pBacPAK8昆虫表达载体说明
提高平末端连接效率的研究

提高平末端连接效率的研究张伟,艾秀莲,王玮,王志方,崔春生,李晨华,谢玉清(新疆农科院微生物所,新疆乌鲁木齐830000)摘 要:在平末端连接反应中,为了提高连接效率,研究载体与片段的分子量比、载体与片段的摩尔比、连接反应体积、连接反应的温度、连接反应时间长短等条件对连接效率的影响,发现这些因素都会直接影响连接效率。
因此在做不同的平末端连接反应时,应该把上述因素协同起来考虑。
此外,反应时间长短也应根据不同情况区别对待。
关键词:平末端连接;连接效率;普适性中图分类号:Q523 文献标识码: 文章编号:1001-4330(2002)05-0300-02The Study to Improve the fficiency of Ligation of B lunt -ended DNAZH ANG Wei ,AI X iu -lian ,W ANG Wei ,W ANG ZHi -fang ,C UI CHun -sheng ,LI CHeng -hua ,XIE Y u -qing(Institute o f Microorganism ,Xinjiang Academy o f Agricultural Sciences ,Urumqi 830000)Abstract :It is important to improve the efficiency of ligation reaction ,especially for joining of blunt -ended DNA which is univer 2sal applicated ,however ,with lower efficiency.In our study ,We draw a conclusion that the m ol -ratio of vector and insert 、the v olume of ligation reaction 、the temperature and time of ligation reaction etc.All can in fluence the efficiency of ligation ,s o we should think of these factors together.K ey w ords :Ligation of blunt -ended DNA ;E fficiency of ligation在分子生物学的研究中,DNA 末端的连接是一极为常用的基本操作,可分为粘末端和平末端连接,粘末端连接效率相对较高,平末端的连接效率一般非常低,但平末端连接具有普适性,被广泛地用于将外源DNA 片段的相互连接当中。
pCoBlast昆虫表达载体说明
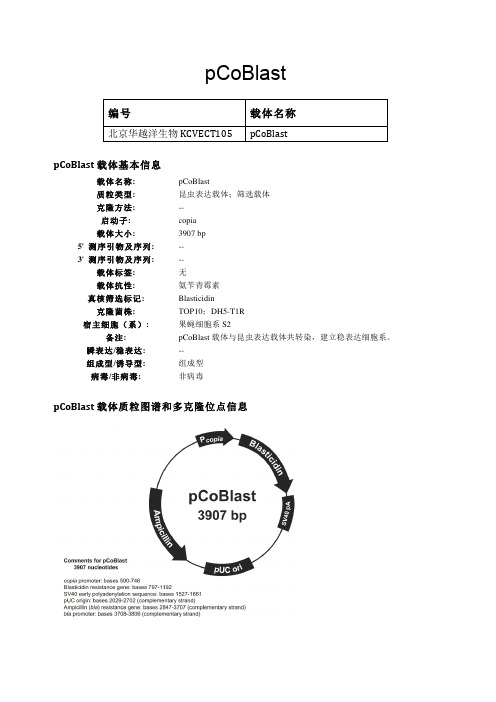
pCoBlast编号 载体名称北京华越洋生物KCVECT105 pCoBlastpCoBlast载体基本信息载体名称: pCoBlast质粒类型: 昆虫表达载体;筛选载体克隆方法: --启动子: copia载体大小: 3907 bp5' 测序引物及序列: --3' 测序引物及序列: --载体标签: 无载体抗性: 氨苄青霉素真核筛选标记: Blasticidin克隆菌株: TOP10;DH5-T1R宿主细胞(系): 果蝇细胞系S2备注: pCoBlast载体与昆虫表达载体共转染,建立稳表达细胞系。
瞬表达/稳表达: --组成型/诱导型: 组成型病毒/非病毒: 非病毒pCoBlast载体质粒图谱和多克隆位点信息pCoBlast载体简介pCoBlast is a 3907 bp selection vector that can be cotransfected with the expression vector of choice to to create stable cell lines in Drosophila. It contains the Streptomyces griseochromogenes bsd gene under control of the Drosophila copia promoter to confer resistance to blasticidin in S2 cells.引用及参考文献A novel allelic variant of the human TSG-6 gene encoding an amino acid difference in the CUB module. Chromosomal localization, frequency analysis, modeling, and expression.Nentwich Hilke A; Mustafa Zehra; Rugg Marilyn S; Marsden Brian D; Cordell Martin R; Mahoney David J; Jenkins Suzanne C; Dowling Barbara; Fries Erik; Milner Caroline M; Loughlin John; Day Anthony J;J Biol Chem (2002) 277:15354-15416Product usage: The full-length coding sequence of human TSG-6 (A431 allele encoding Gln at amino acid 144), including the signal sequence and stop codon,was excised from the pAcCL29-1 vector by digestion with KpnI/XbaI and cloned into the corresponding sites in the Drosophila Expression SystempCoBlast载体序列TCGCGCGTTTCGGTGATGACGGTGAAAACCTCTGACACATGCAGCTCCCGGAGACGGTCACAGCTTGTCT GTAAGCGGATGCCGGGAGCAGACAAGCCCGTCAGGGCGCGTCAGCGGGTGTTGGCGGGTGTCGGGGCTGG CTTAACTATGCGGCATCAGAGCAGATTGTACTGAGAGTGCACCATATGCGGTGTGAAATACCGCACAGAT GCGTAAGGAGAAAATACCGCATCAGGCGCCATTCGCCATTCAGGCTGCGCAACTGTTGGGAAGGGCGATC GGTGCGGGCCTCTTCGCTATTACGCCAGCTGGCGAAAGGGGGATGTGCTGCAAGGCGATTAAGTTGGGTA ACGCCAGGGTTTTCCCAGTCACGACGTTGTAAAACGACGGCCAGTGCCAAGCTTGCATGCCTGCAGGTCG ACTCTAGAGGATCCGGTGCGTGGTGGTTTCATGCTTCTGGGAACGGCAAATGGGTTTAGGATTGGGAACC CCTCATCATCTGTTGGAATATACTATTCAACCTACAAAAATAACGTTAAACAACACTACTTTATATTTGA TATGAATGGCCACACCTTTTATGCCATAAAACATATTGTAAGAGAATACCACTCTTTTTATTCCTTCTTT CCTTCTTGTACGTTTTTTGCTGTGAGTAGGTCGTGGTGCTGGTGTTGCAGTTGAAATAACTTAAAATATA AATCATAAAACTCAAACATAAACTTGACTATTTATTTATTTATTAAGAAAGGAAATATAAATTATAAATT ACAACAGGTTCCCTTTATGCGAAGGGATGGCCAAGCCTTTGTCTCAAGAAGAATCCACCCTCATTGAAAG AGCAACGGCTACAATCAACAGCATCCCCATCTCTGAAGACTACAGCGTCGCCAGCGCAGCTCTCTCTAGC GACGGCCGCATCTTCACTGGTGTCAATGTATATCATTTTACTGGGGGACCTTGCGCAGAACTCGTGGTGC TGGGCACTGCTGCTGCTGCGGCAGCTGGCAACCTGACTTGTATCGTCGCGATCGGAAATGAGAACAGGGG CATCTTGAGCCCCTGCGGACGGTGCCGACAGGTTCTTCTCGATCTGCATCCTGGGATCAAAGCCATAGTG AAGGACAGTGATGGACAGCCGACGGCAGTTGGGATTCGTGAATTGCTGCCCTCTGGTTATGTGTGGGAGG GCTAACCCTTATACGCAAGGGAGTAGATGCCGACCGAACAAGAGCTGATTTCGAGAACGCCTCAGCCAGC AACTCGCGCGAGCCTAGCAAGGCAAATGCGAGAGAACGGCCTTACGCTTGGTGGCACAGTTCTCGTCCAC AGTTCGCTAAGCTCGCTCGGCTGGGTCGCGGGAGGGCCGGTCGCAGTGATTCAGGCCCTTCTGGATTGTG TTGGTCCCCAGGGCACGATTGTCATGCCCACGCACTCGGGTGATCTGACTGATCCCGCAGATTGGAGATC GCCGCCCGTGCCTGCCGATTGGGTGCAGATCAGCCTCGAGGCCAGCTAGCTTGAACTTGTTTATTGCAGC TTATAATGGTTACAAATAAAGCAATAGCATCACAAATTTCACAAATAAAGCATTTTTTTCACTGCATTCT AGTTGTGGTTTGTCCAAACTCATCAATGTATCTTATCATGTCTGGATCGGGTACCGAGCTCGAATTCGTA ATCATGTCATAGCTGTTTCCTGTGTGAAATTGTTATCCGCTCACAATTCCACACAACATACGAGCCGGAA GCATAAAGTGTAAAGCCTGGGGTGCCTAATGAGTGAGCTAACTCACATTAATTGCGTTGCGCTCACTGCC CGCTTTCCAGTCGGGAAACCTGTCGTGCCAGCTGCATTAATGAATCGGCCAACGCGCGGGGAGAGGCGGT TTGCGTATTGGGCGCTCTTCCGCTTCCTCGCTCACTGACTCGCTGCGCTCGGTCGTTCGGCTGCGGCGAG CGGTATCAGCTCACTCAAAGGCGGTAATACGGTTATCCACAGAATCAGGGGATAACGCAGGAAAGAACAT GTGAGCAAAAGGCCAGCAAAAGGCCAGGAACCGTAAAAAGGCCGCGTTGCTGGCGTTTTTCCATAGGCTC CGCCCCCCTGACGAGCATCACAAAAATCGACGCTCAAGTCAGAGGTGGCGAAACCCGACAGGACTATAAA GATACCAGGCGTTTCCCCCTGGAAGCTCCCTCGTGCGCTCTCCTGTTCCGACCCTGCCGCTTACCGGATA CCTGTCCGCCTTTCTCCCTTCGGGAAGCGTGGCGCTTTCTCAATGCTCACGCTGTAGGTATCTCAGTTCG GTGTAGGTCGTTCGCTCCAAGCTGGGCTGTGTGCACGAACCCCCCGTTCAGCCCGACCGCTGCGCCTTAT CCGGTAACTATCGTCTTGAGTCCAACCCGGTAAGACACGACTTATCGCCACTGGCAGCAGCCACTGGTAA CAGGATTAGCAGAGCGAGGTATGTAGGCGGTGCTACAGAGTTCTTGAAGTGGTGGCCTAACTACGGCTAC ACTAGAAGGACAGTATTTGGTATCTGCGCTCTGCTGAAGCCAGTTACCTTCGGAAAAAGAGTTGGTAGCT CTTGATCCGGCAAACAAACCACCGCTGGTAGCGGTGGTTTTTTTGTTTGCAAGCAGCAGATTACGCGCAG AAAAAAAGGATCTCAAGAAGATCCTTTGATCTTTTCTACGGGGTCTGACGCTCAGTGGAACGAAAACTCACGTTAAGGGATTTTGGTCATGAGATTATCAAAAAGGATCTTCACCTAGATCCTTTTAAATTAAAAATGAA GTTTTAAATCAATCTAAAGTATATATGAGTAAACTTGGTCTGACAGTTACCAATGCTTAATCAGTGAGGC ACCTATCTCAGCGATCTGTCTATTTCGTTCATCCATAGTTGCCTGACTCCCCGTCGTGTAGATAACTACG ATACGGGAGGGCTTACCATCTGGCCCCAGTGCTGCAATGATACCGCGAGACCCACGCTCACCGGCTCCAG ATTTATCAGCAATAAACCAGCCAGCCGGAAGGGCCGAGCGCAGAAGTGGTCCTGCAACTTTATCCGCCTC CATCCAGTCTATTAATTGTTGCCGGGAAGCTAGAGTAAGTAGTTCGCCAGTTAATAGTTTGCGCAACGTT GTTGCCATTGCTACAGGCATCGTGGTGTCACGCTCGTCGTTTGGTATGGCTTCATTCAGCTCCGGTTCCC AACGATCAAGGCGAGTTACATGATCCCCCATGTTGTGCAAAAAAGCGGTTAGCTCCTTCGGTCCTCCGAT CGTTGTCAGAAGTAAGTTGGCCGCAGTGTTATCACTCATGGTTATGGCAGCACTGCATAATTCTCTTACT GTCATGCCATCCGTAAGATGCTTTTCTGTGACTGGTGAGTACTCAACCAAGTCATTCTGAGAATAGTGTA TGCGGCGACCGAGTTGCTCTTGCCCGGCGTCAATACGGGATAATACCGCGCCACATAGCAGAACTTTAAA AGTGCTCATCATTGGAAAACGTTCTTCGGGGCGAAAACTCTCAAGGATCTTACCGCTGTTGAGATCCAGT TCGATGTAACCCACTCGTGCACCCAACTGATCTTCAGCATCTTTTACTTTCACCAGCGTTTCTGGGTGAG CAAAAACAGGAAGGCAAAATGCCGCAAAAAAGGGAATAAGGGCGACACGGAAATGTTGAATACTCATACT CTTCCTTTTTCAATATTATTGAAGCATTTATCAGGGTTATTGTCTCATGAGCGGATACATATTTGAATGT ATTTAGAAAAATAAACAAATAGGGGTTCCGCGCACATTTCCCCGAAAAGTGCCACCTGACGTCTAAGAAApCoBlast其他昆虫表达载体:pMT/BioEase-DEST pVL1392 pVL1393 pXINSECT-DEST39 pXINSECT-DEST38 pBacPAK9 pBacPAK8 pIB/V5-His-TOPO pIEX/Bac-1 pFastBacHT B pFastBacHT A pFastBac1pMT/V5-His-TOPO pMT/BiP/V5-His A pCoHygro pAc5.1/V5-His C pAc5.1-V5-His A pAc5/V5-HisA pAc5-V5-HisB pAc5-V5-HisC pCoBlast pAc5.1/V5-His B pIZ/V5-His pIB/V5-HispIZT/V5-His pMIB/V5-His C pMIB/V5-His A pMT/BiP/V5-His C pMT/BiP/V5-His B pIEx/Bac-4 pIEx/Bac-3 pIEXBac-c-EGFP-3 pIEXBac-c-EGFP-1 pIEXBac-c-EGFP-4 pAc5.1B-EGFP pBlueBacHis2 A pBlueBacHis2 C pBlueBacHis2 B pFastBacHT C pMT-Bip-V5-HisA pFBDM pUCDM pFastBac Dual pMIB/V5-His BpMT/V5-His C pMT/V5-His B pMT/V5-His A pAcGP67ApAc5.1b pAc5.1a pVL1392-XyIE control vector pFastBac-c-His-TEV pFastBac-N-GST-TEV pFastBacI-Gus pFastBacHT-CAT pBADZ-His6Cre。
昆虫杆状病毒表达载体系统的应用与展望

昆虫杆状病毒表达载体系统的应用与展望作者:宋姗姗来源:《中国科技纵横》2017年第22期摘要:昆虫杆状病毒表达载体系统具有生物安全性高、能容纳较大外源基因片段并实现多基因共表达、有完备的翻译后加工修饰系统等特点,因而得到广泛的应用。
近年来,随着重组杆状病毒构建技术、昆虫细胞培养技术的不断发展和改进,加大了昆虫杆状病毒表达载体系统在疫苗制备、治疗性抗体、药物研发和基因治疗中的应用力度。
本文综述了杆状病毒表达载体的研究现状和应用范围,讨论了目前杆状病毒表达载体重组蛋白易降解的问题。
随着科技的进步,对杆状病毒载体的研究与优化会更加深入,昆虫杆状病毒表达载体系统的应用研究尤为重要。
关键词:杆状病毒;疫苗;治疗性抗体;基因治疗中图分类号:Q789 文献标识码:A 文章编号:1671-2064(2017)22-0209-01昆虫杆状病毒表达载体系统(Baculovirus expression vector system,BEVS)是当今基因工程四大表达系统之一,具有生物安全性高、能容纳较大外源基因片段、可实现多基因共表达、具有完备的翻译后加工修饰系统、无内毒素污染等优点[1-3]。
同时,由于宿主细胞系的建立和优化、细胞培养基成分的不断改进,使得细胞生存率达到95%,基本可实现大规模培养[1]。
因此,昆虫杆状病毒表达系统越来越广泛的应用于疫苗制备、药物研发、农林牧渔等领域。
1 昆虫杆状病毒表达载体系统的研究进展Kitts等在1990年提出了线性技术,并设计了重组-救活方法,提高了杆状病毒基因组同源重组能力,将其设计的BacPAK6的重组效率提到80%以上,有较强的应用价值[4]。
但由于DNA酶切消化效率影响,重组病毒的获得仍需通过噬斑纯化过程。
Bac-to-Bac技术的出现避免了噬斑纯化步骤,缩短了重组杆状病毒的周期,同时,此方法在大肠杆菌中进行,非常便捷,不需要空斑纯化,较为节省时间。
但由于鳞翅目昆虫细胞系缺乏功能性糖基转移酶和唾液酸生物合成与利用途径,其产生的糖基化修饰缺乏末端唾液酸化,影响糖蛋白的整体性质,因此研究人员设计出转基因昆虫细胞系,使昆虫细胞对重组蛋白的糖基化修饰更加接近哺乳动物细胞,更利于昆虫杆状病毒表达载体系统的研究。
载体及其上下游引物

载体名称上游引物(5'primer)下游引物(3'primer)P3*FLAG-CMV CMV-30(825-854)CMV-24(1129-1152)pAc5.1/V5-His(_A,_B,_C) pAc5-5 BGH revpAcG2T pGEX-5 not availablepACT T7EEV T3/EBVrevpACT2GAL4 ADfor pGAL4-ADrvpACT2p17110p12584pACYC184(BamHI-site) PBRforBam PBRrevBampACYCDuet-1(MCSI)ACYCDuetUP1 Primer DuetDOWN1 pACYCDuet-1(MCSII) DuetUp2 T7-TerminatorpAdEasy-1 not available not availablepAdTrack-CMV not available not availablepAS2-1GAL4 Bdfor/P24990pGAL4-BDrv/P31430pB42AD pB42ADF PB42ADRpBABE pBMN_5' pBacPAK8BAC1BAC2pBAD pBAD-F pBAD-RpBAD/HisABC pBAD-5' pBBR1MCS T3/M13rev T7/M13forpBD-GAL4pBD-GAL4-F PBD-GAL4-RpBI12135S NOSpBIND GAL4-Bdfor T3,EBVrevpBK-CMV M13F/T7M13R/T3 pBluescriptIIKS(+/-) T3/M13rev T7/M13for(优先用M13F)pBluescriptIISK(+/-) T3/M13rev T7/M13for pBluescriptKS(+/-) T3/M13rev T7/M13for pBluescriptSK(+/-) T3/M13rev T7/M13forpBR322(BamHI-site) PBRforBam PBRrevBampBR322(EcoRI/HindIII-sites) pBRforEco pBRrevHindpBR325(BamHI-site) PBRforBam PBRrevBampBR329(BamHI-site) PBRforBam PBRrevBampBridge(MCSI) GAL4-Bdfor3'BDpBridge(MCSII) not available not availablepBS-T II T3/M13rev T7/M13forpBS(AMP)T7T3pBSR T3T7terpBudCE4.1(MCSI) CMV-Profor,T7 c-mycrev,EBVrevpBudCE4.1(MCSII)EF-1α Fwd BGHpBudCE4.1/lacZ/CAT CMV-Profor,T7 c-mycrev,EBVrevpBV220pBV220F PBV220RpCAL-c T7/Tac-Profor T7-TerminatorpCAL-n T7/Tac-Profor T7-Terminator pCAMBIA1301(1300)pCAMBIA1301-5'(ECORI)/M13R pCAMBIA1301-3'(HindIII) pCAMBIA1304pCAMBIA1301-5'(ECORI)pCAMBIA1301-3'(HindIII) pCAMBIA2300M13R(-48)M13F(-47)/M13F(-49) pCANTB5E S1S6pCAT3-enhancer RVP3此载体无反向引物pCDFDuet-1(MCSI)ACYCDuetUP1 Primer DuetDOWN1 pCDFDuet-1(MCSII)DuetUP2T7TERpcDNA1.1 CMV-Profor,T7 pCDM8-3pcDNA2.1 M13for/T7 M13rev/Tac-Profor pcDNA3 T7/CMV-Profor BGH rev/SP6pcDNA3.1(+/-) pcDNA3.1F/T7/CMV-Profor BGH revpcDNA3.1/His(_A,_B,_C) T7/CMV-Profor BGH revpcDNA3.1/myc-His(_A,_B,_C) T7/CMV-Profor BGH rev/c-mycrev pcDNA3.1/V5-His(_A,_B,_C) T7/CMV-Profor BGH rev/V5revpcDNA3.1/Hygro(+,-) T7/CMV-Profor BGH rev/V5revpcDNA3.1/Zeo(+.-) T7BGHrevpcDNA3.3TOPOTA CMV-Profor TKPAreversepcDNA4/HisMax(_A,_B,_C) Xpressfor BGH revpcDNA4/HisMax-TOPO Xpressfor BGH revpcDNA4/HisMax-TOPO/lacZ Xpressfor BGH revpcDNA4/myc-His(_A,_B,_C)T7,CMV-Profor BGH rev,c-mycrev pcDNA4/TO TREfor/CMV-Profor BGH revpcDNA4/V5-His(_A,_B,_C) CMV-Profor,T7 BGH rev,V5revpcDNA4-TET/ON/AMP+CMV-Profor pcDNA5/FRT T7/CMV-Profor BGH revpcDNA6/myc-His(_A,_B,_C) CMV-Profor,T7 BGH rev,c-mycrev pcDNA6/V5-His(_A,_B,_C) CMV-Profor,T7 BGH rev,V5revpCEP4pCEP-F/CMV-Profor EBVrevpCI T7(17BASE) EBVrevpCI-neo T7(17BASE) EBVrev,T3pCMS-EGFP T7 T3pCMS-EGFP T7 EBVrev,T3pCMV-3Tag-4A T3 /PFLAG-CMV-F T7pCMV5pCMV5F PCMV5RpCMV6 CMV-Profor pCMV6-3、HGHrev pCMV6-XL4 CMV-Profor/T7 pCMV6-3、HGHrev pCMV-HA PCMV-F,CMV-Profor,TREfor PCMV-R,EBVrev pCMV-MYC(AMP+)PCMV-F PCMV-R, EBVrev pCMVmyc/nuc CMV-Profor BGH rev,c-mycrevpcmv-scriptEXVector T3 T7pCMV-Sport6 T7/M13for SP6/M13rev pCOLADuet-1(MCSI)ACYCDuetUP1 Primer DuetDOWN1pCOLADuet-1(MCSII)DuetUP2T7TERpCR2.1 T7,M13for M13revpCR2.1-TOPO M13for,T7 M13revpCR3.1 T7 BGH revpCR3.1-Uni T7 BGH revpCR4 T3/M13rev T7/M13forpCR4-Blunt T3/M13rev T7/M13forpCR4-TOPO M13rev,T3 M13for,T7pCR-Blunt M13rev T7,M13forpCR-BluntII M13rev/SP6 T7/M13forpCR-BluntII-TOPO M13rev/SP6 M13for/T7pCRII M13rev/SP6 T7/M13forpCRII-TOPO M13rev/SP6 M13for/T7pCR-ScriptSK(+) T7/M13for T3/M13revpCR-XL-TOPO M13rev T7,M13forpCS2+(MCSI) SP6 EBVrev,T7?pCS2+(MCSII) T3 pSG5-3pDB_GAL4cam T7P pDB_leu PDBLeu-5 PDBLeu-3pDEST22DEST22F DEST22RpDEST15 pGEX-5 T7-TerminatorpDEST20 pGEX-5 pFastBac-3pDEST26 TREfor/CMV-Profor T7/M13forpDisplay T7/CMV-Profor c-mycrevpDNR-LIB(MCS_A) M13for,T7 pSG5-3,M13RpDonar M13F M13RpDONR201PDONR-F PDONR-RpDONR207PDONR-F PDONR-RpDONR223M13F M13RpDrive T7/M13rev SP6/M13forpDsRED1-C1(DsRed1-N-f)DsRed1-C-rpDsRed1-N1 CMV-Profor RFP-Nrev,CMV-R? pDsRed2-N1(DsRed1-N-f)DsRed1-C-rpDsRed2-C1(DsRed1-N-f)DsRed1-C-rpDsRED2-C1(KAN+)pDsRED-ex-C1-F pEGFP-N-3’pDsRed-Express dsRed1_N_primer/CMV-F dsRed1_C_primer/CMV-R pDSRED-monomer-N1(KAN+)dsRed1_N_primer pDSRED-N1(KAN+)pEGFP-N-5’ pDSRED-N-RpEASY-Blunt M13F T7/ M13RpEASY-BluntSimple M13F T7/ M13R(距离插入位点合适,优先安排)pEASY-T1 Simple M13F M13R(距离插入位点合适,优先安排)pEASY-T1M13F/T7M13RpEASY-T3M13F/T7M13R/SP6pECFP-N(C)PECFP-N-5.PECFP-N-3,pEF/myc/cyto/ER/mito/nuc pEF-5Pcdna3.1R/BGH revpEF1(4,6/myc-His) T7/pEF-5Pcdna3.1R/BGH revpEF1(4,6/V5-His) T7/pEF-5Pcdna3.1R/BGH revpEF1(4;6/His_A;_B;_C) T7/Xpressfor Pcdna3.1R/BGH revpEF5/FRT/V5-D(-TOPO) T7/pEF-5Pcdna3.1R/BGH rev pEGFP M13rev EGFP-NrevpEGFP-1 not available EGFP-Nrev/ PEGFP-N-3′pEGFP-C PEGFP-C-5′PEGFP-C-3′pEGFP-C1 EGFP-Cfor SV40-pArevpEGFP-C2 EGFP-Cfor SV40-pArevpEGFP-C3 EGFP-Cfor SV40-pArevpEGFP-F EGFP-Cfor SV40-pArevpEGFP-N PEGFP-N-5′PEGFP-N-3′pEGFP-N1 CMV-Profor EGFP-NrevpEGFP-N2 CMV-Profor EGFP-NrevpEGFP-N3 CMV-Profor EGFP-NrevpENTR M13F M13RpET-11(-a,-b,-c,-d) T7 T7-TerminatorpET-12(-a,-b,-c) T7 T7-TerminatorpET-14b T7 T7-TerminatorpET-15b T7 T7-TerminatorpET-16b T7 T7-TerminatorpET-17b T7 T7-TerminatorpET-17xb T7 T7-TerminatorpET-19b T7 T7-TerminatorpET-20b(+) T7 T7-TerminatorpET-21(-a,-b,-c,-d)(+) T7 T7-TerminatorpET-22b(+) T7 T7-TerminatorpET-23(-a,-b,-c,-d)(+) T7 T7-TerminatorpET-24(-a,-b,-c,-d)(+) T7 T7-TerminatorpET-25b(+) T7 T7-TerminatorpET-26b(+) T7 T7-TerminatorpET-27b(+) T7 T7-TerminatorpET-28a(-b,-c)(+) T7 T7-TerminatorpET-29a(-b,-c)(+)S.tag/T7 T7-TerminatorpET-3(-a,-b,-c,-d) T7 T7-TerminatorpET-30_Ek/LIC T7 T7-TerminatorpET-30_Xa/LIC T7 T7-TerminatorpET-30a(-b,-c)(+)S.tag/T7 T7-TerminatorpET-31b(+) not available T7-TerminatorpET-32_Ek/LIC TRXfor T7-Terminator pET-32_XA/LIC TRXfor T7-Terminator pET-32a(-b,-c)(+)S.tag/TRXfor T7-Terminator pET-33b(+) T7 T7-Terminator pET-34b(+)S.tag T7-Terminator pET-35b(+)S.tag T7-Terminator pET-36b(+)S.tag T7-Terminator pET-37b(+)S.tag T7-Terminator pET-38b(+) not available T7-Terminator pET-39b(+)S.tag T7-Terminator pET-40b(+)S.tag T7-Terminator pET-41_Ek/LIC GST-Cfor T7-Terminator pET-41a(-b,-c)(+) S.tag/pGEX-5 T7-Terminator pET-42a(-b,-c)(+)S.tag/pGEX-5 T7-Terminator pET-43.1a(-b,-c)(+)S.tag coliDOWNpET-43.1a_EK/LIC S.tag not availablepET-44a(-b,-c)(+)S.tag T7-Terminator pET-44a_EK/LIC S.tag not availablepET-45b(+) T7 T7-Terminator pET-46_Ek/LIC T7 T7-Terminator pET-5(-a,-b,-c) T7 T7-Terminator pET-52b(+) T7 T7-Terminator pET-7 T7 T7-Terminator pET-9(-a,-b,-c,-d) T7 T7-Terminator pETcoco-1 T7 T7-Terminator pETDuet-1(MCSI)pETUP1DuetDOWN1 pETDuet-1(MCSII)DuetUp2T7-Terminator pET-T4 T7 T7-Terminator pEYFP-C1pEGFP-C-5’ pEGFP-C-3' pEYFP-N1pEGFP-N-5’,CMV-Profor pEGFP-N-3’ pFLAG-ATS N26for pTrcHis-rv;C24rev pFLAG-CTC N26for pTrcHis-rv;C24rev pFLAG1N26for pTrcHis-rv;C24rev pFLAG-CMV(-2)CMV-30;CMV-Profor CMV-24pFLAG-CMV1CMV-30CMV-24pGAD424GAL4_ADfor3'ADpGADGH GAL4_ADfor3'ADpGADT7T7/GAL4_ADfor3'ADpGADT7-Rec T7/GAL4_ADfor3'ADpGADT-T T73,BDpGAD10GAL4_ADfor3'ADpGAPZa-A a-FACTOR3'AOXpGBKT7T73,BDpGBT9MATCHMAKER5'DNA-BD/GAL4BD MATCHMAKER3'DNA-BDpGEM-1 T7 SP6pGEM-3 T7 SP6pGEM-3Zf(+)(-) SP6/M13rev T7/M13for pGEM-4 T7 SP6pGEM-4Z T7/M13for SP6/M13rev pGEM-5Zf(+)(-) SP6/M13rev T7/M13for pGEM-7Zf(+)(-) SP6/M13rev T7/M13for pGEM-9Zf(+)(-) T7/M13for SP6/M13rev pGEM-T T7/M13for SP6/M13rev pGEM-T_Easy T7/M13for SP6/M13rev pGene/V5-His_A(_B,_C) pGENE-5 BGH rev,V5rev pGEX-2T pGEX-5 pGEX-3 pGEX-2TK pGEX-5 pGEX-3 pGEX-3X pGEX-5 pGEX-3 pGEX-4T-1(-2,-3) pGEX-5 pGEX-3 pGEX-5X-1(-2,-3) pGEX-5 pGEX-3 pGEX-6P-1(-2,-3) pGEX-5 pGEX-3pGL2GLP1 GLP2pGL3RVP3GLP2(RVP4)(没有特别说明用GLP2)pGL3-Basic(MCSI) RVP3(PGL3+)? GLP2(PGL3-)?pGL3-Basic(MCSII) RVP4, PLXSN-3'?pGL4 pGlow(-TOPO) T7 pGlow-3pGM-T T7SP6pGWC SP6T7pHook-2 T7/CMV-Profor not availablepIND/V5-His_A(_B,_C) not available BGH rev,V5rev PinpointTMVector PinpointPrimer SP6pIRES(MCSI) T7 pIRES-3pIRES(MCSII) not available T3,EBVrevpIRES2-EGFP CMV-Profor,pIRES2-EGFP.P5’ IRESrev,pIRES2-EGFP.P3’pIRES-hrGFP-2a CMV-Profor,T3 pIRES-hrGFP-3 pIRESneo2 T7/CMV-Profor pIRES-3pIVEX(all) T7/pBRrevBam T7-TerminatorpLEXA pLEXA-F pLEXA-RpLNCX pLNCX-F pLNCX-RpLNCX2CMV-Profor pRevTRE-3pLVTHM(H1启动子下游)H1primer SP6pLXSN pLXSN-F pLXSN-RpMALc MalE primer M13F(-47)pMAL-c2E MalE primer M13F(-47)pMAL-C2x Pmal-c2x-F(P5’)Pmal-C2X-R(P3’ ) pMALd MalE primer M13F(-47)pMALe MalE primer M13F(-47)pMAL-P4X MalE primer M13F(-47)pMAL-p2X MalE primer M13F(-47)pMET A/B/C AUG1F AUG1RpMD18-T M13R(-48)ECORI M13F(-47)(HINDⅢ) pMD19-T M13F47M13R48pMD20-T M13F47M13R48 pMOSBlue T7/M13rev M13forpMSCV-puro pLXSNfw pMSCVrevpMT/V5-His_A(_B,_C) Mtfor BGH rev,V5rev pNTAP T3T7pCTAP pCTAP5'primer T7pOTB7 M13rev/T7 M13for/SP6 pOptiVEC-TOPO TA CMV-F/CMV-Profor EMCVIRES reverse pPC86PPC86-F PPC86-RpPC97PBD-GAL4-F PBD-GAL4-RpPIC3.5K5'AOX3'AOXpPic9k(A+,Z+)5’AOX(Ppic9k-f)/a-factor3’AOX(Ppic9k-R) pPICZa5'AOX/PPICZa-F3'AOX pPROEXHTA M13R(-48)无反向引物pProEX-HTa(b,c)M13R48/ Tac-Profor pBADrv/pTrcHis-3 pPD129.36 M13F20/M13FpQE30or40or60(AMP+)PQE30-F PQE-30RpRC/CMV2/CAT CMV/T7 pRECEIVER CMV-F 此载体无反向引物pRECEIVER-M03pRECEIVER-M03F pRECEIVER-M03R pREP4pREP4 forward primer EBV reverse primer pRK5 SP6 pSG5-3pRSET T7/Xpressfor T7terpRSFDuet-1(MCSI)DuetUP1DuetDOWN1 pRSFDuet-1(MCSII)DuetUP2T7TERpSecTac2 T7/CMV-Profor BGH rev/c-mycrev pSG5 pSG5-5,/T7PSG5-R(SV40) pSHlox1 T7 SP6pSHUTTLE-CMV PSHUTTLE-CMV-F PSHUTTLE-CMV-R pSI T7 EBVrevpSilencer1.0-U6 T7 T3pSilencer2.0-U6 T7 M13forpSilencer2.1-U6neo T7 M13forpSilencer3.1-H1hygro M13F(-47) 3.0REVpSilencer4_1-CMVneo pSilener4.1-5' CMV-Profor pSILENCEV2.0-U6T7 2.0REVpSIneo T7 EBVrev,T3Psit T7 T7TpSK01-T M13F M13RpSK-CMV(KAN+)T7 T3pSOS Psos5'Psos3'pSP64 SP6 M13revpSP64/65SP6 此载体无反向引物pSP65 SP6 M13revpSP70 SP6 T7pSP71 SP6 T7pSP72 SP6 T7pSP73 SP6 T7pSport1SP6/M13F(-47)T7/M13R pSPORT1 SP6/M13for T7/M13revpSPT18 SP6 T7pSTBlue T7/M13rev SP6/M13for pSUPER.Basic T7/M13F M13RpSUPER T7T3pSURE-T M13F M13RpSURE-Tsimple M13F-47M13R-48 pSuperCos pBRforEco T7pT3T7 M13rev/T3 M13for/T7pT7-7 T7 not availablepT7Blue®T7 M13F(-47)pT7T3_18U M13rev/T7 M13for/T3pT7T3D-PAC M13rev/T7 M13for/T3pTA2M13F/T7(优先使用M13F)T3/M13RpT-Adv M13rev T7,M13forpTAg T7/M13rev M13for pTARGETTM pTarget.F(在T7前面)/T7 pTarget.RpTG19-T M13F(-47)M13R(-48) pTARGETTM T7/M13F PTARGET-SEQ pThioHisA,B,andC Trx Forward Trx Reverse / M13for pTO-T7T7 T7TERpTRC99A PTRC99A-F pTrcHis-rvpTrc99a-c PTRC99C-F PTRC99C-RpTRC99a-c PTRC99C-F PTRC99C-R pTrcHisB PTrcHisA-F/Xpressfor PTrcHisA-R pTrcHisA PTrcHisA-F PTrcHisA-RpT-REX-DEST30 TREfor T7pTriEx1 T7/TriExup TriExdownpTriEx1.1 T7/TriExup TriExdownpTriEx1.1_Hygro T7/TriExup not availablepTriEx1.1_Neo T7/TriExup IRESrevpTriplEX1 TriplEx-LD-5 T7,TriplEx-LD-3pTriplEX2 TriplEx-LD-5;pTR5‘ T7,TriplEx-LD-3pTWIN-1T7 T7TERpTXB1 T7 MxeInteinrevpTXB2 T7 MxeInteinrevpTXB3 T7 MxeInteinrevpTYB1 T7 InteinrevpTYB2 T7 InteinrevpTYB3 T7 InteinrevpTYB4 T7 InteinrevpTZ_18U T7/M13rev M13forpUB6/V5-His UB-F BGH rev,V5revpUC13 M13rev/M13rev(-48) M13for/M13for(-49)pUC18 M13rev/M13rev(-48) M13for/M13for(-49)pUC18(19)/118(119)M13F(-47)M13R(-48)/PUC19(HINDⅢ) pUC19 M13rev/M13rev(-48) M13for/M13for(-49)pUC57 M13rev/M13rev(-48) M13for/M13for(-49)pUC8 M13rev/M13rev(-48) M13for/M13for(-49)pUC9 M13rev/M13rev(-48) M13for/M13for(-49)pUM-T M13F(-20)M13R(-20)pUCm-T(AMP+)M13F/T7M13RpUNI/V5-His not available BGH rev,V5revpVAX1T7 T7/CMV-Profor PCDNA3.1R/BGHRpVP22/mycHis VP22-Cfor BGH rev,c-mycrevpVP22/mycHis2 T7/CMV-Profor VP22-NrevpWE15 pBRrevHind T7pYES2 T7pYes2.RpYES-DEST52 T7/GAL1-Profor V5rev/CYC1-Terminator pYEX_4T_1 Clontech pYEX_4T-3pYEX_4T_2 pGEX-5 pYEX_4T-4pYEX_4T_3 pGEX-5 pYEX_4T-4pYEX_4T_4 pGEX-5 pYEX_4T-4pZeoSV2+/hu-Tyr T7 not availablepZERO-1 M13rev/SP6 T7/M13forpZERO-2 M13rev/SP6 M13for/T7pZome-1-C Zome-F Zome-RpZome-1-N Zome-F Zome-RYCp50(BamHI-site) PBRforBam PBRrevBamYEp13(BamHI-site) PBRforBam PBRrevBamYEp24(BamHI-site) PBRforBam PBRrevBamYIp5(BamHI-site) PBRforBam PBRrevBam RNAi-ReadypSIREN-RetroQ-U6ZsGreen U6 菌PCDB T7BGH天为时代TA载体T7T3m13(+)T7?T3?pMSCV-neo pMSCV-5'pMSCV-3'。
质粒大全 分子大小等

名称长度抗性特殊性能PAO815 7709 bp Amp 酵母表达pAxCAwt 44741 BP Amp 腺病毒表达pAxcw 42410 bp Amp 腺病毒表达pAAV-MCS 4.7 kbp Amp 哺乳动物细胞表达pBacPAK8 5.5 k Amp 杆状病毒表达PBI121 13.0 kbp Kan 植物细胞表达pBV220 3665 bp Amp 原核表达pCAMBIA 1300 植物细胞表达pCAMBIA 1301 11837 bp Kan 植物细胞表达pCAT3-Basic 4047 bp AmppcDNA 3 5446 bp Amp 哺乳动物细胞表达pcDNA 3.1(+) 5428 bp Amp 哺乳动物细胞表达pcDNA 3.1/ mys- HisA 5494 bp Amp 哺乳动物细胞表达pCl-neo 5472 bp Amp 哺乳动物细胞表达pCMV-MCS 4.5 kbp Amp 哺乳动物细胞表达pET-3a 4640 bp Amp 大肠杆菌表达pET-11a 5677 bp Amp 大肠杆菌表达pET-15b(+) 5708 bp Amp 大肠杆菌表达pET-20b(+) 3716 bp Amp 大肠杆菌表达pET-22b(+) 5493 bp Amp 大肠杆菌表达pET-23a(+) 3666 bp Amp 大肠杆菌表达pET-23b(+) 3665 bp Amp 大肠杆菌表达pET-23c(+) 3664 bp Amp 大肠杆菌表达pET-23d(+) 3663 bp Amp 大肠杆菌表达pET-28a(+) 5369 bp Kan 大肠杆菌表达pET-28b(+) 5368 bp Kan 大肠杆菌表达pET-28c(+) 5367 bp Kan 大肠杆菌表达pET-30a(+) 5422 bp Kan 大肠杆菌表达pET-30b(+) 5421 bp Kan 大肠杆菌表达pET-30c(+) 5423 bp Kan 大肠杆菌表达pET-31b(+) 5742 bp Amp 大肠杆菌表达pET-32a(+) 5900 bp Amp 大肠杆菌表达pET-32b(+) 5899 bp Amp 大肠杆菌表达pET-32c(+) 5901 bp Amp 大肠杆菌表达pET-39b 6106 bp Kan 大肠杆菌表达pET-42a 5930 bp Kan 大肠杆菌表达pGAPZ aA 3147 bp Zeo 酵母表达pGBKT7 7.3 kbp Kan 酵母表达pGEM3Zb 原核表达pGEM3Zf(+) 原核表达pGEM7Zf(+) 原核表达pGEX-2T 4969 bp Amp 原核表达pGEX-4T-1 4969 bp Amp 原核表达pGFP-N2 4732 bp Kan 哺乳动物细胞荧光蛋白表达pEGFP-C1 4731 bp Kan 哺乳动物细胞荧光蛋白表达pEGFP-C3 4727 bp Kan 哺乳动物细胞荧光蛋白表达pEGFP-N1 4733 bp Kan 哺乳动物细胞荧光蛋白表达pLEGFP-N1 6892 bp Amp 哺乳动物细胞荧光蛋白表达pGL3-Basic 4818 bp AmppGL36 i3BaUxYEBTpLNCX 6620 bp Amp 反转录病毒表达pLNHX 5.6 kbp Amp 反转录病毒表达pLXSN 5.9 kbp Amp 反转录病毒表达pLXIN 6.1 kbp Amp 反转录病毒表达pMAL-p2x 6721 bp Amp 原核融合蛋白表达pMAL-c2x 6721 bp Amp 原核融合蛋白表达pPIC3.5K 9004 bp Amp/Kan 酵母表达pPIC9 8024 bp Amp 酵母表达pPIC9K 9276 bp Amp 酵母表达pPICZ aA 3593 bp Zeo 酵母表达pQBI PGK 5387 bp Amp 哺乳动物细胞荧光蛋白表达pQE-30 3461 bp Amp 原核表达pQE-9 3439 bp Amp 原核表达pRevTRE 6487 bp Amp 反转录病毒表达pSE420L 4617 bp AmppTac I Amp 原核表达pTac I(BamH I) Amp 原核表达pTAL-Luc 4956 bp Amp 哺乳动物细胞表达pTWIN1 7375 bp AmppTXB1 6706 bp AmppVAX1 2999 bp Kan。
报告基因稳定转化BmN细胞测定BmNPV滴度方法建立

浙江理工大学学位论文独创性声明本人声明所呈交的学位论文是本人在导师指导下进行的研究工作及取得的研究成果。
除了文中特别加以标注和致谢的地方外,论文中不包含其他人已经发表或撰写过的研究成果,也不包含为获得浙江理工大学或其他教育机构的学位或证书而使用过的材料。
与我一同工作的同志对本研究所做的任何贡献均已在论文中作了明确的说明并表示谢意。
浙江理工大学硕士学位论文报告基因稳定转化BmN细胞测定BmNPV滴度方法的建立第—章引言1文献综述1.1病毒滴度的概念病毒滴度是指单位体积病毒样品中所具有感染性病毒粒子的数量,用以衡量单位病毒样品的毒力[1】。
根据检测方法的不同,病毒滴度有多种不同的单位,如病毒粒子数单位(vP)、空斑形成单位(PFU)、半数致死量单位(TCIDso)、克隆形成单位(CFU)、以及基因转导单位(GTU)等,在这些单位之中,VP属于物理学滴度单位,其他均属于生物学滴度单位【2-6】。
1.2测定杆状病毒滴度的意义1983,Smith等‘71利用重组苜蓿丫纹夜蛾核型多角体病毒在昆虫细胞中成功表达了人口.干扰素,从而建立了杆状病毒表达载体系统。
该系统具有安全性好、允许插入片段大、表达效率高及表达产物翻译后修饰完整等特点,目前己成功了表达了数以干计的外源蛋白[8。
lo】o为了最大程度地地提高该表达系统表达外源蛋白质的效率,一个非常重要的因素是要确定感染性杆状病毒粒子数与细胞数的最佳比例,即感染复数(Multiplicityofinfection,MOI)[1t-i31,而为得到表达外源蛋白最合适的MOI值,首先必需准确测定病毒滴度,病毒滴度测定在该系统中是一个必不可少的环节。
1.3测定杆状病毒滴度方法的研究进展蚀斑法【141和终点稀释法嘲是两种经典的杆状病毒滴度测定方法,在目前应用也最为广泛。
但是用这两种方法测滴度均费时费力,且结果重复性差,因此研究人员在这两种方法的基础上做了一些优化,如构建了含3-半乳糖苷酶【16,171,绿色荧光蛋白[1sl报告基因的重组病毒,大大方便了实验人员对发病细胞的辨别并且明显缩短了病毒滴度测定所需要的时间。
载体与引物表(完全)

SP6 PYES2.R
M13R T3
载体抗性 Amp+
Amp+ Amp+ Amp+ Amp+ Amp+ Amp+ Amp+ Amp+
Amp+ Amp+ Amp+ Amp+
共3第3页
PM
PM-F
pMAL-c2E
MalE primer
pMAL-C2x
P5’
pMAL-p2X
MalE primer
pMD18-T
PQE30F/M13R(-48)/RV-M
Pmep4
pMEP4-F
pPIC9K(A+,Z+)
5’AOX(pPIC9K-F)
PPICZa pPROEXHTA
5'AOX/PPICZa-F M13R(-48)
pcDNA3.1R BGH
Amp+
M13F
M13R
pCEP-F pCI-F2/ T7(17base)
EBV-R (此载体没有反向引物)EBR
pCI-F2/ T7(19/17base)
T3
Amp+
T7 2G
T3
pCMV-F
pCMV-R
T3 /PFLAG-CMV-F
T7
Pcmv5f
Pcmv5r
Amp+
Pcmv5f
PDC316.R Primer(R)
载体抗性
pDonar
M13F
M13R
pDONR207
FORWARD
REVERSE
Pdonr221
M13F
pAc5.1a昆虫表达载体说明

pAc5.1a编号 载体名称北京华越洋生物KCVECT126 pAc5.1apAc5.1a载体基本信息载体名称: pAc5.1a质粒类型: 昆虫细胞表达载体高拷贝/低拷贝: --启动子: --克隆方法: 多克隆位点,限制性内切酶载体大小: --5' 测序引物及序列: --3' 测序引物及序列: --载体标签: --载体抗性: --筛选标记: --备注: --稳定性: --组成型: --病毒/非病毒: --pAc5.1a其他昆虫表达载体:pMT/BioEase-DEST pVL1392 pVL1393 pXINSECT-DEST39 pXINSECT-DEST38 pBacPAK9 pBacPAK8 pIB/V5-His-TOPO pIEX/Bac-1 pFastBacHT B pFastBacHT A pFastBac1pMT/V5-His-TOPO pMT/BiP/V5-His A pCoHygro pAc5.1/V5-His C pAc5.1-V5-His A pAc5/V5-HisA pAc5-V5-HisB pAc5-V5-HisC pCoBlast pAc5.1/V5-His B pIZ/V5-His pIB/V5-HispIZT/V5-His pMIB/V5-His C pMIB/V5-His A pMT/BiP/V5-His C pMT/BiP/V5-His B pIEx/Bac-4 pIEx/Bac-3 pIEXBac-c-EGFP-3 pIEXBac-c-EGFP-1 pIEXBac-c-EGFP-4 pAc5.1B-EGFP pBlueBacHis2 A pBlueBacHis2 C pBlueBacHis2 B pFastBacHT C pMT-Bip-V5-HisA pFBDM pUCDM pFastBac Dual pMIB/V5-His B pMT/V5-His C pMT/V5-His B pMT/V5-His A pAcGP67ApAc5.1b pAc5.1a pVL1392-XyIE control vector pFastBac-c-His-TEV pFastBac-N-GST-TEV pFastBacI-Gus pFastBacHT-CAT pBADZ-His6Cre。
- 1、下载文档前请自行甄别文档内容的完整性,平台不提供额外的编辑、内容补充、找答案等附加服务。
- 2、"仅部分预览"的文档,不可在线预览部分如存在完整性等问题,可反馈申请退款(可完整预览的文档不适用该条件!)。
- 3、如文档侵犯您的权益,请联系客服反馈,我们会尽快为您处理(人工客服工作时间:9:00-18:30)。
pBacPAK8编号 载体名称北京华越洋生物KCVECT129 pBacPAK8pBacPAK8载体基本信息载体名称: pBacPAK8质粒类型: 昆虫表达载体;杆状病毒表达载体克隆方法: 限制酶,多克隆位点启动子: Polyhedrin载体大小: 5538 bp5' 测序引物及序列: Bac1 Forward3' 测序引物及序列: Bac2 Reverse载体标签: 无标签载体抗性: 氨苄青霉素真核筛选标记: 无克隆菌株: TOP10宿主细胞(系): 果蝇细胞系 IPLB-Sf21备注: --瞬表达/稳表达: 稳表达组成型/诱导型: 组成型病毒/非病毒: 杆状病毒pBacPAK8载体质粒图谱和多克隆位点信息pBacPAK8载体描述:Available as part of the BacPAK Baculovirus Expression System (Cat. No. 631402). pBacPAK8 is a transfer vector designed for high-‐level expression of a cloned gene driven by the strong AcMNPV polyhedrin promoter. Flanking AcMNPV sequences allow recombination with viral DNA to transfer the expression cassette to the polyhedrin locus of the viral DNA. The polyhedrin coding sequences have been replaced by a multiple cloning s ite w ith 18 u nique s ites t hat f acilitate t he i nsertion o f f oreign g enes i n t he c orrect orientation for expression. The Pac I site at the end of the MCS region provides translational s top c odons i n a ll t hree r eading f rames f or e xpression o f t runcated p roteins. pBacPAK8 has a pUC origin of replication, an M13 origin for single-‐stranded DNA production, a nd a n a mpicillin r esistance g ene f or s election i n E. c oli.1.杆状病毒表达系统简介Baculovirus gene expression is a popular method for producing large quantities of recombinant proteins in insect host cells. In most cases, posttranslational processing of eukaryotic proteins expressed in insect cells is similar to protein processing in mammalian cells. As a result, insect cell-‐processed proteins have comparable biological activities and immunological reactivities to proteins expressed in mammalian cells. Protein y ields f rom b aculovirus s ystems a re h igher, a nd c osts a re s ignificantly l ower t han in m ammalian e xpression s ystems. T he b aculovirus e xpression s ystem c an e xpress g enes from b acteria, v iruses, p lants, a nd m ammals a t l evels f rom 1–500 m g/liter; m ost p roteins are e xpressed i n t he 10–100 m g/liter r ange, a lthough m aking p redictions i s d ifficult.The baculovirus most commonly used to express foreign proteins is Autographa californica nuclear polyhedrosis virus (AcMNPV). AcMNPV can be propagated in certain insect cell lines; the virus enters the cells and replication begins approximately 6 hours post-‐infection (h.p.i.). At approximately 20–48 h.p.i., transcription of nearly all genes ceases. The viral polyhedrin and p10 genes, however, are transcribed at high rates. The polyhedrin p rotein i s e ssential f or p ropagation o f t he v irus i n i ts n atural h abitat; h owever, in cell culture, polyhedrin is not needed, and its coding sequence can be replaced with a sequence for a target protein. Hence, the powerful polyhedrin promoter can drivehigh-‐level transcription of the insert, resulting in expression of a recombinant protein that c an a ccount f or o ver 30% o f t otal c ellular p rotein.The l arge 134 k b-‐size o f t he A cMNPV g enome, m akes d irect m anipulation o f i t d ifficult, s o recombinant b aculovirus e xpression v ectors a re c onstructed i n t wo s teps (Figure 1). F irst, a target gene is cloned into a modified polyhedrin locus contained in a relatively small transfer vector (<10 kb). The polyhedrin coding sequence has been deleted and replaced with a multiple cloning site (MCS). A target gene is inserted into this MCS, between the polyhedrin p romoter a nd p olyadenylation s ignals. T ransfer v ectors a lso c ontain a p lasmid origin of replication and an antibiotic resistance gene for propagation in E. coli, but they are unable to replicate in insect cells. In the second step, the transfer vector and a viral expression v ector a re c otransfected i nto i nsect c ells. D ouble r ecombination b etween v iral sequences in the transfer vector and the corresponding sequences in the viral DNA transfers t he t arget g ene t o t he v iral g enome.The BacPAK Baculovirus Expression System uses BacPAK6, a specially engineered virus that facilitates construction and selection of recombinant expression vectors. BacPAK6 has an essential gene adjacent to the polyhedrin locus that provides selection for recombinant viruses. Sites for Bsu36 I, which does not cut wild-‐type AcMNPV DNA, were introduced i nto t he g enes f lanking t he p olyhedrin e xpression l ocus o f B acPAK6. D igesting BacPAK6 with Bsu36 I releases two fragments. The first carries part of a downstream gene, ORF1629, that is essential for viral replication. If the second large DNA fragment recircularizes by itself, the resulting viral DNA will lack an essential part of the genome and b e u nable t o p roduce v iable v iruses. H owever, t he t ransfer v ector c arries t he m issing ORF1629 sequence, and if the large fragment recombines with it, the resulting circular DNA will contain all the genes necessary for viral replication. This double recombination event r estores t he e ssential g ene a nd t ransfers t he t arget g ene f rom t he t ransfer v ector t o the viral genome. Cotransfections using Bsu36 I-‐digested BacPAK6 viral DNA produce recombinant v iruses a t f requencies a pproaching 100%.This User Manual contains directions for establishing insect cell cultures, as well as for isolating a recombinant baculovirus expression vector using the BacPAK system. More extensive protocols for using baculovirus expression systems are in the baculovirus laboratory m anuals2.实验流程Obtain i nsect c ell m edia a nd e stablish S f21 c ell l ine. T his s tep w ill t ake 3–4 w eeks.Maintain w orking s tocks o f S f21 c ells.When t he s tock o f c ells h as b een p assaged t wice, f reeze a liquots f or l ong-‐term s torage i n liquid nitrogen. Aliquots of frozen cells provide a back-‐up in case the working stock dies or becomes contaminated. Frozen cells are also a source of fresh cells for replacing working s tocks a s t hey b ecome o ld.Isolating pure recombinant virus requires good viral plaques. Therefore, developing a good plaque assay technique before working with recombinant viruses is advisable. Practice a ssaying v iral p laques.Insert t arget g ene i nto t ransfer v ector a nd p repare p lasmid D NA.Produce a recombinant virus by cotransfecting Sf21 cells with BacPAK6 viral DNA and the t ransfer v ector-‐target g ene c lone.Perform plaque assays on the cotransfection supernatant to obtain individual viral plaques.Test t he p utative r ecombinant v iruses t o c onfirm t hat t hey h ave i ncorporated t he t arget gene a nd/or e xpress t he t arget p rotein.Amplify r ecombinant v iruses t o o btain w orking s tocks.Titer a mplified v irus s tock.Perform small-‐scale infections to characterize gene expression and to determine the optimum h arvest t ime a nd i nfection r atio t hat w ill g ive m aximum p rotein y ield.Scale-‐up: produce target protein in large quantities by infecting larger batches of insect cells.pBacPAK8载体序列ORIGIN1 AACGGCTCCG CCCACTATTA ATGAAATTAA AAATTCCAAT TTTAAAAAAC GCAGCAAGAG61 AAACATTTGT ATGAAAGAAT GCGTAGAAGG AAAGAAAAAT GTCGTCGACA TGCTGAACAA121 CAAGATTAAT ATGCCTCCGT GTATAAAAAA AATATTGAAC GATTTGAAAG AAAACAATGT181 ACCGCGCGGC GGTATGTACA GGAAGAGGTT TATACTAAAC TGTTACATTG CAAACGTGGT241 TTCGTGTGCC AAGTGTGAAA ACCGATGTTT AATCAAGGCT CTGACGCATT TCTACAACCA301 CGACTCCAAG TGTGTGGGTG AAGTCATGCA TCTTTTAATC AAATCCCAAG ATGTGTATAA361 ACCACCAAAC TGCCAAAAAA TGAAAACTGT CGACAAGCTC TGTCCGTTTG CTGGCAACTG421 CAAGGGTCTC AATCCTATTT GTAATTATTG AATAATAAAA CAATTATAAA TGCTAAATTT481 GTTTTTTATT AACGATACAA ACCAAACGCA ACAAGAACAT TTGTAGTATT ATCTATAATT541 GAAAACGCGT AGTTATAATC GCTGAGGTAA TATTTAAAAT CATTTTCAAA TGATTCACAG601 TTAATTTGCG ACAATATAAT TTTATTTTCA CATAAACTAG ACGCCTTGTC GTCTTCTTCT661 TCGTATTCCT TCTCTTTTTC ATTTTTCTCC TCATAAAAAT TAACATAGTT ATTATCGTAT721 CCATATATGT ATCTATCGTA TAGAGTAAAT TTTTTGTTGT CATAAATATA TATGTCTTTT781 TTAATGGGGT GTATAGTACC GCTGCGCATA GTTTTTCTGT AATTTACAAC AGTGCTATTT841 TCTGGTAGTT CTTCGGAGTG TGTTGCTTTA ATTATTAAAT TTATATAATC AATGAATTTG901 GGATCGTCGG TTTTGTACAA TATGTTGCCG GCATAGTACG CAGCTTCTTC TAGTTCAATT961 ACACCATTTT TTAGCAGCAC CGGATTAACA TAACTTTCCA AAATGTTGTA CGAACCGTTA1021 AACAAAAACA GTTCACCTCC CTTTTCTATA CTATTGTCTG CGAGCAGTTG TTTGTTGTTA1081 AAAATAACAG CCATTGTAAT GAGACGCACA AACTAATATC ACAAACTGGA AATGTCTATC1141 AATATATAGT TGCTGATATC ATGGAGATAA TTAAAATGAT AACCATCTCG CAAATAAATA1201 AGTATTTTAC TGTTTTCGTA ACAGTTTTGT AATAAAAAAA CCTATAAATA CGGATCCCTG1261 CAGGCCTCGA GTTCGAATCT AGAAGATCTG GTACCGAGCT CGAATTCCCG GGCGGCCGCT1321 TAATTAATTG ATCCGGGTTA TTAGTACATT TATTAAGCGC TAGATTCTGT GCGTTGTTGA1381 TTTACAGACA ATTGTTGTAC GTATTTTAAT AATTCATTAA ATTTATAATC TTTAGGGTGG1441 TATGTTAGAG CGAAAATCAA ATGATTTTCA GCGTCTTTAT ATCTGAATTT AAATATTAAA1501 TCCTCAATAG ATTTGTAAAA TAGGTTTCGA TTAGTTTCAA ACAAGGGTTG TTTTTCCGAA1561 CCGATGGCTG GACTATCTAA TGGATTTTCG CTCAACGCCA CAAAACTTGC CAAATCTTGT1621 AGCAGCAATC TAGCTTTGTC GATATTCGTT TGTGTTTTGT TTTGTAATAA AGGTTCGACG1681 TCGTTCAAAA TATTATGCGC TTTTGTATTT CTTTCATCAC TGTCGTTAGT GTACAATTGA 1741 CTCGACGTAA ACACGTTAAA TAAAGCTTGG ACATATTTAA CATCGGGCGT GTTAGCTTTA 1801 TTAGGCCGAT TATCGTCGTC GTCCCAACCC TCGTCGTTAG AAGTTGCTTC CGAAGACGAT 1861 TTTGCCATAG CCACACGACG CCTATTAATT GTGTCGGCTA ACACGTCCGC GATCAAATTT 1921 GTAGTTGAGC TTTTTGGAAT TATTTCTGAT TGCGGGCGTT TTTGGGCGGG TTTCAATCTA 1981 ACTGTGCCCG ATTTTAATTC AGACAACACG TTAGAAAGCG ATGGTGCAGG CGGTGGTAAC 2041 ATTTCAGACG GCAAATCTAC TAATGGCGGC GGTGGTGGAG CTGATGATAA ATCTACCATC 2101 GGTGGAGGCG CAGGCGGGGC TGGCGGCGGA GGCGGAGGCG GAGGTGGTGG CGGTGATGCA 2161 GACGGCGGTT TAGGCTCAAA TGTCTCTTTA GGCAACACAG TCGGCACCTC AACTATTGTA 2221 CTGGTTTCGG GCGCCGTTTT TGGTTTGACC GGTCTGAGAC GAGTGCGATT TTTTTCGTTT 2281 CTAATAGCTT CCAACAATTG TTGTCTGTCG TCTAAAGGTG CAGCGGGTTG AGGTTCCGTC 2341 GGCATTGGTG GAGCGGGCGG CAATTCAGAC ATCGATGGTG GTGGTGGTGG TGGAGGCGCT 2401 GGAATGTTAG GCACGGGAGA AGGTGGTGGC GGCGGTGCCG CCGGTATAAT TTGTTCTGGT 2461 TTAGTTTGTT CGCGCACGAT TGTGGGCACC GGCGCAGGCG CCGCTGGCTG CACAACGGAA 2521 GGTCGTCTGC TTCGAGGCAG CGCTTGGGGT GGTGGCAATT CAATATTATA ATTGGAATAC 2581 AAATCGTAAA AATCTGCTAT AAGCATTGTA ATTTCGCTAT CGTTTACCGT GCCGATATTT 2641 AACAACCGCT CAATGTAAGC AATTGTATTG TAAAGAGATT GTCTCAAGCT CGGATCGATC 2701 CCGCACGCCG ATAACAAGCC TTTTCATTTT TACTACAGCA TTGTAGTGGC GAGACACTTC 2761 GCTGTCGTCG CCTGATGCGG TATTTTCTCC TTACGCATCT GTGCGGTATT TCACACCGCA 2821 TACGTCAAAG CAACCATAGT ACGCGCCCTG TAGCGGCGCA TTAAGCGCGG CGGGTGTGGT 2881 GGTTACGCGC AGCGTGACCG CTACACTTGC CAGCGCCCTA GCGCCCGCTC CTTTCGCTTT 2941 CTTCCCTTCC TTTCTCGCCA CGTTCGCCGG CTTTCCCCGT CAAGCTCTAA ATCGGGGGCT 3001 CCCTTTAGGG TTCCGATTTA GTGCTTTACG GCACCTCGAC CCCAAAAAAC TTGATTTGGG 3061 TGATGGTTCA CGTAGTGGGC CATCGCCCTG ATAGACGGTT TTTCGCCCTT TGACGTTGGA 3121 GTCCACGTTC TTTAATAGTG GACTCTTGTT CCAAACTGGA ACAACACTCA ACCCTATCTC 3181 GGGCTATTCT TTTGATTTAT AAGGGATTTT GCCGATTTCG GCCTATTGGT TAAAAAATGA 3241 GCTGATTTAA CAAAAATTTA ACGCGAATTT TAACAAAATA TTAACGTTTA CAATTTTATG 3301 GTGCACTCTC AGTACAATCT GCTCTGATGC CGCATAGTTA AGCCAGCCCC GACACCCGCC 3361 AACACCCGCT GACGCGCCCT GACGGGCTTG TCTGCTCCCG GCATCCGCTT ACAGACAAGC 3421 TGTGACCGTC TCCGGGAGCT GCATGTGTCA GAGGTTTTCA CCGTCATCAC CGAAACGCGC 3481 GAGACGAAAG GGCCTCGTGA TACGCCTATT TTTATAGGTT AATGTCATGA TAATAATGGT 3541 TTCTTAGACG TCAGGTGGCA CTTTTCGGGG AAATGTGCGC GGAACCCCTA TTTGTTTATT 3601 TTTCTAAATA CATTCAAATA TGTATCCGCT CATGAGACAA TAACCCTGAT AAATGCTTCA 3661 ATAATATTGA AAAAGGAAGA GTATGAGTAT TCAACATTTC CGTGTCGCCC TTATTCCCTT 3721 TTTTGCGGCA TTTTGCCTTC CTGTTTTTGC TCACCCAGAA ACGCTGGTGA AAGTAAAAGA 3781 TGCTGAAGAT CAGTTGGGTG CACGAGTGGG TTACATCGAA CTGGATCTCA ACAGCGGTAA 3841 GATCCTTGAG AGTTTTCGCC CCGAAGAACG TTTTCCAATG ATGAGCACTT TTAAAGTTCT 3901 GCTATGTGGC GCGGTATTAT CCCGTATTGA CGCCGGGCAA GAGCAACTCG GTCGCCGCAT 3961 ACACTATTCT CAGAATGACT TGGTTGAGTA CTCACCAGTC ACAGAAAAGC ATCTTACGGA 4021 TGGCATGACA GTAAGAGAAT TATGCAGTGC TGCCATAACC ATGAGTGATA ACACTGCGGC 4081 CAACTTACTT CTGACAACGA TCGGAGGACC GAAGGAGCTA ACCGCTTTTT TGCACAACAT 4141 GGGGGATCAT GTAACTCGCC TTGATCGTTG GGAACCGGAG CTGAATGAAG CCATACCAAA 4201 CGACGAGCGT GACACCACGA TGCCTGTAGC AATGGCAACA ACGTTGCGCA AACTATTAAC 4261 TGGCGAACTA CTTACTCTAG CTTCCCGGCA ACAATTAATA GACTGGATGG AGGCGGATAA4321 AGTTGCAGGA CCACTTCTGC GCTCGGCCCT TCCGGCTGGC TGGTTTATTG CTGATAAATC 4381 TGGAGCCGGT GAGCGTGGGT CTCGCGGTAT CATTGCAGCA CTGGGGCCAG ATGGTAAGCC 4441 CTCCCGTATC GTAGTTATCT ACACGACGGG GAGTCAGGCA ACTATGGATG AACGAAATAG 4501 ACAGATCGCT GAGATAGGTG CCTCACTGAT TAAGCATTGG TAACTGTCAG ACCAAGTTTA 4561 CTCATATATA CTTTAGATTG ATTTAAAACT TCATTTTTAA TTTAAAAGGA TCTAGGTGAA 4621 GATCCTTTTT GATAATCTCA TGACCAAAAT CCCTTAACGT GAGTTTTCGT TCCACTGAGC 4681 GTCAGACCCC GTAGAAAAGA TCAAAGGATC TTCTTGAGAT CCTTTTTTTC TGCGCGTAAT 4741 CTGCTGCTTG CAAACAAAAA AACCACCGCT ACCAGCGGTG GTTTGTTTGC CGGATCAAGA 4801 GCTACCAACT CTTTTTCCGA AGGTAACTGG CTTCAGCAGA GCGCAGATAC CAAATACTGT 4861 CCTTCTAGTG TAGCCGTAGT TAGGCCACCA CTTCAAGAAC TCTGTAGCAC CGCCTACATA 4921 CCTCGCTCTG CTAATCCTGT TACCAGTGGC TGCTGCCAGT GGCGATAAGT CGTGTCTTAC 4981 CGGGTTGGAC TCAAGACGAT AGTTACCGGA TAAGGCGCAG CGGTCGGGCT GAACGGGGGG 5041 TTCGTGCACA CAGCCCAGCT TGGAGCGAAC GACCTACACC GAACTGAGAT ACCTACAGCG 5101 TGAGCTATGA GAAAGCGCCA CGCTTCCCGA AGGGAGAAAG GCGGACAGGT ATCCGGTAAG 5161 CGGCAGGGTC GGAACAGGAG AGCGCACGAG GGAGCTTCCA GGGGGAAACG CCTGGTATCT 5221 TTATAGTCCT GTCGGGTTTC GCCACCTCTG ACTTGAGCGT CGATTTTTGT GATGCTCGTC 5281 AGGGGGGCGG AGCCTATGGA AAAACGCCAG CAACGCGGCC TTTTTACGGT TCCTGGCCTT 5341 TTGCTGGCCT TTTGCTCACA TGTTCTTTCC TGCGTTATCC CCTGATTCTG TGGATAACCG 5401 TATTACCGCC TTTGAGTGAG CTGATACCGC TCGCCGCAGC CGAACGACCG AGCGCAGCGA 5461 GTCAGTGAGC GAGGAAGCGG AAGAGCGCCC AATACGCAAA CCGCCTCTCC CCGCGCGTTG 5521 GCCGATTCAT TAATGCAG//pBacPAK8其他昆虫表达载体:pMT/BioEase-DEST pVL1392 pVL1393 pXINSECT-DEST39 pXINSECT-DEST38 pBacPAK9 pBacPAK8 pIB/V5-His-TOPO pIEX/Bac-1 pFastBacHT B pFastBacHT A pFastBac1pMT/V5-His-TOPO pMT/BiP/V5-His A pCoHygro pAc5.1/V5-His C pAc5.1-V5-His A pAc5/V5-HisA pAc5-V5-HisB pAc5-V5-HisC pCoBlast pAc5.1/V5-His B pIZ/V5-His pIB/V5-HispIZT/V5-His pMIB/V5-His C pMIB/V5-His A pMT/BiP/V5-His C pMT/BiP/V5-His B pIEx/Bac-4 pIEx/Bac-3 pIEXBac-c-EGFP-3 pIEXBac-c-EGFP-1 pIEXBac-c-EGFP-4 pAc5.1B-EGFP pBlueBacHis2 A pBlueBacHis2 C pBlueBacHis2 B pFastBacHT C pMT-Bip-V5-HisA pFBDM pUCDM pFastBac Dual pMIB/V5-His B pMT/V5-His C pMT/V5-His B pMT/V5-His A pAcGP67ApAc5.1b pAc5.1a pVL1392-XyIE control vector pFastBac-c-His-TEV pFastBac-N-GST-TEV pFastBacI-Gus pFastBacHT-CAT pBADZ-His6Cre。
