课后习题部分答案(2013版)
逻辑学(第3版)课后练习题答案(部分)人大出版社-图文

逻辑学(第3版)课后练习题答案(部分)人大出版社-图文逻辑学课后练习题答案(部分)王震14年11月8日整理第一章练习题答案二、在下列命题或推理中,哪些具有共同的逻辑形式,用公式表示出来。
1和5:所有S是P2和7:所有P是M,所有S不是M,所以,所有S不是P。
3和8:只有p,才q。
6和9:如果p,那么q;p;所以,q。
三、选择题1.C2.C3.ABCD4.BE第二章练习题答案一、判定下列断定的正误。
1.错误2.错误3.正确4.错误5.错误6.正确7.错误8.错误二、运用本章的相关知识以及相关常识,回答下列问题。
1.错误。
定义过宽。
2.错误。
定义过宽。
3.错误。
“勇敢”和“勇敢的战士”之间不存在属种关系。
4.错误。
“喜马拉雅山”和“珠穆朗玛峰”之间不存在属种关系。
三、在以下各句的括号中填入哪个或哪些选项是适当的?1.C2.ABC3.A4.A5.B6.BC7.B8.B9.BC10.AC四、下列各题中括号内的话,是从内涵方面还是从外延方面来说明标有横线的概念的?1.分别从内涵和外延2.从内涵3.分别从内涵和外延4.分别从内涵和外延5.分别从内涵和外延6.分别从内涵和外延五、从两种概念分类的角度(单独概念与普遍概念、正概念和负概念)说明下列各题中标有横线的概念属于哪一种类。
1.“美术作品”是普遍概念、正概念。
2.“《孔乙己》”是单独概念、正概念;“作品”是普遍概念、正概念。
3.“非司机”是普遍概念、负概念。
4.“中国女子排球队”是单独概念、正概念;“世界冠军”是普遍概念、正概念。
5.“中国工人阶级”是单独概念、正概念。
6.“国家检察机关”是单独概念、正概念。
六、试分析下列各题中标有横线的语词是在集合意义下使用的,还是在非集合意义下使用的?1.集合2.非集合3.非集合4.集合5.1)集合2)非集合3)非集合6.集合7.集合七、下列各组概念是什么关系?1.真包含2.全异(反对)3.交叉4.真包含于5.全异6.全异(矛盾)7.全同8.全异(反对)八、用欧拉图表示下列各题中标有横线的概念之间的关系:十、对下列概念各作一次限制与概括。
遗传学(第3版) 刘祖洞、乔守怡、吴燕华、 赵寿元 高等教育出版社 (2013-01)课后习题答案6

Chapter 6 Circulating Methylated DNA as Biomarkers for Cancer DetectionHongchuan Jin, Yanning Ma, Qi Shen andXian WangAdditional information is available at the end of the chapter/10.5772/514191. IntroductionIn addition to genetic alterations including deletion or point mutations, epigenetic changes such as DNA methylation play an important role in silencing tumor suppressor genes dur‐ing cancer development. By adding a methyl group from S-adenosyl-L-methionine to the cy‐tosine pyrimidine or adenine purine ring, DNA methylation is important to maintain genome structure and regulate gene expression. In mammalian adult tissues, DNA methyla‐tion occurs in CpG dinucleotides that often cluster in the genome as CpG islands in the 5’regulatory regions of the genes. Through recruiting transcriptional co-repressors including methyl-CpG-binding domain proteins (MBDs) and chromatin remodeling proteins like his‐tone deacetylases (HDACs) or impeding the binding of transcriptional activators, DNA methylation could suppress the transcription of many tumor suppressor genes critical to cancer initiation and progression [1-3].More and more results confirmed that cancer is a multi-stage process fuelled by many epige‐netic changes in addition to genetic changes in DNA sequence [4]. Chemical molecules like Trichostatin A (TSA) and 5-aza-2'-deoxycytidine (5-Aza-CdR) targeting epigenetic regula‐tors such as histone modifications and DNMTs (DNA methyltransferases) have been found to inhibit tumor growth both in vitro and in vivo. By reversing the epigenetic silencing of important tumor suppressor genes, an increasing number of epigenetic drugs such as 5-Aza-CdR, 5-Aza-CR and Vorinostat (SAHA) are currently investigated in the clinical trials for cancer treatment as a single drug or in combination with other epigenetic drugs or other ap‐proaches such as chemotherapy and showed very promising activities by offering signifi‐cant clinical benefits to cancer patients [5-13].© 2013 Jin et al.; licensee InTech. This is an open access article distributed under the terms of the CreativeCommons Attribution License (/licenses/by/3.0), which permits unrestricted use,distribution, and reproduction in any medium, provided the original work is properly cited.As one of the major epigenetic changes to inactivate tumor suppressor genes critical to hu‐man cancer development, DNA methylation was recognized as the biomarker for cancer de‐tection or outcome prediction in addition to the identification of novel tumor suppressor genes. DNA mutations will occur randomly in any nucleotides of one particular gene and the comprehensive determination of DNA mutations is thus very difficult and time-consum‐ing. In contrast, aberrant DNA hypermethylation usually takes place in defined CpG Islands within the regulatory region of the genes and it is much more convenient to detect DNA methylation in a quantitatively manner. In addition, DNA methylation can be amplified and is thus easily detectable using PCR-based approaches even when the DNA concentration af‐ter sample extraction is relatively low. Due to such advantages over DNA mutation- or pro‐tein-based biomarkers, DNA methylation-based biomarkers have been intensively investigated in the recent years. A large body of research reports has proved the value of DNA methylations in the prognosis prediction and detection of various cancers. DNAs used for such methylation analyses are usually extracted from tumor tissues harvested after sur‐gical operation or biopsy, thus limiting its wide application as the biomarkers for the early detection or screening of human cancers. Recently, it has been reported that there are certain amount of circulating DNAs in the peripheral blood of cancer patients, providing an ideal source to identify novel biomarkers for non-invasive detection of cancers. Both genetic and epigenetic changes found in the genomic DNAs extracted from primary tumor cells could be detected in the circulating DNAs, indicating that the detection of methylated DNAs in the circulation represents a new direction to develop novel biomarkers for cancer detection or screening in a non-invasive manner.2. Cell free DNA in the circulationAccording to the origin of circulating tumor-related DNA, it could be grouped into circulat‐ing cell free DNA or DNA from cells in the blood such as circulating tumor cells (CTC) in cancer patients (Figure 1).In 1869, the Australian physician Thomas Ashworth observed CTCs in the blood of a cancer patient. Therefore, it was postulated that CTCs were responsible for the tumor metastases in distal sites and should have important prognostic and therapeutic implications [14-16].However, the number of CTCs is very small compared with blood cells. Usually around 1-10CTCs together with several million blood cells could be found in 1 ml of whole blood, mak‐ing the specific and sensitive detection of CTCs very difficult [17-18]. Until recently, technol‐ogies with the requisite sensitivity and reproducibility for CTC detection have been developed to precisely analyze its biological and clinical relevance. The US Food and Drug Administration (FDA) approved the test for determining CTC levels in patients with meta‐static breast cancer in 2004. Currently, it has been expanded to other cancer types such as advanced colorectal cancer and prostate cancer. Although CTCs-counting based test have proven its value in predicting prognosis and monitoring therapeutic effects, the number of CTCs per ml of blood limited its sensitivity greatly [19]. With the development of high-sen‐sitive PCR-based methods, the detection of gene mutations or epigenetic changes such asMethylation - From DNA, RNA and Histones to Diseases and Treatment138DNA methylation within small amount of CTCs could be the next generation of CTC-based test for cancer detection. However, the cost of such tests will be greatly exacerbated, thuslimiting its wide application in the clinic [20-22].Figure 1. Circulating tumor cells and cell free DNA. Circulating Tumor cells (CTC) escape from primary sites and spread into the vessel to form metastases in the distal organs with. Cell free DNAs (cf-DNAs) are released into the circulation from dead cancer cells or proliferating tumor cells. RBC: red blood cell; WBC: white blood cell.Although its origin and biological relevance remains unknown, circulating cell free DNA (cf-DNA) is supposed to be valuable source to identify cancer markers with ideal sensitivity and specificity for non-invasive detection of cancer [23-24]. Early in 1948, two French scientists Mandel and Metais firstly reported the presence of cf-DNAs in human plasma [25]. Such an important discovery has been unnoticed for a long time until cell-free circulating nucleic acid was found to promote the spread and metastasis of crown gall tumor in plants [26]. Subse‐quently, increased level of cf-DNAs was found in patients with various diseases such as lupus erythematosus and rheumatoid arthritis cancer [27-28]. In 1977, Leon et al. reported that higher level of circulating DNA in the plasma of cancer patients when compared to healthy con‐trols. Moreover, greater amounts of cf-DNA were found in the peripheral blood of cancer patients with tumor metastases and cf-DNA levels decreased dramatically after radiothera‐py while persistently high or increasing DNA concentrations were associated with a lack of response to treatment [29], clearly revealing the potential value of cf-DNA as biomarker for cancer detection. Following studies confirmed that cf-DNAs in the plasma contains genetic and epigenetic changes specific to DNAs within the tumor cells from primary tissues, indicat‐ing that tumor specific cf-DNAs are originated from tumor cells rather than lymphocytes reacting towards the disease [30-31]. For example, K-Ras mutation was found in cf-DNA from 17 out of 21 patients with pancreatic adenocarcinoma and mutations were similar in corre‐sponding plasma and tissues samples. Importantly, such DNA alterations were found inCirculating Methylated DNA as Biomarkers for Cancer Detection/10.5772/51419139patients with pancreatitis who were diagnosed as pancreatic cancer 5-14 months later, indi‐cating that release of tumor-specific DNA into the circulation is an early event in cancer development and cf-DNA could be used as the biomarkers for early cancer detection [32].Treatment resulted in disappearance of K-Ras mutations in plasma DNA in six of nine pa‐tients. Three patients with a persistently positive K-Ras gene mutation in plasma samples from patients before and after treatment showed early recurrence or progression and pancreatic carcinoma patients with the mutant-type K-ras gene in plasma DNA exhibited a shorter survival time than patients with the wild-type gene, indicating the cf-DNA could be of value in monitoring disease progression or evaluating treatment response [31, 33].Through quantitatively analyzing plasma DNAs from patients with organ transplantation,Lo et al found that the majority of plasma DNAs was released from the hematopoietic sys‐tem. However, donor DNA could be detected in the plasma of recipients suffering from the graft rejection because of the large amount of cell death which promotes the release of donor DNAs into the peripheral blood of the recipients [34]. Therefore, it was postulated that cell-free tumor related DNA could originate from the apoptotic tumor cells since high-rate of apoptosis indeed occurs in primary and metastatic tumor tissues. However, cf-DNA quanti‐ties are significantly reduced in cancer patients after radiotherapy when a great number of tumor cells were believed to undergo apoptotic cell death and cf-DNAs in supernatants of cultured cancer cells increases with cell proliferation rather than apoptosis or necrosis, indi‐cating that proliferating tumor cells could actively release cf-DNA into the tumor microen‐vironment and circulation.In contrast to labile RNAs that were included into the actively secreted exosomes, the nature of cf-DNAs remains to be clarified. As negatively charged molecules, cf-DNA was bound by plasma proteins to escape from endonuclease-mediated degradation. Unfortunately, plasma proteins bound to cf-DNAs was not well characterized yet. Meanwhile, secreted exosomes could remodel microenviroments and promote tumor metastasis since RNAs within exo‐somes especially microRNA with high stability may influence gene expression in neighbor cells. The biological relevance of cf-DNAs remains unknown. DNA was believed to be more structural rather than functional. However, it was supposed that cf-DNA could play a role as vaccine in tumor microenvironment.3. Methods for the detection of methylated DNAIt is unclear so far whether serum or plasma is better for cf-DNA extraction. Although the DNA amount is significantly higher in the serum, the majority of the increase was due to the release of nuclear acids from destroyed blood cells during blood clotting [35]. In addition,the time gap between blooding drawing and DNA extraction as well as the methodologies used for DNA isolation contribute greatly to the amount of cf-DNA harvested. On an aver‐age, around 30 ng cf-DNA could be extracted from one ml of blood sample [36]. Therefore,in order to determine the quantity of potential cf-DNA-based biomarkers precisely and pro‐mote its wide application for cancer detection, it is very important to unify the source asMethylation - From DNA, RNA and Histones to Diseases and Treatment140well as the methodologies for cf-DNA extraction and use various internal controls to adjustpossible inter-laboratory variations.Figure 2. Schematic introductions of various methods for methylation analyses. MSP, BGS and COBRA are based on bisulfite-mediated conversion of unmethylated cytosines into uracils. CpG methylation could block DNA digestion by some restriction enzymes, making it possible to determine methylation status independent of bisulfite treatment by analyzing digestion products. Alternatively, DNA fragments containing methylated CpG sites could be enriched by an‐ti-methylcytosine antibody or methylation binding proteins. Advances in next generation genome sequencing tech‐nology led to the development of noel techniques such as SMRT which can specially analyze 5-methylcytosines with genome wide coverage.In general, the detection of DNA methylation could be bisulfite-dependent or -independent (Figure 2).The chemical reaction of sodium bisulfite with DNA could convert unmethylated cytosine of CpG into uracil or UpG but leave methylated cytosine of CpG unchanged. The following analyses such as methylation-and unmethylation specific polymerase chain reaction (M- and U-SP), bisulfite genome sequencing (BGS) or combined bisulfite restriction analysis (CO‐BRA) could determine the conversion of CpG sites of interest, thus reflecting their methyla‐tion status as methylated or unmethylated [37]. With varied resolution levels, different bisulfite-dependent DNA methylation analysis methods detect the conversion after bisulfite treatment of genomic DNA, which could have certain artificial effects such as incomplete conversion of unmethylated CpG into UpG, leading to high rate of false negative conclusion of DNA methylation status.Recently, some new modifications of cytosine in CpG dinucleotides have been discovered such as 5-hydoxymethylcytosine which was called the sixth base since 5-methylcytosine was named as the fifth base [38]. Generated from the oxidation of 5-methylcytosine by the Tet family of enzymes, 5-hydoxymethylcytosine was first found in bacteriophages and recentlyCirculating Methylated DNA as Biomarkers for Cancer Detection/10.5772/51419141shown to be abundant in human and mouse brains as well as in embryonic stem cells [39-40]. Although the exact relevance of 5-hydoxymethylcytosine in the genome is still not fully clarified, it has been found to regulate gene expression or promote DNA demethyla‐tion. The in vitro synthesized artificial oligonucleotides containing 5-hydoxymethylcyto‐sines can be converted into unmodified cytosines when introduced into mammalian cells,indicating that 5-hydoxymethylcytosine might be one of intermediate products during ac‐tive DNA demethylation [41]. Therefore, the increase of 5-hydoxymethylcytosine might re‐flect the demethylation of CpG dinucleotides. Unfortunately, 5-hydoxymethylcytosines,similar to 5-methylcytosines, appear to be resistant to bisulfite-mediated conversion and PCR could amplify DNA fragments containing 5-hydoxymethylcytosines or 5-methylcyto‐sines with similar efficiency [42-43]. Therefore, bisulfite-dependent methylation analyses could produce false positive results by counting 5-hydoxymethylcytosines into 5-methylcy‐tosines. In addition to 5-hydroxymethylcytosines, some forms of DNA modifications such as the seventh base, 5-formylcytosine and the eighth base, 5-carboxylcytosine, have been found in mammalian cells recently [44-47]. As the products of 5-hydoxymethylcytosine oxidation through TET hydroxylases, both 5-formylcytosine and 5-carboxylcytosine will be read as the uracil after bisulfite conversion, thus making it impossible for bisulfite-dependent analyses to distinguish unmodified cytosines from 5-formylcytosines and 5-carboxylcytosines.Bisulfite independent analyses such as MedIP (methylated DNA immunoprecipitation)could more or less detect DNA methylation specifically. In bisulfite independent analyses, 5-methylcytosines are differentiated from unmethylated cytosine by either enzyme digestion or affinity enrichment. DNA methylation analysis using restriction enzyme digestion is based on the property of some methylation-sensitive and -resistant restriction enzymes such as HpaII and MspI that target CCGG for digestion. HpaII fails to cut it once the second cyto‐sine was methylated while MspI-mediated digestion is not affected by DNA methylation,thus making it possible to determine the methylation status of CpG in the context of CCGG tetranucleotides by analyzing the products of DNAs digested by HpaII and MspI respective‐ly. As a primary method to analyze DNA methylation, it can only determine the methyla‐tion of CpG in the context of CCGG tetranucleotides and will overlook the majority of CpG dinucleotides in the genome.The development of monoclonal antibody specific to 5-methylcytosines revolutionized the analyses of DNA methylation [48-49]. Immunoprecipitated DNA by this antibody could be subject to DNA microarray or even deep sequencing to reveal novel sequences or sites con‐taining 5-methylcytosines [50]. This antibody specifically recognizes 5-methylcytosines but not 5-hydoxymethylcytosines. However, 5-methylcytosines could present not only in CpG dinucleotides but also in CHH or CHG trinucleotides, especially in plants, human embryon‐ic stem cells and probably cancer cells as well. CHH methylation indicates a 5-methylcyto‐sine followed by two nucleotides that may not be guanine and CHG methylation refers to a 5-methylcytosine preceding an adenine, thymine or cytosine base followed by guanine. Such non-CpG DNA methylations were enriched at transposons and repetitive regions, although the exact biological relevance remains unknown. However, antibody against 5-methylcyto‐Methylation - From DNA, RNA and Histones to Diseases and Treatment142sine may precipitate methylated CHH and CHG trinucleotide containing DNA fragments in addition to DNA sequences with methylated CpG sites.DNA methylation functions as the signal for DNA-interacting proteins to maintain genome structure or regulate gene expression. The proteins such as MBD1 (methyl-CpG binding do‐main protein 1), MeCP2 (methyl CpG binding protein 2) and MBD4 (methyl-CpG binding domain protein 4) bind methylated CpG specifically to regulate gene expression [51-52].Therefore, methyl-CpG binding domain could specifically enrich differentially methylated regions (DMRs) of physiological relevance [53]. Similar to MeDIP, MBD capture specifically enrich methylated CpG sites rather than hydroxymethlated CpG sites. The detailed analysis to compare MeDIP and MBD capture revealed that both enrichment techniques are sensitive enough to identify DMRs in human cancer cells. However, MeDIP enriched more methylat‐ed regions with low CpG densities while MBD capture favors regions of high CpG densities and identifies the greater proportion of CpG islands [49].Recently, the advance of next generation sequencing led to the development of several novel techniques, making it possible to quantitatively analyze DNA methylation at single nucleo‐tide resolution with genome wide coverage. Both the single molecule real time sequencing technology (SMRT) and the single-molecule nanopore DNA sequencing platform could dis‐criminate 5-methylcytosines from other DNA bases including 5-hydroxymethylcytosines even methyladenine independent of bisulfite conversion [54-55]. With many advantages such as less bias during template preparation, lower cost and better accuracy, such new techniques could offer more methods to detect DNA methylation with high specificity and sensitivity in addition to more potential DNA methylation based biomarkers for cancer de‐tection and screening.4. Potential DNA methylation biomarkers for cancer detectionIt has been questioned whether the methylated DNA in the circulation is sensitive to detect cancers early enough for curative resection. However, the development of sensitive detection methods confirmed the potential value of DNA methylation in cancer detection (Table 1).Most of DNA methylation biomarkers are well-known tumor suppressor genes silenced in primary tumor tissues. However, the biomarks do not have to be functional relevant. For ex‐ample, currently well-used biomarkers such as AFP (Alpha-Fetal Protein), PSA (Prostate-specific antigen) and CEA (Carcinoembryonic antigen) are not tumor suppressor genes with important biological functions. Profiling of methylated DNA in the circulation instead of primary tumor tissues with MeDIP or MBD capture or other methylation specific analyses methods would identify more potential biomarks rather than functional important tumor suppressor genes.Circulating Methylated DNA as Biomarkers for Cancer Detection/10.5772/51419143Cancer Markers Sensitivity Specificity Methods Ref.Bladder cancer CDKN2A (ARF) CDKN2A(INK4A)CDKN2A (INK4A)13/27 (48%)2/27 (7%)19/86 (22%)N/AN/A31/31 (100%)MSPMSPMSP[58][59]Breast cancer CDKN2A (INK4A)CDKN2A (INK4A)5/35 (14%)6/43 (14%)N/AN/AMS-AP-PCRMS-AP-PCR[56][57]Colorectal cancerMLH1CDKN2A (INK4A) CDKN2A(INK4A) CDKN2A (INK4A)ALX4CDH4NGFRRUNX3SEPT9TMEFF23/18 (17%)14/52 (27%)13/94 (11%)21/58 (36%)25/30 (83%)32/46 (70%)68/133 (51%)11/17 (65%)92/133 (69%)87/133 (65%)N/A44/44 (100%)N/AN/A36/52 (70%)17/17 (100%)150/179 (84%)10/10 (100%)154/179 (86%)123/179 (69%)MSPMSPMSPMSPMSPMSPMSPMSPMSPMSP[60][61][62][63][64][65][66][67][66]Esophageal cancer APCAPCCDKN2A (INK4A)13/52 (25%)2/32 (6%)7/38 (18%)54/54 (100%)54/54 (100%)N/AMSPMSPMSP[68][69]Gastric cancer CDH1CDKN2A (INK4A)CDKN2B (INK4B)DAPK1GSTP1Panel of five 31/54 (57%)28/54 (52%)30/54 (56%)26/54 (48%)18/54 (15%)45/54 (83%)30/30 (100%)30/30 (100%)30/30 (100%)30/30 (100%)30/30 (100%)30/30 (100%)MSPMSPMSPMSPMSPMSP[70]Head and neck cancer CDKN2A (INK4A)DAPK1MGMTPanel of threeDAPK18/95 (8%)3/95 (3%)14/95 (15%)21/95 (22%)N/AN/AN/AN/AN/AN/AMSPMSPMSPMSPMSP[71][72]Liver cancer CDKN2A (INK4A) CDKN2B(INK4B)13/22 (45%)4/25 (16%)48/48 (100%)35/35 (100%)MSPMSP[73][74]Lung cancer CDKN2A (INK4A)DAPK1GSTP1MGMTPanel of fourCDKN2A (INK4A)APC 3/22 (14%)4/22 (18%)1/22 (5%)4/22 (18%)11/22 (50%)N/A42/89 (47%)N/AN/AN/AN/AN/AN/A50/50 (100%)MSPMSPMSPMSPMSPMSPMSP[75][76][77]Methylation - From DNA, RNA and Histones to Diseases and Treatment 144Cancer Markers Sensitivity Specificity Methods Ref.CDKN2A (INK4A)CDKN2A (INK4A)77/105 (73%)12/35 (34%)N/A15/15 (100%)MSP MSP [78][79]Prostate cancer GSTP1GSTP123/33 (70%)25/69 (36%)22/22 (100%)31/31 (100%)MSP MSP[80][81]Table 1. Methylated DNA biomarkers in the literature.Most of the methods used for methylation biomarkers analyses are still bisulfite dependent.Few reports used MS-AP-PCR (methylation-sensitive arbitrarily primed PCR) which takes the advantage of methylation sensitive restriction endonucleases to distinguish methylated CpG from unmethylated form, although the sensitivity seems to be lower than MSP [56-57].Interestingly, combination of more than one methylated DNA as a methylation panel could great increase the sensitivity for cancer detection without significant reduction of specificity.Unfortunately, most of studies were performed in a retrospective manner. More prospective studies with large sample sizes will be warranted to compare different approaches especial‐ly bisulfite-independent methods in addition to confirm the value of DNA methylation for cancer detection.5. Conclusion and PerspectivesWith the development of the next generation genome sequencing as well as single molecular PCR, it became possible to analyze trace amount of DNAs including circulating cell-free DNA. Circulating tumor cells have been proven its value in prognosis predication even ear‐ly detection of various cancers. The analyses of methylated DNAs in the circulating will be the next promising epigenetic biomarkers for cancer detection. As one of the intermediate products of DNA demethylation, 5-hydroxymethlcytosines are resistant to bisulfite conver‐sion. Therefore, it should be carefully to interpret the data of methylation analyses based on bisulfite treatment due to potentially high rate of false positive results. Although some me‐thylated DNAs were found to valuable as a single biomarker for cancer detection, more po‐tential DNA methylations will be found after the wide application of SMRT and other sequencing platforms with high speed, depth and accuracy. DNA methylation signatures in‐cluding a panel of methylated DNAs will show the potential in the early diagnosis or screening and prognosis or therapy response prediction of many cancers. In addition, such DNA methylation biomarkers could be more sensitive and specific for cancer detection when combined with well-used biochemical biomarkers. However, unified methods with gold standards will be warranted to promote the development and clinical application of DNA methylation biomarkers.Circulating Methylated DNA as Biomarkers for Cancer Detection/10.5772/51419145AcknowledgementsThis work was supported by the National Natural Science Foundation of China (81071963;81071652), Program for Innovative Research Team in Science and technology of Zhejiang Province (2010R50046) and Program for Qianjiang Scholarship in Zhejiang Province (2011R10061; 2011R10073).Author detailsHongchuan Jin, Yanning Ma, Qi Shen and Xian Wang **Address all correspondence to: wangx118@Department of Medical Oncology, Laboratory of Cancer Epigenetics, Biomedical Research Center, Sir Runrun Shaw Hospital, Zhejiang University, ChinaReferences[1]Jones, P. A., & Baylin, S. B. (2007). The epigenomics of cancer. Cell , 128, 683-692.[2]Jones, P. A., & Baylin, S. B. (2002). The fundamental role of epigenetic events in can‐cer. Nat Rev Genet , 3, 415-428.[3]Baylin, S. B., Esteller, M., Rountree, M. R., Bachman, K. E., Schuebel, K., & Herman, J.G. (2001). Aberrant patterns of DNA methylation, chromatin formation and gene ex‐pression in cancer. Hum Mol Genet , 10, 687-692.[4]Baylin, S. B., & Herman, J. G. (2000). DNA hypermethylation in tumorigenesis: epige‐netics joins genetics. Trends Genet , 16, 168-174.[5]Oki, Y., & Issa, J. P. (2006). Review: recent clinical trials in epigenetic therapy. Rev Re‐cent Clin Trials , 1, 169-182.[6]Kelly, T. K., De Carvalho, D. D., & Jones, P. A. (2010). Epigenetic modifications astherapeutic targets. Nat Biotechnol , 28, 1069-1078.[7]Ramalingam, S. S., Maitland, M. L., Frankel, P., Argiris, A. E., Koczywas, M., Gitlitz,B., Thomas, S., Espinoza-Delgado, I., Vokes, E. E, Gandara, D. R., & Belani,C. P.(2010). Carboplatin and Paclitaxel in combination with either vorinostat or placebo for first-line therapy of advanced non-small-cell lung cancer. J Clin Oncol , 28, 56-62.[8]Braiteh, F., Soriano, A. O., Garcia-Manero, G., Hong,D., Johnson, MM, Silva Lde, P.,Yang, H., Alexander, S., Wolff, J., & Kurzrock, R. (2008). Phase I study of epigenetic modulation with 5-azacytidine and valproic acid in patients with advanced cancers.Clin Cancer Res , 14, 6296-6301.Methylation - From DNA, RNA and Histones to Diseases and Treatment146/10.5772/51419 [9]Font, P. (2011). Azacitidine for the treatment of patients with acute myeloid leukemiawith 20%-30% blasts and multilineage dysplasia. Adv Ther, 3(28), 1-9.[10]Fu, S., Hu, W., Iyer, R., Kavanagh, J. J., Coleman, R. L., Levenback, C. F., Sood, A. K.,Wolf, J. K., Gershenson, D. M., Markman, M., Hennessy, B. T., Kurzrock, R., & Bast, R. C., Jr. (2011). Phase 1b-2a study to reverse platinum resistance through use of a hypomethylating agent, azacitidine, in patients with platinum-resistant or platinum-refractory epithelial ovarian cancer. Cancer, 117, 1661-1669.[11]Silverman, L. R., Fenaux, P., Mufti, G. J., Santini, V., Hellstrom-Lindberg, E., Gatter‐mann, N., Sanz, G., List, A. F., Gore, S. D., & Seymour, J. F. (2011). Continued azaciti‐dine therapy beyond time of first response improves quality of response in patients with higher-risk myelodysplastic syndromes. Cancer.[12]Sonpavde, G., Aparicio, A. M., Zhan, F., North, B., Delaune, R., Garbo, L. E., Rousey,S. R., Weinstein, R. E., Xiao, L., Boehm, K. A., Asmar, L., Fleming, M. T., Galsky, M.D., Berry, W. R., & Von Hoff, D. D. (2011). Azacitidine favorably modulates PSA ki‐netics correlating with plasma DNA LINE-1 hypomethylation in men with chemo‐naive castration-resistant prostate cancer. Urol Oncol, 29, 682-689.[13]Keating, G. M. (2012). Azacitidine: a review of its use in the management of myelo‐dysplastic syndromes/acute myeloid leukaemia. Drugs, 72, 1111-1136.[14]Alix-Panabieres, C., Schwarzenbach, H., & Pantel, K. (2012). Circulating tumor cellsand circulating tumor DNA. Annu Rev Med, 63, 199-215.[15]Zhe, X., Cher, M. L., & Bonfil, R. D. (2011). Circulating tumor cells: finding the needlein the haystack. Am J Cancer Res, 1, 740-751.[16]Fidler, I. J. (2003). The pathogenesis of cancer metastasis: the ‘seed and soil’ hypothe‐sis revisited. Nat Rev Cancer, 3, 453-458.[17]Ghossein, RA, Bhattacharya, S, & Rosai, J. (1999). Molecular detection of micrometa‐stases and circulating tumor cells in solid tumors. Clin Cancer Res, 5, 1950-1960. [18]Pelkey, TJ, Frierson, H. F., Jr, & Bruns, D. E. (1996). Molecular and immunological de‐tection of circulating tumor cells and micrometastases from solid tumors. Clin Chem, 42, 1369-1381.[19]Mocellin, S., Keilholz, U., Rossi, C. R., & Nitti, D. (2006). Circulating tumor cells: the‘leukemic phase’ of solid cancers. Trends Mol Med, 12, 130-139.[20]Chimonidou, M., Strati, A., Tzitzira, A., Sotiropoulou, G., Malamos, N., Georgoulias,V., & Lianidou, E. S. (2011). DNA methylation of tumor suppressor and metastasis suppressor genes in circulating tumor cells. Clin Chem, 57, 1169-1177.[21]Garcia-Olmo, D. C., Gutierrez-Gonzalez, L., Ruiz-Piqueras, R., Picazo, M. G., & Gar‐cia-Olmo, D. (2005). Detection of circulating tumor cells and of tumor DNA in plas‐ma during tumor progression in rats. Cancer Lett, 217, 115-123.。
自动控制原理课后习题答案

• 20世纪40年代,Evans提出并完善了根轨迹法。
• 20世纪50年代末,最优控制系统设计。
• 20世纪50年代末,基于时域分析的现代控制理 论。
• 60年代~80年代:最优控制、随机系统的最优控 制、复杂系统的自适应控制和学习控制得到了研 究。
5. 干扰量(Disturbance):引起被控量偏离预定运 行规律的量。除给定值之外,凡能引起被控量变 化的因素,都是干扰。干扰又称扰动
6.反馈(Feedback):将系统输出量引回输入端,并 与参考输入进行比较的过程。
7.前向通路 (Forward Channel):从给定量到被控 量的通道。
缺点: 闭环控制系统的参数如果匹配得不好,会造成被控量的 较大摆动,甚至系统无法正常工作。
例: 飞机自动驾驶控制
被控对象: 飞机
被控量: 飞机的俯仰角 θ
控制任务:系统在任何扰动作用下,保持飞机俯仰角不变。
仰俯角控制系统方块图
IV 复合控制
开环控制和闭环控制相结合的一种控制。实质上,它是在 闭环控制回路的基础上,附加了一个输入信号或扰动作用 的顺馈通路,来提高系统的控制精度。
an
d
n n
c(t
)
dt n
+
an-1
d n-1n-1c(t ) dt n-1
+"+
a1
dc(t) dt
+
a0c(t )
=
bm
d m m r (t ) dt m
+ bm-1
d m-1m-1r (t ) dt m-1
+" + b1
中国科学院大学现代信息检索课后习题答案

《信息检索导论》课后练习答案王斌最后更新日期 2013/9/28第一章布尔检索习题1-1 [*] 画出下列文档集所对应的倒排索引(参考图1-3中的例子)。
文档 1 new home sales top forecasts文档 2 home sales rise in july文档 3 increase in home sales in july文档 4 july new home sales rise习题1-2 [*] 考虑如下几篇文档:文档1 breakthrough drug for schizophrenia文档2 new schizophrenia drug文档3 new approach for treatment of schizophrenia文档4 new hopes for schizophrenia patientsa. 画出文档集对应的词项—文档矩阵;解答:breakthrough 1 0 0 0drug 1 1 0 0for 1 0 1 1hopes 0 0 0 1new 0 1 1 1patients 0 0 0 1schizophrenia 1 1 1 1treatment 0 0 1 0b. 画出该文档集的倒排索引(参考图 1-3中的例子)。
解答:参考a。
习题1-3 [*] 对于习题1-2中的文档集,如果给定如下查询,那么返回的结果是什么?a.schizophrenia AND drug解答:{文档1,文档2}b.for AND NOT (drug OR approach)解答:{文档4}习题1-4 [*] 对于如下查询,能否仍然在O(x+y)次完成?其中x和y分别是Brutus和Caesar所对应的倒排记录表长度。
如果不能的话,那么我们能达到的时间复杂度是多少?a.Brutus AND NOT Caesarb.Brutus OR NOT Caesar解答:a.可以在O(x+y)次完成。
传感器课后习题答案2013

返回
上页
下页
图库
第2章
2.5 解:①图 2-32(c)
6 2 2 A R r 59 . 7 10 ②圆桶截面积
应变片 1,2,3,4 感受的是纵向应变,有
1 2 3 4 x
应变片 5,6,7,8 感受的是纵向应变,有
5 6 7 8 y
返回
上页
下页
图库
第2章
2.5 一个量程为10kN的应变式测力传感器,其弹 性元件为薄壁圆筒轴向受力,外径20mm,内径18mm, 在其表面粘贴八各应变片,四个沿周向粘贴,应 变片的电阻值均为120Ω ,灵敏度为2.0,波松比为 0.3,材料弹性模量E=2.1×1011Pa。要求: 1) 绘出弹性元件贴片位置及全桥电路; 2) 计算传感器在满量程时,各应变片电阻变化; 3) 当桥路的供电电压为10V时,计算传感器的输出 电压。
nxi yi xi yi nxi (xi ) 2
2
2
6 182.54 21 42.23 208.41 1.98 6 91 21 21 105
91 42.23 21 182.54 9.59 0.09 6 91 21 21 105
第1章
1.5
某传感器为一阶系统,当受阶跃函数作用时,在 t=0时,输出为10mV;t→∞时,输出为100mV;在t=5s时, 输出为50mV,试求该传感器的时间常数。
解:此题与炉温实验的测飞升曲线类似:
y t 10 90(1 e t / T )
5 8.51 由 y 5 50 T 5 ln 9
返回 上页
解:
2.10题图
中级财务会计课后习题答案(全部)教材习题答案(全部).docx

教材练习题参考答案第二章货币资金【参考答案】(1)①出差借支时借:其他应收款一一张某1000贷:银行存款1000②归来报销时借:管理费用850库存现金150贷:其他应收款1000(2)①开立临时采购户吋借:其他货币资金一一外埠存款80 000贷:银行存款80 000②收到购货单位发票时借:原材料60 000应交税费一一应交增值税(进项税额)10 200贷:其他货币资金一一外埠存款70 200③将多余资金转回原来开户行时借:银行存款9 800贷:其他货币资金一一外埠存款9 800(3)①收到开户银行转来的付款凭证吋借:其他货币资金一一信用卡3 000贷:银行存款3 000②收到购物发票账单时借:管理费用2 520贷:其他货币资金一一信用卡25 20(4)拨出备用金时借:备用金1000贷:银行存款10 00(5)总务部门报销时借:管理费用900贷:库存现金9 00(6)①期末盘点发现短缺时借:待处理财产损溢一一待处理流动资产损溢50贷:库存现金50②经批准计入损益吋借:管理费用5 0贷:待处理财产损溢一一待处理流动资产损溢50第三章应收款项【参考答案】1.(1)办妥托收银行收款手续时:借:应收账款11700贷:主营业务收入10 000应交税费一应交增值税(销项税额)17 0 00(2)如在10天内收到货款时借:银行存款11 466财务费用23 4贷:应收账款11700(3)如在30内收到货款时借:银行存款11700贷:应收账款117002.(1)收到票据时借:应收票据93 6 00贷:主营业务收入80 000应交税费一应交增值税(销项税额)13 600(2)年终计提票据利息借:应收票据15 60贷:财务费用1560(3 )到期收回货款借:银行存款98 280贷:应收票据95 160财务费用3 1203.(1)第一年末借:资产减值损失5 000贷:坏账准备5 000(2)第二年末借:资产减值损失7 500贷:坏账准备7 500(3 )第三年末借:坏账進备1500贷:资产减值损失1500(3)第四年6月发生坏账时借:坏账准备18 000贷:应收账款18 00010月收回己核销的坏账时借:应收账款5 00 0贷:坏账准备5 000借:银行存款5 000贷:应收账款5000年末计提坏账准备时借:资产减值损失1 2 000贷:坏账准备12 00 0第四章存货【参考答案】1.(1)实际成本核算:该批甲材料的实际总成本=20 000+200=2 0 200 (元)借:原材料•甲材料20 2 00应交税费•应交增值税3 400贷:银行存款23 600(2)计划成本核算①购进借:材料采购■甲材料20 200应交税费•应交增值税3 400贷:银行存款23 600②入库材料成本差异=20 200-990X18 =2380元,超支差异借:原材料--- 甲材料17 820 (=990X18)材料成本差异2380贷:材料采购——甲材料20 20 02.(1)先进先出法6月7日①发出A材料的成本=200X 60+20 0X 66=25 20 0 (元)6月18日②发出A材料的成本=300X 66 +500X 70=54 800 (元)6月29日③发出A材料的成本=100 X70+200X68 =20 600 (元)期末结存A材料成本=300X68=20 400 (元)(2)月末一次加权平均法加权平均单位成本二(12 000+109 0 00) 4- (200+1 600) ^67.22 (元/公斤)期末结存A材料的成本=300 X67.22=201 66 (元)本月发出A 材料的成本二(12 000+109 00 0) -20166=1 00834 (元)(3 )移动加权平均法6月5日①购进后移动平均单位成本二(12000+33 000)一(200+500) =64.29 (元/公斤)6月7日结存A材料成本=300X64 .29=19287 (元)6 月7 日发出A 材料成本=(12000+330 00) -19287=25713 (元)6月16日②购进后移动平均单位成本二(19287+42 000) 4- (300+600) =68.10 (元/公斤)6月18日结存A材料成本=100X68 .10=6810 (元)6 月18 日发出A 材料成本二(19287+4200 0) -6810=54 477 (元)6月27日③购进后移动平均单位成本二(6810+34000 )0 (100+500 )=68.02 (元/公斤)6月29日结存A材料成本=300X68.02 =20406 (元)6月29日发出A材料成本二(68 10+34000)・20406=2040 4 (元)期末结存A材料成本=300X68 .02=20406 (元)3.A产品:有销售合同部分:A产品可变现净值=40X (1105)=4 20(万元),成本=40X10=400 (万元),这部分存货不需计提跌价准备。
《线性代数》(经科社2013版)习题解答

5. A2 − 2A − 4E = O ⇒ A2 − 2A − 3E = E ⇒ (A + E )(A − 3E ) = E , 故(A + E )−1 = (A − 3E ).
3(A − E )−1 A = 3(A−1 (A − E ))−1 = 3(E − A−1 )−1 , 其中A−1 = 9. AA∗ = |A|E ⇒ 10.
−1 1
2
1 (4)A31 + A32 + A33 + A34 = 3 1
2 3 1
−3 6 3 1 3 1 .
3 4 1 8 3.(1)第i行减去末行的ai 倍(i = 1, 2, · · · , n), 再按末列展开. (2)仿教材例1.4.4. (3)从第一行开始, 上一行的x倍加到下一行, 再按末行展开. (4)按末列展开. 4.(1)见《线性代数学习指导》P25例25. (2)见《线性代数学习指导》P26例26. 或: 第一行减去第二行, 按第一行展开, 得递推关系式; 列同样 处理. 联立解之. 注: ::::::::: 此题较难,::::::::::: 可不作要求. (3)从第一行开始, 用上一行消下一行, 化为上三角行列式. 1 5. M11 + M21 + M31 + M41 = A11 − A21 + A31 − A41 = −1 1 −1 1 A11 + A12 + A13 + A14 = 1 −1 1 1 3 1 0 1 1 −5 3 −3 . −5 1 3 −4 2 0 1 1 −5 3 .
i=1 i=1 i=1
注: :::::::::::::::::::::::::::::::::: 要牢记矩阵乘法的口诀“前行乘后列”.
大学物理学(第三版上) 课后习题3答案详解
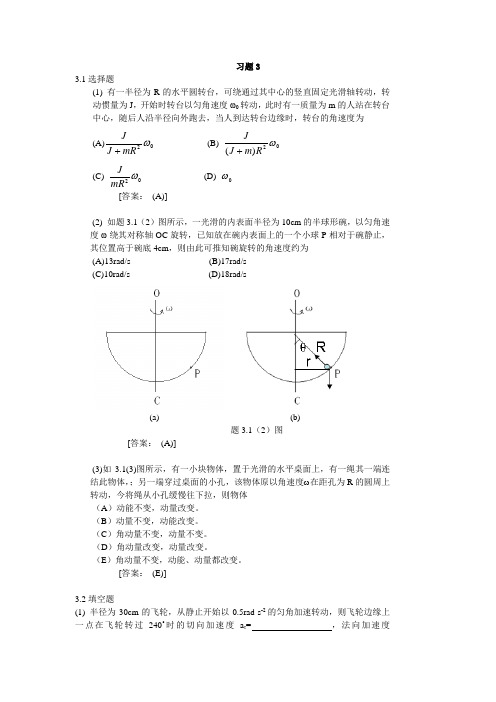
习题33.1选择题(1) 有一半径为R 的水平圆转台,可绕通过其中心的竖直固定光滑轴转动,转动惯量为J ,开始时转台以匀角速度ω0转动,此时有一质量为m 的人站在转台中心,随后人沿半径向外跑去,当人到达转台边缘时,转台的角速度为(A)02ωmRJ J+ (B) 02)(ωR m J J + (C)02ωmRJ(D) 0ω [答案: (A)](2) 如题3.1(2)图所示,一光滑的内表面半径为10cm 的半球形碗,以匀角速度ω绕其对称轴OC 旋转,已知放在碗内表面上的一个小球P 相对于碗静止,其位置高于碗底4cm ,则由此可推知碗旋转的角速度约为 (A)13rad/s (B)17rad/s (C)10rad/s (D)18rad/s(a) (b)题3.1(2)图[答案: (A)](3)如3.1(3)图所示,有一小块物体,置于光滑的水平桌面上,有一绳其一端连结此物体,;另一端穿过桌面的小孔,该物体原以角速度ω在距孔为R 的圆周上转动,今将绳从小孔缓慢往下拉,则物体 (A )动能不变,动量改变。
(B )动量不变,动能改变。
(C )角动量不变,动量不变。
(D )角动量改变,动量改变。
(E )角动量不变,动能、动量都改变。
[答案: (E)]3.2填空题(1) 半径为30cm 的飞轮,从静止开始以0.5rad·s -2的匀角加速转动,则飞轮边缘上一点在飞轮转过240˚时的切向加速度a τ= ,法向加速度a n=。
[答案:0.15; 1.256](2) 如题3.2(2)图所示,一匀质木球固结在一细棒下端,且可绕水平光滑固定轴O转动,今有一子弹沿着与水平面成一角度的方向击中木球而嵌于其中,则在此击中过程中,木球、子弹、细棒系统的守恒,原因是。
木球被击中后棒和球升高的过程中,对木球、子弹、细棒、地球系统的守恒。
题3.2(2)图[答案:对o轴的角动量守恒,因为在子弹击中木球过程中系统所受外力对o轴的合外力矩为零,机械能守恒](3) 两个质量分布均匀的圆盘A和B的密度分别为ρA和ρB (ρA>ρB),且两圆盘的总质量和厚度均相同。
- 1、下载文档前请自行甄别文档内容的完整性,平台不提供额外的编辑、内容补充、找答案等附加服务。
- 2、"仅部分预览"的文档,不可在线预览部分如存在完整性等问题,可反馈申请退款(可完整预览的文档不适用该条件!)。
- 3、如文档侵犯您的权益,请联系客服反馈,我们会尽快为您处理(人工客服工作时间:9:00-18:30)。
1、结合唯物辩证法的基本观点分析科学发展观是关于发展的观点和方法论的集中体现
马克思主义是我们认识和改造世界的强大思想武器,也是指导我们发展的世界观和方法论的基础。
科学发展观坚持马克思主义的基本原理,紧密结合中国特色社会主义的伟大实践,吸收人类文明进步的新成果,站在历史和时代的高度,进一步明确了新世纪新阶段我国社会主义发展的一系列重大问题,用新的思想理论观点丰富了马克思主义关于发展的理论,是与时俱进的马克思主义发展观。
科学发展观坚持以经济建设为中心,把发展生产力作为首要任务,把经济发展作为一切发展的前提,体现了历史唯物主义关于生产力是人类社会发展基础的基本观。
科学发展观强调人民群众是发展的主体力量,把实现和维护好人民群众的根本利益作为发展的出发点和落脚点,切实保障人民群众的经济、政治和文化权益,体现了历史唯物主义关于人民群众是历史发展的主体和人的全面发展的基本观点。
科学发展观提出全面、协调、可持续发展,强调既要按照经济社会发展规律全面推进经济建设、政治建设、文化建设和社会建设,又要遵循自然规律推动人与自然和谐发展,实现经济发展与资源、人口、环境相协调,注重城乡发展、区域发展、经济社会发展、人与自然和谐发展、国内发展和对外开放的统筹协调;强调发展是相互推进、系统协调的过程,要正确处理中心与全面、重点与非重点、平衡与不平衡的关系,实现经济社会又快又好发展,充分体现了唯物辩证法关于事物之间普遍联系、辩证统一的基本原理。
科学发展观关于和谐社会与和谐世界的思想观点,既坚持矛盾无处不在、无时不有,是一切事物发展的内在动力,又强调通过解决矛盾,求得协调和统一,实现人与人、人与社会、人与自然之间的和谐,实现不同国家、不同文明之间的友好相处与共同繁荣,进一步深化了对辩证唯物主义对立统一规律的认识。
总的看,科学发展观贯穿了马克思主义的立场、观点、方法,体现了世界观和方法论的统一。
它集中反映了社会主义建设的内在规律,创造性地回答了什么是发展、为什么发展和怎样发展的重大问题,进一步丰富发展了中国特色社会主义理论,开辟了马克思主义发展的新境界,是全面推进社会主义经济建设、政治建设、文化建设、社会建设必须长期坚持的指导方针。
在新的历史条件下,树立和落实科学发展观,就是坚持运用马克思主义世界观和方法论指导新的发展实践。
2、为什么说马克思主义认识论是能动的反映论?
之所以说马克思主义认识论是能动的反映论,是因为:
1、马克思主义认识论坚持实践的观点,认为人的认识是在实践的基础产生和发
展的。
马克思主义认识论是实践论,它把科学的实践观引入认识论。
马克思主义认识论把实践观点作为首要的基本的观点,阐明了认识对实践的依赖性,人们只有从实践中获得“材料”,才能通过头脑这个“加工厂”产生认识。
人们在社会实践中变革客观世界的同时,也不断改造着自身的认识能力。
从认识论角度讲,主体和客体之间的关系,是反映和被反映的关系。
所以,正是人们改造客观事物的社会实践活动,才使得人们的认识得以形成和发展。
2、马克思主义认识论坚持辩证法的观点,认为人的认识不是一次完成的,而是一
个辩证发展的过程。
揭示了人类认识的发展规律。
在马克思主义认识论看来,人的认识与照相和镜子照东西有某些相似之处,但又有本质的不同。
照相和镜子照东西是消极
被动的反映,人的认识是经过头脑加工改造,有一个对客体信息的选择、加工整理和创造的过程,是能动的反映,不是机械的直观。
这不仅体现在对客观事物认识的初级阶段,而且体现在理性观念的形成过程中。
认识主体在感知客体的过程中,根据自身已有的知识背景和认知图式,从不同层面上选择、整理和解释客体信息,将客体的存在方式转变为主体能够理解、说明的方式;而且更重要的是对客体信息进行建构,将客体信息同化到既有的观念体系中,或者将客体信息建构成为新的概念理论体系。
在这方面,辩证思维方法就是思维对来自客体的感性信息进行加工制作,将其升华为表征客体的理性观念及其体系。
由此可见,马克思主义认识论在坚持反映论的前提下,肯定主体在认识中的选择、建构等能动作用。
人的认识不仅反映事物的表面现象,而且能动地揭示出事物的本质和规律。
人的认识是随着实践的发展而发展的,是一个由不知到知,由浅入深的辩证发展过程。
3、马克思主义认识论的目的不是为了揭示和说明世界,而是为了更好的改造世界。
3、为什么说真理是客观的,绝对又是相对的?把握这一观点对于我们坚持和发展马克思主
义有什么重要意义?
1、把握真理的绝对性和相对性相统一的原理,对于我们正确对待马克思主义有重
要意义。
马克思主义是真理,它也是绝对性和相对性的统一。
它正确地反映了自然、社会和思维发展的普遍规律,因而具有绝对性的一面。
但是,它又没有穷尽一切事物及其规律,需要随着社会实践的发展而发展,因而又具有相对性的一面。
正因为马克思主义真理具有绝对性,所以我们必须坚持它并以它作为我们的指导思想;又因为它具有相对性,所以我们又必须在实践中丰富它、发展它。
既坚持又发展,才是对待马克思主义的正确态度。
2、马克思主义真理的生命力在于理论创新。
与时俱进,最主要的就是进行理论创
新。
实践始终在向前发展,理论必须随着实践的不断前进而发展,实践没有止境,创新也没有止境。
在我国社会主义市场经济建设中,思想解放和理论创新能对制度创新、文化科技创新、经济领域创新提供能动的反作用,必须不断加强理论创新工作。
3、强调马克思主义的品质是与时俱进,不是抛弃马克思主义哲学基本原理,而是
要科学地反映人类在实践领域和认识领域所取得的最新成果,及时回答和解决时代提出的重大问题,从中得出新的结论,提出新的观点,发展新的理论。
马克思主义哲学的历史进程有力地证明,与时俱进是马克思主义哲学的内在要求。
4、绝对真理和相对真理辩证关系的原理,指导我们树立对马克思主义的科学态度,
即一要坚持,二要发展。
马克思主义是在无产阶级革命实践中产生并接受实践的检验,然后又指导无产阶级革命实践的普遍真理。
马克思主义是对社会发展规律的正确认识,是客观真理,也就是包含着绝对真理的因素的正确认识。
从这个意义上说,马可思主义是绝对真理。
因此,必须坚持,任何时候都不能违背。
要反对否定马可思主义的错误倾向。
这种观点怀疑和否定马可思的基本原理的正确性,因此,也就是怀疑和否定其真理的客观性和绝对性。
马克思主义又是在一定的历史条件下产生的。
它没有也不可能穷尽我们对社会发展规律的认识。
马克思主义也有一个不断发展、不断完善的过程,它的个别观点也会随着实践的发展而改变。
这说明,马克思义主同时也是相对真理
4、如何理解辩证唯物主义认识论与党的思想路线的关系?
两者是统一的。
辩证唯物主义认识论是“实事求是”思想路线的哲学基础,而“实事求是”思想路线则辩证唯物主义认识论在实际工作中的具体运用。
思想路线与认识路线的一
致性,充分体现了实践中知与行的统一。
①一切从实际出发:就是坚持客观决定主观的唯物主义认识路线,这是确定正确思想路线的前提;②理论联系实际:强调实践必须有理论的指导,而理论只有从实践中来,并用以指导实践才有意义;③实事求是:强调规律的客观性,指明认识的任务是探索客观事物的内部联系,以指导实践;④在实践中检验真理和发展真理:坚持认识论关于实践是检验真理的唯一标准的观点、真理性的认识只有经过实践的检验才能得到进一步补充、丰富和发展等观点。
5、商品的价值量是由生产商品所耗费的劳动量决定的,但为什么不源于私人劳动和社会劳
动的矛盾?
答:商品的价值量就是由生产商品所耗费的劳动量决定的。
劳动的量是用劳动的持续时间来衡量的,因而商品价值的大小也就取决于生产商品的劳动时间的多少。
但是,商品的价值量,不能由各个商品生产者生产商品所实际耗费的个别劳动时间所决定,这是因为:
第一,各个商品生产者生产商品的主观条件和客观条件不同,他们生产同种商品所耗费的个别劳动时间也就不同。
如果由各个商品生产者生产商品所实际耗费的个别劳动时间所决定,同种商品就不会有同一的价值量,不利于交换的进行。
第二,个别劳动时间决定商品的价值量不利于技术进步。
如果那些生产技术差、生产工具落后、工作懒惰、劳动熟练程度低的商品生产者所生产的商品,因为消耗的个别劳动时间长,所生产的每件商品的价值量就大,而且消耗的劳动时间越长,每件商品的价值量就越大;而那些生产技术好、生产工具先进、工作勤勉、劳动熟练程度高的商品生产者所生产的商品,因为消牦的个别劳动时间短,所生产的每件商品的价值反而小,那么,这种情况实际上是在鼓励落后,显然是极不合理的,商品的价值不可能这样来决定。
所以,商品的价值不是由生产商品的个别劳动时间决定的,而是由生产商品的社会必要劳动时间决定的。
商品价值量和市场的劳动生产率成反比,生产率高了,产品多了,这样商品供大于求,导致商品价格降低;生产率低了,产品少了,商品供不应求,商品价格上升..
商品价值量和个人的劳动生产率成正比,当个人的生产率大于市场的生产率时,你所供应的商品多了,但商品价格没变,所以你的商品的价值总量增加了。
