生物英文文献分享
微生物专题英文文献

班级:生物工程 学生:马春玲 2013年12月13日
LOGO
试验内容
1. Purpose and meaning 2. Introduction 3. Materials and methods
4.5 正交试验结果
Table 5. Results of ortho.1 Trend curve
Fig.2 Relationship between xylanase and time of fermentation in Aspergillus niger N212
通过对出发菌株注入不同剂量的氮离子,低能氮离子 束对菌体细胞均有一定程度的致死和损伤作用,细胞及其 损伤DNA又在其修复系统的作用下得到不同程度的修复, 从而导致黑曲霉孢子的存活率先下降,后上升,然后又下 降,并且菌种的修复出错会使其突变率大大提高,从而提 高了菌株的正突变率,从而确定了氮离子最佳注入参数。 以上试验可以得出最优培养基的组成(即各组分的最 适浓度),而且在以上培养得到了黑曲霉N212(表2),当 它发酵60个小时后酶活达到600IU/ml,比之前未优化的菌 株减少了12个小时,而且相对于原出发菌株酶活增加了100 %。 试验证明离子注入对微生物进行诱变改良是一种行之 有效的诱变技术。
木聚糖酶是植物细胞壁的主要之一,属 于非淀粉多糖。可作为生物漂白剂用于造纸工 业,也可用于生物转化等等。目前木聚糖酶的 生产主要还依靠真菌。
对于产酶微生物的育种,国外多采用基因工程手段 构建高产菌,而国内多采用传统的诱变方法,如紫外辐 射、化学诱变剂处理等,这些诱变手段获得的突变株一 般稳定性差、容易产生回复突变且负突变较多及诱变选 育的工作量很大,而20世纪80年代末,人们发现离子束 可以引起靶物质原子移位和重排,使细胞表面刻蚀和穿 孔,并能影响和改变细胞电性等现象,提出了离子束可 以用于细胞加工和基因转移的设想,并陆续得到了研究 证实,由此产生了国内外普遍关注的离子束生物技术工 程学,而且离子束育种是一项具有我国自主知识产权且 被国际所承认的定向遗传改良的集物理诱变和化学诱变 于一身的综合诱变方法,具有损伤小、突变谱广、突变 率高的特点。
关于生物学的书籍英语作文

关于生物学的书籍英语作文Biology is a vast and fascinating field of study that delves into the intricate workings of living organisms, from the smallest microbes to the most complex multicellular life forms. As an area of scientific inquiry, biology encompasses a wide range of sub-disciplines, each with its own unique focus and set of research methodologies. Whether one's interest lies in the realm of cell biology, evolutionary theory, ecology, or any of the numerous other branches of this discipline, there is a wealth of informative and engaging literature available to explore the myriad aspects of the living world.One of the most valuable resources for those seeking to deepen their understanding of biology is the abundance of high-quality books on the subject. These publications, written by leading experts in the field, offer readers a comprehensive and authoritative exploration of the various concepts, theories, and discoveries that have shaped our knowledge of the biological sciences. From introductory textbooks designed for students to specialized monographs tackling the latest advancements in research, the diversity of biology-focused literature is truly remarkable.For those new to the field, introductory biology books serve as an excellent starting point, providing a solid foundation in the fundamental principles and processes that underpin the study of living organisms. These works often cover a broad range of topics, including the structure and function of cells, the mechanisms of inheritance and genetic expression, the principles of evolution, and the complex interactions between organisms and their environments. By presenting complex scientific concepts in a clear and accessible manner, these introductory texts help to demystify the subject matter and inspire a genuine curiosity in the reader.As one delves deeper into the study of biology, more specialized and advanced books become increasingly valuable. These works delve into the nuances of specific sub-disciplines, offering a more detailed and comprehensive exploration of the research, theories, and methodologies that define these specialized areas of study. For instance, books on cellular and molecular biology might delve into the intricate workings of the cell, examining the various organelles, signaling pathways, and genetic mechanisms that govern the life and function of these fundamental units of life. Similarly, works on evolutionary biology might explore the historical development of life on Earth, the mechanisms of natural selection and adaptation, and the ongoing debates and controversies surrounding the origin and evolution of species.One of the key advantages of biology-focused books is their ability to provide readers with a more in-depth and contextualized understanding of the subject matter, compared to the often-limited scope of scientific articles or online resources. By presenting information in a cohesive and comprehensive manner, these books allow readers to gain a deeper appreciation for the interconnectedness of the various concepts and principles that underlie the biological sciences. Moreover, many biology books also include detailed illustrations, diagrams, and case studies that help to reinforce the key ideas and facilitate a more engaging and interactive learning experience.Beyond the realm of academic and research-oriented literature, there is also a wealth of popular science books that explore the fascinating world of biology in a more accessible and engaging manner. These works, written by talented science communicators, aim to bring the wonders of the living world to a broader audience, often by highlighting the latest discoveries, groundbreaking research, and the profound implications of our understanding of biology. From books that delve into the intricacies of the human body to those that explore the remarkable diversity of life on our planet, these popular science publications serve to inspire a sense of wonder and curiosity in readers of all backgrounds.Regardless of one's level of expertise or specific area of interest within the field of biology, the abundance of high-quality books on the subject provides a valuable resource for learning, exploration, and personal growth. Whether one is seeking to deepen their understanding of the fundamental principles of life or to delve into the latest advancements in a particular sub-discipline, the wealth of biology-focused literature offers something for every curious mind. By engaging with these informative and engaging works, individuals can not only expand their knowledge but also cultivate a deeper appreciation for the incredible complexity and beauty of the living world.。
关于微生物的英文作文

关于微生物的英文作文The World of Microorganisms: Tiny Creatures with Enormous Impact.Microorganisms, often overlooked due to their minute size, play a pivotal role in various aspects of our lives. These tiny creatures, ranging from bacteria and viruses to fungi and protozoa, exist everywhere, from the depths of the ocean to the highest peaks of the mountains. Their influence extends across all ecosystems, influencing everything from the health of our bodies to the fertility of our soils.One of the most remarkable features of microorganisms is their diversity. There are millions of species of bacteria, each with its unique characteristics and functions. Some bacteria are beneficial to humans, aiding in digestion, synthesizing vitamins, and even helping to fight off harmful invaders. Others, however, can cause diseases ranging from minor infections to life-threateningconditions.Viruses, another type of microorganism, are even more enigmatic. They consist of genetic material enclosed within a protein capsid and lack the ability to replicate independently. Instead, they depend on infecting host cells and hijacking their replication machinery to produce more virus particles. This dependence on hosts makes viruses extremely adaptable and able to infect a wide range of organisms, from bacteria to plants and animals.Fungi and protozoa are other significant groups of microorganisms. Fungi, known for their role in bread making and wine fermentation, also play crucial roles in decomposition and nutrient cycling. Protozoa, on the other hand, are single-celled animals that can be eitherparasitic or free-living. Some protozoa, such as amoebas, are known for their unique cell structure and movement patterns.The impact of microorganisms on human health cannot be overstated. The human microbiome, which consists of themicroorganisms that live on and within our bodies, plays a crucial role in maintaining our health. These microorganisms help digest food, synthesize vitamins, and even regulate our immune systems. Dysbiosis, or an imbalance in the microbiome, can lead to various diseases, including inflammatory bowel disease, obesity, and even some types of cancer.Microorganisms also play a vital role in the environment. They are responsible for decomposing organic matter, releasing nutrients back into the soil and water, and supporting the growth of plants and other organisms. Additionally, microorganisms play a key role in the carbon cycle, helping to convert carbon dioxide into organic matter and vice versa.In recent years, the field of microbiology has seen significant advancements, thanks to the development of new technologies such as next-generation sequencing and microscopy. These advances have allowed scientists to explore the microbial world in unprecedented detail, revealing the incredible complexity and diversity of thesetiny creatures.As we delve deeper into the microbial world, it becomes increasingly apparent that these tiny creatures play a crucial role in shaping our planet and our lives. It is essential that we continue to study and understand them,not only to improve our health and environment but also to harness their potential for biotechnological applications and sustainable development.In conclusion, microorganisms, despite their small size, are巨大的影响力on our world. They are essential for maintaining the health of our bodies, the fertility of our soils, and the balance of our ecosystems. As we continue to explore and understand these tiny creatures, we will discover even more remarkable ways that they shape our world.。
关于生物技术的英文文献

关于生物技术的英文文献Biotechnology is such a fascinating field, really bringing the wonders of science to life. You know, I've always been fascinated by how we can manipulate the tiniest building blocks of life to create amazing things.Take genetic engineering, for instance. It's like having a magic wand that can change the DNA of a plant or animal to give it new traits. Imagine growing crops that are resistant to drought or insects, or animals that produce milk with higher nutritional value. It's like science fiction becoming reality.And then there's biopharmaceuticals. Using biotechnology, we can produce drugs and vaccines that are more effective and targeted. It's like having a precision tool to fight diseases, giving us a better chance of curing or preventing them.But it's not just about medicine and agriculture.Biotechnology is also revolutionizing industries like energy and environmental protection. We can now use microbes to produce biofuels, reducing our dependence on fossil fuels and their impact on the environment.Plus, the advances in biotechnology are happening so fast. It's exciting to think about what the future holds. Maybe one day, we'll be able to regenerate organs for transplant using stem cells, or even create synthetic biology systems that can perform complex tasks.The possibilities are endless, and it's truly remarkable to be a part of this era where biotechnology is changing the world for the better. It's not just about scientific discovery; it's about improving lives and making a difference.。
微生物英文文献及翻译—原文

微生物英文文献及翻译—原文本期为微生物学的第二讲,主要讨论炭疽和蛔虫病这两种既往常见而当今社会较为罕见的疾病。
炭疽是由炭疽杆菌所致的一种人畜共患的急性传染病。
人因接触病畜及其产品及食用病畜的肉类而发生感染。
临床上主要表现为皮肤坏死、溃疡、焦痂和周围组织广泛水肿及毒血症症状;似蚓蛔线虫简称蛔虫,是人体内最常见的寄生虫之一。
成虫寄生于小肠,可引起蛔虫病。
其幼虫能在人体内移行,引起内脏幼虫移行症。
案例分析Case 1:A local craftsman who makes garments from the hides of goats visits his physician because over the past few days he has developed several black lesions on his hands and arms. The lesions are not painful, but he is alarmed by their appearance. He is afebrile and his physical examination is unremarkable.案例1:一名使用鹿皮做皮衣的当地木匠来就医,主诉过去几天中手掌和手臂上出现几个黑色皮肤损害。
皮损无痛,但是外观较为骇人。
患者无发热,体检无异常发现。
1. What is the most likely diagnosis?Cutaneous anthrax, caused by Bacillus anthracis. The skin lesions are painless and dark or charred ulcerations known as black eschar. It is classically transmitted by contact with thehide of a goat at the site of a minor open wound.皮肤炭疽:由炭疽杆菌引起,皮损通常无痛、黑色或称为焦痂样溃疡。
生物英文文献.doc

Application of α-amylase and Researchα-amylase to be widely distributed throughout microorganisms to higher plants. The International Enzyme classification number is EC. 3.2.1.1, acting on the starch from the starch molecules within the random cut α a 1,4 glycosidic bond to produce dextrin and reducing sugar, because the end product of carbon residues as Α configuration configuration, it is called α-amylase. Now refers to α-amylase were cut from the starch molecules within the α-1,4 glycosidic bond from the liquefaction of a class of enzymes.α-amylase is an important enzyme, a large number of used food processing, food industry, brewing, fermentation, textile industry and pharmaceutical industries, which account for the enzyme about 25% market share. Currently, both industrial production to large-scale production by fermentation α-amylase. α-amylase in industrial applications1.1 The bread baking industry, as a preservative enzymes used in baking industry, production of high quality products have been hundreds of years old. In recent decades, malt and microbial α-amylase, α-amylase is widely used in baking industry. The enzymes used for making bread, so that these products are much larger, better colors, more soft particles.Even today, baking industry have been α-amylase from barley malt and bacterial, fungal leaf extract. Since 1955 and after 1963 in the UK GRAS level validation, fungal amylase, has served as a bread additive. Now, they are used in different areas. Modern continuous baking process, add in f lour α-amylase can not only increase the fermentation rate and reduce dough viscosity (improving product volume and texture) to increase the sugar content in the dough, improved bread texture, skin color and baking quality, but also to extend the preservation time for baked goods. In the storage process, the bread particles become dry, hard, not crisp skin, resulting in deterioration of the taste of bread. These changes collectively referred to as degenerate. Each year simply because the losses caused by deterioration of bread more than 100 million U.S. dollars. A variety of traditional food additives are used to prevent deterioration and improve the texture and taste of baked goods. Recently, people started to pay attention enzyme as a preservative, preservative agent in improving the role of the dough, as amylopectin, amylase enzyme and a match can be effectively used as a preservative. However, excessive amylase causes a sticky bread too. Therefore, the recent trend is the use of temperature stability (ITS) α a amylase activity are high in starch liquefaction, but the baking process is completed before the inactivation. Despite the large number of microbes have been found to produce α-amylase, but with the temperature stability of the nature of the α-amylase only been found in several microorganisms.1.2 starch liquefaction and saccharification of the main α-amylase starch hydrolysis product market, such as glucose and fructose. Starch is converted into high fructose corn syrup (HFCS). Because of their high sweetness, are used in the soft drink beverage industry sweeteners. The liquefaction process is used in thermal stability at high temperature α-amylase. α-amylase in starch liquefaction ofthe application process is already quite mature, and many relevant reports.1.3 fiber desizing modern fiber manufacturing process in knitting yarn in the process will produce large amounts of bacteria, to prevent these yarn faults, often increase in the surface layer of the yarn can remove the protective layer. The surface layer of the material there are many, starch is a very good choice because it is cheap and easy to obtain, and can be easily removed. Starch desizing α-amylase can be used, it can selectively remove the starch without harming the yarn fibers, but also random degradation of starch dextrin soluble in water, and are easily washed off. 1.4 Paper Industry amylase used in the paper industry mainly to improve the paper coating starch. Paste on the paper is primarily to protect the paper in the process from mechanical damage, it also improved the quality of finished paper. Paste to improve the hardness and strength of paper, enhanced erasable paper, and is a good paper coating. When the paper through two rolls, the starch slurry is added the paper. The process temperature was controlled at 45 ~ 6O ℃, need a stable viscosity of starch. Grinding can also be controlled according to different grades of paper starch viscosity. Nature of the starch concentration is too high for the sizing of paper, you can use part of α-amylase degradation of starch to adjust.1.5 Application of detergents in the enzyme is a component of modern high-efficiency detergents. Enzymes in detergents in the most important function is to make detergents more modest sound. Automatic dishwasher detergents early is very rough, easy to eat when the body hurts, and on ceramics, wood tableware can also cause damage. α-amylase was used from 1975 to washing powder. Now, 90% of the liquid detergents contain an amylase, and automatic dishwasher detergents α-amylase on demand is also growing. α 1 amylase ca2 + is too sensitive to low ca2 + in the stability of poor environment, which limits an amylase in the remover in. And, most of the wild-type strains produced an amylase on raw materials as one of the oxidants detergents are too sensitive. Keep household detergents, this limitation by increasing the number of process steps can be improved. Recently, two major manufacturers of detergents NovozymesandGcncncoreInternational enzyme protein technology has been used to improve the stability of amylase bleaching. They leucine substitution of Bacillus licheniformis α-amylase protein in the first 197 on the methionine, resulting in enzymes of the oxidant component of resistance increased greatly enhanced the oxidation stability of the enzyme stability during storage better. The two companies have been pushing in the market these new products.1.6 Pharmaceutical and clinical chemical analysis with the continuous development of biological engineering, the application of amylase involved in many other areas, such as clinical, pharmaceutical and analytical chemistry. Have been reported, based on the liquid α-amylase stability of reagents have been applied to automatic biochemical analyzer (CibaComingExpress) clinical chemistry system. Amylase has been established by means of a method of detecting a higher content of oligosaccharides, is said this method is more than the effective detection method of silver nitrate.2.1 Research amylase α-amylase enzymes in domestic production and application in 1965, China began to apply for a 7658 BF Bacillus amyloliquefaciens amylase production of one, when only exclusive manufacturing plant in Wuxi Enzyme. 1967 Hangzhou Yi sugar to achieve the application of α-amylase production of caramel new technology can save 7% ~ 10% malt, sugar, increase the rate of 10%.1964, China began a process of enzymatic hydrolysis of starch production of glucose. In September l979 injection of glucose by the enzyme and identification of new technology and worked in North China Pharmaceutical Factory, Hebei Dongfeng Pharmaceutical Factory, Zhengzhou Songshan applied pharmaceutical units and achieved good economic benefits. Compared with the traditional acid to improve the yield of 10% Oh, cost more than 15%. In addition to enzyme for citric acid production in China, glutamic acid fermentation system for beer saccharification, fermentation, rice wine, soy sauce manufacture, vinegar production also has been studied and put into production successfully.2.2 Overseas Researc h α-amylase, present, and in addition a large number of T for conventional mutation breeding, the overseas production has been initially figure out the regulation of α-amylase gene, the transduction of the transformation and gene cloning techniques such as breeding. The Bacillus subtilis recombinant gene into the production strain to increase α-amylase yield of 7 to 10 times and has been used in food and the wine industry, for breeding high-yield strains of α-amylase to create a new way.2.3 domestic and foreign research institutions and major research direction as α-amylase is an important value of industrial enzymes, weekly discussion group and outside it was a lot of research. Representative of the domestic units: Sichuan University, major research produc tion of α-amylase strains and culture conditions; Jiangnan University, the main research structure of α-amylase and application performance, such as heat resistance, acid resistance; Northwest universities, major research denatured α-amylase and the environment on the mechanism of α-amylase; South China University of Technology, the main α-amylase of immobilization and dynamic nature; there Huazhong Agricultural University, Chinese Academy of Sciences Institute of Applied Ecology in Shenyang, Tianjin University, Nankai University, College of Life Sciences, Chinese Academy of Agricultural Sciences, Chinese Academy of Sciences Institute of Microbiology and a number of research institutions on a variety of bacterial α-amylase production of a amylase gene cloning and expression. Representative of foreign research units are: Canada UniversityofBritishColumbia, they were a pancreatic amylase structure and mechanism of in-depth research; Denmark's Carlsberg Research Laboratory of the main structure of barley α-amylase domain and binding sites; U.S. WesternRegionalResearchCenter major study α-amylase in barley and the role of antibiotics and the barley α-amylase active site.3, α-amylase conclusion has become the industrial application of one of the most important enzymes, and a large number of micro-organisms can be used for efficient production of amylase, but large-scale commercial production of the enzyme is still limited to some specific fungi and bacteria. For the effective demandfor α-amylase more and more, this enzyme by chemical modification of existing or improved technology through the white matter are. Benefit from the development of modern biotechnology, α an amylase in the pharmaceutical aspects of growing importance. Of course, the food and starch indust ries is still the main market, α amylase in these areas, a demand is still the largest.Journal of Southeast University(English Edition)2008 24(4)。
生物科学中英文对照外文翻译文献

中英文资料外文翻译文献译文标题:传统意大利榛子的体外繁殖用于当地遗传资源库的稳定和保存译文:关键词:欧洲榛,榛属,传统种质,体外繁殖摘要:在地中海盆地,榛子(欧洲榛)是非常重要的一种作物。
体外繁殖能够有效的稳定当地遗传资源库。
为了提高榛子微组织繁殖实验记录的精确性,各种不同的研究已经在进行。
这些研究通常以重要的品种为材料,然而,微组织繁殖实验记录应用在这些幼小品种上比起传统方法通常会产生相反的结果,这种技术在幼小品种上很少取得成功。
本实验的目的是为重要品种微组织繁殖的操作积累相关的知识和信息。
实验过程中需要设计不同成分的培养基,灭菌时间和培养时间都要进行详细的讨论。
传统意大利品种植株茎芽中的N6-异戊烯腺嘌呤的作用是改善这种状态。
生根阶段是榛属微组织繁殖应用于大型商业生产的关键步骤。
欧洲榛在欧洲特别是生物地理分布区地中海盆地代表一种重要的经济类林木。
榛子主要产于土耳其,意大利,美国和西班牙(分别是每年55,000, 110,000, 25,000, 18,000+吨),其次是法国,希腊,葡萄牙。
大约90%的产品被去皮并且以树芯的形式卖出,然而剩余的10%则作为树苗消费。
极好的营养成分和营养制品的特性也使该物种产生很高的利润。
此外,在一些特有的栽培地区,传统和文化身份严重受榛子产量的影响,文化身份常常会促进贫瘠土地的回收和利用。
即使这样,在一些地区,这种林业作物仍然不是重要的农业资源,然而,就当地足够维持的生产式系统和作为宝贵的食物的传统而言,它却是一种有趣的收入来源。
世界第二大生产商意大利说一些传统的品种主要种植在Campania ,Latium, Piedmont,在西西里岛有大量的属典型种。
近几年,一些主要品种由于质量和传统特性获得了欧洲质量印模。
此外,这些品种还被引进其他国家特定的果园中以增大他们的生长范围。
没有经过检验的物质可能会传播疾病,也可能会导致原因不明的物质的出现。
微组织繁殖法等生物技术的应用会促进健康的合乎本性的物质的产生(Nas et al.,2004),并且提高这种林木的经济价值。
分子生物学英文文献1
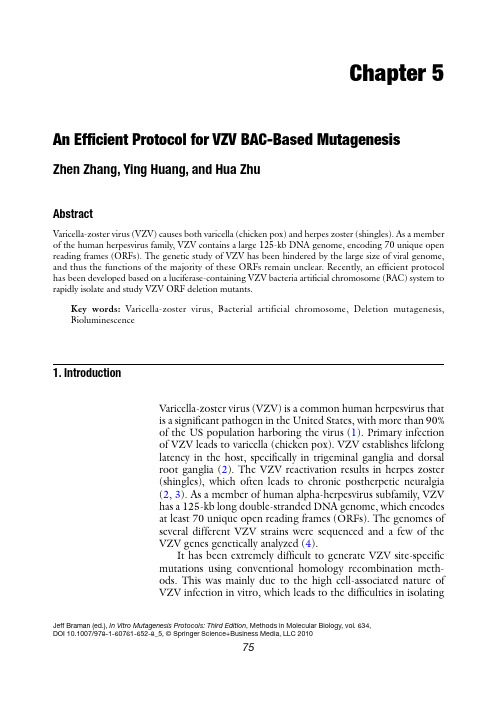
Chapter 5An Efficient Protocol for VZV BAC-Based MutagenesisZhen Zhang, Ying Huang, and Hua ZhuAbstractVaricella-zoster virus (VZV) causes both varicella (chicken pox) and herpes zoster (shingles). As a member of the human herpesvirus family, VZV contains a large 125-kb DNA genome, encoding 70 unique open reading frames (ORFs). The genetic study of VZV has been hindered by the large size of viral genome, and thus the functions of the majority of these ORFs remain unclear. Recently, an efficient protocol has been developed based on a luciferase-containing VZV bacteria artificial chromosome (BAC) system to rapidly isolate and study VZV ORF deletion mutants.Key words:Varicella-zoster virus, Bacterial artificial chromosome, Deletion mutagenesis, Bioluminescence1. I ntroductionVaricella-zoster virus (VZV) is a common human herpesvirus thatis a significant pathogen in the United States, with more than 90%of the US population harboring the virus (1). Primary infectionof VZV leads to varicella (chicken pox). VZV establishes lifelonglatency in the host, specifically in trigeminal ganglia and dorsalroot ganglia (2). The VZV reactivation results in herpes zoster(shingles), which often leads to chronic postherpetic neuralgia(2, 3). As a member of human alpha-herpesvirus subfamily, VZVhas a 125-kb long double-stranded DNA genome, which encodesat least 70 unique open reading frames (ORFs). The genomes ofseveral different VZV strains were sequenced and a few of theVZV genes genetically analyzed (4).It has been extremely difficult to generate VZV site-specificmutations using conventional homology recombination meth-ods. This was mainly due to the high cell-associated nature ofVZV infection in vitro, which leads to the difficulties in isolatingJeff Braman (ed.), In Vitro Mutagenesis Protocols: Third Edition,Methods in Molecular Biology, vol. 634,DOI 10.1007/978-1-60761-652-8_5, © Springer Science+Business Media, LLC 20107576Zhang, Huang, and Zhuviral DNA and purifying recombinant virus away from wild-type virus. In the last few years, a popular method for VZV in vitro mutagenesis involves a four-cosmid system covering the entire viral genome (5–7). Using the cosmid system to generate recom-binant VZV variants involves technically challenging steps such as co-transfection of four large cosmids into permissive mammalian cells and multiple homologous recombination events within a single cell to reconstruct a full-length viral genome. The highly cell-associated nature of VZV also makes the downstream appli-cations of traditional virology methods such as plaque assay-based titering and plaque purification difficult. To date, the functions of the majority of VZV ORFs remain uncharacterized (8).In order to create recombinants of VZV more efficiently, the full-length VZV (P-Oka strain, a cloned clinical isolate of VZV) genome has been successfully cloned as a VZV bacteria artificial chromosome (BAC) (9, 10). This VZV BAC combined with a highly efficiently E. coli homologous recombination system allows quick and easy generation of recombinant VZV. To further ease the downstream virus quantification assays, a firefly luciferase reporter gene, was inserted into the VZV BAC to generate a novel luciferase-expressing VZV (10). In this protocol, we show the generation and analyses of VZV full-length ORF deletion mutants and genetic revertants as examples to demonstrate the utility and efficiency of this versatile system for VZV mutagenesis in vitro. Furthermore, this protocol can be easily modified to broaden its applications to a variety of genetic maneuvers including making double ORF deletions, partial ORF deletions, insertions, and point mutations.1. Human melanoma (MeWo) cells were grown in DMEM supplemented with 10% fetal bovine serum, 100 U penicillin–streptomycin/ml, and2.5 m g amphotericin B/ml at 37°C in a humidified incubator with 5% CO 2. All tissue culture reagents were obtained from Sigma (St. Louis, MO).2. VZV luc was recently developed in the laboratory (10). It con-tains a full-length VZV P-Oka genome with a firefly luciferase cassette (see Note 1). The BAC vector was inserted between VZV ORF60 and ORF61, which includes a green fluorescent protein (GFP) expression cassette and a chloramphenicol resistance cassette (Cm R ).3. pGEM-oriV/kan was previously constructed (11) in the lab-oratory and used as a PCR template to generate the expres-sion cassettes for the kanamycin or ampicillin resistance genes (Kan R and Amp R ).2. M aterials2.1. Cells, VZV luc , Plasmids, and E. coliStrain77An Efficient Protocol for VZV BAC-Based Mutagenesis 4. pGEM-lox-zeo was derived from pGEM-T (Promega, Madison, WI) (12) and was used to generate the rescue clones of VZV ORF deletion mutants. 5. E. coli strain DY380 was obtained from Neal Copeland and Craig Stranthdee and used for mutagenesis (13). 6. A cre recombinase expression plasmid pGS403 was a gift from L. Enquist (Princeton University, NJ). 1. All primers were synthesized by Sigma-Genosys (Woodlands, TX) and stored in TE buffer (100 m M). 2. HotStar Taq DNA polymerase (Qiagen, Valencia, CA) was used for PCR reactions and Platinum Taq DNA polymerase (Invitrogen, Carlsbad, CA) could be used for optional hi-fidelity PCR reactions (see Note 2). 3. PCR purification was carried out using a PCR purification kit (Qiagen, Valencia, CA). 4. The amplified linear DNAs were suspended in sterile ddH 2O and w ere q uantified b y s pectroscopy (NanoDrop T echnologies, Wilmington, DE). 5. DpnI (New England Biolabs, Ipswich, MA) restriction treat-ment following PCR was carried out in order to eliminate circular template DNA. 6. Electroporation was carried out with a Gene Pulser II Electroporator (Bio-Rad, Hercules, CA). 1. All antibiotics were obtained from Sigma (St. Louis, MO). LB plates containing specific antibiotics were used for appro-priate selections (Table 1).2. A 37°C air shaker and a 37°C water bath shaker were used for bacterial culturing.2.2. Primers, PCR, PCRpurification, DpnITreatment, andElectroporation2.3. AntibioticsSelection and BACDNA Purification Table 1Antibiotics concentrations for selectionFor BACs (single or low copy numbers)For plasmids (high copynumbers)78Zhang, Huang, and Zhu3. NucleoBond Xtra Maxi Plasmid DNA purification kits (Clontech Laboratories, Inc., Palo Alto, CA) were used to purify VZV BAC DNA from E. coli .4. Kimwipes (Kimberly-Clark Global Sales, Inc., Roswell, GA) were used as small filters in BAC DNA Mini-preparations.5. Phenol/chloroform, isopropanol, and ethanol were obtained from Sigma (St. Louis, MO) and were used as additional reagents in BAC DNA preparations.6. Hin dIII (New England Biolabs, Ipswich, MA) digestions were performed to check the integrity of BAC DNA.1. FuGene6 transfection kit (Roche, Indianapolis, IN) was used for transfecting viral BAC DNA into MeWo cells (ATCC).2. An inverted fluorescent microscope was used to observe and count plaque numbers.3. Tissue culture media containing 150 m g/ml d -luciferin (Xenogen, Alameda, CA) was used as substrate for in vitro bioluminescence detection.4. An IVIS Imaging 50 System (Xenogen, Alameda, CA) was used to record bioluminescence signal from virally infected cells.5. Bioluminescence data were quantified by using Living Image analysis software (Xenogen, Alameda, CA).In order to generate VZV ORF deletion mutants using this new VZV luc system, we took advantage of an efficient recombination system for chromosome engineering in E. coli DY380 strain (13). A defective lambda prophage supplies the function that protects and recombines linear DNA. This system is highly efficient and allows recombination between homologies as short as 40 bp. The experimental design is summarized in Fig. 1.1. The first step in making any specific VZV ORF deletion was to amplify a Kan R cassette containing 40-bp flanking sequences of the targeted ORF.2. Primers were stored in TE buffer (100 m M). The Kan R expres-sion cassette was amplified from pGEM-oriV/Kan using a HotStar DNA polymerase kit following a standard protocol.3. PCR product was purified using a PCR purification kit fol-lowing the manufacturer’s protocol.2.4. Transfectionand SubsequentVirological Assays(Tittering and Growth Curve Analysis)3. M ethods3.1. Generation of VZVORF Deletion BACClones 3.1.1. Making a Kan R Cassette Targeting a Specific VZV Open Reading Frame79An Efficient Protocol for VZV BAC-Based Mutagenesis 4. The purified PCR product was treated with DpnI in order to eliminate the template DNA. This step greatly reduces the background in later selections.5. PCR product was purified again as above (step 3) and the amplified linear DNA was suspended in sterile ddH 2O and was quantified by spectroscopy (see Note 3).1. DY380 cells were grown at 32°C until the OD 600nm measure-ment reached 0.5 (see Note 5).2. The culture was shifted to 42°C by placing the flask into a 42°C water bath with vigorous shaking for 10–15 min (see Note 4).3.1.2. Induction of theLambda RecombinationSystem and Preparationof Electroporation-Competent DY380Fig. 1. Generating ORF deletion mutants (ORFD). (a ) The E. coli DY380 strain provides a highly efficient homologous recombination system, which allows recombination of homologous sequences as short as 40 bp. The homologous recombination system is strictly regulated by a temperature-sensitive repressor, which permits transient switching on by incubation at 42°C for 15 min. VZV luc BAC DNA is introduced into DY380 by electroporation. Electro-competent cells are prepared with homologous recombination system activation. (b ) Amplification of the Kan R expression cassette by PCR using a primer pair adding 40-bp homologies flanking ORFX. (c ) About 200 ng of above PCR product are transformed into DY380 carrying the VZV luc BAC via electroporation. (d ) Homologous recombination between upstream and downstream homologies of ORFX replaces ORFX with the Kan R cassette, creating the ORFX deletion VZV clone. The recombinants are selected on LB agar plates containing kanamycin at 32°C. (e ) The deletion of ORFX is confirmed by testing antibiotic sensitivity and PCR analysis. The integrity of viral genome after homologous recombination is examined by restriction enzyme Hin dIII digestion. (f ) VZV luc BAC DNA with ORFX deletion is propagated and isolated from DY380. (g ) Purified BAC DNA is transfected into MeWo cells. (h ) 3–5 days after transfection, the ORFX deletion virus is visualized under a fluores-cent microscope due to EGFP expression given nonessentiality of ORFX.Select for kan Rat 32°Cseqs. (40 bp)ORFX kan R ORFE. coli 32°C ts λ cI repressorVZV-BAC Defective l prophage D BkanR E BAC DNATransfect MeWo cells ProducerecombinantVZV (givenORFX is notessential)x G Hx MR WTORFXDORFXR Confirm recombinant VZV by antibiotic sensitivity, PCR and HindIII digestion80Zhang, Huang, and Zhu3. The culture was immediately transferred to an ice–water slurry for 30 min. (see Note 6).4. After incubation on ice, the culture was then pelleted at 6,000 × g for 10 min 4°C, washed with ice-cold sterile ddH 2O, and repelleted.5. Prechilled 10% glycerol (use about 1% of original volume of culture) was used to resuspend cells, and a 40-m l aliquot (>1 × 1010 cells) was used for each electroporation reaction. 1. Two microliters of Kan R cassette DNA (greater than 200 ng) were electroporated into competent DY380 cells harboring the VZV luc BAC. Homologous recombination took place between the 40-bp ORF flanking sequences and the targeted BAC ORF was replaced by the linear Kan R cassette creating the expected VZV ORF deletion clones. 2. Electroporation was carried out at 1.6 kV, 200 W , and 25 m F in a Gene Pulser II electroporator. Two microliters of con-centrated linear DNA cassette (greater than 200 ng) were used in each reaction. 3. The bacteria were immediately transferred to 1 ml LB medium after electroporation and incubated at 32°C for 1 h before plating. The resultant recombinants were selected on LB agar plates containing kanamycin at 32°C for 16–24 h (see Note 7). 4. Antibiotic sensitivity: it is important to further test that kanamycin-resistant colonies are resistant to kanamycin but not to ampicillin because the circular pGEM-oriV/Kan R (containing Amp R ) was used as the PCR template. This can be tested by re-streaking single colonies on mul-tiple LB agar plates containing different antibiotics. VZV ORFX deletion clones should be resistant to chloram-phenicol (from BAC vector), hygromycin (from luciferase cassette), and kanamycin (VZV ORF replacement cassette), but sensitive to ampicillin (potentially from pGEM-oriV/Kan R ; see Note 8). 1. Mini-BAC DNA preparations.(a) A single DY380 clone containing the recombinant VZV BAC was inoculated in 5 ml LB supplemented with the appropriate antibiotics and cultured at 32°C overnight.(b) BAC DNA was isolated by pelleting the bacteria, resus-pending in 1 ml resuspension buffer supplemented with RNase A (Buffer RES), lysing in 1 ml NaOH/SDS lysis buffer (Buffer LYS), and neutralizing in 1 ml potassium acetate neutralization buffer (Buffer NEU) for 5 min for each step (NucleoBond Xtra Maxi Plasmid DNA purifi-cation kit).3.1.3. Electroporation andRecombinant Screening3.1.4. BAC DNAPurification and BACClone Verification81An Efficient Protocol for VZV BAC-Based Mutagenesis (c) The cloudy solution was centrifuged at 4,500 × g for 15 min at 4°C. The supernatant was filtered through a small piece (cut to 4 × 4 cm) of Kimwipe tissue (Kimberly-Clark Global Sales, Inc., Roswell, GA).(d) The filtered solution was extracted with an equal volume of phenol/chloroform and the BAC DNA precipitated with two volumes of ethanol.(e) After the final spin at 4,500 × g for 30 min at 4°C, the DNA pellet was air-dried and resuspended in 20 m l sterile ddH 2O. 2. PCR verification: multiple colonies with the correct antibiotic sensitivities were picked for the mini-BAC DNA preparations. The ORF deletions with Kan R replacements were confirmed by PCR using a HotStar DNA polymerase kit following a standard protocol. The target ORF should be absent in ORF deletion clones while the adjacent ORFs should remain intact as positive controls. 3. Maxi-BAC DNA preparations: the large-scale BAC DNA preparations using the NucleoBond Xtra Maxi Plasmid DNA purification kit (Clontech Laboratories, Inc., Palo Alto, CA) started with 500 ml of overnight cultures. The standard man-ufacturer’s protocol for BAC DNA purification was followed. The final DNA products were resuspended in 250 m l sterile ddH 2O and quantified by spectroscopy (see Note 9). 4. Hin dIII digestion profiling: PCR verified clones were selected for maxi-BAC DNA preparations. To confirm that no large VZV genomic DNA segment is deleted, Hin dIII digestion profiling was routinely carried out (see Note 10). Three micrograms of BAC DNA from maxi-preparations were digested with 20 U of Hin dIII in a 20-m l reaction at 37°C overnight. Hin dIII digestion patterns were compared by electrophoresis on ethidium bromide stained 0.5% agarose gels. As shown in Fig. 1, Hin dIII digestion patterns of each VZV ORF deletion clone were highly comparable with the parental wild-type VZV luc clone (see Note 11).The generation of VZV ORF deletion revertants is necessary to prove that the deleted ORF is responsible for any phenotype (usu-ally a growth defect) observed in analyses of the deletion mutants. The viral revertants should be able to fully restore the wild-type phenotype. As an example, generating the VZV ORFX deletion rescue virus is described to demonstrate the approach (see Fig. 2).1. VZV ORFX was amplified from wild-type VZV luc BAC DNAby PCR. Two unique restriction enzyme sites and two addi-tional 6-bp random sequences were added to the ends of the PCR product. A hi-fidelity PCR kit could be used in order to minimize unwanted mutations during PCRs (see Note 2).3.2. Generation of VZVORF DeletionRevertant BAC Clones82Zhang, Huang, and Zhu2. The ORFX gene was directionally cloned into pGEM-zeo to form pGEM-ORFX-zeo. The cloned ORFX was verified by sequencing analysis.3. ORFX-zeoR cassette was made by PCR using pGEM-ORFX-zeo as template (Fig. 2). The PCR product contained 40-bp homologies of flanking sequences of Kan R cassette, which was also used to generate the ORFX deletion mutant.4. The subsequent procedures are similar to producing the ORFX deletion mutant. Briefly, the linear ORFX-zeoR cas-sette was treated with DpnI and electroporated into compe-tent DY380 cells harboring VZV luc ORFX deletion BAC. Similarly, homologous recombination functions were tran-siently induced by switching the culture temperature from 32 to 42°C for 10–15 min when electroporation-competent cells were prepared. The recombinants were selected on LB agar plates containing zeocin. After verification, the ORFX dele-tion rescue BAC DNA was isolated from E. coli .Because of VZV’s highly cell-associated nature in cell culture,conventional virology techniques, including plaque purification and plaque assay, become troublesome. By developing andexploiting the new luciferase VZV BAC system, the resulting virus has a removable EGFP expression cassette and a built-in 3.3. Transfectionand Subsequent Virological Assayszeo R lox mcs mcskan lox zeo R ORFX lox lox E. ORFXR D. ORFXR-zeo B. C. ORFXD zeo R ORFX zeo ORFX ORFX Fig. 2. Generating an ORFX deletion rescue clone (ORFXR). (a ) To generate the ORFXR clone, ORFX was amplified by PCR from the wild-type VZV BAC DNA. The ORFX was directionally cloned into plasmid pGEM-lox-zeo to form pGEM-zeo-ORFX. (b ) Amplification of the ORFX-Zeo R cassette by PCR using a primer pair adding 40 bp homologies flanking ORFX. (c ) Such PCR product was transformed into DY380 carrying the VZV luc ORFXD BAC via electroporation. (d ) Homologous recombination between upstream and downstream homologies of ORFX replaced Kan R with the ORFX-Zeo R cassette, creating the ORFXR clone. (e ) Zeo R was removed while generating virus from BAC DNA by co-transfecting a Cre recombinase expressing plasmid.83An Efficient Protocol for VZV BAC-Based Mutagenesis luciferase reporter. In this protocol, an alternative biolumines-cence quantification approach has been provided to significantly increase the reproducibility of results. This approach has also been successfully used in monitoring VZV growth in vivo (10). 1. VZV BAC DNA from maxi-preparations was transfected into MeWo cells using the FuGene6 transfection kit according to the manufacturer’s standard protocol. 2. One and a half micrograms of BAC DNA and 6 m l of transfec-tion reagent were used for a single reaction in one well of 6-well tissue culture plates (see Note 12). 3. As an option, 0.5 m g of Cre expression plasmid was co-transfected with the VZV BAC DNA to remove the BAC sequence flanked by two loxP site from the viral genome (see Note 13). 4. In order to prevent the precipitation of BAC in solution, 1.5 m g BAC DNA were diluted in serum-free tissue culture medium, and the volume of DNA solution was adjusted to 50 m l (see Note 14). 5. The DNA solution was slowly added to the transfection reagent with gentle stirring using pipet tips. 6. Because of GFP expression from the BAC vector, VZV plaques were usually visually discernable using a fluorescent microscope within 3–5 days after transfection given deleted ORF is dispensable (see Note 15). If a VZV ORF is essential for viral replication, no plaque will be observed. 7. Since VZV is highly cell-associated in tissue culture, mutant VZV-infected MeWo cells were harvested and stored in liquid nitrogen for future studies.Recombinant viruses were titered by infectious focus assay. MeWo cells were seeded in 6-well tissue culture plates and inoculated with serial dilutions of VZV-infected MeWo cell suspensions. Plaques were counted by fluorescent microscopy 3 days after inoculation and viral titer was determined. 1. MeWo cells were infected with 100 PFU of infected MeWo cell suspensions in 6-well tissue culture plates. 2. After every 24-h interval, cell culture media was replaced with media containing 150 m g/ml d -luciferin.3. After incubation at 37°C for 10 min, the bioluminescent sig-nal was quantified and recorded using an IVIS ImagingSystem following the manufacturer’s instructions. 4. Fresh tissue culture medium was added to replace the luciferin-containing medium for further incubation at later time points.3.3.1. Transfection of BACDNA into MeWo Cells3.3.2. Titering by InfectiousFocus Assay3.3.3. Growth CurveAnalyses Based onBioluminescence Imaging(See Fig. 3 and Note 16)84Zhang, Huang, and Zhu5. Measurements from the same plate were repeated every day for 7 days.6. Bioluminescence signal data from each sample was quantified by manual designation of regions of interest and analyzed using Living Image analysis software (see Note17).1. The luciferase expression cassette, driven by an SV40 early pro-moter, was inserted between VZV ORF65 and ORF66. The cassette also contains a hygromycin B resistance gene (Hyg R ).2. Platinum Taq DNA polymerase can be used alternatively if a hi-fidelity PCR product is preferred.3. In order to achieve optimum results, the final concentration of the linear DNA cassette for the subsequent electroporation was adjusted to at least 100 ng/m l.4. The 42°C temperature shift is critical for the success of the homologous recombination. The temperature needs to be adjusted accurately to 42°C and remain constant. Too much recombination system activity is detrimental to E. coli and harm the integrity of BAC DNA. On the other hand, inade-quate induction of the recombination system in DY 380 leads to inefficient recombination. Ten to fifteen minutes might need to be adjusted carefully in order to achieve optimized efficiency of homologous recombination.5. E. coli DY380 strain needs to be cultured at 32°C all the time except when the recombination system is transiently activated and expressed by shifting the culture to 42°C.6. Beyond this point, every step needs to be carried out at a low temperature (0–4°C). All reagents, centrifuge rotor and glass-ware need to be prechilled.4. N otes Growth curve analysisVZVluc infectedMeWo cells / animal. a b c d Bioluminescenceimaging Image acquisition Fig. 3. Growth curve analyses based on bioluminescence imaging. (a ) Small animals/tissue culture can be infected with VZV luc . (b ) After administration of an enzyme substrate, luciferin, bioluminescence emitting from living animals/cultured cells can be detected and monitored by using a bioluminescence imaging system (a CCD camera mounted on top of a light-tight imaging dark chamber). (c ) Data can be stored in a connected PC and quantified by using region-of-interest analysis. (d ) Viral growth kinetics can be analyzed based on quantification of bioluminescence signals.85 An Efficient Protocol for VZV BAC-Based Mutagenesis7. Recombinants often have multiple antibiotic resistances. For instance, VZV ORFX/Kan clone will have Kan R, Cm R (from BAC vector), and Hyg R (from luciferase cassette). Screening for recombinants with more than one antibiotic is optional. However, the growth rate under such conditions could be much slower than selection under one antibiotic.8. If a clone also has Amp R, it should count as a false positive result.9. Due to the large size, handling BAC DNAs needs to avoid any harsh physical sheering force including vortexing or quickly passing through fine pipette tips. Freeze and thaw should also be avoided. BAC DNA solutions should always be stored at 4°C.10. Although it has been shown that VZVluc DNA is highly stablein E. coli (10) under the conditions described in this protocol, large undesirable deletions in the BAC clones were observed if homo l ogous recombination system in DY380 was over-induced.11. Since many large DNA fragments are generated by a Hin dIIIdigestion of the VZV genome, smaller genetic alterations, including replacement of an ORF by a Kan R cassette, would be difficult to recognize by this assay.12. The ratio of BAC DNA and FuGene6 reagent might need tobe adjusted to maximize transfection efficiency.13. The ORFX rescue clone was generated by introducing the wild-type ORFX back into the deletion viral genome along with a Zeo R cassette flanked by two loxP sites. By following this optional step in transfection, Zeo R will be removed from the genome by Cre-mediated recombination. The resulting virus will have a wild-type copy of ORFX restored in the same direction and loca-tion as the parental wild-type strain except a remaining loxP site(34 bp) in the 3¢ noncoding region of ORFX.14. Highly concentrated (greater than 250 m g/m l) BAC DNAsolutions are viscous and BAC DNA molecules easily precipi-tate out of solution when added to transfection reagent solu-tions. When such precipitation becomes visible, it is irreversible and the result of the transfection assays is often poor.Therefore, we predilute each BAC DNA in media before gen-tly mixing with the transfection reagent.15. Transfection efficiency was easy to monitor because of theresulting GFP expression from the BACs.16. Growth curve analyses were traditionally carried out by aplaque assay-based method.17. See ref. 14 for more detailed methods and more applicationof in vivo bioluminescence assay.86Zhang, Huang, and Zhu References1. Abendroth A, Arvin AM (1999) Varicella-zoster virus immune evasion. Immunol Rev 168:143–1562. Gilden DH, Kleinschmidt-DeMasters BK,LaGuardia JJ, Mahalingam R, Cohrs RJ (2000) Neurologic complications of the reactivation of varicella-zoster virus. N Engl J Med 342:635–6453. Arvin AM (2001) Varicella-zoster virus. In:Knipe DM, Howley PM (eds) Fields virology, vol 2. Lippincott Williams & Wilkins, Philadelphia, PA, pp 2731–27674. Davison AJ, Scott J (1986) The completeDNA sequence of varicella zoster virus. J Gen Virol 67:1759–18165. Cohen JI, Seidel KE (1993) Generation ofvaricella-zoster virus (VZV) and viral mutants from cosmid DNAs: VZV thymidylate syn-thetase is not essential for replication in vitro.Proc Natl Acad Sci USA 90:7376–73806. Mallory S, Sommer M, Arvin AM (1997)Mutational analysis of the role of glycoproteinI in varicella-zoster virus replication and itseffects on glycoprotein E conformation and trafficking. J Virol 71:8279–82887. Niizuma T, Zerboni L, Sommer MH, Ito H,Hinchliffe S, Arvin AM (2003) Construction of varicella-zoster virus recombinants from P-Oka cosmids and demonstration that ORF65 protein is dispensable for infection of human skin and T cells in the SCID-hu mouse model. J Virol 77:6062–60658. Cohen JI, Straus SE, Arvin AM (2007)Varicella-zoster virus replication, pathogene-sis, and management. In: Knipe DM, HowleyPM (eds) Fields virology, vol 2. Lippincott Williams & Wilkins, Philadelphia, PA, pp 2773–28189. Nagaike K, Mori Y, Gomi Y, Yoshii H,Takahashi M, Wagner M, Koszinowski U, Yamanishi K (2004) Cloning of the varicella-zoster virus genome as an infectious bacterial artificial chromosome in Escherichia coli.Vaccine 22:4069–407410. Zhang Z, Rowe J, Wang W, Sommer M, ArvinA, Moffat J, Zhu H (2007) Genetic analysis of varicella zoster virus ORF0 to 4 using a novel luciferase bacterial artificial chromosome sys-tem. J Virol 81:9024–903311. Wang W, Patterson CE, Yang S, Zhu H(2004) Coupling generation of cytomegalovi-rus deletion mutants and amplification of viral BAC clones. J Virol Methods 121:137–143 12. Netterwald J, Yang S, Wang W, Ghanny S,Cody M, Soteropoulos P, Tian B, Dunn W, Liu F, Zhu H (2005) Two gamma interferon-activated site-like elements in the human cyto-megalovirus major immediate-early promoter/enhancer are important for viral replication.J Virol 79:5035–504613. Yu D, Ellis HM, Lee EC, Jenkins NA,Copeland NG, Court DL (2000) An efficient recombination system for chromosome engi-neering in Escherichia coli. Proc Natl Acad Sci USA 97:5978–598314. Tang QY, Zhang Z, Zhu H (2010) Bioluminesc-ence imaging for herpesvirus studies in vivo.In: Gluckman TR (ed) Herpesviridae: viral structure, life cycle and infections. Nova Science, Huntington, in press。
- 1、下载文档前请自行甄别文档内容的完整性,平台不提供额外的编辑、内容补充、找答案等附加服务。
- 2、"仅部分预览"的文档,不可在线预览部分如存在完整性等问题,可反馈申请退款(可完整预览的文档不适用该条件!)。
- 3、如文档侵犯您的权益,请联系客服反馈,我们会尽快为您处理(人工客服工作时间:9:00-18:30)。
High Mobility Group Box-1 Protein
Tethered particle motion
— 2—
背景知识
V(D)J Recombination
V(D)J重组
V(D)J代表可变区V,多样区D,连接区J。重组是在T和B细胞成熟的早期阶段 仅发生在淋巴细胞中的基因重组的独特机制。它是B和T细胞上发现的免疫球蛋 白(Ig)和T细胞受体(TCR)的高度多样性的组成部分。 V(D)J重组负责组装在B和T淋巴细胞发育期间抗原受体的可变区。 在V(D)J 重组期间,同时位于特定染色体上的V,D和J片段被重排到功能性V(D)J或者 有特异性决定簇的B细胞受体或免疫球蛋白(Ig)和 T细胞受体(TCR)VJ等位 基因中。 与V,D和J基因区段相邻的是由高度保守的七聚体(共有序列5‘CACAGTG-3’)和九聚体(共有序列5‘-ACAAAAACC-3’)组成的重组信号序列 (RSS),被相对不保守的间隔序列12bp 或23bp(分别称为12RSS和23RSS)分 开。
— 3—
背景知识
Recombination activating gene
RAG
重组激活基因(RAG)是在编码免疫球蛋白和T细胞受体分子的基因 的重排和重组中起重要作用的酶。它有两种称为RAG-1和RAG-2的重组激 活基因产物,其细胞表达在其发育阶段限于淋巴细胞。RAG-1和RAG-2对 于产生成熟B和T淋巴细胞是两种细胞类型是必需的。 RAG蛋白启动V(D)J重组,这是pre-B和pre-T细胞的成熟所必需的。 研究表明RAG-1和RAG-2必须以协同方式工作以激活VDJ重组。当分离和 转染成纤维细胞样品时,显示RAG-1低效诱导VDJ基因的重组活性。当 RAG-1与RAG-2共转染时,重组频率增加了1000倍,这一发现促进了新修 订的理论,RAG基因不仅可以帮助VDJ重组,而是直接诱导VDJ基因的重 组。
In the presence of 5 nM RAG1/2c and 80 nM HMGB1 for 1 h。
结果
— 13 —
Results
C
(i) paired complex formation and excision of the looped DNA faster than we can resolve with our method (ii) a pathway for cutting that does not require the two RSS sites to synapse
讨论
— 19 —
Discussion
我们发现HMGB1增加RAG-RSS复合物中DNA长度的减少量。 这种 观察结果与在12RSS-RAG-HMGB1或23RSS-RAG-HMGB1复合物中检测 到的大弯曲的HMGB1依赖性一致,尽管那些研究使用短的寡核苷酸底 物。 我们还使用这种DNA链系长度的减少量来确定在RAG1 / 2c和 HMGB1存在下12RSS和23RSS的Kd,并且发现HMGB1与单独的RAG1 / 2c相比降低了两个Kd值,并且它们是相似的,与前面的观察一致。我们 研究HMGB1依赖过程的单分子实验可以扩展到研究除RAG1 / 2c以外的 其它可以与DNA结合并且可以被HMGB1促进的蛋白质(例如,肿瘤抑 制蛋白p53)。
讨论
— 22 —
Thank You
其中B box是发挥炎症的功能区域,A box是B box的拮抗位点。A box和B box都能够与DNA结合, 并参与DNA双链的折叠与扭曲。
HMGB1
— 5—
背景知识
Tethered particle motion
TPM
Tethered particle motion (TPM)是用生物物理方法研究各种聚合物如DNA及其与其他物质( 例如蛋白质)的相互作用。
讨论
— 20 —
Discussion
在我们实验中使用的RAG1 / 2c和HMGB1的浓度下,在HMGB1存在 (或不存在)下与RAG1 / 2c相关的结合事件相对频繁,但是该结合 (配对复合物形成)的下游产物相对稀少 ; 在我们的实验中,珠子结合 到DNA上在配对的复合物状态中仅花费总观察时间的4%。 我们直接观 察到在DNA底物上存在12RSS和23RSS时配对复合物的形成。 当仅存在 12RSS(或不存在RSS)时,我们没有看到复合物形成的证据,并且我 们确认当使用1,200和1,800-bp间距时配对复合物时的表观长度与已经被 缩短了12/23的信号间距的系链一致。我们无法确定是否一个或两个 RSSs在配对的复杂形成之前被占领, 我们需要进行更多的研究来解决 这个重要机制问题。
实时检测单个RAG-RSS复合物。
结合动力学显示RAG-RSS复合物的平均停留时间。
HMGB1改变RAG-RSS复合物的结合性质。
直接观察平行复合物形成。
12/23规则调节珠释放作为RAG1 / 2c和HMGB1的函数的动力学。
讨论
— 18 —
Discussion
我们观察到在没有HMGB1的情况下通过RAG的RSS结合的DNA长度 的明显减少,这与先前的工作一致,显示单独的RAG蛋白在RSS处弯曲 DNA。这种计算提供了RAG-RSS交互的Kd。我们期望,我们的单分子方 法可用于测量非共有RSS的Kd值,从而提供对RSS和RAG活性之间的关 系的更完整的理解。 该方法还允许首先测定RAG1 / 2c复合物结合到12RSS或23RSS的平 均停留时间。 我们发现RAG1 / 2c-RSS复合物是相当稳定的,停留时间在 几分钟的量级。 特别地是,我们发现,与23RSS(〜230s)相比,复合 物在12RSS(〜430s)处保持更长的结合时间,与两个位点之间的Kd的 差异一致;
5 nM RAG1/2c-----the red line is the rms trajectory of the bead and the black dashed line is the resulting segmentation between long and short states of the DNA tether.
结果
— 14 —
fig.7
Dynamics of 12/23 Rule-Regulated Bead Release as a Function of RAG1/2c and HMGB1.
Results
结果
— 15 —
Results
结果
— 16 —
fig.8
Results
结果
— 17 —
Discussion
结果
— 12 —
fig.6
2900bp
Results
Direct Observation of Paired Complex Formation.
Display distinctly different DNA lengths of 1,360 bp for the 1,200bp intersignal substrate and 880 bp for the 1,800-bp intersignal substrate
结果
— 8—
fig.3
Detecting Single RAG–RSS Complexes in Real Time.
As such, we interpret these periods of reduced rms motion to be binding events that change the effective tether length by 64 ± 5 bp for a single 12RSS binding site and 57 ± 5 bp for a single 23RSS binding site
475 ± 5 bp
结果
482 ± 5 bp
— 10 —
fig.5
Results
HMGB1 Alters the Binding Properties of RAG–RSS Complexes.
结果
— 11 —
Results
We observe that HMGB1 alters the reduction in DNA tether length for RAG1/2c alone from 64 ± 5 bp and 57 ± 5 bp for a single 12RSS and 23RSS to 101 ± 8 bp and 132 ± 4 bp, respectively.
讨论
— 21 —
Discussion
最后,我们通过观察珠子释放推断发夹结构的形成作为研究RAGHMGB1-RSS介导的DNA上的切割。在这些实验中,我们证实了珠释放 和RAG1 / 2c和HMGB1的浓度之间的关系。我们发现珠粒释放在含有 12RSS / 23RSS对的底物上被显着增强,需要催化活性RAG,并且当信 号间距离缩短超过以前显示抑制RAG介导的切割时V(D) J重组。利 用这些工具,现在可以对V(D)J反应过程进行大量的系统研究是有可 能的。我们的结果已经显示,大量的RSS和它们的序列可能改变重组动 力学,并且对确定在淋巴发育期间产生Ig和TCR基因的各种12/23规则调 节的V,D和J反应的突触和发夹形成的定量规则是有很大意义的。
Single-molecule analysis of RAG-mediated V(D)J DNA cleavage
PNAS 2015 112 (14) 4193-4194; doi:10.1073/pnas.ss11214
• reporter:
背景知识
V(D)J Recombination
Recombination Activating Gene
— 9—
fig.4
Results
