Eppendorf Research
Proflex PCR System使用说明书

USER GUIDEFor Research Use Only. Not for use in diagnostic procedures.ProFlex ™PCR System User GuideInstallation, Use, and Maintenancefor use with: PCR reagents from Invitrogen ™ and Applied Biosystems ®Catalog Number 4483636, 4483637, 4483638, 4484071, 4484073, 4484074, 4484075, 4484076, 4484078,4484072, and 4484077Publication Number MAN0007697RevisionA.0For Research Use Only. Not for use in diagnostic procedures.The information in this guide is subject to change without notice.DISCLAIMERLIFE TECHNOLOGIES CORPORATION AND/OR ITS AFFILIATE(S) DISCLAIM ALL WARRANTIES WITH RESPECT TO THIS DOCUMENT, EXPRESSED OR IMPLIED, INCLUDING BUT NOT LIMITED TO THOSE OF MERCHANTABILITY, FITNESS FOR A PARTICULAR PURPOSE, OR NON-INFRINGEMENT. TO THE EXTENT ALLOWED BY LAW, IN NO EVENT SHALL LIFE TECHNOLOGIES AND/OR ITS AFFILIATE(S) BE LIABLE, WHETHER IN CONTRACT, TORT, WARRANTY, OR UNDER ANY STATUTE OR ON ANY OTHER BASIS FOR SPECIAL, INCIDENTAL, INDIRECT, PUNITIVE, MULTIPLE OR CONSEQUENTIAL DAMAGES IN CONNECTION WITH OR ARISING FROM THIS DOCUMENT, INCLUDING BUT NOT LIMITED TO THE USE THEREOF.Important Licensing InformationThis product may be covered by one or more Limited Use Label Licenses. By use of this product, you accept the terms and conditions of all applicable Limited Use Label Licenses.TrademarksAll trademarks are the property of Thermo Fisher Scientific and its subsidiaries unless otherwise specified. AmpliTaq and AmpliTaq Gold are registered trademarks of Roche Molecular Systems, Inc. Clorox is a registered trademark of The Clorox Company, Bio-Rad, MyCycler, and C1000 Touch are trademarks of Bio-Rad Laboratories, Inc. Eppendorf, Mastercycler, and MJ Research are trademarks of Eppendorf AG.©2014 Thermo Fisher Scientific Inc. All rights reserved.ContentsAbout this Guide (6)Revision History (6)Purpose (6)■CHAPTER 1 About the System (7)About the system (7)Use the Touchscreen (8)Enter text (9)Enter numerals (10)Touchscreen menu overview (11)■CHAPTER 2 Initial Set-up (14)Site requirements (14)Environmental requirements (14)Temperature and humidity requirements (14)Pollution (14)Altitude (14)Materials (15)Required materials (15)Optional protective hardware (15)Unpack the product (15)Set up the system (16)Set up the wired connection (18)Set up the wireless connection (20)Recommended instrument settings (21)Change sample blocks (30)■CHAPTER 3 Operating the ProFlex™ PCR System (31)Load samples into the instrument (31)Load samples (31)Load samples in the ProFlex™ 3x32-Well PCR System (33)Create a new run method (34)Edit a run method (36)ProFlex™ PCR System User Guide 3 For Research Use Only. Not for use in diagnostic procedures.ContentsManage methods and folders (41)Perform a run (43)Perform a run on multiple instruments (45)Connect the instruments (45)Perform a run on multiple instruments (45)Monitor a run (46)Monitor via the Home screen (46)Monitor via the run monitor screen (46)If the power fails (48)View and export the run report (48)Remove the samples from the instrument (48)■CHAPTER 4 Routine Maintenance (49)As-needed maintenance (49)Clean the instrument (49)Replace the fuses (51)Upgrade the system firmware (51)Self Verification test (53)Order kits and replacement parts (54)■APPENDIX A Ordering Information (55)Instrument part numbers (55)Consumables (55)■APPENDIX B Troubleshooting (59)Troubleshoot problems (59)Return an instrument for service (60)■APPENDIX C Instrument specifications (62)Technical specifications (62)System specifications (64)Location of power point and ports on the instrument (65)■APPENDIX D Predefined Run Methods (66)■APPENDIX E Safety (81)Symbols on this instrument (81)Safety alerts on this instrument (82)Location of safety labels on this instrument (83)Safety information for instruments not manufactured by Thermo Fisher Scientific (85)4ProFlex™ PCR System User GuideFor Research Use Only. Not for use in diagnostic procedures.Contents Instrument safety (86)General (86)Physical injury (86)Electrical (86)Cleaning and decontamination (86)Laser (87)Safety and electromagnetic compatibility (EMC) standards (87)Safety (87)EMC (87)Environmental design (88)Chemical safety (88)Biological hazard safety (89)Documentation and support (90)Related documentation (90)Obtaining SDSs (90)Obtaining support (91)Limited product warranty (91)ProFlex™ PCR System User Guide 5 For Research Use Only. Not for use in diagnostic procedures.About this Guide Revision HistoryPurposeThe ProFlex™ PCR System User Guide provides information about installing, using,and maintaining the ProFlex™ PCR System.6ProFlex™ PCR System User GuideFor Research Use Only. Not for use in diagnostic procedures.1About the SystemThe ProFlex™ PCR System is an end-point thermal cycler, specifically designed for theamplification of nucleic acids using the Polymerase Chain Reaction (PCR) process.The user interface consists of a touchscreen with a graphical display that shows thetime, status, and temperature for each run. A touchscreen keypad allows you to enterinformation into fields on the display screen.About the systemThere are five models of the ProFlex™ PCR System:Five interchangeable ProFlex™ Sample Blocks are available:The ProFlex™ PCR System allows you to:•Run three experiments at a time with the 3x32-well, 0.2-mL block•Interchange the blocks (e.g. exchange the ProFlex™ 3x32-Well Sample Block withthe ProFlex™ 96-Well Sample Block, the ProFlex™ Dual 96-Well Sample Block, theProFlex™ Dual Flat Sample Block, the ProFlex™ Dual 384-Well Sample Block, andvice-versa)•Access the system remotely through a mobile application•Program the instrument from the touchscreen interface•Simulate other PCR instruments with Thermal Simulation modesProFlex™ PCR System User Guide 7 For Research Use Only. Not for use in diagnostic procedures.•Optimize your PCR with the help of the Veriflex ™blocksFigure 1 ProFlex ™3x32-Well PCR SystemFigure 2 ProFlex ™ 96-Well PCR SystemThis user guide provides information on how to maximize the capabilities of your ProFlex ™ PCR System. This user guide provides unique instructions for the different ProFlex ™ block types at the beginning of each section.Detailed specifications for the ProFlex ™ PCR System are found in Appendix C,“Instrument specifications“.Use the TouchscreenYou interact with the instrument using a touchscreen. Table 1 describes the several buttons common to many of the screens in the ProFlex ™ System.Table 1 Buttons in the touchscreenChapter 1 About the System Use the Touchscreen18ProFlex ™ PCR System User Guide For Research Use Only. Not for use in diagnostic procedures.and buttons.When you touch a field that requires the input of text, the text editor, as seen in the following figure, opens. Table 2 displays the buttons to touch to enter different information types.Table 2 Buttons to enter different informationEnter text Chapter 1 About the System Use the Touchscreen 1ProFlex ™ PCR System User Guide 9For Research Use Only. Not for use in diagnostic procedures., then the symbol(s). When you are done, touch again. to accept the entry and close the editor.When you touch a field that requires a numerical input, the numeric editor, as seen in the following figure, opens. Table 3 displays the buttons to touch to perform different functions while entering numbers.Table 3 Functions performed by entering numbersEnter numerals Chapter 1 About the System Use the Touchscreen110ProFlex ™ PCR System User Guide For Research Use Only. Not for use in diagnostic procedures.Touchscreen menu overviewOn powering on the instrument, you will first see the following Splash screen.Figure 3 ProFlex ™ PCR System Splash screenAfter the Splash screen, the Home screen appears.The instrument has interchangeable blocks, and detects which module is present on the system automatically. After the block completes initializing, depending on the block in use, the Home screen displays status dials; three for the ProFlex ™ 3x32-Well Sample Block and one for the ProFlex ™ 1x96-Well Sample Block, the ProFlex ™ Dual 96-Well Sample Block, the ProFlex ™ Dual Flat Sample Block, and the ProFlex ™ Dual384-Well Sample Block.Figure 4 Home screenThe Home screen includes the following features:1.Status dial : The number of dials appearing on the Home screen varies with the block module in use. While the instrument is in use, the status dial displays the current temperature of the block, the time elapsed for a run, and the run status.When the instrument is not in use, the status dial/s will show the 'Set Up Run'display. You can start a run by touching the status dial where it says Set Up Run.For instructions on starting a run by this method, see “ Perform a run“ on page 43.Chapter 1 About the System Touchscreen menu overview 12.Sign In : Touch Sign In to create an account or to enter information to sign in to your account.•To create a new account:a.In the Sign In screen, touch Sign Up .b.In the Create An Account screen, enter your personal information,create a user name and password, and enter a name for the defaultfolder that will contain the account information.c.TouchCreate Account•To sign-in to an existing account:1.Enter your user name and password.Note: To sign-in to an existing account as an administrator, enter youruser name as "Administrator" and password as "password".2.Touch Sign In .Chapter 1 About the System Touchscreen menu overview1Note: Once you have signed into your account, the Sign In button on the Home screen turns to My Account. The user name (custom or administrator) appears inthe bottom-left of the Home screen.3.New Method : Touch New Method to create a new run method with the help of the default templates that are pre-loaded with the instrument. Touching New Method takes you to the Setup Run screen that allows you to create a new run method by using the default templates or existing methods. You can also create a run method for incubation from the Setup Run screen.4.Open Method : Touch Open Method to select an existing run method to start a run. Touching Open Method takes you to the Select Method screen that displays the existing run methods. The run methods are sorted and stored in various folders. Select a folder to display the run methods within the folder. Touch a run method under the Method name to edit that run method in the edit mode.5.Incubate : Touch Incubate to use the instrument as a precise incubator for non-PCR workflows.6.Settings : Touch Settings to configure the instrument.Chapter 1 About the System Touchscreen menu overview 1Initial Set-up This chapter includes the procedures for installing the ProFlex ™PCR System.Site requirements The ProFlex ™ PCR System is for indoor use. Ensure that the installation site:•Meets the spatial and weight requirements (see “System specifications“ on page 64)•Meets environmental requirements (see “Environmental requirements“ on page 14)•Is within 1 m (3 ft.) of an AC power source receptacle•Is away from waterEnvironmental requirementsEnsure that the installation site is maintained under the following conditions:Table 4 Temperature and humidity requirementsAvoid placing the instrument adjacent to heaters, cooling ducts, or in direct sunlight.Fluctuations between day and night temperatures can cause system instability. Place away from any equipment that vibrates, such as a refrigerator or centrifuge.The ProFlex ™ PCR System has a Pollution Degree rating of 2. It may be installed in an environment that has non-conductive pollutants only, such as dust particles or wood chips. Typical environments with a Pollution Degree II rating are laboratories and sales and commercial areas.The safety of ProFlex ™ PCR System use was tested for altitudes up to 6000 ft.2Temperature and humidity requirements Pollution AltitudeMaterials•Scissors, pocket knife, or box cutter •Compressed air •Protective hardware, as appropriate (see below)Life Technologies supports the use of the following devices to protect the ProFlex ™PCR System from damage resulting from electrical hazards and the resultant loss of data. Before installing the system, decide what additional hardware (if any) you want to install.•Power line regulator (1.5-kVA)•Surge protector/line conditioner (10-kVA)•Uninterruptible power supply (1.5-kVA)Unpack the productIMPORTANT! Save the packing materials and box in case you need to ship the instrument to Life Technologies for service.1.To unpack the ProFlex ™ System:a.Cut the straps securing the instrument box.b.Cut the tape securing the top flaps of the instrument crate, then open the flaps.c.Remove the ProFlex ™ System Accessories from the instrument and set them aside.d.Lift and remove the cover from the instrument crate.e.Remove the packing material from the ProFlex ™ System, then inspect the instrument for shipping damage.IMPORTANT! If the ProFlex ™ System is damaged, note the location and appearance of the damage, then contact Life Technologies Technical Support or your service representative (see “Obtaining support“ on page 91).2.Move the ProFlex ™ System to the desired installation site. Follow these guidelines for lifting and moving:•Make sure that you have a secure, comfortable grip.•Keep your spine in a neutral position.•Bend at the knees and lift with your legs.•Do not lift and twist your torso at the same time.Required materialsOptional protective hardware Chapter 2 Initial Set-up Materials 23.Verify that the package containing the ProFlex ™ System Accessories includes the ProFlex ™ System Starter Kit :•Power cord cable•ProFlex ™ System Starter Kit•USB-enabled Wi-Fi Card (Part no. 4483658)Set up the system1.Remove the packing material:a.Open the ProFlex ™ System heated cover.b.Remove the packing material from the ProFlex ™ sample block(s) (ProFlex ™96-Well Sample Block, ProFlex ™ 3x32-Well Sample Block, ProFlex ™ Dual 96-Well Sample Block, ProFlex ™ Dual Flat Sample Block, or ProFlex ™ Dual 384-Well Sample Block) .ing a can of compressed air, deliver a blast of air into each well of the sample block(s) to remove any particles that may have collected during transportation.2.Install the sample block. To place the sample block into the instrument base:a.Facing the ProFlex ™ Base unit away from you, pull the lever all the way tothe right hand side.Figure 5 ProFlex ™ System Base unit and the ProFlex ™ 3x32-Well Sample Block)b.Place the sample block onto the ProFlex ™ Base with the latching mechanismto the right.Figure 6 ProFlex ™ System Base unit with the latching mechanism to the rightChapter 2 Initial Set-up Set up the system2c.Push the latching mechanism all the way to the left to secure the block ontothe base.Figure 7 ProFlex ™ System Base unit with the latching mechanism to the left Note: If the sample block module is not seated in place correctly, the instrument will not be able to detect the block type.Note: To change the sample block from one type to the other, see Change sample blocks.3.Close the heated cover.4.Connect the power cable to the ProFlex ™System.Figure 8 Power cable inlet in the ProFlex ™ System5.(Optional) Install any of the recommended protective devices (see “Optional protective hardware“ on page 15).6.Connect the power cable to the AC power source receptacle.7.Connect the instrument to the network by inserting the ethernet cable into the ethernet port at the back of the ProFlex ™ System. For instructions on setting up the wired connection, see “Set up the wired connection“ on page 18.In the absence of the ethernet cable or the ethernet port, you can connect the instrument to the network via the USB-enabled Wi-Fi Card (Cat. no. 4483658) as shown in the following graphic.Insert the Wi-Fi Card into the USB port at the back of ProFlex ™ System. For instructions on setting up the wireless connection, see “Set up the wireless connection“ on page 20Chapter 2 Initial Set-up Set up the system 2Figure 9 USB port in the ProFlex ™ System unit 8.Toggle the power switch, then wait for the instrument to start up. The touchscreen displays the Main Menu, indicating that the ProFlex ™ System startup is complete. When you power on the instrument, the instrument may require about 45 seconds to start up.Note: First time users will be prompted with the End User License Agreement (EULA) screen. See “Recommended instrument settings“ on page 21 for moreinformation on the EULA.Figure 10 ProFlex ™ System Touchscreen Home See “Set up the system“ on page 16for instructions on connecting the ProFlex ™ System to the network. To set up the wired connection:1.On the Home screen, touch Settings.Figure 11 ProFlex ™ PCR System Settings screen Set up the wired connectionChapter 2 Initial Set-up Set up the system22.In the Settings screen, touch Instrument Settings.Figure 12 ProFlex ™ PCR System Instrument Settings screen3.In the Instrument Settings screen, touch Network Connection.Figure 13 ProFlex ™ PCR System Network Connection screen4.In the Network Connection screen, touch Wired .The Network Configuration screen opens up.Figure 14 ProFlex ™ PCR System Network Configuration screenIn the Network Configuration screen, you can choose to connect to the network by obtaining the IP address either automatically (using DHCP) or manually. If your instrument is not on a network, you do not need to set the IP address.Chapter 2 Initial Set-up Set up the system 2Note: Ask your system administrator if the IP address is assigned statically or dynamically. For static addresses, you need to know the IP address for the instrument, the subnet mask, and the default gateway.•Automatically:Touch Obtain an IP address automatically (using DHCP). A check mark appears when DHCP is selected.or •Manually:Touch Use the following IP Address , then enter the appropriate IP addresses for the instrument, the Subnet Mask, and, optionally, the Default Gateway, the Primary DNS Server, and the Secondary DNS Server using the numeric editor. Addresses are in the form of X.X.X.X, where each X is a 3-digit number, from 001 to 255.5.Touch OK to save the changes and go back to the Network Connection screen.Touch Cancel to exit the screen without saving the changes.See “Set up the system“ on page 16on connecting the USB-enabled Wi-Fi Card into the ProFlex ™ System. To set up the wireless connection:1.Refer to “Set up the wired connection“ on page 18 through step 3 to arrive at the Network Connection screen.2.In the Network Connection Type screen, touch Wireless.Figure 15 ProFlex ™ PCR System Network Connection screen The Choose Network screen will come up and the wireless symbol will appear active. Touch Cancel to exit the screen.Note: During initial setup, if you selected the Wired option in the Network Connection Type screen, you will be required to enter your IP address if you selected the Static IP wired option. If you selected the Dynamic IP wired option,the IP address is automatically populated.3.Once a wireless connection has been detected, a list of the available networks will be displayed in the Choose Network screen. Touch the network name of your choice or touch Join Other Network .If you choose Join Other Network, the Configure and Join Network screen opens.Set up the wireless connectionChapter 2 Initial Set-up Set up the system 24.In the Configure and Join Network screen, enter the name and security type of the network.Note: When you touch in the Network Name field, a keypad will come up to facilitate making the entry.5.From the Security type drop-down menu, touch to select the security type and enter the relevant information in the screen.Note: Contact your IT Systems Administrator for information on security type.You can select from the following options:•Open•WEP•WPA Personal•WPA2 Personal•WPA Enterprise•WPA2 EnterpriseNote: The above options are available only if you selected Join Other Network in step 3. You cannot change the security type if you selected an existing network.6.Touch Join to continue or Cancel to exit from the Find and Join a Network screen.7.Depending on the security type you have selected, enter the appropriate passwords and touch Join .If you entered the correct information, the Network Connection Complete screen will appear. Touch OK to continue. If you entered incorrect information, the Network Connection Failed screen will come up and you will require to go back to security type screen to resolve the issue. Touch OK to continue.You can configure the following instrument settings by touching Settings on theHome screen:Note: The Manage Users button is visible only when you sign in as an administrator.•Instrument Settings : Touch Instrument Settings to set various additional parameters:Set the following in the Instrument Settings screen:Recommended instrument settings Chapter 2 Initial Set-up Set up the system 2Note: The Backup/Restore and Restore Factory Settings are available only when you sign in as an administrator.–Instrument Name : Touch the Instrument Name field and, using the text editor, enter up to 25 alphanumeric characters to identify the instrument.Note: The instrument name cannot have spaces. Separate consecutive characters with a hyphen or underscore; for example, MyInstrument.–Date/ Time : Touch Date/ Time to set the date and time for a run.1.Enter the date and time in the Date/Time field using the numerical editor that comes up when you place the cursor in the respective fields.You can toggle between AM and PM by touching in the field directly.2.Touch OK to save the date and time changes or Cancelto exit.–Sleep Mode : In the Sleep Mode screen, touch the Off and On toggle button to disable or enable, respectively, the sleep mode.Chapter 2 Initial Set-up Set up the system 2In the 'On' mode, you can edit the time (in minutes) after which theinstrument will go from the idling state into standby mode.–Network Connection : Touch Network Connection to select the type of network connection. You can select from the Wireless and Ethernet options.For details on using the Wireless and Ethernet options, see “Set up the wired connection“ on page 18 and “Set up the wireless connection“ on page 20,respectively.–Heated Cover :1.In the Heated Cover screen, touch the On and Off toggle button to enable or disable, respectively, the heated cover idling temperature.2.Edit the idling temperature if the button is in the 'On' mode.Note: The temperature for the heated cover must be between 30°C and 110°C.3.Click OK to save the changes or Cancelto exit the screen.Chapter 2 Initial Set-up Set up the system 2–Remote Service : In the Remote Service screen, use the On and Off toggle button to enable or disable, respectively, remote service. The feature allows the instrument to upload data, periodically, to a remote server. For more information on remote services, visit /proflex .Note: To connect to the ProFlex ™ PCR System with your mobile device,Remote Services must be switched on.Note: Ensure that you are connected to a network if you want to enable the Remote Service feature.After you have set the toggle button, touch OK to save the change or Cancelto exit the screen.–Printer Configuration : In the Printer Configuration screen, enter the IP address of the printer in the Remote Printer IP Address After you have entered the IP address, touch OK to save the change or Cancelto exit the screen.–Multi-Instrument Setup : Use this feature to allow multi-instrument runs on visible instruments. In the Multi-Instrument Setup screen, touch:1.The Off and On toggle button to make the instrument on which you are running an experiment invisible or visible, respectively, to other instruments on the network.2.The second set of Off and On toggle buttons to ignore or find,respectively, other instruments on the network.Chapter 2 Initial Set-up Set up the system 2–Backup/Restore (for Admin only): In the Backup/ Restore screen, touch:–Backup Instrument to back up the instrument settings, user accounts,and methods on the instrument you are working to a USB drive.–Restore a Backup to view the files that you have backed up on a USBdrive.This feature can be useful in event of a hardware failure or while setting up multiple instrument runs.–Restore Factory Settings (for Admin only): Touch Restore Factory Settings to remove all the data and customized settings and revert to factory settings.Touch Yes to confirm if you want to restore factory settings or Cancel to exit the screen.At the end of the restoration process, the message, "Your instrument has been restored." will be displayed and the instrument will automatically reboot after 30 seconds.Note:All data and settings will be erased once factory settings are restored.Chapter 2 Initial Set-up Set up the system 2•About Instrument : Touching About Instrument will take you to the About Instrument screen where you can find out more information about the instrument as well as view the End User License Agreement (EULA).–Touch About Instrument to find out more about the ProFlex ™ PCR System.In the About Instrument screen, you can view information like firmware version and instrument statistics.–Touch EULA to view the End User License Agreement. You can also save theEULA document to a USB drive.•Run History : The Run History screen displays the entire list of runs performed using a particular ProFlex ™ PCR System.a.In the Run History screen, touch a particular Run ID to view the details of that run.b.Touch Export to save the run details to a USB device or Print to print the rundetails.Chapter 2 Initial Set-up Set up the system2Note: If you sign in as an Administrator, you can manage the Run History. Forexample, deleting a run history.•Maintenance & Services : You can perform the following in the Maintenance &Services screen:–Software Update : Touch Software Updates to update the System firmware.See “ Upgrade the system firmware“ on page 51 for instructions on updating the firmware.–Service Reminders : Touch Service Reminders to set the time interval for the service reminder. Use the Off and On toggle button to turn off and on,respectively, the service reminder. If you select On, the time interval field becomes visible. Touch the time interval field to choose from 12 Months, 9Months, 6 Months, 3 months, and 1 Month. Touch OK to save the change or Cancel to exit the screen.–Self Verification Test : Touch Self Verification Test for the instrument to conduct a check on the instrument hardware. The check includes testing the block, heated cover, and other components. See Chapter 4, “ Routine Maintenance“ for instructions on conducting the self-verification test.–Export Instrument Log : Touch Export Instrument Log to export the instrument logs to a USB. Insert the USB into the USB drive before using this feature.–Block Verification Test : Touch Block Verification Test to perform a block verification test. Ensure that you have the Temperature Verification Kit (TVK) (Cat. no. 4377669) (need corresponding TVK for ProFlex) before performing this test. The TVK kit is available at /temperatureverificationkits . Touch:–Verify Block Temperature to carry out the block temperature test.1.Choose the test type from:–Heated Cover–Temperature Verification–Temperature Non-Uniformity2.Touch Next .Connect the TVK and depending on the test type perform insert the TVK probe into a specific zone.3.Touch Start Test .–Verify Cycle Performance to check the cycle performance of the instrument.Chapter 2 Initial Set-up Set up the system 2。
ITS序列分析。

摘要关于水稻与近缘稻种关系的研究方法,生物学上已有多种学说,由于目前国内外的近缘水稻rDNA的(internal transcribed spacers,ITS)以及他们的二级结构并未进行很多研究,所以本研究拟选取ITS序列作为一个分子分析指标,对他们的亲缘关系进行探索。
我们用PCR扩增的方法获得了ITS,并进行PCR产物的克隆测序。
材料选取包括广陆矮四号稻、药用野生稻、宽叶野生稻、高杆野生稻四种。
对以上四种稻的rDNA内IT S(ITS1+ITS2)以及5.8s rDNA序列进行测定和分析。
最后,本文还用软件对栽培稻与这几种野生稻ITS2的二级结构进行了预测。
关键词野生稻;ITS序列;PCR1 前言稻属(Oryza)是种子植物门,单子叶植物纲,禾本目,包括20余个野生种。
中国是世界上水稻栽培历史最悠久的国家,据浙江余姚河姆渡发掘考证,早在六七千年以前这里就已种植水稻,比泰国还早千余年。
目前,我国水稻播种面占全国粮食作物的1/4,而产量则占一半以上。
栽培历史已有6000~7000年。
为重要粮食作物;不仅如此,我国的稻种类型繁多。
目前中国收集保存的水稻种植中,来自国内的就占87.84%[1],其中地方品种占81.26%[1]。
在如此众多的品种中,如何区分判断不同稻种之间的亲缘关系,利用更有效的方法研究优良遗传性状,这是水稻资源研究的重要内容之一。
目前人们对植种亲缘关系的研究方法有很多,例如非常成熟的杂交法。
从整体上看,遗传多样性的研究方法从个体形态学水平、细胞学水平、生理生化水平发展到了分子水平,研究层次也随之深入。
论述了植物遗传变异的来源,总结并分析比较了不同水平的遗传多样性研究方法[12]。
进入21世纪现代生物学基因技术飞速发展,从分子水平认识和了解水稻间的亲缘关系成为一种潮流,成为生物学的新的研究领域。
在基因方面的研究中,人们发现植物的rDNA中的ITS(internal transcribed spacers)序列有着非常丰富的遗传学信息。
219402627_HR催化剂在PP_EP548R生产中的应用

工业技术CHINA SYNTHETIC RESIN AND PLASTICS合 成 树 脂 及 塑 料 , 2023, 40(2): 47DOI:10.19825/j.issn.1002-1396.2023.02.11聚丙烯(PP)EP548R是一种具有中高流动性的抗冲共聚聚丙烯,具有优异的刚韧平衡性和加工性能,适用于家电、汽车、办公、医疗、日常消费用品等的生产[1]。
在300 kt/a的Spheripol-Ⅱ工艺装置进行生产的过程中,用进口催化剂生产的PP EP548R存在轴流泵功率波动、装置运行不稳定的问题。
同时,近年来市场对塑料制品轻量化的需求与日俱增,对EP548R的刚韧平衡性提出了更高的要求,提升PP的刚性有利于减少材料厚HR催化剂在PP EP548R生产中的应用王子强(中国石化催化剂有限公司北京奥达分公司,北京 100029)摘要:采用HR催化剂在300kt/a的Spheripol-Ⅱ工艺装置上生产了中高流动、高刚性抗冲共聚聚丙烯EP548R,考察了HR催化剂的活性、立构定向能力、氢调敏感性、抗冲共聚能力及装置运行稳定性,并测试了EP548R的力学性能。
结果表明:与进口催化剂相比,HR催化剂活性提高约20%,外给电子体用量减少约30%(w),氢气加入量降低约10%(y),说明HR催化剂具有高活性、高立构定向性和高氢调敏感性。
在生产负荷相当的情况下,轴流泵功率波动仅为应用进口催化剂的10%,且细粉含量仅为应用进口催化剂时的1/2。
在气相反应釜乙烯浓度相同的条件下,采用HR催化剂生产的EP548R具有更高的弯曲模量和相似的悬臂梁缺口冲击强度,说明HR催化剂具有更好的抗冲共聚能力。
关键词:丙烯聚合 抗冲共聚聚丙烯 HR催化剂 高刚性中图分类号:TQ 325.1+4文献标志码:B 文章编号:1002-1396(2023)02-0047-05Application of HR catalysts in PP EP548R productionWang Ziqiang(SINOPEC Catalyst Co.,Ltd. Beijing AUDA Division,Beijing 100029,China)Abstract:EP548R grade impact copolymerized polypropylene(PP) with medium-high melt flow rate (MFR) and high stiffness was produced by HR catalysts at a 300 kt/a Spheripol plant. The activity,stereospecificity,hydrogen sensitivity and impact copolymerization capability of HR catalyst as well as the stability of plant operation when utilizing HR catalyst were investigated. The mechanical properties of the produced resin were characterized. The results show that compared to imported catalysts,the activity of HR catalysts is increased by 20%,the mass fraction of external electron donor is decreased by 30% and the molar fration of hydrogen is reduced by 10%,which means the activity,stereospecificity and hydrogen sensitivity of HR catalyst are higher than those of imported catalysts. The fluctuation of axial pump power is decreased by 90% when HR catalyst is used than that when imported catalyst is used at the same operation conditions. The content of fine powders is halved by utilizing HR catalysts as well. EP548R grade resin produced by HR catalysts has higher flexural modulus and same Izod notched impact strength at the same ethylene to propylene ratio in the gas-phase reactor,indicating that HR catalysts perform better in impact copolymerization.Keywords:propylene polymerization; impact copolymerized polypropylene; HR catalyst; high stiffness收稿日期:2022-11-26;修回日期:2023-01-13。
材料类的国外期刊以及投稿经验

英文材料期刊简介Journal of Alloys and Compounds《合金与化合物杂志》瑞士ISSN:0925-8388,1959年创刊,全年36期,Elsevier Science出版社出版,SCI收录期刊,SCI2003年影响因子1.080。
国际性材料科学和固体化学与固体物理学杂志。
刊载稀有金属及其化合物、合金的实验和理论研究论文、会议报告、简讯与书评。
文章多用英文发表。
Journal of Composites for Construction《建筑复合材料杂志》美国ISSN:1090-0268,1997年创刊,每年4期,American Society of Civil Engineers,USA出版。
刊载有关建筑用合成纤维增强复合材料的研究论文。
SCI、EI收录期刊,SCI2003年影响因子1.234,被引频次249、即年指标0.125、年载文量40。
2003年EI收录91篇。
EI收录期刊,EI2001年收录25篇。
Journal of Materials in Civil Engineering《土木工程材料杂志》美国ISSN:0899-1561,1989年创刊,每年6期,American Society of Civil Engineers,USA出版。
刊载土建材料的开发、加工与现场生产、特性评价、应用和性能等方面的研究论文。
SCI、EI收录期刊,2002年SCI影响因子0.346、被引频次193、即年指标0.015、年载文量66、被引半衰期4.9。
EI2002年收录66篇。
Journal of the European Ceramic Society《欧洲陶瓷学会志》英国ISSN:0955-2219,1985年创刊,全年16期,Elsevier Science出版社出版,SCI、EI收录期刊,SCI2003年影响因子1.248,2003年EI收录396篇。
主要发表研究陶瓷材料结构、特性和加工的原始论文。
华中科技大学学术期刊分类目录(T-D)_最新权威版
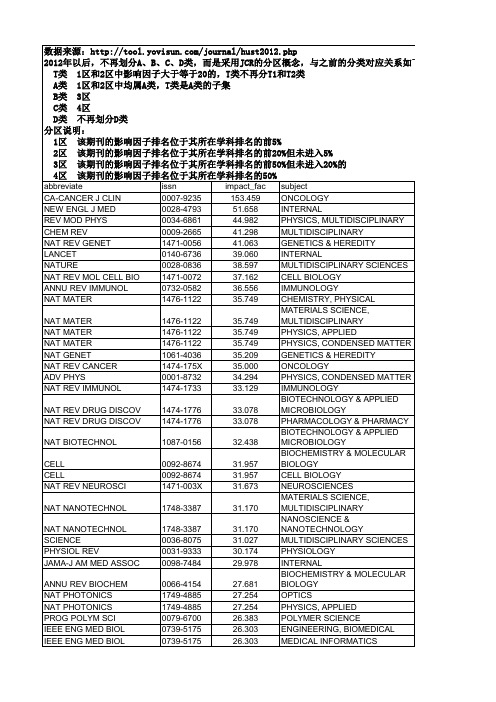
24.755 23.917 23.654 23.565 23.333 23.194 22.929 22.864 22.864 22.864 22.490 22.345 22.333 21.757 21.543 21.543 20.833 20.761 20.614 19.966 19.795 19.547 19.352 18.571 18.571 18.038 17.983 17.983 17.949 17.689 17.689 17.689 17.436 17.313 17.215 16.417 16.238 16.179 16.008 16.008 15.766 15.575 15.518 15.389 15.389 15.389 15.333 15.333 15.280 15.280 15.265 15.253 15.251 15.202
ONCOLOGY CLINICAL NEUROLOGY PLANT SCIENCES BIOCHEMICAL RESEARCH METHODS ASTRONOMY & ASTROPHYSICS MATERIALS SCIENCE, MULTIDISCIPLINARY PHYSICS, MULTIDISCIPLINARY BIOCHEMISTRY & MOLECULAR BIOLOGY CELL BIOLOGY MEDICINE, RESEARCH & EXPERIMENTAL MICROBIOLOGY PHARMACOLOGY & PHARMACY PHYSICS, PARTICLES & FIELDS CHEMISTRY, MULTIDISCIPLINARY PHARMACOLOGY & PHARMACY TOXICOLOGY CHEMISTRY, MULTIDISCIPLINARY CELL BIOLOGY NEUROSCIENCES INFECTIOUS DISEASES IMMUNOLOGY PHYSIOLOGY PHYSICS, MULTIDISCIPLINARY BEHAVIORAL SCIENCES NEUROSCIENCES ONCOLOGY CELL BIOLOGY DEVELOPMENTAL BIOLOGY ECOLOGY CHEMISTRY, MULTIDISCIPLINARY MAபைடு நூலகம்ERIALS SCIENCE, MULTIDISCIPLINARY NANOSCIENCE & NANOTECHNOLOGY GENETICS & HEREDITY MICROBIOLOGY MEDICINE, GENERAL & INTERNAL MICROBIOLOGY ASTRONOMY & ASTROPHYSICS MATERIALS SCIENCE, MULTIDISCIPLINARY BEHAVIORAL SCIENCES NEUROSCIENCES NEUROSCIENCES PSYCHOLOGY CLINICAL NEUROLOGY ECOLOGY EVOLUTIONARY BIOLOGY GENETICS & HEREDITY CHEMISTRY, PHYSICAL PHYSICS, CONDENSED MATTER BIOCHEMISTRY & MOLECULAR BIOLOGY CELL BIOLOGY PSYCHOLOGY MEDICINE, GENERAL & INTERNAL NEUROSCIENCES CARDIAC & CARDIOVASCULAR SYSTEMS
高压法聚乙烯管式反应器模拟研究

高压法聚乙烯管式反应器模拟研究何加强(中国石化股份有限公司齐鲁分公司塑料厂,山东淄博255411)Simulation of a High-pressure Ethylene Polymerization Tubular ReactorHe Jiaqiang(Plastics Complex of Qilu Branch Co.,SINOPEC,Zibo255411,China)Abstract:A tubular polyethylene reactor model was established in Aspen Plus.The effects of the reactor temperature peak value,initiator ratios and reactor cooling water temperature on the molecular chain structure,ethylene conversion rate and initiator consumption were investigated respectively.The results show that the number average molecular weight of LDPE is mainly affected by the reactor temperature peak value when the modifier is constant,and the molecular weight distribution of LDPE is mainly affected by the cooling water temperature and the reactor temperature peak values.Meanwhile,the ethylene single-pass conversion is mainly affected by the cooling water temperature,and the initiator consumption is mainly affected by the reactor temperature peak values of the upstream reaction zone and the initiator ratios.Keywords:free radical polymerization;polyethylene;tubular reactor;simulation;molecular chain structure高压聚乙烯工艺生产的低密度聚乙烯(LDPE)是三大聚乙烯通用树脂中最早实现工业化生产的聚乙烯塑料产品,具有优良的透明性和加工性,其薄膜制品、电气绝缘材料、注塑、吹塑制品、涂层、板材、管材等在工农业生产和日常生活中得到普遍应用。
Advanced materials research
Keywords: Konjac glucomannan; Poly(acrylic acid); Interpenetrating polymer network; Enzymatic degradation; Molecular weight
Introduction KGM is a high molecular weight water-soluble non-ionic polysaccharide found in tubers of
Amorphophallus konjac[1-2]. KGM with various molecular weights can show different physicochemical properties[3-4] and physiological function[5]. Polymer molecular weight has an influence on their efficacy in vitro and in vivo when they are used as the drug delivery systems. For example, polymeric nanoparticles based on low molecular weight (MW) chitosan exhibit higher transfection efficiency in vitro and in vivo than those made from high molecular weight chitosan[6].
β-mannanase and were hydrolyzed for different times. The enzymatic hydrolysis solution was precipitated with ethanol, and then the precipitate was dried at room temperature and ground into powders. Consequently, a series of powdered KGM with different molecular weights were obtained and were coded as KGM1, KGM2 and KGM3.
美国能源部未来工业材料研究进展
美国能源部未来工业材料研究进展作者:马廷灿冯瑞华姜山黄可未来工业材料(IMF)研究计划是由美国能源部能效与可再生能源办公室领导的一个全国性的研究计划,致力于研究、设计、开发、制造和测试新型与改良材料,同时积极探索对现有材料的更为有效的利用,提升工业生产和制造过程的能源效率,满足铝、玻璃、钢铁、金属铸造、化学制品、石油、林产业、农业和采矿等美国未来九大工业的需求。
本文对这一研究计划进行了介绍,并重点总结了其2006年完成的一些项目。
美国能源部未来工业材料研究计划未来工业材料(Industrial Materials for the Future,IMF)研究计划是美国能源部能效与可再生能源办公室(EERE)的工业技术计划(Industrial Technologies Program,ITP)的一项子计划,于2000年由美国能源部工业技术办公室(DOE’s Office of Industrial Technologies,OIT)的“先进工业材料(Advanced Industrial Materials,AIM)”和“连续纤维陶瓷复合材料(Continuous Fiber Ceramic Composites,CFCC)”两个计划合并而成。
IMF的使命是引领全国的工作,研究、设计、开发、制造和测试新型和改良材料,同时积极探索对现有材料的更为有效的利用,提升工业生产和制造过程的能源效率,满足未来美国工业对更强、更轻且抗高温疲劳、抗腐蚀、抗磨损等性能更加优良的材料的需求。
IMF重点关注以下四个领域并优先开展相关研究:(1)抗衰退材料(材料开发与生产、涂层与表面改善、耐火材料);(2)热物理学数据库与模型;(3)分离材料;(4)工程应用材料。
根据委托兰德公司(RAND)进行的一项独立审查,IMF确定了针对铝、玻璃、钢铁等9种未来工业的优先研究方向,如表1所示。
2004年是IMF开始取得重大进展的第一年,开展的课题多达近30项,结题10项。
chip 的操作指南(中英文)
Fast chromatin immunoprecipitation assayJoel D.Nelson 1,2,Oleg Denisenko 2,Pavel Sova 2and Karol Bomsztyk 1,2,*1Molecular and Cellular Biology Program and 2UW Medicine Lake Union Research,University of Washington,Seattle,WA 98109,USAReceived November 15,2005;Revised and Accepted December 12,2005ABSTRACTChromatin immunoprecipitation (ChIP)is a widely used method to explore in vivo interactions between proteins and DNA.The ChIP assay takes several days to complete,involves several tube transfers and uses either phenol–chlorophorm or spin columns to purify DNA.The traditional ChIP method becomes a chal-lenge when handling multiple samples.We have developed an efficient and rapid Chelex resin-based ChIP procedure that dramatically reduces time of the assay and uses only a single tube to isolate PCR-ready DNA.This method greatly facilitates the probing of chromatin changes over many time points with several antibodies in one experiment.INTRODUCTIONChromatin is composed of DNA,proteins and RNA (1–3).Chromatin structure is dynamic,responds to extracellular sig-nals,and controls gene expression,cell division and DNA repair (3–5).Chromatin is one of the most intensely studied structures in biology and ChIP assays have proven to be a powerful means to investigate a host of DNA-dependent pro-cesses (3,6,7).Along with other techniques (8,9),Chromatin immunoprecipitation reveals an extraordinarily rich and dynamic chromatin environment (3,9).The traditional method has several limitations,it takes several days to complete,requires DNA precipitations and involves multiple tube transfers (6).It is becoming increasingly clear that chromatin is a very dynamic structure (1,3,8,10–13).Thus,the traditional method may not be suffi-cient enough to explore the rich environment of chromatin.In the traditional ChIP protocol (6),DNA extractions and precipitations,are not only time consuming and tedious but also introduce steps for potential sample loss and contam-ination.This becomes a bigger problem in experiments invol-ving multiple chromatin samples.We set out to simplify the standard ChIP protocol by eliminating the multiple tube transfers to purify DNA.MATERIALS AND METHODS Mammalian cellsRat mesangial cells were grown in 150mm plastic cell culture dishes in RPMI 1640media supplemented with 10%FBS,2mM glutamine,penicillin (100U/ml),streptomycin (0.01%)and humidified with 7/93%of CO 2/air gas mixture (14).Yeast strainsMedia used for the growth of Saccharomyces cerevisiae were previously described (15);cells were grown at 30 C.The strain used in this study was MAT a HML a HMRa ade2-101his3-D 200leu2-D 1lys2-801trp1D -1ura3-52(16).ReagentsAntibodies and blocking peptides.The antibody to the C-terminal peptide of K protein was raised in rabbits as described before (17).Anti-Sir2p serum was a gift from Jasper Rine,University of California,Berkeley (18).The other antibodies used,anti-RNA polymerase II,anti-histone H3and rabbit IgG were from Santa Cruz (cat#sc-899),Abcam (cat#1791),and Vector Laboratories (cat#I-1000),respectively.The blocking peptide to K protein antibody was previously described (17)and the blocking peptide to the RNA polymerase II antibody was obtained from Santa Cruz (cat#sc-899p).Chelex-100was purchased from BioRad (cat#142–1253)and proteinase K from Invitrogen (cat#25530–015).In vivo cross-linking and immunoprecipitationsCell cross-linking was done by adding 0.8ml of 37%formaldehyde to 20ml of overlaying media for 15min at RT,followed by the addition of glycine to a final concentration of 125mM (6).After cross-linking,cells were harvested and then washed twice with 10ml phosphate-buffered saline (PBS).Cells from one dish were lysed with 1.0ml IP buffer [150mM NaCl,5mM EDTA,1%Triton X-100,0.5%NP-40,50mM Tris–HCl (pH 7.5)and 0.5mM DTT]containing the following inhibitors;10m g/ml leupeptin,0.5mM phenyl-methlysulfonyl fluoride (PMSF),30mM p -nitrophenyl phos-phate,10mM NaF,0.1mM Na 3VO 4,0.1mM Na 2MoO 4and*To whom correspondence should be addressed at UW Medicine Lake Union,Box 358050,University of Washington,Seattle,WA 98109,USA.Tel:+12066167949;Fax:+12066168591;Email:karolb@ÓThe Author 2006.Published by Oxford University Press.All rights reserved.The online version of this article has been published under an open access ers are entitled to use,reproduce,disseminate,or display the open access version of this article for non-commercial purposes provided that:the original authorship is properly and fully attributed;the Journal and Oxford University Press are attributed as the original place of publication with the correct citation details given;if an article is subsequently reproduced or disseminated not in its entirety but only in part or as a derivative work this must be clearly indicated.For commercial re-use,please contact journals.permissions@Nucleic Acids Research,2006,Vol.34,No.1e2doi:10.1093/nar/gnj004Published online January 5, 2006 at Library of the Third School of Clinical Medical of Peking University on September 22, 2012/Downloaded from10mM b-glycerophosphate.After one wash with1.0ml IP buffer the pellet was resuspended in1ml IP buffer(containing all inhibitors)and sheared with a sonicator microprobe for 4rounds of15,1s pulses at output level7(Misonix3000). Sheared chromatin was cleared by centrifugation(10min at 17000g,Eppendorf5403),split into two0.5ml fractions,and was used immediately or stored atÀ70 C.After adding anti-body pre-incubated(30min at room temp)with or without blocking peptide,0.5ml of the sheared chromatin fraction was incubated in an ultrasonic water bath(15min,4 C)(Bronson 3510).Tubes were centrifuged(10min at17000g,Eppendorf 5403)and the supernatant was transferred to fresh tubes con-taining20m l of washed protein A beads(Pharmacia)(19).The slurry was rotated for45min(4 C)and then the beads were washedfive times with1ml cold IP buffer containing no inhibitors.Yeast chromatin was prepared as previously described(6).Isolation of DNA using conventionalphenol–chlorophorm methodThis procedure is based on previously described methods (6,20).Briefly,DNA is eluted twice from the protein A beads with250m l of elution buffer(1%SDS and0.1M NaHCO3)for15min with periodic vortexing at room tem-perature.Cross-linking is reversed by adding20m l of5M NaCl and incubating the eluate overnight at65 C.After add-ing5m g linear acrylamide,as a carrier(21),DNA is precipi-tated with1.0ml100%EtOH.The pellet is washed with1ml 70%EtOH,and then dissolved in100m l TE buffer(pH8.0). Proteins are digested by adding11m l10·proteinase K buffer [0.1M Tris(pH7.8),50mM EDTA and5%SDS]and1m l of 20m g/m l proteinase K at50 C for30min.DNA is extracted using phenol/chlorophorm and then chloroform,precipitated with ethanol and thefinal DNA pellet is dissolved in200m l TE buffer.Isolation of PCR-ready DNA using the new methodA total of100m l of10%Chelex(10g/100ml H2O)is added directly to the washed protein A beads and vortexed.After 10min boiling,the Chelex/protein A bead suspension is allowed to cool to room temperature.Proteinase K (100m g/ml)is then added and beads are incubated for 30min at55 C while shaking,followed by another round of boiling for10min.Suspension is centrifuged and super-natant is collected.The Chelex/protein A beads fraction is vortexed with another100m l water,centrifuged again,and thefirst and the second supernatants are combined.Eluate is used directly as a template in PCR and makes up to25%of the final reaction volume.Real-time PCRThe reaction mixture contained5m l2·SYBR Green PCR Master Mix(Applied Biosystems),2.5m l DNA template and 0.3m M primers(10m lfinal volume)in384-Well Optical Reaction Plate(Applied Biosystems).Amplification(three step,40cycles),data acquisition and analysis were done using the7900HT Real-Time PCR system and SDS Enterprise Database(Applied Biosystems).CalculationsFactor density from ChIP assays are expressed as a signal ratio, R,using the following formula,R¼exp2(CT mockÀCT specific), where CT mock and CT specific are mean threshold cycles of PCR done in triplicates on DNA samples from specific and mock immunoprecipitations.RESULTS AND DISCUSSIONMethod developmentChelex-100resin has been used previously for DNA extraction from forensic specimens(22).We reasoned that addition of Chelex resin to immunoprecipitated chromatin samples could facilitate DNA extraction.To test if the Chelex-based method can efficiently extract DNA from immunoprecipitated chro-matin we used antibodies to hnRNP K(K protein).K protein is a conserved DNA/RNA-binding protein involved in gene expression including transcription(23–27).Using the tradi-tional ChIP assay,we have previously shown the binding of K protein to multiple gene loci(20).Recruitment of this factor to DNA was estimated by comparing the DNA signal obtained with the specific antibody to that obtained in mock immuno-precipitation where the antibody is blocked with a specific peptide(20).Control experiment(western blot,Figure1A) shows that K protein immunoprecipitation is blocked with the specific peptide.Sheared chromatin from3H-thymidine-labeled cells was pulled down with protein A beads and anti-K protein antibody pre-incubated with or without blocking pep-tide.Chelex-100suspension was added to the washed protein A immunoprecipitates and after boiling,the Chelex/protein A bead suspensions were treated with proteinase K.3H counts in the bead and supernatant fractions were measured by liquid scintillation.As shown in Figure1B most of the3H counts were recovered in the supernatants indicating that the DNA is efficiently extracted from the immunoprecipitated chromatin. In agreement with previous studies(20),these results also show that a fraction of chromatin binds to the antibody-loaded protein A beads non-specifically.The DNA eluted from protein A beads is typically treated with proteinase K to remove associated proteins.After proteo-lysis DNA is purified to remove the proteinase and peptide fragments(6).To avoid this DNA purification step,after treat-ment with proteinase K,we boiled the Chelex/protein A bead suspension containing genomic DNA.After centrifugation, the supernatant was used as template in real-time PCR with primers to the egr-1and b-globin genes.Figure1C illustrates that this procedure eliminated the potential inhibitory effects of proteinase K on PCR.We next estimated the concentration of proteinase K needed to isolate DNA from the chromatin precipitated with antibod-ies to hnRNP K(Figure1D).We found that DNA can be extracted even without proteinase K,however,addition of the enzyme increases DNA yield.The enhancing effect of proteinase K digestion was greater for the silent b-globin locus(Figure1D,lower panel)compared with the active egr-1 locus(Figure1D,upper panel),with a2.4-compared with 1.5-fold increase in specific DNA yield,respectively.In sub-sequent experiments,we used100m g/ml concentration of proteinase K.In addition,we found that30min incubatione2Nucleic Acids Research,2006,Vol.34,No.1P AGE2OF7at Library of the Third School of Clinical Medical of Peking University on September 22, 2012/Downloaded fromwith proteinase K is as effective as 4h (data not shown).In subsequent experiments we used 30min proteinase K treatment.We also introduced an improvement in the immunoprecipi-tation step itself.In the ChIP assay,immunoprecipitation is typically carried out over several hours (6).We found that pre-incubating sheared chromatin with the antibody in an ultra-sonic water bath (15min at 4 C)first and then binding to protein A beads (45min at 4 C)is sufficient to immunopre-cipitate chromatin.This faster method for chromatin IP works well with different antibodies (20,28).Verification of the fast ChIP assay in mammalian cells To verify the new method,we examined kinetics of the recruit-ment of RNA polymerase II and K protein to the PMA-inducible ing the traditional ChIP assay,we have previously demonstrated transient recruitment of hnRNP K to the inducibly transcribed egr-1locus (20).The recruitment of RNA polymerase II is a highly regulated key step control-ling the rate of mRNA synthesis from target gene loci (4,29).Treatment of rat mesangial cells with mitogens potently acti-vates the immediate-early egr-1gene (20).We compared kinetics of PMA-induced recruitment of RNA polymerase II and K protein to the egr-1locus to the induction of egr-1mRNA measured by real-time RT–PCR.This comparison revealed that the kinetics of RNA polymerase II recruitment to the egr-1gene parallels the PMA-induced changes in the transcript (Figure 2A).The nearly identical changes in mRNA levels (Figure 2A,top panel,blue lines)and recruitment of RNA polymerase II (Figure 2,middle panel,blue lines)vali-date our ChIP protocol.Note reproducibility of the results of two independent ChIP is similar to that of two separate RT–PCR parison of the new (Figure 2A,middleFigure 1.A Chelex-100-based method to isolate PCR-ready DNA from immunoprecipitated chromatin.(A )Anti-K protein antibody (10m g)was pre-incubated with (+)or without (À)blocking peptide (4m g)(30min,RT)(17).Lysates were incubated with the antibodies,complexes were pulled down with protein A beads (IP),and after washing,proteins were eluted by boiling in SDS–PAGE loading buffer.Proteins were resolved by SDS–PAGE and,after transfer to PVDF membranes,immunostained (IS)with anti-K protein antibody.(B )Cells were labeled with 3H-thymidine overnight (10m Ci in 10ml media).After cross-linking with formaldehyde,cells were lysed and sonicated.Half of the sonicated chromatin was incubated with antibody blocked with the peptide [(+)blocking peptide]and the other half with antibody that was not blocked [(À)blocking peptide].After five washes with 1ml of IP buffer,Chelex was added and the mixture was boiled.After cooling,proteinase K (200m g/ml)was added and the tubes were incubated at 55 C for 60min,mix was again boiled and beads were centrifuged.3H counts in the supernatant and the beads were measured using a liquid scintillation counter.(C )Purified rat genomic DNA (Total DNA)was boiled with the Chelex/protein A beads suspension.After cooling to room temperature the suspension was treated with [(+)proteinase K]or without [(À)proteinase K]proteinase K and then the mix was boiled again for 10min.The suspension was centrifuged and the supernatant was used as a template in real-time PCR using primers to the egr-1and b -globin genes.Three step real-time PCR was run for 40cycles.Results are expressed as 40-CT,(Threshold Cycle,Applied Biosystems,ABI7900manual),which directly reflects levels of amplicons.(D )Sonicated chromatin was incubated with anti-K protein antibody as before.After five washes with 1ml of IP buffer,Chelex was added and the mixture was boiled.After cooling the mix was treated without or with proteinase K (100or 200m g)for 60min (55 C),suspension was boiled again and the released DNA was used as a template in real-time PCR.Plots show values mean ±SD n ¼3.P AGE 3OF7Nucleic Acids Research,2006,Vol.34,No.1e2at Library of the Third School of Clinical Medical of Peking University on September 22, 2012/Downloaded fromFigure 2.Verification of the new ChIP protocol.(A )Serum-deprived rat mesangial cells were treated with PMA (10À7M)for indicated time points.Whole cell RNA was used in RT with random hexamer primers.Real-time PCR was carried out with primers to either egr-1(exon 1)or LAMC1(exon 28)genes.Results normalized to b -actin mRNA are shown as fold induction,mean ±SD,two experiments,PCR done in triplicates (top panel).Serum-deprived mesangial cells were treated with PMA as above (top panel).After cross-linking with formaldehyde,cells were lysed,pelleted and sonicated.Chromatin IPs were prepared with either RNA polymerase II (4m g)(Middle panel)or K protein (10m g)(Bottom panel)antibody with or without blocking peptides (4m g).Equal amounts of chromatin fraction were used in the IPs.DNA purified with either the new (solid blue line)or the conventional (dotted blue line)ChIP protocols was used as a template in real-time PCR.The results are expressed as a ratio of the level of PCR products obtained without (À)and with (+)blocking peptide.The graphs show results from two independent IPs done with the new ChIP protocol and results of one representative experiment is shown for the traditional method.PCR was done in triplicates (middle and bottom panel).(B )New ChIP protocol was used to assesses PMA-induced kinetics of RNA polymerase II and K protein recruitment to the different regions (I–VII)along the LAMC1gene in rat mesangial cells.The graph with results of RT–PCR analysis of mRNA is shown in the right panel (V).Results are are shown as mean values of two independent experiments.Diagram above the graphs represents LAMC1transcribed (rectangle)and flanking regions (lines).The arrows point at the sites of the respective pair of primers (I and II are 20and 5kb 50to the start of transcription,respectively,III is the promoter region,IV is exon 2,V is exon 28and VI and VII are 5and 20kb 30to the end of the last exon,exon 28).(C )Comparison of the density of RNA polymerase II,K protein and histone H3in the 50flanking (I)and transcribed (II)regions of egr-1,and at the silent b -globin (III)locus.Equal aliquots of sheared chromatin were used in the new ChIP assay with either anti-H3(4m g),anti-RNA polymerase II or K antibodies.For H3ChIP,purified rabbit IgG fraction (4m g)was used as a mock IP control.Diagram above the graphs represents egr-1and b -globin genes (rectangle)and flanking regions (lines).The arrows represent the sites of the respective pair of primers (I–III).e2Nucleic Acids Research,2006,Vol.34,No.1P AGE 4OF7at Library of the Third School of Clinical Medical of Peking University on September 22, 2012/Downloaded frompanel,solid blue line)and the conventional(Figure2A,dotted blue line)ChIP protocols done on the same extracts reveals similar PMA-induced kinetics of RNA polymerase II recruit-ment to the egr-1locus.Both methods also revealed similar kinetics for K protein recruitment to this locus(Figure2A, lower panel).Thus,results obtained with the new and the traditional methods were very similar.In comparison to the rapid and robust activation of the short egr-1gene(3.8kb),induction of the long laminin g1(LAMC1) (128kb)gene is slow and of small magnitude(Figure2A, upper panel,red lines);2to3-fold induction for LAMC1 mRNA levels compared with30–50-fold induction for egr-1 transcript.Results of ChIP analysis show that recruitment of RNA polymerase II and K protein to the last exon of LAMC1 (exon28,primer V,diagram in Figure2B)is much lower than to the egr-1locus and parallels low levels of mRNA induction (Figure2A,compare the red and blue lines).We next examined the inducible recruitment of hnRNP K and RNA polymerase II to the long and weakly induced LAMC1gene in greater details(Figure2B).Results of the ChIP analysis revealed that there was PMA-inducible increase in hnRNP K and RNA polymerase II recruitment to the LAMC1gene and50and30flanking regions.The highest levels were observed in the promoter region with the density not only decreasing along the transcribed region but also exhibiting different kinetics(Figure2B).Likewise,in the case of the egr-1locus(Figure2A),at most of the examined LAMC1 sites the recruitment of K protein resembled but was not ident-ical to that of RNA polymerase II(II–VI).At the intergenic sites20kb50and30from the gene there were low constitutive and PMA-inducible levels of K protein with little or no RNA polymerase II detected.Thefinding here that K protein binding differs from that of RNA polymerase II is consistent with the notion that hnRNP K is involved not only in tran-scription but also in chromatin remodeling(24).The higher density of hnRNP K at regions that encode laminin g1protein (IV and V)than at the intergenic sites(I and VII)indicates preferential K protein recruitment to domains that include LAMC1open reading frame(ORF).The analysis of the LAMC1gene illustrates that the new method has a sufficient signal to noise ratio to study genes that exhibit low levels of induction.The ability to simultaneously monitor DNA-binding of sev-eral factors enhances chromatin studies.We used equal ali-quots of chromatin from the same time course experiment to compare density of RNA polymerase II,histone H3and K protein at50-flanking and transcribed region of e gr-1gene and the silenced b-globin locus(Figure2C).These experiments revealed that at zero time point the histone H3density was comparable at transcribed(Figure2C,upper panels,II)and non-transcribed(I)regions of egr-1and at the silent b-globin locus(III).In the transcribed region of egr-1(II)there was large PMA-inducible loss of H3-DNA contact associated with RNA polymerase II recruitment to this site(Figure2C,center top and middle panel).The kinetics of K protein recruitment (Figure2C,bottom panel)is similar but not identical to that of RNA polymerase II.In the intergenic region5kb50to the egr-1gene and at the silent b-globin locus RNA polymerase II was not detected(Figure2C,I and III)and the level of H3 decreased slightly or remained the same after PMA treatment (Figure2C,top panels I and III).As in the case of the regions flanking the LAMC1gene(Figure2B,I and VII)there were low levels of K protein(Figure2C,bottom panels I and III). Finding K protein in the intergenic regions is consistent with the previous suggestion that its binding is genome-wide(20). These data confirm previous observations of the inverse rela-tionship between the density of RNA polymerase II and histones(30–36).These results also further validate the new ChIP method and show the ability to simultaneously monitor DNA-binding of several factors.The presence of RNA polymerase II in the transcribed and promoter regions but not in the intergenic(Figure2B and C) domains and the differential loss of H3-DNA contact (Figure2C,top panels,I–III)confirms the specificity of the fast ChIP assay.The high reproducibility of the results obtained with the new ChIP method is illustrated by close kinetics of changes in the density of RNA polymerase II,histone H3and hnRNP K protein at egr-1locus observed in two separate experiments (Figure2C).Also,note that in these experiments(Figure2C) the kinetics of RNA polymerase II and hnRNP K protein recruitment to the transcribed region of egr-1is similar to the set of two other experiments shown in Figure2A. These experiments illustrate that the new method yields repro-ducible results and allows probing of multiple factors in chro-matin preps from one experiment.We use one15cm($107cells)dish to prepare chromatin sufficient to IP with one antibody,with and withoutblocking Figure3.Verification of the new ChIP assay in yeast.The new and traditional ChIP methods were used to assess recruitment of the yeast Sir2p to HMR and an adjacent genomic locus.(A)Diagram of the yeast HMR locus(37).Primers for PCR analysis were designed to the indicated regions.(B)Results of ChIP analysis with either antibodies to Sir2p or no antibodies(mock IP).IPs and DNA purification using either the new(blue)or traditional(purple)methods were done in parallel with equal amounts of yeast chromatin.Purified DNA samples were analyzed as above with real-time PCR.Results are expressed as signal ratios of anti-Sir2p IP to mock IP.PCR were done in triplicates,data represented as mean±SD.P AGE5OF7Nucleic Acids Research,2006,Vol.34,No.1e2at Library of the Third School of Clinical Medical of Peking University on September 22, 2012/Downloaded frompeptide.DNA purified with this method from one chromatin IP is sufficient for80–100PCR.Verification of the fast ChIP assay in yeastS.cerevisiae has proven to be a valuable system to study chro-matin processes(6)and many heterochromatin factors were initially characterized in yeast[reviewed in(37)].Among these factors,Sir2p is a very conserved protein that binds to all major silenced domains in the yeast nucleus,including HML and HMR mating type loci(37).We estimated the den-sity of Sir2at HMR and at a control locus that does not bind this protein using the new and traditional ChIP methods.Equal amounts of extracts and antibodies were used in IPs.The results of real-time PCR analysis demonstrate that both meth-ods reveal Sir2p recruitment to the silenced HMR region but not to the control locus(Figure3).These results demonstrate that the fast ChIP method can be also used to study chromatin processes in yeast.In summary,we have developed a simple and efficient ChIP assay that is fast,and allows using of multiple antibodies in one experiment to simultaneously process many samples. The new ChIP assay will greatly facilitate experiments designed to study chromatin dynamics from yeast to mammals.ACKNOWLEDGEMENTSWe thank Dr J.Rine for antibodies,and members of KB lab for valuable discussions of the method.This work was supported by NIH DK45978,GM45134and Juvenile Diabetes Research Foundation(K.B.).Funding to pay the Open Access publica-tion charges for this article was provided by NIH DK45978. Conflict of interest statement.None declared. REFERENCES1.Bernstein,E.and Allis,C.D.(2005)RNA meets chromatin.Genes Dev.,19,1635–1655.2.Schubeler,D.and Elgin,S.C.(2005)Defining epigenetic states throughchromatin and RNA.Nature Genet.,37,917–918.3.Felsenfeld,G.and Groudine,M.(2003)Controlling the double helix.Nature,421,448–453.4.Sims,R.J.,IIIrd,Mandal,S.S.and Reinberg,D.(2004)Recent highlights ofRNA-polymerase-II-mediated transcription.Curr.Opin.Cell Biol.,16, 263–271.5.Thiriet,C.and Hayes,J.J.(2005)Chromatin in need of a fix:phosphorylation of H2AX connects chromatin to DNA repair.Mol.Cell, 18,617–622.6.Kuo,M.H.and Allis,C.D.(1999)In vivo cross-linking andimmunoprecipitation for studying dynamic Protein:DNA associations in a chromatin environment.Methods,19,425–433.7.Impey,S.,McCorkle,S.R.,Cha-Molstad,H.,Dwyer,J.M.,Yochum,G.S.,Boss,J.M.,McWeeney,S.,Dunn,J.J.,Mandel,G.and Goodman,R.H.(2004)Defining the CREB regulon:a genome-wide analysis oftranscription factor regulatory regions.Cell,119,1041–1054.8.Dundr,M.,Hoffmann-Rohrer,U.,Hu,Q.,Grummt,I.,Rothblum,L.I.,Phair,R.D.and Misteli,T.(2002)A kinetic framework for a mammalian RNA polymerase in vivo.Science,298,1623–1626.9.Cheutin,T.,McNairn,A.J.,Jenuwein,T.,Gilbert,D.M.,Singh,P.B.andMisteli,T.(2003)Maintenance of stable heterochromatin domains bydynamic HP1binding.Science,299,721–725.10.Spector,D.L.(2003)The dynamics of chromosome organization and generegulation.Annu.Rev.Biochem.,72,573–608.11.Bannister,A.J.and Kouzarides,T.(2005)Reversing histone methylation.Nature,436,1103–1106.12.Belotserkovskaya,R.,Oh,S.,Bondarenko,V.A.,Orphanides,G.,Studitsky,V.M.and Reinberg,D.(2003)FACT facilitates transcription-dependent nucleosome alteration.Science,301,1090–1093.13.Festenstein,R.,Pagakis,S.N.,Hiragami,K.,Lyon,D.,Verreault,A.,Sekkali,B.and Kioussis,D.(2003)Modulation of heterochromatinprotein1dynamics in primary Mammalian cells.Science,299,719–721.14.Suzuki,H.,O’Neill,B.C.,Suzuki,Y.,Denisenko,O.N.and Bomsztyk,K.(1996)Activation of a nuclear DNA-binding protein recognized by a transcriptional element,bcn-1,from the laminin B2chain gene promoter.J.Biol.Chem.,271,18981–18988.15.Adams,A.,Gottschling,D.E.,Kaiser,C.A.and Stearns,T.(1997)Methodsin Yeast Genetics.CSHL Press.16.Brachmann,C.B.,Davies,A.,Cost,G.J.,Caputo,E.,Li,J.,Hieter,P.andBoeke,J.D.(1998)Designer deletion strains derived fromSaccharomyces cerevisiae S288C:a useful set of strains and plasmids for PCR-mediated gene disruption and other applications.Yeast,14, 115–132.17.Van Seuningen,I.,Ostrowski,J.and Bomsztyk,K.(1995)Description of anIL-1-responsive kinase that phosphorylates the K protein.Enhancement of phosphorylation by sequence-selective DNA and RNA motifs.Biochemistry,34,5644–5650.18.Rusche,L.N.,Kirchmaier,A.L.and Rine,J.(2002)Ordered nucleation andspreading of silenced chromatin in Saccharomyces cerevisiae.Mol.Biol.Cell,13,2207–2222.19.Ostrowski,J.,Schullery,D.S.,Denisenko,O.N.,Higaki,Y.,Watts,J.,Aebersold,R.,Stempka,L.,Gschwendt,M.and Bomsztyk,K.(2000)Role of tyrosine phosphorylation in the regulation of the interaction ofheterogenous nuclear ribonucleoprotein K protein with its protein and RNA partners.J.Biol.Chem.,275,3619–3628.20.Ostrowski,J.,Kawata,Y.,Schullery,D.S.,Denisenko,O.N.andBomsztyk,K.(2003)Transient recruitment of the hnRNP K proteinto inducibly transcribed gene loci.Nucleic Acids Res.,31,3954–3962.21.Gaillard,C.and Strauss,F.(1990)Ethanol precipitation of DNA withlinear polyacrylamide as carrier.Nucleic Acids Res.,18,378.22.Walsh,P.S.,Metzger,D.A.and Higuchi,R.(1991)Chelex-100as amedium for simple extraction of DNA for PCR-based typingfrom forensic material.Biotechinques,10,506–513.23.Tomonaga,T.and Levens,D.(1995)Heterogeneous nuclearribonucleoprotein K is a DNA-binding transactivator.J.Biol.Chem.,270, 4875–4881.24.Bomsztyk,K.,Denisenko,O.and Ostrowski,J.(2004)hnRNP K:oneprotein multiple processes.Bioessays,26,629–638.25.Stains,J.P.,Lecanda,F.,Towler,D.A.and Civitelli,R.(2005)Heterogeneousnuclear ribonucleoprotein K represses transcription from a cytosine/thymidine-rich element in the osteocalcin promoter.Biochem.J., 385,613–623.26.Lynch,M.,Chen,L.,Ravitz,M.J.,Mehtani,S.,Korenblat,K.,Pazin,M.J.and Schmidt,E.V.(2005)hnRNP K binds a core polypyrimidine element in the eukaryotic translation initiation factor4E(eIF4E)promoter,and its regulation of eif4e contributes to neoplastic transformation.Mol.Cell Biol.,25,6436–6453.27.Da Silva,N.,Bharti,A.and Shelley,C.S.(2002)hnRNP-K and Pur(alpha)act together to repress the transcriptional activity of the CD43genepromoter.Blood,100,3536–3544.28.Denisenko,O.and Bomsztyk,K.(2002)Yeast hnRNP K-like genes areinvolved in regulation of the telomeric position effect and telomere length.Mol.Cell Biol.,22,286–297.29.Ptashne,M.and Gann,A.(1997)Transcriptional activation byrecruitment.Nature,386,569–577.30.Zhang,H.,Roberts,D.N.and Cairns,B.R.(2005)Genome-wide dynamicsof Htz1,a histone H2A variant that poises repressed/basal promoters for activation through histone loss.Cell,123,219–231.31.Lee,C.K.,Shibata,Y.,Rao,B.,Strahl,B.D.and Lieb,J.D.(2004)Evidencefor nucleosome depletion at active regulatory regions genome-wide.Nature Genet.,36,900–905.32.Katan-Khaykovich,Y.and Struhl,K.(2005)Heterochromatin formationinvolves changes in histone modifications over multiple cell generations.EMBO J.,24,2138–2149.33.Reinke,H.and Horz,W.(2003)Histones are first hyperacetylated and thenlose contact with the activated PHO5promoter.Mol.Cell,11,1599–1607.e2Nucleic Acids Research,2006,Vol.34,No.1P AGE6OF7at Library of the Third School of Clinical Medical of Peking University on September 22, 2012/Downloaded from。
大连化学物理研究所科研成果介绍异丁烯高附加值下游产品 甲基丙烯膳和甲基丙烯酸甲酯制备工艺研究
28辽宁化工2021年1月尽管如此,MOFs所具备的结构灵活和成分可调等固有优点,对其在储能和转换之外的潜在性能和应用的探索将是一个持久的发展方向。
而随着对其复杂反应过程背后的基础科学的不断研究,表征和合成技术的快速发展,MOFs基衍生物将在催化、气体存储/分离、光电子和药物传递等领域迎来另一个飞跃。
参考文献:[1]CHU S,MAJUMDAR A.Opportunities and challenges for a sustainableenergy future[J].Nature、2012,488:294-303.[2]MARTIREZ J M P,CARTER E.A.Unraveling oxygen evolution oniron-doped卩-nickel oxyhydroxide:The key role of highly active molecular-like sites[J].J.Am.Chem.Soc.2018,141:693-705.[3]JAMES S L.Metal-organic Frameworks[J].Chem.Soc.Rev.2003,32:276.[4]LIU S W,ZHANG H M,ZHAO Q,et al.Metal-organic frameworkderived nitrogen-doped porous carbon@graphene sandwich J ike structured composites as bifunctionai electrocatalysts for oxygen reduction and evolution reactions[J].Carbon,2016,106:74-83.[5]QIAN Y,HU Z,GE X,et al.A metal-free ORR/OER bifunctionalelectrocatalyst derived from metal-organic frame-works for rechargeable Zn-Air batteries[J].Carbon,2016,111:641.[6]XU X,CAO R,JEONG S,et al.Spindle-like mesoporous a-FepChanode material prepared from MOF template for high-rate lithium batteries卩].Nano厶e".,2012,12:4988-4991[7]WEI L,KARAHAN H E,ZHAI S L,et al.Amorphous bimetallicoxide-graphene hybrids as bifunctional oxygen electrocatalysts for rechargeable Zn-Air batteries[J].A c h'.Mater.,2017,29:1701410.[8]ZHENG Y,JIAO Y,QIAO S Z.Engineering of carbon-basedelectrocatalysts for emerging energy conversion:from Fundamentality to functionality[J].Jt/v.Mater.,2015,27:5372-537&[9]ZHOU J,DOU Y B,ZHOU A,et al.MOF template-directed fabricationof hierarchically structured electrocatalysts for efficient oxygen evolution reaction[J].Adv.Energy Mater.,2017,7:1602643.[10]XU A N C J,WANG J,XIA W W,et al.Heteroatom(P,B,or S)incorporated NiFe-based nanocubes as efficient electrocatalysts for the oxygen evolution reaction[J].J.Mater.Chem.A,201&6:7062-7069.[11]ZHOU L.SHAO M G,LI J B,et al.Two-dimensional ultrathin arraysof CoP:Electronic modulation toward high performance overall water splitting[J].Nano Energy.2017,41:583-590.[12]YU X Y,FENG Y,GUAN B Y,et al.Carbon coated porous nickelphosphides nanoplates for highly efficient oxygen evolution reaction[J].Energy Environ.Sci.,2016,9:1246-1250.[13]HAN S,FENG Y L,ZHANG F,et al.Metal-phosphide-containingporous carbons derived from an Ionic-polymer framework and applied as highly efficient electrochemical catalysts for water splitting[J].Adv.Fund.Mater.,2015,25:3899-3906.Metal-organic Frameworks-based Catalyst for Oxygen Evolution ReactionWANG YuZHANG Shu,LI Zi-yi,YU Zhou(College of Chemistry and Chemical Engineering,Shenyang Normal University,Shenyang Liaoning110034,China)Abstract:Oxygen evolution reaction(OER)plays an important role in electrochemical hydrocracking,providing a more feasible way for sustainable hydrogen production.The derivatives derived from metal organic framework(MOFs)have the advantages of controllable structure/composition,high surface area and ordered pore structure.In this paper,the research progress of MOFs-based derivatives in OER applications in recent years was reviewed.The examples presented can provide some reference for the preparation of highly active MOFs-based derivatives.Finally,the challenges and prospects of the engineering of MOF-based electrocatalysts were prospected.Key words:Metal organic framework;Oxygen evolution reaction;Non-precious metal catalysts大连化学物理研究所科研成果介绍异丁烯高附加值下游产品甲基丙烯膳和甲基丙烯酸甲酯制备工艺研究负责人:高爽电话:84379248联络人:王连月Email:************.cn学科领域:精细化工项目阶段:实验室开发项目简介及应用领域我国石油化工催化裂解装置副产大量G资源,主要成分为丁二烯、叔丁醇、异丁烯等:经过分离丁二烯等其他成分后,可得到大啟叔丁醇、异丁烯,而R在催化剂作用下叔丁醇可脱水得到异丁烯,因此以异丁烯为原料制备其高附加值下游产品甲基丙烯睛和甲基丙烯酸甲酯是充分利用C4的有效途径目前,工业上以异丁烯为原料直接制备甲基內烯睛是高温气相反应工艺甲基丙烯酸甲酯的制备匚艺主要是三步法,并且国外垄断现有的主要匸艺技术:由异丁烯催化氧化得到甲基丙烯醛,再由甲基丙烯醛直接氧化氨(或酯化)为甲基丙烯睛(或者甲基内烯酸甲酯);该工艺路线简化了甲基丙烯醛氧化过程及分离中间产物的设备,原子经济性高,可实现甲基丙烯睛和甲基丙酸甲酯的绿色化生产目前.异丁烯制备甲基丙烯醛技术已经成熟,本项目为甲基丙烯醛一步液相催化氧化氨化(或者酯化)为甲基丙烯睛(或者甲基丙烯酸甲酯)甲基丙烯睛:甲基丙烯睛是一种重要的有机合成原料,尤其是制备聚甲基丙烯酰亚胺(PMI)的原料——重要的芯层材料.用于航空航天、车辆、船舶等高科技领域研发了一种禽链催化剂材料用于制造型料、涂料、粘合剂、PVC改性剂、高档轿车漆、纺织浆料、髙级酯类油品添加剂等精细化学品研发由一种甲基丙烯醛分钟氧选择氧化同时和甲醇发生酯化反应生成甲基内烯酸甲酯的新型纳米金催化剂材料.与已有T.业化催化材料相比,本项冃催化材料制备过程简单.催化剂成分简单.反应条件温和.催化剂效率高等特点合作方式:合作开发;投资规模:100万~500万。
- 1、下载文档前请自行甄别文档内容的完整性,平台不提供额外的编辑、内容补充、找答案等附加服务。
- 2、"仅部分预览"的文档,不可在线预览部分如存在完整性等问题,可反馈申请退款(可完整预览的文档不适用该条件!)。
- 3、如文档侵犯您的权益,请联系客服反馈,我们会尽快为您处理(人工客服工作时间:9:00-18:30)。
Eppendorf Research®可调量程移液器和可调量程移液器价格
Eppendorf Research®
可调量程移液器 标题:Eppendorf Research®可调量程移液器
这套高质量的可调量程移液器受到全世界许多
实验室的欢迎。
由于其操作的高度舒适性以及
符合人体工程学的完美设计,Research®
可调量程移液器使得繁重的实验操作变得非常
简单轻松且易于掌握。
Research®移液器
的量程设定单手可调,快速简单,在操作过程
中随时可观察到量程的数值 量程范围
Eppendorf 吸头 容量 准确度*1 精确度*1 0.1-2.5 µl 10 µl (深灰色) 0.25 µl ±12.0% ≤ 6.0% 1.25
µl ±2.5% ≤1.5% 2.5
µl ±1.4% ≤ 0.7% 0.5-10
µl 20 µl (灰色) 1 µl
±2.5% ≤ 1.8% 5 µl
±1.5% ≤ 0.8% 10 µl
±...
厂家:上海政泓 市场价格: 优惠价格:百度搜索联系
Eppendorf
Reference®固定量程
移液器 标题:Eppendorf Reference®固定量程移液器
Eppendorf Reference 系列移液器除具有平滑
的活塞、紧凑坚固的设计、操作舒适外,更是
移液器精度的象征,其单一的多功能按钮既可
进行移液、打出液体、也可进行脱卸吸头操作。
Reference 系列提供从0.1 µl 至2,500
µl 范围的9种可调量程的移液器;同时也提供16种固定量程的移液器,其移液量程范围从1 µl 到2,500 µl 。
产品特性 "pipette/tip"系统确保了超凡
的准确性和精确性 移液器下部套筒可脱卸;脱
卸后吸头脱卸功能将失效 活塞移动平滑,且抗
化学腐蚀 坚固耐用的新型材料,可整支高压消
毒、耐紫外线处理,且不影响移液器准确度和厂家:上海政泓 市场价格: 优惠价格:百度搜索联系
精密度使用附件工具,方便快捷进行简单维修
保养提供相关的保证书可单手调节移液体积
可调校适用于不同密度的液体量...
Eppendorf
Reference®可调量程
移液器
标题:Eppendorf Reference®可调量程移液器
Eppendorf Reference 系列移液器除具有平滑
的活塞、紧凑坚固的设计、操作舒适外,更是
移液器精度的象征,其单一的多功能按钮既可
进行移液、打出液体、也可进行脱卸吸头操作。
Reference 系列提供从0.1 µl至2,500
µl范围的9种可调量程的移液器;同时
也提供16种固定量程的移液器,其移液量程范
围从1 µl到2,500 µl。
产品特
性单独的标记使用附件工具,方便快捷进行
维修保养和刻度校准配有量程调节锁定钮,保
证所选择的液体体积固定不变量程准确度
精确度 0.1-2.5 µl,烟灰色 0.25
µl ±12.0% ≤6.0% 1.25
µl ±2.5% ≤1.5% 2.5
µl ±1.4% ≤0.7% 0.5-10
µl, 灰色 ...
厂家:上海政泓
市场价格:
优惠价格:百度
搜索联系
Eppendorf
BioSpcetrometer®紫
外/可见光分光光度
计
标题:Eppendorf BioSpcetrometer®紫外/可见
光分光光度计
全新Eppendorf BioSpectrometer 紫外/可见
光分光光度计系列产品,体积小巧,适用于紫
外和可见光范围的分光检测。
该系列产品可以
记录数据结果,并在200nm至830nm进行波长
扫描。
此外,BioSpectrometer kinetic动力
学款具备温控比色杯滑盖,可在 +20°C 至
+42°C 范围内进行温度控制。
无需外连任
何温控设备,即可直接在仪器上进行酶动力学
和底物动力学检测。
产品特性:紫外线/可见
光光谱检测范围:220 nm 至 830 nm 氙灯光源,
厂家:上海政泓
市场价格:
优惠价格:百度
搜索联系。
