Domain mutagenesis kit随机突变试剂盒说明书
定点诱变技术

How DNA shuffling works ?
一、单基因和基因家族的重组装
Single gene shuffling
. .. . ....... library of point mutants
Similar mutants generated by error-prone PCR, random and site-directed mutagenesis
在正常情况下,尿嘧啶N-糖基化酶(ung+)可以去除掺 入DNA中的尿嘧啶残基。但 在ung-的菌株中,此酶失活。
在大肠杆菌dut- ung-菌株 中生长的M13噬菌体的单链基 因组DNA中将含有20-30个尿 嘧啶残基。用这些噬菌体感染 ung+菌株,尿嘧啶被迅速去 除,DNA链遭到破坏,感染 力下降约5个数量级。
牛乳糖酶到岩藻糖苷酶的直接进化(JI-HU ZHANG等,1997)
How DNA shuffling works ?
二、随机引物PCR(RPR)和重组装
多功能氧化酶的定向进化 (Hikaru Suenaga等,2001)
How DNA shuffling works ?
三、交错延伸PCR突变法(StEP)
枯草杆菌蛋白酶E热稳定性的分子进化 (Huimin Zhao等,1999)
进化的热稳定性枯草芽孢杆菌蛋白酶E的突变谱系
正面
反面
进化后的枯草杆菌蛋白酶E
芽孢杆菌脲酸酶的定向 进化 (Su-Hua Huang等,2004)
定点突ቤተ መጻሕፍቲ ባይዱ的研究意义
1.对调控区进行突变
研究基因结构与功能之间的关系
2. 对编码基因进行突变
PCR介导的基因突变
在基因5’和3’末端产生突变 重叠延伸PCR 大引物PCR法
First Strand cDNA Synthesis Kit 说明书

GeneCopoeia Inc.19520 Amaranth DriveGermantown, Maryland 20874USATel: 301-515-6982; 1-866-360-9531Fax: 301-515-6983Web: First Strand cDNA Synthesis Kit产品套装编号:C0210A 储存条件:-20℃保存产品内容M-MLV Reverse Transcriptase (RNase H -)5×RT Reaction Buffer RNase Inhibitor 25mM dNTP60 µM Oligo(dT)18250 µM Random PrimerddH 2O (DNase/RNase Free)试剂组分total RNA 或Poly(A) mRNA60 µM Oligo(dT)18 或250 µM Random Primer 或10 µM Sequence-Speci fic Primer ddH 2O (DNase/RNase Free)产品编号C02010A C02011A C10200A C10011A C10210A C10220A C10230A体积1 µl 1 µl 1 µl包装规格50 μl (200 U/μl)250 μl50 μl (25 U/μl)50 μl 50 μl 50 μl 1 ml终浓度1 µg 10 ng 2.4 µM 10 µM 0.4 µM至总体积13 µl■ 产品概述:RNA 逆转录成的单链cDNA 是基因克隆和RNA 研究的主要材料。
本产品是采用莫洛尼鼠白血病病毒(Moloney Murine Leukemia Virus) 逆转录酶(M-MLV Reverse Transcriptase)来合成单链cDNA 的专用试剂盒。
第一链cDNA合成试剂盒Transcriptor First Strand cDNA Synthesis Kit 中文说明书

第一链cDNA合成试剂盒Transcriptor First Strand cDNA Synthesis Kit中文说明书2013-12-07 13:20:16Transcriptor First Strand cDNA Synthesis Kit试剂盒包装与含量小瓶/瓶盖标签适用于a) 04 379012001b)04896866001c************1红色TranscriptorReverseTranscriptase(逆转录酶)a) 1瓶,25 μl (20 U/μl)b) 1瓶,50 μl (20 U/μl)c) 2瓶,各50 μl (20 U/ μl)•储存缓冲液:200 mM 磷酸钾,2 mM 二硫苏糖醇,0.2% Triton X-100(v/v),50% 甘油(v/v),pH 约为7.22无色Transcriptor RTReaction Buffer(5×)(逆转录缓冲液)a) 1瓶,1 mlb) 1瓶,1 mlc) 2瓶,各1 ml• 5×浓度:250 mM Tris/HCl,150 mM KCl,40 mM MgCl2,pH约为8.5(25°C)3无色ProtectorRNase Inhibitor(RNase抑制剂)a) 1瓶,50 μl(40 U/μl)b) 1瓶,100 μl(40 U/μl)c) 2瓶,各100 μl(40 U/μl)•储存缓冲液:20 mM Hepes-KOH,50 mM KCl,8 mM 二硫苏糖醇,50 % 甘油(v/v),pH 约为7.6 (4°C)4黄色/紫色Deoxynuc-leo-tideMix(dNTP)a) 1瓶,100 μl(黄色瓶盖)b) 1瓶,200 μl(紫色瓶盖)c) 2瓶,各200 μl(紫色瓶盖)• dATP, dCTP, dGTP, dTTP各10 mM5蓝色Anchored-oligo(dT)18Primer(锚定oligo(dT)18引物)a) 1瓶,100 μl(50 μM)b) 1瓶,200 μl(50 μM)c) 2瓶,各200 μl(50 μM)6Random Hexamera) 1瓶,100 μl(600 μM)蓝色Primer(随机引物)b) 1瓶,200 μl(600 μM)c) 2瓶,各200 μl(600 μM)7绿色Control RNA(对照RNA)a) 1瓶,20 μl(50 ng/μl)•包含提取于永生细胞系(K562)的总RNA片段稳定溶液8绿色Control Primer Mix PBGD(对照基因引物)a) 1瓶,40 μl•5 μM 人类PBGD特异性正向与反向引物9(b和c为瓶7)无色Water, PCR-gradea) 1瓶,1 mlb) 2瓶,各1 mlc) 3瓶,各1 ml注意:货号为***********和***********的产品不含有control试剂(瓶7和瓶8),因此瓶7即为Water, PCR-grade。
碧云天 Human IgE ELISA Kit 产品说明书
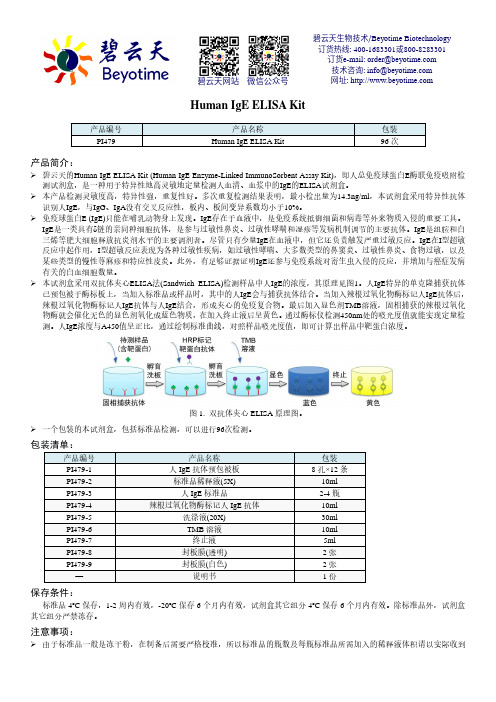
Human IgE ELISA Kit产品编号 产品名称包装 PI479Human IgE ELISA Kit96次产品简介:碧云天的Human IgE ELISA Kit (Human IgE Enzyme-Linked ImmunoSorbent Assay Kit),即人总免疫球蛋白E 酶联免疫吸附检测试剂盒,是一种用于特异性地高灵敏地定量检测人血清、血浆中的IgE 的ELISA 试剂盒。
本产品检测灵敏度高,特异性强,重复性好。
多次重复检测结果表明,最小检出量为14.3ng/ml ,本试剂盒采用特异性抗体识别人IgE ,与IgG 、IgA 没有交叉反应性,板内、板间变异系数均小于10%。
免疫球蛋白E (IgE)只能在哺乳动物身上发现。
IgE 存在于血液中,是免疫系统抵御细菌和病毒等外来物质入侵的重要工具。
IgE 是一类具有δ链的亲同种细胞抗体,是参与过敏性鼻炎、过敏性哮喘和湿疹等发病机制调节的主要抗体。
IgE 是组胺和白三烯等肥大细胞释放抗炎剂水平的主要调剂者。
尽管只有少量IgE 在血液中,但它还负责触发严重过敏反应。
IgE 在I 型超敏反应中起作用,I 型超敏反应表现为各种过敏性疾病,如过敏性哮喘、大多数类型的鼻窦炎、过敏性鼻炎、食物过敏,以及某些类型的慢性荨麻疹和特应性皮炎。
此外,有足够证据证明IgE 还参与免疫系统对寄生虫入侵的反应,并增加与癌症发病有关的白血细胞数量。
本试剂盒采用双抗体夹心ELISA 法(Sandwich ELISA)检测样品中人IgE 的浓度,其原理见图1。
人IgE 特异的单克隆捕获抗体已预包被于酶标板上,当加入标准品或样品时,其中的人IgE 会与捕获抗体结合。
当加入辣根过氧化物酶标记人IgE 抗体后,辣根过氧化物酶标记人IgE 抗体与人IgE 结合,形成夹心的免疫复合物。
最后加入显色剂TMB 溶液,固相捕获的辣根过氧化物酶就会催化无色的显色剂氧化成蓝色物质,在加入终止液后呈黄色。
Kras基因突变检测试剂盒说明书

人类K-ras基因突变检测试剂盒(PCR-熔解曲线法)说明书【产品名称】通用名:人类K-ras基因突变检测试剂盒(PCR-熔解曲线法)英文名:Diagnostic kit for Mutations of Human K-ras Gene(PCR-Melting Curve Analysis)【包装规格】20测试/盒【预期用途】K-ras基因位于12号染色体短臂上,是重要的癌基因之一,编码一种21kD 的kras蛋白,参与细胞内的信号传递,主要包括PI3K/PTEN/AKT 和RAF/MEK/ERK信号转导途径,这些转导途径是当前肿瘤靶向药物研究的热点,靶向药物通过抑制这些途径发生药理作用。
K-ras基因第12和13密码子发生突变,将导致kras蛋白变异并处于持续激活状态,使药物失效。
据中国2010版《肿瘤学临床实践指南》,在一项包含101例肺腺癌亚型细支气管肺泡癌患者的回顾性研究中,所有患者均接受厄洛替尼单药一线治疗。
K-ras突变者无一例缓解(0/18),而无K-ras突变者则有20例缓解(20/62,32%),差别有统计学意义(P <0.01)。
因此,指南建议,非小细胞肺癌和结直肠癌患者使用靶向药物前应进行K-ras基因突变状态的检测。
本试剂盒以人非小细胞肺癌、结直肠癌肿瘤组织切片提取的基因组DNA为检测样本,用于检测肿瘤组织K-ras基因第12,13密码子的12种体细胞突变(表1),提供突变状态的定性结果。
为临床肿瘤靶向药物的个体化用药提供辅助诊断依据,本品适用于进入个体化靶向治疗疗程前的患者使用。
表1 本品可检测的K-ras基因突变K-ras基因12密码子突变K-ras基因13密码子突变Gly12Ser GGT > AGT Gly13Ser GGC > AGCGly12Arg GGT > CGT Gly13Arg GGC > CGCGly12Cys GGT > TGT Gly13Cys GGC > TGCGly12Asp GGT > GAT Gly13Asp GGC > GACGly12Ala GGT > GCT Gly13Ala GGC > GCCGly12Val GGT > GTT Gly13Val GGC > GTC【检验原理】本试剂盒基于实时PCR平台,结合了特异引物、荧光探针和熔解曲线技术,定性检测DNA样品中K-ras 基因12,13密码子是否存在突变。
东盛生物 RT-PCR Kit(逆转录试剂盒) 说明书

0.6 μl RNasin , M-MLV ; 按下列条件进行反转录反应 42℃ 温浴 60 min(随即引物 37℃ 温浴 60 min) ; 5 终止反应 70℃ 温浴 5 min 终止反应,置冰上进行后续实验或-20℃ 保存。 6 用 RNase-free ddH2O 将反应体系稀释到 50 μl ,取 2-5 μl 进行 PCR 扩增反应。
20 次逆转录反应,R1012 可进行 100 次逆转
录反应(20 μl 标准 PCR 反应体系,每次使用 M-MLV 1 μl)。
M-MLV 储存液 20 mM Tris-HCl (pH 7.5) 200 mM NaCl 0.1 mM EDTA 1 mபைடு நூலகம் DTT 0.01% NP-40 50% glycerol 5xfirst-strand buffer 成分 250 mM Tris-HCl (pH 8.3 at 25 ℃) 375 mM KCl 15 mM MgCl2 50 mM DTT 保存条件 各组分均-20℃保存,避免反复冻融。
●
在逆转录反应中经常加入 RNase 抑制剂以增加 cDNA 合成的长度和产量。RNase 抑制剂要在第一链合成反应中,在缓冲
液和还原剂(如 DTT)存在的条件下加入,因为 cDNA 合成前的过程会使抑制剂变性,从而释放结合的可以降解 RNA 的 RNase。蛋白 RNase 抑制剂仅防止 RNase A,B,C 对 RNA 的降解,并不能防止皮肤上的 RNase,因此尽管使用了这些抑 制剂,也要小心不要从手指上引入 RNase。
----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------广州东盛生物科技有限公司 电话:020-87791356 传真:020-87791381 网址:
Random Primer DNA Labeling Kit Ver.2 商品说明书.pdf_171
Certificate of AnalysisProduct Name/Description: Random Primer DNA Labeling Kit Ver.2Cat. #: 6045Lot #: AK4601Storage Conditions: -20 degrees CShipping Conditions: -20 degrees CExpiration Date: Specified on product label or packagePackage Size:30 RxnsPackage Contents: 1. Random Primer (9 mer)60 µl 2. 10X Buffer75 µl 3. dNTP Mixture (dGTP, dATP, dTTP)0.2 mM each75 µl 4. Exo-free Klenow Fragment 2 U/µl30 µl 5. Control DNA (λ-Hind III Fragment)25 ng/µl10 µl Product Documents: Documents for Takara Bio products are available for download at Quality Control Data:1) The specific radioactivity was ≥1.2E+09 dpm/μg when the incorporation test was performedusing 25 ng of Control DNA according to the user manual.2) The expected pattern of Control DNA was confirmed by agarose gel electrophoresis.It is certified that this product meets above specifications.Manager, Quality AssuranceTAKARA BIO INC.Safety Information :Please refer to our website for safety information :Notice To Purchaser :This product is for research use only. It is not intended for use in therapeutic or diagnostic procedures for humans or animals. Also, do not use this product as food, cosmetic, or household item, etc.Takara products may not be resold or transferred, modified for resale or transfer, or used to manufacture commercial products without written approval from TAKARA BIO INC.If you require licenses for other use, please contact us by phone at +81 77 543 7247 or from our website.Your use of this product is also subject to compliance with any applicable licensing requirements described on the product web page. It is your responsibility to review, understand and adhere to any restrictions imposed by such statements.All trademarks are the property of their respective owners. Certain trademarks may not be registered in all jurisdictions.。
Braf基因突变检测试剂盒说明书
人类B-raf基因V600E突变检测试剂盒(荧光PCR法)说明书【产品名称】通用名:人类B-raf基因V600E突变检测试剂盒(荧光PCR法)英文名:Diagnostic kit for V600E Mutation of Human B-raf Gene(Fluorescence PCR Analysis)【包装规格】20测试/盒【预期用途】B-raf基因位于7号染色体长臂上,是一种癌基因,属RAF基因家族,有18个外显子,编码一种含783个氨基酸的B-raf蛋白,是EGFR通路RAS/RAF/MEK/MRK/MAPK中重要的转导因子,参与调控细胞生长、分化和凋亡等多种生理过程。
针对EGFR的肿瘤靶向药物通过抑制该途径发生药理作用。
研究表明在非小细胞肺癌、结直肠癌等多种恶性肿瘤中存在B-raf基因突变,其中第15外显子上V600E点突变最常见,约占所有突变的90%以上,该突变导致B-raf蛋白被异常激活,从而使患者接受EGFR-TKI药物和EGFR单抗类药物治疗失效。
据中国2010版《肿瘤学临床实践指南》建议,对K-raf基因检测正常的非小细胞肺癌和结直肠癌患者,应进一步检查B-raf基因的突变状态,以指导靶向药物治疗方案。
本试剂盒以人非小细胞肺癌、结直肠癌肿瘤组织切片提取的基因组DNA为检测样本,用于肿瘤组织B-raf 基因V600E点突变的定性检测,为临床肿瘤靶向药物的个体化用药提供依据。
本公司尚无临床实例证实B-raf 基因突变与靶向药物的相关性,其相关性主要来自文献报道,因此本试剂盒检测结果仅用于辅助临床医生对肿瘤患者制定用药方案。
【检验原理】本试剂盒基于实时荧光PCR平台,结合等位基因特异性扩增(ARMS)技术、野生型基因扩增抑制技术和多重PCR技术检测B-raf基因V600E突变。
ARMS技术是指PCR引物的3’端末位碱基必须与其模板DNA互补才能有效扩增,通过设计特异性ARMS引物,对存在V600E突变的B-raf基因靶序列进行PCR扩增放大,并利用FAM基团标记的Taqman 探针对扩增产物进行检测。
点突变
定点突变定点突变原理图定点突变是指通过聚合酶链式反应(PCR)等方法向目的DNA片段(可以是基因组,也可以是质粒)中引入所需变化(通常是表征有利方向的变化),包括碱基的添加、删除、点突变等。
定点突变能迅速、高效的提高DNA所表达的目的蛋白的性状及表征,是基因研究工作中一种非常有用的手段。
意义体外定点突变技术是研究蛋白质结构和功能之间的复杂关系的有力工具,也是实验室中改造/优化基因常用的手段。
蛋白质的结构决定其功能,二者之间的关系是蛋白质组研究的重点之一。
对某个已知基因的特定碱基进行定点改变、缺失或者插入,可以改变对应的氨基酸序列和蛋白质结构,对突变基因的表达产物进行研究有助于人类了解蛋白质结构和功能的关系,探讨蛋白质的结构/结构域。
而利用定点突变技术改造基因:比如野生型的绿色荧光蛋白(wtGFP)是在紫外光激发下能够发出微弱的绿色荧光,经过对其发光结构域的特定氨基酸定点改造,现在的GFP能在可见光的波长范围被激发(吸收区红定点突变移),而且发光强度比原来强上百倍,甚至还出现了黄色荧光蛋白,蓝色荧光蛋白等等。
定点突变技术的潜在应用领域很广,比如研究蛋白质相互作用位点的结构、改造酶的不同活性或者动力学特性,改造启动子或者DNA作用元件,提高蛋白的抗原性或者是稳定性、活性、研究蛋白的晶体结构,以及药物研发、基因治疗等等方面。
单点突变对于单点突变,Stratagene公司的QuikChange Site-directed Mutagenesis kit是不错的选择,通过巧妙设计,将质粒定点突变技术变得简单有效。
准备突变的质粒必须是从常规E.coli 中经纯化试剂盒(Miniprep)或者氯化铯纯化抽提的质粒。
设计一对包含突变位点的引物(正、反向),和模版质粒退火后用PfuTurbo聚合酶“循环延伸”,(所谓的循环延伸是指聚合酶按照模版延伸引物,一圈后回到引物5’端终止,再经过反复加热褪火延伸的循环,这个反应区别于滚环扩增,不会形成多个串联拷贝。
Stratagene突变解决方案
• 为多位点突变和饱和突变提供建议 • 将导入的 DNA 序列翻译为氨基酸序列 • 支持我们所有 QuikChange 定点突变
试剂盒
访问 /mutagenesis, 体验 QuikChange 引物设计程序
7
和高效的定点突变;其它试剂盒组成可促
聚合酶和 Dpn I 筛选酶
进大质粒(大于 8 kb)或复杂质粒的突变
• 采用 QuikSolution 以增强扩增
• 包含我们最高效的 XL-10 Gold® 超级 感受态细胞
多位点定点突变
QuikChange II-E 试剂盒 提供超高保真定点突变,随后即可将突变 • 包含我们的超高保真 PfuUltra DNA
5
定点突变领域的创新技术
Stratagene 意识到定点突变在您研究中的重要 作用。因此我们持续研发功能更强大的新工 具,帮助您在更少的时间里更轻松地开展实 验,成功应对条件最为挑战的应用。
QuikChange Lightning 定点突变试剂盒: 功能强大的新工具 在三小时内即可引入点突变、插入突变 或缺失突变,随后便可进行过夜转化, 同时保持了 QuikChange 试剂盒预期所能 达到的高准确性和高效率(图 2)。由于 采用了我们独有的三步 QuikChange 实验 方案及催化速度更快的酶,扩增和筛选 步骤所需时间得到了缩短。这些新型酶 类仅为我们的 QuikChange Lightning 定 点突变试剂盒所独有。
2
突变用产品选择指南
应用
产品
说明
特点
页码
定点突变
QuikChange Lightning
- 1、下载文档前请自行甄别文档内容的完整性,平台不提供额外的编辑、内容补充、找答案等附加服务。
- 2、"仅部分预览"的文档,不可在线预览部分如存在完整性等问题,可反馈申请退款(可完整预览的文档不适用该条件!)。
- 3、如文档侵犯您的权益,请联系客服反馈,我们会尽快为您处理(人工客服工作时间:9:00-18:30)。
GeneMorph II EZClone Domain Mutagenesis KitINSTRUCTION MANUALCatalog #200552Revision C.01For In Vitro Use Only200552-12L IMITED P RODUCT W ARRANTYThis warranty limits our liability to replacement of this product. No other warranties of any kind,express or implied, including without limitation, implied warranties of merchantability or fitness fora particular purpose, are provided by Agilent. Agilent shall have no liability for any direct, indirect,consequential, or incidental damages arising out of the use, the results of use, or the inability to usethis product.O RDERING I NFORMATION AND T ECHNICAL S ERVICESUnited States and CanadaAgilent TechnologiesStratagene Products Division11011 North Torrey Pines RoadLa Jolla, CA 92037373-6300Telephone(858)424-5444Order Toll Free (800)Technical Services(800) 894-1304Internet techservices@World Wide Web EuropeServices Location Telephone Fax TechnicalAustria 0800 292 499 0800 292 496 0800 292 498Belgium00800 7000 7000 00800 7001 7001 00800 7400 74000800 15775 0800 15740 0800 15720France00800 7000 7000 00800 7001 7001 00800 7400 74000800 919 288 0800 919 287 0800 919 289 Germany00800 7000 7000 00800 7001 7001 00800 7400 74000800 182 **** **** 182 **** **** 182 8234 Netherlands00800 7000 7000 00800 7001 7001 00800 7400 74000800 023 0446 +31 (0)20 312 5700 0800 023 0448 Switzerland00800 7000 7000 00800 7001 7001 00800 7400 74000800 563 080 0800 563 082 0800 563 081 United Kingdom00800 7000 7000 00800 7001 7001 00800 7400 74000800 917 3282 0800 917 3283 0800 917 3281 All Other CountriesPlease contact your local distributor. A complete list of distributors is available at .C ONTENTSMaterials Provided (1)Storage Conditions (1)Additional Materials Required (1)Notice to Purchaser (2)Introduction (3)Eliminate Bias and Easily Control Mutation Rate (5)EZClone Reaction (9)GeneMorph II EZClone Domain Mutagenesis Control (10)XL10-Gold Ultracompetent Cells (10)Preprotocol Considerations (11)Mutant Megaprimer Synthesis Considerations (11)EZClone Reaction Considerations (13)Protocol (14)Mutant Megaprimer Synthesis (14)EZClone Reaction (17)Expected Results (21)Transformation Guidelines (22)Troubleshooting (23)Preparation of Media and Reagents (25)Appendix: How to Calculate Mutation Frequency (26)References (28)MSDS Information (28)Quick-Reference Protocol (29)M ATERIALS P ROVIDEDQuantity Materials provided a ConcentrationUU/μl 25 Mutazyme II DNA polymerase b 2.510× Mutazyme II reaction buffer 10× 150 μl40 mM dNTP mix c10 mM each dNTP 10 μl2× EZClone enzyme mix 2× 250 μlU Dpn I restriction enzyme 10 U/μl 100EZClone solution — 30 μlng Positive control plasmid 10 ng/μl 210ng Positive control primer mix 250 ng/μl 750μl1.1-kb gel standard 20 ng/μl 50XL10-Gold ultracompetent cells d,e (yellow tubes) — 4 × 135 μlXL10-Gold β-mercaptoethanol mix (β-ME) — 50μlng/μl 10μlpUC18 control plasmid (0.1 ng/μl in TE buffer f) 0.1a Sufficient reagents are provided for 10 reactions, which includes 3 control reactions.b Mutazyme II DNA polymerase is not sold separately.c Thaw the dNTP mix once, prepare single-use aliquots, and store the aliquots at –20°C. Do not subject the dNTP mixto multiple freeze-thaw cycles.d Genotype: Tet rΔ (mcrA)183 Δ(mcrCB-hsdSMR-mrr)173 endA1 supE44 thi-1 recA1 gyrA96 relA1 lac Hte[F’ proAB lacI q ZΔM15 Tn10 (Tet r) Amy Cam r]e The XL10-Gold ultracompetent cells must be stored at the bottom of a –80°C freezer immediately on receipt.The ultracompetent cells are very sensitive to small variations in temperature. Transferring tubes from one freezer toanother may result in a loss of efficiency.f The pUC18 control plasmid is stored in TE buffer (see Preparation of Media and Reagents).S TORAGE C ONDITIONSXL10-Gold ultracompetent cells: Store the cells immediately at –80°C.Do not place the cells in liquid nitrogen.All other components: Store at –20°C upon receipt. Store the 2× EZClone enzyme mix at4°C after thawing. Once thawed, full activity is guaranteed for 3 months.A DDITIONAL M ATERIALS R EQUIREDTemperature cyclerPCR tubes llPCR primers14-ml BD Falcon polypropylene round-bottom tubes (BD Biosciences Catalog #352059)ll Thin-walled PCR tubes are highly recommended for use with Stratagene thermal cyclers. These PCR tubes ensure idealcontact with the multiblock design to permit more efficient heat transfer and to maximize temperature cyclingperformance.Revision C.01 © Agilent Technologies, Inc. 2009.N OTICE TO P URCHASERNotice to Purchaser: Limited LicensePurchase of this product includes an immunity from suit under patents specified in the product insert to use only the amount purchased for the purchaser’s own internal research. No other patent rights (such as 5’ Nuclease Process patent rights) are conveyed expressly, by implication, or by estoppel.Further information on purchasing licenses may be obtained by contacting the Director of Licensing, Applied Biosystems, 850 Lincoln Centre Drive, Foster City, California 94404, USA.I NTRODUCTIONRandom mutagenesis is a powerful tool for elucidating protein structure-function relationships and for modifying proteins to improve or alter their characteristics. E rror-prone PCR is a random mutagenesis technique for generating amino acid substitutions in proteins, domains or promoter elements by introducing mutations during PCR. However, cloning amplicons generated by error-prone PCR can be difficult and labor-intensive due to low product yields, mutations at the ends which interfere with restriction-based cloning, and/or inefficient synthesis of 3´ dA overhangs or blunt ends which reduces the efficiency of TA- or blunt-end cloning strategies. In addition, targeting specific functional domains for random mutagenesis can also be difficult and inefficient using current methods. To address the need for efficient and flexible cloning methods, the GeneMorph II E ZClone domain mutagenesis kit* offers an easy and fast cloning method to perform targeted random mutagenesis on protein domains and promoter elements, while delivering a uniform mutational spectrum. Utilizing a unique cloning method, the GeneMorph II E ZClone kit allows you to target specific protein domains or promoter elements without the need for restriction sites or sub-cloning (see Figure 1).1, 2The kit contains Mutazyme II DNA polymerase, which deliberately introduces mutations during PCR with a more uniform mutational spectrum compared to other error-prone PCR enzymes. Additionally, this kit includes an optimized reaction buffer for the error-prone PCR step so that you need only vary the input DNA amount added to the reaction to produce the desired mutational frequency. The resulting purified mutated PCR products serve as megaprimers for the E ZClone reaction** during which they are denatured and annealed to the original donor plasmid and extended with a specialized enzyme mix containing a high fidelity DNA polymerase. Usinga high-fidelity polymerase minimizes unwanted secondary mutations during the cloning process, which can affect downstream results. The E ZClone reaction is temperature cycled several times before being treated with a unique enzyme to remove parental DNA prior to transformation into competent E. coli . Our XL10-Gold ultracompetent cells are included to maximize library size and diversity. Screening libraries created by random mutagenesis allows researchers to identify beneficial mutations in the absence of structural information, or when such mutations are difficult to predict from protein structure.3* U.S. Patent Nos. 6,803,216, 5,489,523, and patent pending.** U.S. Patent Nos. 6,713,285, 6,391,548, 5,789,166 and 5,932,419, and patents pending.变性的退火亲代在之前F IGURE 1 GeneMorph II EZClone domain mutagenesis kit method.变性退火保真度Eliminate Bias and Easily Control Mutation RateThe mutational bias exhibited by error-prone PCR enzymes undoubtedly skews representation of random mutant libraries, diminishing the effective size of the collection produced by error-prone PCR. Mutazyme II DNA polymerase is a novel error-prone PCR enzyme blend, formulated to provide useful mutation rates with minimal mutational bias. Mutazyme II is a blend of two error-prone DNA polymerases—Mutazyme I DNA polymerase (from the GeneMorph Random Mutagenesis Kit) and a novel Taq DNA polymerase mutant that exhibits increased misinsertion and misextension frequencies compared to wild type Taq . For the Mutazyme II polymerase formulation, the Mutazyme I polymerase and the Taq polymerase mutant have been combined to produce a less biased mutational spectrum with equivalent mutation rates at A’s and T’s vs. G’s and C’s. Therefore, libraries created with Mutazyme II exhibit greater mutant representation compared to libraries generated with other enzymes.With the GeneMorph II E ZClone kit, mutation rates of 1–16 mutations per kb can be achieved using the provided buffer, which is optimized for high product yield. The desired mutation rate can be controlled simply by varying the initial amount of target DNA in the reaction or the number of amplification cycles performed.How Mutation Frequency is ControlledMutation frequency is the product of DNA polymerase error rate and number of duplications (see Appendix ). In the GeneMorph II EZClone kit, a sufficiently high error rate is achieved through use of Mutazyme II DNA polymerase. A low, medium or high mutation frequency is produced by adjusting the initial target DNA amounts in the amplification reactions. For the same PCR yield, targets amplified from low amounts of target DNA undergo more duplications than targets amplified from high concentrations of DNA. The more times a target is replicated, the more errors accumulate. Therefore, higher mutation frequencies are achieved simply by lowering input DNA template concentration. Conversely, lower PCR mutation frequencies can be achieved by using higher DNA template concentrations to limit the number of target duplications. Mutation rates can also be decreased by lowering the number of cycles to achieve fewer target duplications. For targets that produce high product yields after 30 cycles, lower mutation rates can be achieved by amplifying lower target amounts for 20–25 cycles. Selecting the Appropriate Mutation FrequencyThe GeneMorph II EZClone kit allows researchers to choose the mutation frequency that is most appropriate for a particular application. For analyzing protein structure-function relationships, the desired mutation frequency is one amino acid change (1–2 nucleotide changes) per gene.4 In directed evolution studies, mutation frequencies of 1–4 amino acid changes (2–7 nucleotide changes) per gene are commonly employed.5–8 Proteins with improved activities have also been isolated from highly mutagenized libraries exhibiting 20 mutations per gene.3无疑地歪斜地减少经历重复重复Achieving the Desired Mutation FrequencyTable I presents the initial amount of target DNA required to produce low, medium, or high mutation frequencies. An initial target amount of 500–1000 ng is recommended to achieve low mutation frequencies of 0–4.5 mutations/kb. Low mutation frequencies can also be achieved by using 100–500 ng of target DNA with a lower number of PCR cycles (see Cycle Number in Preprotocol Considerations). Initial target amounts ranging from 100–500 ng are recommended for producing mutation frequencies of 4.5–9 mutations/kb (medium mutation frequency range). High mutation frequencies (>9 mutations/kb) are obtained by using 0.1–100 ng of input target DNA, where the highest mutation rates can be achieved using the lowest recommended target amounts. Mutation rates up to 16 mutations per kb have been achieved using 0.01 ng of target DNA, although PCR product yields tend to decrease at amounts below 0.1 ng. The predicted mutation frequencies shown in Table I are accurate for amplification reactions producing the indicated approximate fold amplification. The actual number of mutations in individual clones may differ as the values in Table I represent the average mutation frequency for the entire pool of clones.T ABLE IMutation Frequency vs. Initial Target QuantityMutation rate Mutation frequency(mutations/kb)aInitial targetamount (ng)b,cRecommended foldamplificationLow 0–4.5 500–1000 1.5–10 Medium 4.5–9 100–500 10–100High 9–16 0.1–100 100–10,000 a These values are accurate for reactions achieving the approximate fold amplification (total yield/input DNA) indicated. The actual number of mutations in each clone may differ as these values represent the average frequency for all clones.b The amount of template indicated is the amount of target DNA to be amplified, not the total amount of DNA template to add to the reaction. See Initial Amount of Target in Preprotocol Considerations for an example on how to calculate initial target amount.c The recommended DNA target amounts are higher for Mutazyme II compared to Mutazyme I since Mutazyme II exhibits a ~3-fold higher error rate compared to Mutazyme I.Mutational Spectrum of the GeneMorph II KitThe mutational spectra of Mutazyme II DNA polymerase, Mutazyme I DNA polymerase, and Taq DNA polymerase (with Mn 2+-containing buffer and unbalanced dNTP concentrations) are compared in Table II. These error-prone PCR enzymes introduce all possible nucleotide substitutions, however, Mutazyme II DNA polymerase exhibits less mutational bias compared to Mutazyme I and Taq DNA polymerases.There are several ways to assess bias in an enzyme’s mutational spectrum.Bias can be examined by analyzing the ratio of transition (Ts) to transversion (Tv) mutations produced. Transition mutations are purine (A and G) to purine changes and pyrimidine (C and T) to pyrimidine changes, while transversions are purine to pyrimidine and pyrimidine to purine changes. There are eight possible transversions and four possible transitions, and an enzyme completely lacking bias would exhibit a Ts/Tv ratio of 0.5. Secondly, mutational bias has been assessed by calculating the ratio of AT →GC to GC →AT transition mutations (AT →GC/GC →AT ratio), which would equal 1 for a completely unbiased enzyme. Thirdly, mutational bias can be assessed by comparing the frequency of mutating A’s and T’s vs. the frequency of mutating G’s and C’s (AT →NN/GC →NN ratio), which should be equal for an unbiased DNA polymerase.ll The Taq DNA polymerase was used in the PCR with Mn 2+-containing buffer andunbalanced deoxynucleotide concentrations, which are mutagenic conditions for Taq DNA polymerase.转换范围 频谱颠换T ABLE IIMutational Spectra of Mutazyme and Taq DNA PolymerasesType(s) of mutations Mutazyme IIDNA polymerase aMutazyme IDNA polymerase aTaq DNA polymerase(Reference 5) bBias IndicatorsTs/Tv 0.91.20.8 AT→GC/GC→AT 0.6 0.2 1.9A→N, T→N 50.7% 25.6% 75.9%G→N, C→N 43.8% 72.5% 19.6% TransitionsA→G, T→C 17.5% 10.3% 27.6%G→A, C→T 25.5% 43.7% 13.6% TransversionsA→T, T→A 28.5% 11.1%40.9%A→C, T→G 4.7% 4.2% 7.3%G→C, C→G 4.1% 8.8% 1.4%G→T, C→A 14.1% 20.0% 4.5% Insertions and DeletionsInsertions 0.7%0.8%0.3% Deletions 4.8%1.1%4.2% Mutation FrequencyMutations/kb (per PCR)c3–16 (per PCR) <1 to 7 (per PCR) 4.9 (per PCR)a The Mutazyme DNA polymerases were used with the corresponding GeneMorph random mutagenesis kits.b The Taq DNA polymerase was used with Mn2+-containing buffer and unbalanced dNTP concentrations, which are mutagenic conditions for Taq DNA polymerase.c Initial target amounts of 16 pg to 1 μg (Mutazyme II DNA polymerase), 1 pg to 100 ng (Mutazyme I DNA polymerase), and 0.01 nM template (Taq DNA polymerase) were used to generate data.As shown in Table II, error-prone enzymes generally favor transitions overtransversions, as shown by Ts/Tv ratios greater than 0.5, with Mutazyme IIand Taq exhibiting a somewhat higher tendency to create transversions overtransitions and Mutazyme I exhibiting a greater tendency for introducingtransitions over transversions. E xamining transition mutation frequenciesshows that Mutazyme II produces AT→GC and GC→AT mutations withsimilar rates (AT→GC/GC→AT ratio = 0.6), while Mutazyme I is 4 timesmore likely to generate GC→AT transitions over AT→GC transitions, andTaq is 2 times more likely to introduce AT→GC transitions over GC→ATtransitions. In addition, Mutazyme II DNA polymerase introduces mutationsat A’s and T’s only slightly more frequently than G’s and C’s. In contrast,Mutazyme I is nearly 3 times more likely to mutate G’s and C’s, while Taqunder error-prone conditions is 4 times more likely to mutate A’s and T’sthan G’s and C’s.The spectrum of mutations produced by the GeneMorph II EZClone kit is the same at all mutation frequencies. With the GeneMorph II EZClone kit, low, medium, and high mutation frequencies are achieved using a single set of buffer conditions (MgCl 2, balanced dNTPs) optimized for high product yield. The only parameter varied is the initial amount of target DNA in the reaction or the number of cycles employed. In contrast, Taq DNA polymerase–based mutagenesis methods typically employ different sets of reaction conditions to vary mutation levels. Varying the buffer conditions (e.g., different Mn 2+ concentrations) and/or the concentrations of one or more nucleotides to alter mutation frequency can lead to changes in Taq ’s mutational spectrum and increased mutational bias.Furthermore, mutational hotspots have not been observed in any of the mutagenized genes generated by Mutazyme II DNA polymerase that have been sequenced.9EZClone ReactionThe E ZClone reaction utilizes (1) a supercoiled double-stranded DNA (dsDNA) vector containing the same region targeted for mutagenesis in the mutagenesis reaction and (2) two megaprimers generated in the mutagenesis reaction containing random mutations. The E ZClone enzyme mix is a 2× formulation containing a high fidelity DNA polymerase to minimize unwanted second site errors, an optimized PCR reaction buffer, magnesium, and dNTPs. The megaprimers, each complementary to opposite strands of the vector, are extended during temperature cycling by the EZClone enzyme mix, without primer displacement. Extension of the megaprimers generates a mutated plasmid containing staggered nicks. Following temperature cycling, the product is treated with Dpn I restriction enzyme. The Dpn I endonuclease (target sequence: 5´-Gm 6ATC-3´) is specific for methylated and hemimethylated DNA and is used to digest the parental DNA template. Because DNA produced during PCR is not methylated, Dpn I selects for the mutation-containing synthesized DNA.10 The nicked vector DNA incorporating the desired mutations is then transformed into competent cells (provided), where the nicks are repaired by endogenous enzymes in the cell. Note While plasmid DNA isolated from almost all of the commonly usedE. coli strains (dam +) is methylated and is a suitable template formutagenesis, plasmid DNA isolated from the exceptional dam – E. coli strains, including JM110 and SCS110, is not suitable. 超螺旋的突变易发生的大引物变异发生高保真充分利用的镁充足的延伸缺口甲基化半甲基 消化亲本带切口的意外的G ENE M ORPH II EZC LONE D OMAIN M UTAGENESIS C ONTROLTo demonstrate the effectiveness of the GeneMorph II EZClone method the3.0-kb positive control plasmid, which contains the lacZ gene, is used to testthe efficiency of mutant megaprimer synthesis and the E ZClone reaction.XL10-Gold ultracompetent cells* transformed with this control plasmidappear blue on LB–ampicillin agar plates (see Preparation of Media andReagents), containing IPTG and X-gal. Following random mutagenesis andE ZCloning, >25% of the colonies plated appear white on LB–ampicillinplates containing IPTG and X-gal because β-galactosidase activity has beenobliterated.XL10-G OLD U LTRACOMPETENT C ELLSStratagene XL10-Gold ultracompetent cells, a derivative of the highest-efficiency Stratagene competent cell line, XL2-Blue MRF´, possess the Htephenotype, which increases transformation efficiency of ligated DNA.11XL10-Gold cells are both endonuclease deficient (endA1) andrecombination deficient (recA). The endA1 mutation greatly improves thequality of plasmid miniprep DNA,12 and the recA mutation helps ensureinsert stability. In addition, the McrA, McrCB, McrF, Mrr, and HsdRsystems have been removed from XL10-Gold ultracompetent cells. ThemcrA, mcrCB and mrr mutations prevent cleavage of cloned DNA thatcarries cytosine and/or adenine methylation, which is often present ineukaryotic DNA and cDNA.13–15 The McrA and McrCB systems recognizeand restrict methylated cytosine DNA sequences, and the Mrr systemrecognizes and restricts methylated adenine DNA sequences. The Mrrsystem also restricts methylated cytosine DNA sequences with a specificitydiffering from that of McrA and McrCB. This activity has been namedMcrF. This McrF activity against methylated cytosines has been shown tobe equal to or greater than the restriction activity of the McrA and McrCBsystems.16 The hsdR mutation prevents the cleavage of cloned DNA by theEco K (hsdR) endonuclease system. XL10-Gold cells grow faster than XL1or XL2-Blue cells, resulting in larger colonies. To permit blue-white colorscreening, the XL10-Gold ultracompetent cells contain the lacI q ZΔM15 geneon the F´ episome.Host strain References GenotypeXL10-Gold ultracompetent cells 11, 17, 18 Tet RΔ(mcrA)183 Δ(mcrCB-hsdSMR-mrr)173endA1 supE44 thi-1 recA1 gyrA96 relA1 lac Hte[F´ proAB lacI q ZΔM15 Tn10 (Tet R) Amy Cam R]It is important to store the XL10-Gold ultracompetent cells at –80°C to prevent a loss of efficiency. For best results, please follow the directions outlined in the following sections.* U.S. Patent Nos. 6,706,525, 5,512,468 and 5,707,841 and patents pending and equivalentforeign patentsP REPROTOCOL C ONSIDERATIONSMutant Megaprimer Synthesis ConsiderationsAmplification TargetsThe GeneMorph II EZClone kit has been used to mutagenize targets up to3.5 kb in length from plasmid DNA.Genomic DNA templates are not generally recommended for error-pronePCR as researchers are limited to medium-to-high mutation levels due to thelow copy number of genomic DNA targets. If genomic DNA is the onlysource of the target gene, we recommend amplifying the target with ahigh-fidelity DNA polymerase, such as PfuUltra high-fidelity DNApolymerase, followed by re-amplification of the diluted PCR product withMutazyme II DNA polymerase.Initial Amount of Target DNAThe mutation frequency depends upon the initial amount of target DNAemployed in the reaction. The amount of target to add to a reaction can bedetermined using Table I.The initial amount of target DNA required to achieve a particularmutation frequency refers to the amount of target DNA to amplify, notthe total amount of plasmid DNA template to add to the reaction. As anexample, to mutagenize a 1.0-kb target gene at a low mutation frequency, aninitial target amount of 500 ng is recommended. For a 1.0-kb target genethat is an insert in a 3.0-kb plasmid (the total construct is 4.0 kb), 2 μg of theplasmid construct should be added to the reaction to provide 500 ng of targetDNA.Cycle NumberIn addition to using higher target DNA amounts, mutation rates can also be lowered by decreasing the number of cycles employed to achieve fewertarget duplications. For targets that produce high product yields after30 cycles, lower mutation rates can be achieved by amplifying lower target amounts for 20 to 25 cycles (see Table III).T ABLE IIIAchieving Low Mutation Frequency Using Fewer Cycle Numbers Mutation frequency(mutations/kb) a Cycle Number Initial target amount b0–4.5 (low range)ng20–25 100ngng–100030 500a These values are accurate for reactions achieving the approximate 1.5–10 fold amplification (total yield/input DNA). The actual number of mutations in each clonemay differ as these values represent the average frequency for all clones.b The amount of template indicated is the amount of target DNA to be amplified, not thetotal amount of DNA template to add to the reaction. See Initial Amount of Target in Preprotocol Considerations for an example on how to calculate initial target amount. Primer DesignStandard PCR primers flanking the region targeted for mutagenesis are usedin the mutant megaprimer synthesis reaction. For best results, PCR primers should be designed with similar melting temperatures ranging from 55 to72°C. The use of primers with melting temperatures within this rangereduces false priming and ensures complete denaturation of unextended primers at 94–95°C.PCR Product YieldThe PCR product yield should be within the recommended range to obtainthe predicted mutation frequencies listed in Tables I and III. To ensuresufficient product yield, sample PCR reactions are electrophoresed adjacentto a DNA standard provided in the kit. PCR product yields are quantified by被量化comparing the staining intensity of PCR product bands to the DNAstandard.Achieving High Mutation FrequenciesThe highest mutation frequency that can be achieved in one round of PCR islimited by the minimum amount of target DNA that can be amplified in high product yield. In the GeneMorph II kit, we recommend using 0.1–100 ng of target DNA, which is sufficient to produce high product yields after30 cycles and mutation frequencies up to 9–16 mutations per kb of target. Higher mutation frequencies can be achieved by amplifying from<0.1ng target DNA, although product yields may be noticeably lower.Alternatively, mutation frequencies > 20 mutations per kb can be achievedby performing sequential PCRs, in which a small aliquot of the first PCRreaction is re-amplified in a second PCR reaction.EZClone Reaction ConsiderationsRequired Host StrainE nsure that the plasmid DNA template is isolated from a dam+ E. colistrain. The majority of the commonly used E. coli strains are dam+.Plasmid DNA isolated from dam– strains (e.g. JM110 and SCS110) is notsuitable.Plasmid DNA Template GuidelinesThe plasmid DNA template used in the EZClone reaction may be the sameor different than the original plasmid DNA used as template in the mutantmegaprimer synthesis reaction, provided that the region targeted in themutagenesis reaction is present in the plasmid.The plasmid DNA template used in the E ZClone reaction should be lessthan 10 kb in length to ensure full extension during temperature cycling.P ROTOCOLMutant Megaprimer SynthesisNote Gently mix and centrifuge each component before use. Prepare allreactions on ice.1. Refer to Table I to determine the initial amount of target to use in eachreaction.Note Target DNA refers to the DNA sequence to be amplified,not the total amount of plasmid DNA in the reaction(see Initial Amount of Target in PreprotocolConsiderations).2. Prepare the control reaction as indicated below:5 μl of 10× Mutazyme II reaction buffer2 μl of 0.5 ng/μl positive control plasmid (dilute the controlprovided 1:20 in high-quality water for a final concentrationof 0.5 ng/μl)1 μl of positive control primer mix (125 ng/μl of each primer)1 μl of 40 mM dNTP mix (200 μM each final)40 μl of ddH2O1 μl of Mutazyme II DNA polymerase (2.5 U/μl)3. Prepare the sample reaction(s) as indicated below:5 μl of 10× Mutazyme II reaction bufferx μl template (see Table I for recommended amount)1 μl of sample primers (125 ng/μl of each primer)1 μl of 40 mM dNTP mix (200 μM each final)x μl of ddH2O for a final reaction volume of 50 μl1 μl of Mutazyme II DNA polymerase (2.5 U/μl)4. Centrifuge each reaction briefly.5. If the thermal cycler does not have a heated lid, overlay each reactionwith a few drops of mineral oil.。
