levels of lipids and lipoproteins, obesity,hypertension,and diabetes are major risk fac-tors for atherosclerotic cardiovascular disease.Genetic and environmental factors are known to affect the inci-dence of these conditions.Although monogenic condi-tions can result in abnormalities,it is now thought that much of the population variance is due to the contribu-tion of many genes and gene-environment interactions. Genes involved in lipid metabolic pathways have been investigated as candidates in many general population association studies,but the results from these studies are far from conclusive and often inconsistent,partly due to the common pitfalls in these types of association studies,such as inadequate sample size and population stratification.Attempts to identify genes responsible for these risk factors by association studies should be easier in studies of populations that are more homogeneous, both genetically and with regard to life style,assuming that significant phenotypic variation still exists.With this idea in mind we have initiated a comprehensive epidemiological and genetic study on the Pacific island of Kosrae,Federated States of Micronesia.Grant sponsor:National Institutes of Health(N.I.H.);Grant numbers:HL33714-17,GM58757-01,GM07982,DK56208; Grant sponsor:National Human Genome Research Institute (N.H.G.R.I.);Grant number:HG00008.*Correspondence to:Prof.Jan L.Breslow,The Rockefeller University,Director,Laboratory of Biochemical Genetics and Metabolism,1230York Avenue,Box179,New York,NY10021. E-mail:breslow@Received5June2001;Accepted18February2002DOI10.1002/ajmg.10445ß2002Wiley-Liss,Inc.Kosrae is located2,500miles northeast of Australia and was originally settled by a small number of founders(estimated to be50),originating from Poly-nesia around50A.D.Westernersfirst visited Kosrae in 1824.During the19th century the combined effects of typhoons and exposure of the native population to Western communicable diseases reduced the indigen-ous population from3,000–6,000to about300indivi-duals by1910.Historical and genealogical records indicate that these300survivors were the result of extensive admixture between native Kosraean females and male Caucasian whalers from New England and Europe who visited the island in the mid to late19th century.As late as1945Kosraeans consumed a diet consisting mostly offish,fruits,and vegetables,and the average individual was noted to be lean.After WWII Kosrae was designated a U.S.Trust Territory and this led to drastic lifestyle changes,with islanders becom-ing more sedentary and consuming large quantities of high-fat foods,such as Spam,turkey tails,and hamburgers,supplied through U.S.aid.These changes resulted in a large increase in the Kosraean population, accompanied by a dramatically increased prevalence of obesity,an outcome similar to that of other indigenous populations,such as the Pima Indians of Arizona and the Nahruans[Shmulewitz et al.,2001].Concern over the epidemic of obesity led in1994to a screening of practically all adult Kosraeans not only for obesity,but also for other related conditions such as dyslipidemia,diabetes,and hypertension.In addition, blood was obtained for DNA analysis from each parti-cipant and an extensive family tree was constructed for the entire island.In the current study,eight markers in or near the following candidate genes previously impli-cated in lipoprotein abnormalities,apolipoprotein AII (apo AII),apolipoprotein AI-CIII-AIV gene cluster(apo AI-CIII-AIV),apolipoprotein E(apo E),cholesteryl ester transfer protein(CETP),cholesterol7a-hydro-xylase(CYP7a),hepatic lipase(HL),and microsomal triglyceride transfer protein(MTP),were tested in a family-based association study for correlation to lipid and lipoprotein,body mass index(BMI),blood pressure (BP),leptin,and fasting blood sugar(FBS)levels.We also assessed the existence of population stratification on these marker/trait pairs and its implications on the association results.MATERIALS AND METHODSSample CollectionA total of2,188adult Kosraeans(ages,20–85years) were studied for dyslipidemia,obesity,diabetes,and hypertension as part of a public health campaign to identify risk factors for noncommunicable diseases, begun in1994.The study protocol was approved by the Institutional Review Board of Rockefeller Univer-sity and the Kosrae Department of rmed consent was obtained for all participants.Recruitment was through open meetings and radio announcements, and over90%of eligible Kosraeans participated.Parti-cipantsfilled out questionnaires that included family data that were subsequently used to construct a family tree of the population of the island.A detailed descrip-tion of the study protocol;the measurement of lipids and lipoproteins,BMI,systolic blood pressure(SBP), diastolic blood pressure(DBP),leptin,and FBS levels; the prevalence of risk factors;the effects of covariates; and a factor analysis of these traits has been recently published[Shmulewitz et al.,2001].Of relevance to the study,standard lipoprotein quantification requires ultracentrifugation of either fresh specimens or speci-mens kept overnight at48C,and cannot be performed on frozen plasma or serum.Due to the lack of adequate facilities on the island and the inability to ship serum overnight at48C,it was not possible to determine low-density lipoprotein(LDL)and high-density lipopro-tein(HDL)cholesterol levels directly.Instead,their major apolipoprotein components,apolipoproteinB (apo B)and apolipoprotein AI(apo AI),respectively, were measured.The levels of apo AI and apo B are highly correlated with HDL cholesterol and LDL cho-lesterol,respectively,making them excellent surro-gates[Bachorik et al.,1997].Total cholesterol(TC) (Boehringer-Mannheim,Indianapolis,IN)and trigly-ceride(Tg)(Sigma Diagnostics,St.Louis,MO)levels were measured with commercially available enzymatic kits.Apo AI,apolipoprotein AII(apo AII),and apo B levels were determined using standard enzyme-linked immunosorbent assay(ELISA)techniques.FBS was measured by afinger stick with a glucometer.Leptin levels were determined by a commercially available radioimmunoassay kit(Linco,St.Louis,MO),with a lower detection limit of0.5ng/m l and5%interassay coefficient of variation.Genomic DNA was extracted from whole blood that was previously frozen on dry ice by standard techniques and kept atÀ208C until ana-lysis.Genotyping was done on1,102Kosraeans,who comprised one major large branch of the family tree.GenotypingAll10single nucleotide polymorphism(SNP)sites were genotyped by PCR amplification of specific frag-ments of genomic DNA followed by digestion with the respective restriction enzymes.The candidate genes and their related polymorphic sites are listed in columns1and2in Table I.The primers for DNA ampli-fication are listed in column3.A typical PCR reaction was done in a20-m l volume consisting of10mM Tris-HCl(pH9.0at258C),50mM KCl,3mM MgCl2,0.1% Triton X-100,300m M deoxyribonucleoside tripho-sphates(dNTPs),10ng of genomic DNA,50pmol of each primer,and0.5unit of Taq polymerase.For apo E/HhaI,apo CIII/SstI,and CETP/TaqIB,10%dimethyl sulphoxide(DMSO)was included in the reaction solution.The reaction was performed byfirst denatur-ing at948C for4min,followed with35rounds of denaturing for30sec at948C,annealing for30sec at an experimentally optimized annealing temperature and elongating for30sec at728C,and afinal step of elongation at728C for7min.Restriction enzyme and buffer were subsequently added into the mix and in-cubated at378C for4hr.The products were separated on agarose gel,except for apo E,which was separated Genes for Cardiovascular Risk Factors235T A B L E I .L i s t a n d C h a r a c t e r i s t i c s o f T e s t e d P o l y m o r p h i c S i t e s i n K o s r a e a n P o p u l a t i o nG e n e c l u s t e r P o l y m o r p h i s mA m p l i fic a t i o n p r i m e r s (50t o 30)L o c a t i o nG e n e f u n c t i o n R e f e r e n c eA p o A I I A p o A I I /M s p lC T T T G C A T T G C A C A A G G A A C T A G a n d T T C T T T T C T T C A T A G A A T G A C T A A C A G A p o A I I (C 2777A .30-u n t r a n s l a t e d )HD L c o m p o n e n t [T s a o e t a l .,1985]A p o A I I (C A )nA C T G C T G T G G A C C C A G C T G A A A A G a n d C C T G T C T C C G A A C C A A A G C T C C T G A p o A I I i n t r o n 2[T s a o e t a l .,1985]A p o A I -C I I I -A I V A p o C I I I /S s t IG A T T C C T G C C T G A G G T C T C A G G G C T G T C G T a n d C C T G G A G T C T G T C C A G T G C C C A C C C A C A A p o C I I I (G 3147C .30-U T R )A p o C I I I i s V L D L a n d H D L c o m p o n e n t [D a m m e r m a n e t a l .,1993]A p o A I /X m n IG G A A A C A G G G G C C T A C A C T G T G a n d G T C T G C A G C C T T T G C A G T C T G A T C A p o A I (C 2500T )A p o A I i s m a j o r H D L c o m p o n e n t[S h o u l d e r s e t a l .,1993]D 11S 1998A G C C A T C A A C T A G C T T T C C C a n d G G G A G G C A C C A A C A G A T G W i t h i n 3c m o f A p o A I[w w w .g d b .o r g ]A p o E A p o E /H h a IG C G G C G C A G G C C C G G C T G G G C G C G a n d C G C T C G G C G C C C T C G C G G G C C C C G A p o E 1112(C 3745T )A p o E 158(C 3883T )V L D L ,I D L ,c h y l o m i c r o n r e m n a n t c l e a r a n c e [H i x s o n a n d V e r n i e r ,1990]C E T PC E T P /T a q I BC A C T A G C C C A G A G A G A G G A G T G a n d C T G A G C C C A G C C G C A C A C T A A CI n t r o n 1o f C E T PL i p i d e x c h a n g e b e t w e e n H D L a n d T g -r i c h l i p o p r o t e i n s [F u m e r o n e t a l .,1995]C Y P 7a C y p 7a /B s a IA A T G T T T T T C C C A G T T C T C T T T C a n d A A T T A G C C A T T T G T T C A T T C T A T T A G C Y P 7a (A 278C )R a t e -l i m i t i n g s t e p i n b i l e a c i d s y n t h e s i s [W a n g e t a l .,1998]H e p a t i c l i p a s e H L /D r a IG G G G G A A G A A G T G T G T T T A C T C T A G G A T C A C C a n d C A C A G G G G A C T T G T G T C C A T T T C T C C G H L (G -250A )T g h y d r o l y s i s[G u e r r a e t a l .,1997]M T P M T P /H h p IG G A T T T A A A T T T A A A C T G T T A A T T C A T A T C A C a n d A G T T T C A C A C A T A A G G A C A A T C A T C T A M T P (G -493T )A s s e m b l y a n d s e c r e t i o n o f V L D L [K a r p e e t a l .,1998]L P LL P L /R s a IG C C G A G A T A C A A T C T T G G T G a n d C T G C T T C T T T T G G C T C T G A C T G T AL p l 291,A s n t o S e rH y d r o l y s i s o f T g i n c h y l o m i c r o n a n d V L D L[R e y m e r e t a l .,1995]L P L /M n l IC T C T G A T T C T G A T G T G G C C T G A G T G a n d C T C C C T T A G G G T G C A A G C T C A G GL p l 471,S e r t o s t o p [H a t a e t a l .,1990]on6%acrylamide gel.The common alleles were defined as allele1,except in apo E/HhaI,where allele3was the most common of the three alleles(E2,E3,and E4).The sizes of DNA fragments for all of the alleles are listed in Table II,column2.The two microsatellite markers,the apo AII intra-genic dinucleotide repeat and D11S1998near apo AI-CIII-AIV,were genotyped withfluorescent primers on the373system(Applied Biosystems,Foster City,CA). PCR was carried out in condition similar to that described above,except that the primer concentration was reduced to5pmol.Allele calling was done by two individuals independently with Genotyper;afterwards the data were exported to a database where discrepan-cies were resolved.Statistical AnalysisIn the family-based association study the quantita-tive traits were corrected for age and sex by linear regression,and outliers(values that fell more than3SD from the mean)were excluded.All of the quantitative variables had an approximately symmetric distribu-tion,except for Tg,leptin,and FBS.To reduce skewness in the latter,a natural logarithm transformation was applied.However,in order to facilitate comparisons with other studies,we still reported the analysis result using Tg,since this has been done customarily.There was no essential difference between the results from Tg and log Tg.A standard way to estimate association between geno-type and trait is to regress the trait on the genotype and obtain estimates of the phenotypic means.In this pro-cess,the phenotypic means for the various genotypes are calculated,and assuming a normal distribution for the means and environmental noise,their significance is estimated.A prerequisite for this type of analysis is that the samples are independent.Although the familial relationship of the Kosraean samples violates this assumption,the regression analysis is valid as long as the following two conditions are met:1)the cor-relation among relatives is solely due to the genotype of the locus being tested,and2)the genotype for every pedigree member is known[Ewens and Spielman, 1995].Since the Kosraeans lived in a remarkably homo-genous environment,we can assume a uniform envir-onment and we assumed that thefirst condition was met.For the second condition,since only about45%of the population was typed,we used the known familial relationships in the Loki program[Heath,1997]to estimate the genotype of the individuals.In doing so, we corrected for the familial(genetic)correlation be-tween relatives.The Loki program uses a Markov chain Monte Carlo method to sample the unknown genotypes and gives an accurate description of the genotype distribution profile.In this study,1million sets of mis-sing genotypes were generated,proportional to the probability given the observed genotype data and the pedigree structure.A regression analysis was perfor-med as described earlier for each set of samples,and the estimates obtained from each regression were then averaged over the1million sets of samples.This pro-vides estimates of the phenotypic means that have been averaged over the missing genotype data.For simpli-city,the phenotypic mean of the most common genotype (11)was always set at zero,while the phenotypic means of the other genotypes(12or22)were estimated as deviations from that of11.In addition to a point estimate,this process also allows estimation of the dis-tribution of the phenotypic means.From this,95% confidence intervals and the probability that a parti-cular phenotypic mean differs from zero can be empiri-cally estimated,without relying on assumptions of normality.For example,to estimate the P value of the phenotypic mean of genotype12,which is smaller than the phenotypic mean of11,we simply counted the proportion of samples in which the regression estimate of the phenotypic mean of12is<0.The Loki method takes advantage of the whole set of trait data and is robust in estimating P values in the sense that it does not rely on the normality of the estimates.The method easily allows for the addition of other factors into the regression model,and in this study,age and sex were added to the model.Individuals with Mendelian incon-sistencies were excluded prior to the analysis. MAPMAKER/SIBS0.9was used in the sibpair ana-lysis[Kruglyak and Lander,1995].The Kosraean pedi-gree was too large to be analyzed with existing linkage software,so we split the Kosrae pedigree into two-generation nuclear families.Those families with two or more genotyped children were used in a sibpair analy-sis for the quantitative traits using the MAPMAKER/ SIBS0.9program.Prior to running the MAPMAKER/ SIBS0.9,PEDMANAGER was used to check Mende-lian inconsistencies.For each marker,we excluded those families whose member(s)showed any inconsis-tency.In general,the numbers of inconsistent indivi-duals were very low.The QTDT program by Abecasis et al.[2000]was used to examine the evidence of population stratifica-TABLE II.Allelic Characteristics of the Genotyped Markers inKosraean PopulationPolymorphism Allele size(bp)Allele frequencyApo AII/MspI1:175and2960.832:4710.17Apo CIII/SstI1:5490.622:232and3170.38Apo AI/XmnI1:3920.742:173and2190.26CETP/taq-1B1:174and3610.622:5350.38Cyp7a/BsaI1:3930.842:90and3030.16HL/DraI1:4620.722:3500.28MTP/HhpI1:20and890.692:1090.31Apo E/HhaI2:83and910.023:35and48and910.814:39and48and720.17LPL/RsaI1:2380.9952:23and2150.005LPL/MnlI1:32512:26and2580Genes for Cardiovascular Risk Factors237tion.The test evaluates whether there are subpopula-tion groupings where the following quantity varies in the absence of linkage:(p–q)Âm.The p and q are allele frequencies and m is the phenotypic mean,so it reports stratification that affects the traits only.The same two-generation nuclear families used in the sibpair analysis were used in this analysis,since the complexity of the Kosraean pedigree exceeded the limit of the QTDT. This should give a good estimation,although it tends to overestimate the existence of stratification,since each sibship would have unique allele frequencies(deter-mined by parental alleles)and phenotypes(determined by shared genetic and environmental components).RESULTSFamily-Based AssociationsWe genotyped10polymorphisms and tested whether there were significant differences in phenotypic means for Tg,log Tg,TC,apo AI,apo AII,apo B,BMI,SBP, DBP,log leptin,and log FBS among all the genotypes in the eight polymorphisms that were common.These common polymorphisms included the apo AII/MspI, apo CIII/SstI,apo AI/XmnI,apo E/HhaI,CETP/TaqIB, CYP7a/BsaI,HL/DraI,and MTP/HhpI.Allele frequen-cies of these polymorphisms in the Kosraean popula-tion are listed in Table II.In Tables III and IV,we summarize all of the polymorphisms that displayed sig-nificantly different genotype-specific means at the signi-ficance level of P<0.01.For the apo AII/MspI polymorphism,compared to individuals with two common alleles(M1/M1),the mean Tg level in individuals with homozygous minor alleles(M2/M2)was increased by56%.The difference was highly significant(P<0.0001).The result was similar when the analysis was done using log Tg(P¼0.008),thus not caused by outliers.The Tg-raising effect of the M2allele seemed to be recessive,since the M1/M2 individuals displayed a Tg level similar to that in M1/ M1.The M2allele was also associated with a decreased level of apo AI.This effect was dosage dependent(3% decrease in the heterozygous M1/M2and7%decrease in the homozygous M2/M2)and reached significance for the homozygous M2individuals(P¼0.01).Interest-ingly,both M1/M2and M2/M2subjects also demon-strated statistically significant decreases in DBP and SBP,compared to that of M1/M1subjects.These results suggest that the M2allele is a recessive allele associa-ted with higher Tg levels,a codominant allele asso-ciated with lower apo AI levels,and a dominant allele associated with decreased BP.For the apo CIII/SstI polymorphism,compared to individuals with two common alleles(S1/S1),the mean Tg level was10%higher in S1/S2individuals(P¼0.004) and27%higher in S2/S2individuals(P¼0.0002).A similar result was obtained when the analysis was done using log Tg,whereas log Tg was4.5%higher in S1/S2 individuals(P¼0.0008)and9.5%higher in S2/S2indi-viduals(P<0.0001).The mean TC level was 2.0% higher in S1/S2individuals and6.4%higher in S2/S2 individuals(P¼0.0006).The apo B level displayed the same trend as the TC level.These results suggest that the S2allele is a codominant allele associated with increased Tg levels and a recessive allele associated with increased TC and apo B levels.For the CETP/TaqIB polymorphism,compared to individuals carrying two common alleles(T1/T1),the heterozygous individuals(T1/T2)showed a2.7%in-crease in apo AI level(P¼0.1),and individuals carrying two rare alleles(T2/T2)showed a6.1%increase in apo AI level(P¼0.002).This indicates that the rare allele is associated with increased apo AI levels.For the CYP7a/BsaI polymorphism,compared to homozygous common allele carriers(B1/B1),individualsTABLE III.Significant Genotype/Phenotype Associations,Part I*Genotype/phenotypic mean 111222Publishedbefore? 1112P-value22P-valueApo AII/MspITg98.73100.480.3152.68<0.0001YesLog Tg 1.94 1.970.2 2.110.008YesApo AI115.99112.450.02107.790.01NoDBP78.4674.220.00474.600.002NoSBP121.70116.270.009116.180.006NoApo CIII/SstITg94.57104.010.004119.820.0002YesLog Tg 1.93 2.010.0008 2.11<0.0001YesTC173.70177.260.06184.730.0006YesApo B85.8987.710.0992.270.001YesCETP/Taq1BApo AI112.76115.850.01119.640.002YesCYP7a/BsaIApo AI114.26114.740.4128.290.0008NoBMI31.0631.820.0233.360.004No *For all the polymorphisms,1refers to the common allele and2refers to the rare allele.Tg,TC,apo AI,and apo Bare reported as mg/dl.DBP and SBP are reported as mmHg.The unit for BMI was kg/m2.The phenotypic meansof12and22were compared with those of11,and the P-values at which differences happened by chance wereshown.The analyses were done with the Loki method that corrected for age and gender,and took into account ofthe family.238Han et al.with two rare alleles(B2/B2)displayed a12%in-crease in apo AI levels(P¼0.0008).Also,compared to B1/B1individuals,BMI was increased by2.4%for B1/B2individuals(P¼0.02)and7.4%for B2/B2 individuals(P¼0.004).For the apo AI/XmnI,HL/DraI,and MTP/HhpI polymorphisms,no associations were found at the sig-nificance level we set(P¼0.01).For the apo E/HhaI polymorphism,since there were not enough E2/E2and E4/E2individuals(0and6, respectively),we examined the effects of genotypes E3/ E2,E3/E3,E4/E3,and E4/E4on the lipid and other traits.We observed,in the above order,stepwise in-creases in apo B and TC levels,and stepwise decreases in apo AI levels(see Table IV).Compared to individuals of E3/E3,the mean apo B level was16%lower in E3/E2 individuals,2.7%higher in E4/E3individuals,and10% higher in E4/E4individuals.Similarly,the mean TC level was the lowest in E3/E2individuals and the highest in E4/E4individuals.For the Apo AI level the trend was pared to E3/E3individuals, the mean apo AI level was1.1%higher in E3/E2indi-viduals,4.7%lower in E4/E3individuals(P<0.0001), and6.4%lower in E4/E4individuals.The mean Tg level was the lowest in E3/E3individuals and was signifi-cantly higher in E4/E3individuals(P¼0.01).Similar results were obtained when analysis was done with log Tg.Overall,these results are consistent with the gene-rally established effects of E4,E3,and E2on apo B levels and further clarified their influence on TC,apo AI,and Tg levels.The N291S and S447X polymorphisms in lipoprotein lipase have been shown to influence the lipid and lipo-protein profile[Hata et al.,1990;Reymer et al.,1995; Fisher et al.,1997]and were chosen initially for the study.Yet the rare allele of N291S was not detected, and the rare allele frequency of S447X was0.5%.As a result,these polymorphisms were not informative enough to be included in the study.LinkageIn order to corroborate our results in the association analyses,we performed linkage analysis on all the traits for the apo AII and apo AI-CIII-AIV loci,using micro-satellite markers of the apo AII dinucleotide repeat (intragenic;heterozygosity,0.57)and D11S198(within 3cm of the apo AI-CIII-AIV;heterozygosity,0.69).Out of the223nuclear families used in the sibpair analysis,68had2siblings,contributing68sibpairs;46had3 siblings,contributing138sibpairs;36had4siblings, contributing216sibpairs;34had5siblings,contribut-ing340sibpairs;16had6siblings,contributing240 sibpairs;17had7siblings,contributing357sibpairs; and6had8siblings,contributing168sibpairs.The total number of sibpairs was1,527.For the intragenic apo AII dinucleotide repeat,a single-point nonpara-metric analysis yielded a Z-score of1.86for Tg levels, corresponding to P¼0.0314;and a Z-score of2.22for log Tg,corresponding to P¼0.0132.For the D11S1998, no significant linkage was found.The result from lin-kage analysis further confirms that the apo AII locus or a gene in its vicinity is important for Tg levels.Population StratificationOur family-based association analysis can correct fully for population stratification,on the condition that all individuals were genotyped.Since we genotyped only approximately45%of the individuals,it was possible that residual effects of population stratification still existed.To guard against this possibility,we used the QTDT program[Abecasis et al.,2000]to check the exis-tence of population stratification on all the80possible marker/trait combinations(8markers and10traits). The following two marker/trait combinations gave indi-cations of population stratification at a significance level of P<0.01:the apoA II/MspI vs.SBP(P¼0.007)and the HL/DraI vs.apo AII levels(P¼0.004).There was no evidence of population stratification for most of the associations we observed,such as the apo AII/MspI with Tg and apo AI levels,the apo CIII/SstI with Tg,TC,and apo B levels,the CETP/TaqIB with apo AI levels,and the apo E/HhaI with TC,apo AI,and apo B levels.In conclusion,the associations we found were not a result of stratification.DISCUSSIONIn this study we have selected common polymorph-isms that have been shown to influence lipid and lipo-protein levels and tested their roles in modulating lipid, BMI,BP,leptin,and FBS levels in a family-based asso-ciation study.We observed the following significant associations:apo AII/MspI to Tg,apo AI,and BP;apo AI-CIII-AIV to Tg,TC,and apo B;apo E/HhaI to Tg,TC, and apo B;CETP/TaqIB to apo AI;and CYP7a/BsaI to apo AI and BMI levels.Our study is thefirst to report that the apo AII/MspI polymorphism is associated withTABLE IV.Significant Genotype/Phenotype Associations,Part II*Genotype/phenotype mean 33344432Agree withpreviousreports? 3334P-value44P-value32P-valueApo E/HhaITg97.15105.050.0198.810.4102.260.3YesLog Tg 1.94 2.020.003 2.050.1 1.970.3YesTC175.12178.110.1187.310.04159.730.002Yes Apo AI116.54111.07<0.0001109.040.04117.780.4Yes Apo B86.2688.680.0495.420.0272.16<0.0001Yes*The details for this table are similar to those of Table III,except that,for the apo E/HhaI polymorphism,the E3is the common allele,and the phenotypic means of34,44,and32were compared to those of33.Genes for Cardiovascular Risk Factors239。
男男同性恋人群治疗前HIV-1流行毒株及其变异研究
•短篇论著•男男同性恋人群治疗前HIV-1流行毒株及其变异研究路新利张玉琪邱延超李保军李岩王莹莹刘萌安宁050021石家庄,河北省疾病预防控制中心艾滋病所,河北省传染病病原学鉴定分析与流行病学重点实验室(路新利、张玉琪、李保军、李岩、王莹莹、刘萌、安宁);050011石家庄,石家庄市疾病预防控制中心艾滋病所(邱延超)通信作者:路新利,E-mail:lxlii2009@ DOI:10.16462/ki.zhjbkz.2020.12.019【摘要】目的了解掌握男男同性恋者(men who have sex with men, MSM)人群治疗前人类免疫缺陷病毒I型(human immunodeficiency virus type one, HIV-1)流彳了毒株与基因变异特点。
方法使用In-house方法进行耐药基因型检测,通过系统进化分析和H IV-1耐药数据库比对确定基因型和基因变异情况。
结果石家庄市M SM人群中共有5种H IV-1基因型毒株在流行,CRF01_AE是最主要流行毒株,占到 56. 8%(25/44),其次是 CRF07_BC 29. 5%( 13/44) ,URFs 9. 1%(4/44),B 亚型 2. 3%(1/44)和 CRF55_01B 2.3%( 1/44)。
URFs 主要包括 CRF01—A E/B和 CRF01_AE/C 两种重组模式。
f检验显示不同基因型毒株在婚姻状况、年龄和首次CD4+ T淋巴细胞中的分布差异均有统计学意义(均有P<〇.〇5)。
发现1例M S M在川V-l N N R T编码区225位点处发生了耐药基因突变(P225H),原发耐药率为2. 3%( 1/44),可导致依非韦伦(E F V)和奈韦拉平(N V P)中度耐药。
结论加强M SM人群中H IV-1流行和耐药监测研究对控制本市H IV疫情发展具有重要意义。
【关键词】人类免疫缺陷病毒I型;基因型;男男同性恋者;变异【中图分类号】R373 【文献标识码】A 【文章编号】1674-3679(2020) 12-1465-05基金项目:河北省自然科学基金(H2016303006)HIV-1 epidemic strains and genetic variation among therapy-naive men who have sex with menLU X in-li,Z H A N G Y u-q i,Q IU Y a n-c h a o,U B a o-ju n,LI Y a n,W ANG Y in g-y in g,LIU M e n g,A NN in gD epartm ent o f A ID S R esearch , H ebei K ey Laboratory o f P a th o g en a n dE p idem iology o f Infectious D isease,Hebei P rovincial Center fo r D isease Control a n d P revention,S h ijia zh u a n g050021, C hina ( Lu X L,Z h a n g Y Q, Li B J, Li F,W a n g Y Y, Liu M, A n N);D epartm ent o f A ID S R esearch, S h ijia zh u a n g Centerfo r Disease Control a n d P re ve n tio n, Sh ijia zh u a n g050011, C hina( Q iu Y C)Correspongding a u th o r:LU X in-li^ E-m a il:Lxlii2009@ 【Abstract】Objective To explore HIV-1 genotypes and genetic variation among therapy-naivemen who have sex with men (M SM) in Shijiazhuang. Methods HIV-1 genotypic resistance assay wascarried out using In-house method. HIV-1 genotypes and drug resistance were analyzed by phylogenetic a-nalysis and submitting the sequences to the Stanford University Network HIV-1 database. Results FiveHIV-1 subtypes were found among MSM in Shijiazhuang. Of them, CRF01_AE was the most frequent, accounting for 56. 8% (25/44) , followed by CRF07_BC 29. 5% ( 13/44) , URFs 9. 1%(4/44) , B 2. 3%(1/44) and CRF55_01B 2. 3% ( 1/44). URFs mainly included CRF01_AE/B and CRF01_AE/C. %2-test verified that HIV-1 genotype distribution showed significant difference (P<0. 05) in marital status,age and first CD4 groups, respectively. The rate of HIV-1 primary drug resistance was 2. 3% ( 1/44).P225H mutation was found in NNRT gene coding region, and one MSM with P225H occurred intermediate resistance to EFV and NVP. Conclusion It is critical for us to take measures to monitor HIV-1 epidemic and drug resistance.【Key words】Human immurmdenriency virus type one; Gen〇l\[H*;M<m i who have sex with m en;MutationFund Program:Provincial Natural Sc ienrt1Foundation ol Hehei ( H2016303006).(Chin J DLs Control Prev2020,24( 12):1465-1469)自1981年在美国男男同性恋者(men who have sex with men,MSM)中发现艾滋病以来,人类免疫缺 陷病毒I型(human immunodeficiency virus type one,H I V-丨)已在全球肆虐近四十年,对全球经济和社会发 展造成成严重挑战高效抗病毒治疗能够有效抑制 病毒复制,防控H I V传播•1但是,H1V-1具有快速 的复制能力和高度异质性,易产生耐药基因突变,使 得抗病毒药物和治疗方案的选择范围更加狭窄》 因此,在当前“发现即治”政策下,评估高危人群新诊 断未治疗感染者中H I V-1流行和基因变异,对防控 m v-1传播具有重要意义。
- 1、下载文档前请自行甄别文档内容的完整性,平台不提供额外的编辑、内容补充、找答案等附加服务。
- 2、"仅部分预览"的文档,不可在线预览部分如存在完整性等问题,可反馈申请退款(可完整预览的文档不适用该条件!)。
- 3、如文档侵犯您的权益,请联系客服反馈,我们会尽快为您处理(人工客服工作时间:9:00-18:30)。
A Large Maize(Zea mays L.)SNP Genotyping Array: Development and Germplasm Genotyping,and Genetic Mapping to Compare with the B73Reference Genome Martin W.Ganal1,Gregor Durstewitz1,Andreas Polley1,Aure´lie Be´rard2,Edward S.Buckler3,Alain Charcosset4,Joseph D.Clarke5,Eva-Maria Graner1,Mark Hansen6,Johann Joets4,Marie-Christine Le Paslier2,Michael D.McMullen7,Pierre Montalent4,Mark Rose5,Chris-Carolin Scho¨n8,Qi Sun3,Hildrun Walter8,Olivier C.Martin4,Matthieu Falque4*1TraitGenetics GmbH,Gatersleben,Germany,2Etude du Polymorphisme des Ge´nomes Ve´ge´taux,INRA–CEA–Institut de Ge´nomique–Centre National de Ge´notypage, Evry,France,3Cornell University,Ithaca,New York,United States of America,4UMR de Ge´ne´tique Ve´ge´tale,INRA–Universite´Paris-Sud–CNRS–AgroParisTech,Gif-sur-Yvette,France,5Syngenta Biotechnology Inc.,Research Triangle Park,North Carolina,United States of America,6Illumina Inc.,San Diego,California,United States of America,7Plant Genetics Research Unit,USDA-Agricultural Research Service,Columbia,Missouri,United States of America,8Department of Plant Breeding,Technische Universita¨t Mu¨nchen,Freising,GermanyAbstractSNP genotyping arrays have been useful for many applications that require a large number of molecular markers such as high-density genetic mapping,genome-wide association studies(GWAS),and genomic selection.We report the establishment of a large maize SNP array and its use for diversity analysis and high density linkage mapping.The markers, taken from more than800,000SNPs,were selected to be preferentially located in genes and evenly distributed across the genome.The array was tested with a set of maize germplasm including North American and European inbred lines,parent/ F1combinations,and distantly related teosinte material.A total of49,585markers,including33,417within17,520different genes and16,168outside genes,were of good quality for genotyping,with an average failure rate of4%and rates up to8% in specific germplasm.To demonstrate this array’s use in genetic mapping and for the independent validation of the B73 sequence assembly,two intermated maize recombinant inbred line populations–IBM(B736Mo17)and LHRF(F26F252)–were genotyped to establish two high density linkage maps with20,913and14,524markers respectively.172mapped markers were absent in the current B73assembly and their placement can be used for future improvements of the B73 reference sequence.Colinearity of the genetic and physical maps was mostly conserved with some exceptions that suggest errors in the B73assembly.Five major regions containing non-colinearities were identified on chromosomes2,3,6,7and9, and are supported by both independent genetic maps.Four additional non-colinear regions were found on the LHRF map only;they may be due to a lower density of IBM markers in those regions or to true structural rearrangements between lines.Given the array’s high quality,it will be a valuable resource for maize genetics and many aspects of maize breeding.Citation:Ganal MW,Durstewitz G,Polley A,Be´rard A,Buckler ES,et al.(2011)A Large Maize(Zea mays L.)SNP Genotyping Array:Development and Germplasm Genotyping,and Genetic Mapping to Compare with the B73Reference Genome.PLoS ONE6(12):e28334.doi:10.1371/journal.pone.0028334Editor:Lewis Lukens,University of Guelph,CanadaReceived September14,2011;Accepted November5,2011;Published December8,2011This is an open-access article,free of all copyright,and may be freely reproduced,distributed,transmitted,modified,built upon,or otherwise used by anyone for any lawful purpose.The work is made available under the Creative Commons CC0public domain dedication.Funding:This work has been supported by a grant from the Agence Nationale de la Recherche http://www.agence-nationale-recherche.fr/(ANR-10-GENOMIQUE-SingleMeiosis)to MF and OCM,a grant from the German Federal Ministery for Education and Research http://www.bmbf.de/en/(AgroclustER Synbreed–Synergistic Plant and Animal Breeding,grant no.0315528)to MWG and C-CS,and funds provided by USDA-ARS /main/main.htm to MDM and ESB.The funders had no role in study design,data collection and analysis,decision to publish,or preparation of the manuscript.Competing Interests:The authors M.W.Ganal,G.Durstewitz,A.Polley,and E-M.Graner have competing commercial interests as members of TraitGenetics, which is a commercial company that performs molecular marker analysis with the maize SNP50array.TraitGenetics has also a commercial interest in the data generated with the chip since it increases the value of their services to their customers.The author M.Hansen has competing commercial interests as a member of Illumina Inc.which produces and sells the MaizeSNP50array.The authors J.D.Clarke and M.Rose have competing commercial interests as members of Syngenta Biotechnology Inc.,which has provided a number of their SNPs for the chip(these data are already public and released in dbSNP).This does not alter the authors’adherence to all the PLoS ONE policies on sharing data and materials.There are no further products in development or marketed products or patents to declare.*E-mail:falque@moulon.inra.frIntroductionMaize(Zea mays ssp.mays),along with wheat and rice,is one of the most important crop plants.Being widely grown around the world in tropical and temperate climatic zones,it is important both as a food and feed plant but it has also recently gained additional interest as a renewable energy plant due to its high biomass potential.The cultivated maize was domesticated from the grass teosinte(Zea mays ssp.parviglumis).Current maize morphology differs from teosinte mainly because of a few major genes for which specific alleles were selected during domestication [1].Through selfing,homozygous inbred lines have been developed and in current hybrid varieties a high level of heterosis is achieved through the combination of lines from different heterotic groups.The genome of maize is approximately2.3Gbp which makes it comparable in terms of size to the human genome.The maize inbred B73has been used as a reference line for sequencing[2].The current version of the B73genome assembly covers a‘‘golden path’’of2066Mb.This assembly is based on the most recent physical mapping data(BAC contig assembly through FPC fingerprinting,optical mapping),integration of molecular markers from genetic maps,and within-BAC sequence assembly[3–5]. Like many other plant genomes,the maize genome has undergone several duplication events of which the most recent is a whole genome duplication resulting in a haploid chromosome number of n=10approximately12million years ago[6,7].Maize in traditional populations is a highly heterozygous plant that displays an extremely high level of sequence polymorphism. Through whole genome sequencing of maize inbreds[8], sequencing of genomic fractions with reduced complexity(i.e. through the elimination of highly repeated DNA sequences)or transcriptome sequencing,large numbers of SNP markers have been identified.SNP polymorphisms appear,on average,every 44–75bp[9].This level of polymorphism is10to20times higher than in most animal species.Furthermore,it has been found that individual maize lines have extensive structural differences such as copy number variations and presence/absence polymorphisms [10].To date,over180genetic mapping studies have been performed in maize(/),based on different map-ping populations such as F2[11],recombinant inbred lines(RILs) [12],and high-resolution Intermated Recombinant Inbred Lines (IRILs)which include several generations of random intermating starting from F2plants to increase the number of effective meioses before repeated selfing to obtain inbred lines,thus increasing the resolution of the map[13–15].The current biparental reference genetic map for maize is the IBM map based on IRILs obtained from the cross B736Mo17.An additional population named LHRF that is more relevant to studying European maize material was produced from the cross F26F252[16]using exactly the same scheme.More recently,a star-shaped multi-parental mapping experiment called Nested Association Mapping(NAM)[17]was developed using the B73inbred as the pivotal line.The analysis of very large numbers of SNP markers in precisely located single copy sequences is a prerequisite towards the elucidation of the detailed genome structure and precision breeding.Arrays with many thousands of SNPs genotyped in a highly parallel fashion[18,19]as well as new genotyping by sequencing methods[20],are approaches towards this goal.As originally demonstrated for humans,large genotyping arrays with several million SNPs are useful for the analysis of many individuals at a very high genetic resolution.They permit the analysis of traits that are inherited as single locus(qualitative)traits as well as traits that are influenced by multiple loci(QTLs or quantitative traits)to a resolution that leads directly to the identification of candidate genes,using linkage mapping in very large sets of recombinant individuals or genome wide association studies(GWAS).In contrast to the genetic analysis in segregating populations,GWAS studies are based on the precise phenotypic analysis of a given trait in a large set of individuals that are widely unrelated(i.e.have no or little family structure)but are derived from a common gene pool.GWAS with large SNP genotyping arrays containing several million SNP markers are now routinely used to identify loci that are associated with many complex traits in human and other organisms[21].In important domesticated animal species[22,23], the analysis of large numbers of SNP markers has opened the door to new breeding schemes such as genomic selection(GS).For GS, the effect of all markers present on the array is estimated in precisely phenotyped reference populations through a variety of statistical approaches[24,25].Breeding values are subsequently calculated for newly generated,not yet phenotyped progenies based on their genotyping and marker effects estimated in the reference population(s).In cattle,this approach has been so successful that genomic breeding values are now used as reliable predictors for progeny individuals.Recent data suggest that GS is also promising in maize and other plant species[26].The objectives of the work presented here are(1)to use SNPs previously identified in maize to develop a first reliable and standardized large scale SNP genotyping array;(2)to genotype a set of several hundred maize lines in order to define the functionality over a wide set of maize germplasm and(3)to produce two high-density linkage maps based on biparental IRIL populations for an independent comparison with the B73genome sequence to identify assignment and ordering discrepancies between the genetic and physical maps.ResultsEstablishment of an accurate genotype calling procedure through cluster definitionFrom the57,838synthesized SNP markers,56,110markers passed bead representation and decoding quality metrics.When the cluster distribution of the genotypes from the274maize lines was analyzed,it was found that the distribution of the genotype calls in the two-dimensional analysis was producing mainly four distinct patterns(Figure1).Pattern Type1was represented by three clearly defined clusters representing the three possible genotypes(AA,AB and BB).Such markers did not require any significant adjustment for the genotype calling as the genotyping software algorithm ually this pattern was observed with markers that could be scored in all or nearly all274 maize lines.Pattern Type2was obtained with the majority of the markers on the array and was similar to the Pattern Type1. However,frequently the cluster corresponding to the heterozygous situation was not as compact as in Type1and often a significant number of lines were not automatically scored due to lower than expected signal intensities.With the appropriate adjustment of the area for the respective genotype,such a marker could be scored accurately with the genotypes of the samples producing very weak signal intensities set to failed.The Pattern Type3also produced three defined clusters but two of those clusters were shifted to normalized theta space along the X axis of the SNP graph (normalized theta ranges from0–1corresponding to an angle relative to the‘‘A’’allele intensity of0–90degrees,respectively). Such a marker could not be easily interpreted with the GenomeStudio software[27].In Pattern Type3,the genotype clusters required manual adjustment in a significant manner so that the three genotype classes could be called accurately.In cases when the three clusters were very close and shifted strongly to one side of the theta space,the GenomeStudio software did not permit the definition of the three different genotypes and the marker had to be scored as failed.Such markers often occurred in groups when the markers were positioned along the maize genome sequence. The final Pattern Type4resulted in five instead of three clusters. These markers could not be scored with the GenomeStudio v2009 software and were thus set to failed.After evaluating all56,110 markers,49,585passed the analytical phase.Figure2shows the final attribution of the49,585functional markers to different groups used for the SNP selection procedure,as well as the individual functionality rates for each group.Among the49,585 scorable SNPs,34,182came from Panzea,13,037from Syngenta, 1,816from INRA,400from TraitGenetics,and150from other sources.These markers covered a total of17,520different genes, with sometimes numerous markers in the same gene,which may increase resolution for gene haplotyping.Quality control of the markers for pedigree consistency,reproducibility and call rate in the different maize samplesIn addition to the definition of the genotype clusters based on the distribution on the two-dimensional plane of the genotyping software,further quality control steps were performed such as the analysis of the markers in a number of parent/F1triplets for pedigree consistency and in DNA duplicates for technical reproducibility.35triplet combinations including 25parent/F1combinations for the NAM populations,9parent/F1combina-tions from European maize material and the parent/F1combi-nation of B73/Mo17(IBM population)were used for determining the pedigree consistency of the genotype calls based upon Mendelian expectations.The consistency of the genotype calls was also determined in technical (same DNA analyzed twice)duplicates (three samples each from B73,Mo17and the F1)and sample duplicates from different seed sources.In summary,the allele calls were highly reproducible (Table S1)with no or negligible inconsistencies observed between samples with respect to genotype calls in technical replicates.A slightly higher levelofFigure 1.Representative samples from the observed cluster types with maize SNPs based on the GenomeStudio software.The three highlighted clusters display the area where the three different genotypes with homozygous allele A (red),heterozygous AB (purple)and homozygous allele B (blue)are called.Allele calls that fall in the lighter colored areas in between or below these areas are set to ‘failed’.Ellipses are used to adjust the position of the allele calling areas.A)Cluster Type 1:Accurate genotype calling of all three genotypes in essentially all 274maize lines with clearly defined clusters;B)Cluster Type 2:Three clusters with a number of failed samples at the bottom of the analysis plane that are not called;C)Cluster Type 3:Three clusters with a shift towards one side which is indicative for a duplicated locus.Clusters have to be shifted to the left for accurate allele calling;D)Cluster Type 4:Five clusters which are indicative of two simultaneously scored polymorphic loci in a duplicated sequence.Such markers cannot be scored accurately,so they were deleted from the data set.doi:10.1371/journal.pone.0028334.g001inconsistency (up to 1%)was observed with duplicated samples from different seed samples and sources.Similarly,a pedigree inconsistency of less than 1%was observed in the analyzed triplets further confirming the quality of the genotype calling with the array.Also,for the three DNA replicates of B73,Mo17and the hybrid,highly similar but not identical numbers of markers (B73:49,546–49,560out of 49,585;Mo17:48,719–48,737out of 49,585;B736Mo17F1:48,795–48,889out of 49,585)could be scored.In the next step,the cluster file was used to score all 274maize samples and the quality of the genotyping data was assessed with respect to the failure rate on a marker by marker basis (Figure 3and Table S2).The 49,585markers that could be scored with the established cluster had an average failure rate of 4.0%corre-sponding to an average of 11samples per marker.However,8,628(17.4%)of the markers had a failure rate of more than 5%although these markers produced the correct genotypes in the analyzed triplets as far as they could be fully analyzed there.ToFigure 2.Distribution and success rates of SNP markers on the Infinium array.A)Numbers and percentage of SNP markers from the different marker groups on the array,showing that most of the SNPs coming from sources other than Panzea were located in genes;B)Success rates for the individual marker classes.Numbers are based on all 57,838SNP markers manufactured.doi:10.1371/journal.pone.0028334.g002further elucidate the reasons for the relatively high failure rate for the maize SNP markers,a more detailed analysis was performed for individual groups of maize lines (Figure 4).These data revealed that the highest call rate (0.9987)was obtained for B73and the lowest (0.9187)for the teosinte samples with additional major groups such as the INRA material with European inbreds and the expired PVP inbred material from the US [28]in the middle,suggesting a correlation with increased sequence divergence relative to B73and other materials used to discover intital polymorphisms.Detailed information for each SNP,including source and genotypes across all 274lines is given as Table S3.Level of polymorphism between lines and potential ascertainment biasThe SNP markers were selected from different sources:many via the B73-Mo17pair (especially the Syngenta SNPs,see alsoMaterial and Methods),many as well from the NAM material which represented a more comprehensive sample of the maize germplasm,and still others from polymorphisms between key sets of lines (Table S4).A very high number of polymorphisms was observed for the combination of B736Mo17with approximately 25,325SNPs (numbers differ slightly between individual samples due to variation between the samples,see above).A dendrogram generated using all markers illustrated that Mo17was one of the most distant lines compared to the other analyzed samples and that the unadapted teosinte material was slightly less distant in the dendrogram than Mo17.This was mainly due to the Syngenta SNPs which were specifically selected for their high value in detecting polymorphism between B73and Mo17.When only the less biased Panzea markers (derived from the diverse set of NAM parents)were considered,the position of Mo17in the dendrogram changed significantly and to a more expected position (FigureS1).Figure 3.Number of SNPs with their corresponding failure rate.Failure rates are presented in %for the 49,585markers analyzed in the 274maize samples based on the MaizeSNP50_B.egt cluster file.doi:10.1371/journal.pone.0028334.g003Figure 4.Success rate for the 49,585markers in the different sample groups.doi:10.1371/journal.pone.0028334.g004Analysis of the functional markers in comparison to the maize genome sequenceTo include markers in as many maize genes as possible,one of the criteria during the original marker selection was their presence in maize genes.Originally,for19,350maize genes at least one SNP was put on the array.After the elimination of the markers that could not be scored,the49,585SNPs consisted of33,417 SNPs located in17,520genes and16,168SNPs located in intergenic regions.The minimum number of SNPs in a gene was1 and the maximum number was15SNPs(Table S5).Altogether 17,520genes could be analyzed with the established cluster file for the49,585markers,whereby the vast majority(15,404or88%)of these genes contained from one to three SNPs.The details about the number of SNPs per gene and their correspondence to the respective filtered gene or other maize genes are displayed in the Table S6.Because the remaining16,168SNPs were not in known maize genes,the full set of the49,585SNPs were located onto the maize B73reference sequence(AGPv2).In general,markers were well-distributed over the chromosomes with lower numbers of markers per megabase in many centromeric regions and a slightly higher number of markers per megabase near the telomeres.While typically there was an uncovered region of approximately 1Mbp on most of the chromosomes,on maize chromosome6, there was a region of more than2Mbp that did not contain a single SNP.The distribution of distances between adjacent markers shows that most distances are a few kilobases,but nevertheless for a significant number of regions the distance between adjacent markers was more than100kb.The IBM and LHRF framework mapsAll polymorphic markers were first analyzed for segregation bias in the allele frequencies within the two mapping populations. Some regions were skewed(Figure S2),with close markers generally distorted towards the same parent.On chromosome3, the IBM and LHRF populations showed similar distortion patterns,suggesting the presence of common sources of segrega-tion bias in both populations.Some markers are clear outliers, falling outside of the patterns,confirming the need for filtering out markers with high fractions of missing data or extreme distortion. After removing low-quality markers,24,816SNPs were used for mapping on the IBM population and17,047on the LHRF population.8,883SNPs were common to both populations.The genomic distribution of mapped markers for each population is given in Figure S3.The IBM and LHRF linkage maps were constructed independently using precisely the same procedures and parameters.To generate maps that are independent of the B73genome sequence,the map construction was based only on the SNP genotyping data.The theoretical resolution of the IBM and LHRF maps,based on populations composed of239and226 IRIL individuals respectively,should be better than0.1cM if there are enough markers.Neglecting heterogeneity in the spacings of our24,816or17,047marker,such a resolution seems accessible.However,for such high numbers of markers,especially if they are heterogeneously spaced,many will be indistinguishable, preventing one from building maps with robust marker orders. Thus,as a first step,a scaffold map was constructed with a limited number of markers,but whose order is very reliable.The IBM (respectively LHRF)scaffold obtained contained311markers separated by4.7to11.4cM(respectively345markers separated by4.5to10.0cM).The scaffolds were then augmented by adding as many markers as possible while keeping the robustness of marker order above a threshold.This produced an IBM framework map with1,934markers separated by0.2to12.3cM and a LHRF framework map with1,785markers separated by0.2 to14.5cM.The total map length was1,689cM for IBM and 2,168cM for LHRF(Figure S4and Table S7).Mapping of all additional polymorphic SNPsThe two framework maps were limited in their marker number by the constraint of maintaining high statistical robustness of the ordering.Given a framework map,additional markers can be placed by determining their positions without including them explicitly into the map.This approach is mandatory for mapping high numbers of markers using relatively small populations.For the IBM population,this placement led to a high density map with 20,913markers of total length1,725cM with a largest gap size of 11.6cM.For the LHRF population,we obtained a high density map with14,524markers,a total length of2,208cM,and a largest gap size of12.1cM.Furthermore,a total of7,368SNPs monomorphic on IBM could be mapped on LHRF,so the use of LHRF increased the overall number of markers that could be genetically mapped by35%.The results are presented in Figure S5and detailed in Table S8for all mapped markers. Genome-wide and chromosome-specific comparison between the genetic maps and the B73genome sequenceAmong the49,585markers used,over400markers,or approximately1%could not be unambiguously located on the B73genome sequence using a BLAST search.The majority of these unassigned markers were not found at all in the B73 reference sequence indicating that the corresponding genomic sequence was missing in the current assembly.Among such markers,172could be mapped on one or both of the genetic maps,indicating a potential location of the associated genomic sequence on a maize chromosome(Table S9).Figure5shows the relationship between genetic and physical positions on all chromosomes,along with the first derivative of this relationship providing an estimate of the meiotic recombination rate.In all chromosomes(less for chromosome6),recombination occurred predominantly near both telomeres whereas very large pericentromeric regions were almost devoid of recombination. Variations in recombination rate along the chromosomes were similar in both genetic maps except for some highly recombining regions that were specific for one cross such as in the middle of chromosome10for the LHRF map.Among20,788(respectively14,432)markers genetically mapped on the IBM(respectively LHRF)map and which were physically placed on the B73sequence,23(respectively24)did not show conserved chromosomal assignments(Figure6).Some of these non-syntenic markers were singletons,but in other cases they formed clusters.There were cases of singletons and clusters where both the IBM and LHRF genetic maps provided the same chromosomal assignments,yet were non-syntenic with the B73 genome.For instance,on chromosome10of the B73sequence, there was a cluster of seven markers that were mapped to chromosome2in both genetic maps strongly suggesting that these markers were erroneously positioned on the B73genome sequence.In another case,there were six markers forming a cluster in the B73sequence on chromosome8that the IBM map assigned to chromosome2whereas the LHRF map showed no assignment discrepancy with the B73genome on chromosome8. Table S10lists all markers that were non-syntenic between genetic and physical maps,as well as their inferred physical position on the B73genome based on their genetically determined chromosome assignment and positions.Colinearity within chromosomes was determined through the genome-wide comparative analysis of physical and genetic positions of shared markers(Figure5),based on the complete genetic maps.Some physical segments were devoid of markers on the two genetic maps(e.g.the centromeric region of chromosome 1)or specifically in one of the two maps.For instance,a large segment of approximately15Mb was completely lacking any markers on chromosome8of the IBM map.Otherwise,the coverage was quite dense and the maps mainly colinear.A first class of exceptions within chromosomes consisted of individual markers that lied far away from the common pattern of the other markers.These were quite rare,but interestingly they were often outliers for both the IBM and the LHRF maps(e.g.on chromosome5).A second class of exceptions consisted of groups of markers generally corresponding to small inversions between the genetic and physical map(e.g.on chromosome3at position 85Mb in both IBM and LHRF,see Figure5).Since small inversions would require larger populations to reach a high confidence level,these were not considered further.Based on the framework maps,we then defined regions containing major non-colinearities with the B73sequence (Figure6and Table S11).These regions involved several markers of one of the genetic maps,or involved one marker but overlapped with a non-colinear region of the other genetic map.This led to five regions for the IBM genetic map(2.3I,3.1I,6.2I,7.1I,and 9.1I)and nine(2.1L,2.2L,2.3L,3.1L,5.1L,6.1L,6.2L,7.1L,and 9.1L)for the LHRF genetic map.Interestingly,all five IBM regions closely overlapped with LHRF regions(see Figure6and Table S11):this concordance strongly points to probable errors in the B73sequence assembly.On the other hand,the remaining regions(2.1L, 2.2L, 5.1L,and 6.1L)could be suggestive of structural rearrangements between parental lines of the two populations.Using framework maps alone led to order robustness.However, their coverage is low and potentially misses small regions of non-colinearities or lacks power in revealing the fine structure of non-colinearities.Thus,the complete maps were used to further analyze these regions(e.g.in Figure7;all regions are shown in Figure S6);this analysis reinforced the evidence for non-colinearities and refined their structure.The extra resolution also revealed a non-colinearity between the IBM genetic map and the B73sequence in the region5.1L compatible with that observed on LHRF,suggesting in this case an error in the B73assembly rather than a structural rearrangement between maize lines. DiscussionArray development and array characterizationStarting from over800,000SNPs that were identified in a number of SNP discovery projects,a set of57,838SNPs was selected for synthesis and manufacture.The marker selection was based on the fact that maize is a highly polymorphic plant species with a low level of linkage disequilibrium(LD).Especially in unadapted or wild maize material,LD extends only from several hundred base pairs to several kilobases[29].Because polymor-phism within genes or their close vicinity are expected to be the main basis of phenotypic variation,in the SNP selection process a first priority was given to SNPs located in genes.SNPs in genes are potentially more informative in GWAS studies and thus it was attempted to cover as many genes as possible based on the filtered gene or high confidence gene set[2].With17,520genes containing at least one SNP,more than50%of these high confidence genes could be covered.Another16,168SNPs were used to populate other regions of the maize genome with markers and to obtain a relatively even marker distribution.In theory,an even distribution of50,000markers would result in an average distance between markers of40–50kilobases.In reality and with the focus on one or more SNPs in maize genes,the distance between many adjacent SNP markers was much lower(in the range of a few kilobases).In other regions,the distance between markers went up to hundreds of kilobases because the maize genome contains large stretches of highly repeated sequences that cannot be used for SNP analysis on arrays.Compared to animal species for which large genotyping arrays have been developed[22,23],the MaizeSNP50array contains a relatively large number of markers that were difficult to score or had to be dropped from the analysis altogether.Furthermore, compared to mammalian species where significantly more than 99%of all marker/individual combinations could be scored,in maize on average,only approximately96%of all marker/ individual combinations could be scored.Also,in parent/offspring triplets,more than300markers did not produce the correct genotype in the F1compared to the two inbred parents.This relatively low marker functionality has probably two causes:The first is that maize,compared to animals,has a much higher level of genetic variation.While in animals and humans[30,31],on average one SNP is observed about every kilobase,in maize there is a10–20times higher genetic variation.Depending on the analyzed germplasm,on average,one SNP appears every44–75 base pairs.With such a high level of SNP polymorphism,it is very likely that in different maize lines there will be adjacent SNPs within the approximately20base pairs that are necessary for the Infinium assay primer.If this is the case,then it is very likely that the respective SNP will fail in the respective line(s).The percentage of generated SNP data points for a given line is roughly correlated with its genetic distance to B73.Indeed the reference sequence represented by B73[2]has the highest success rate over all assays, reaching99.87%while the genetically highly diverse teosinte had only a success rate of,on average,91.87%.A second source of low marker functionality is the fact that maize is an ancient polyploid species with large genomic regions that have been duplicated in its evolutionary past.Due to this genome duplication,many maize genes have a second copy at another position in the maize genome that differs by a varying extent.A considerable number of these duplicated genes have a sequence diversity of significantly less than 10%.Thus,it is very likely that a number of SNP assays detect not one locus but multiple highly similar paralog(s).This is confirmed by the observation that in maize,a considerable number of SNP markers show a pattern(Pattern Types3and4as described in the Results)that are indicative of detecting more than one locus(shift of the clusters to one side or five clusters)as it is found in true tetraploid species[27].Markers with such a pattern occurred frequently in groups at specific positions that correspond to the duplicated regions identified in the maize genome sequence.As aFigure5.Relationship between physical and genetic positions,and corresponding recombination rates.X-axis:physical position(in Mbp)of the SNPs on the B73physical map.Left Y-axis:genetic positions of SNP markers on IBM(red triangles)and LHRF(blue circles)linkage maps. Right Y-axis:recombination rate in centiMorgan per Mega base pair for the IBM(solid red line)and LHRF(dashed blue line)maps.Recombination rates were obtained as the first derivative of the smoothed curve representing genetic versus physical positions.The thick arrow indicates approximate centromere position according to MaizeGDB.doi:10.1371/journal.pone.0028334.g005。
