Chapter 2 Nucleotide(2009)
核酸的结构与功能

目录
1979年 Rich提出了左手螺旋的Z DNA结构模型。 1979年,Rich提出了左手螺旋的Z-DNA结构模型。 提出了左手螺旋的 结构模型 左手螺旋Z-DNA只是右手螺旋结构模型的一个 左手螺旋Z DNA只是右手螺旋结构模型的一个 补充发展。 补充发展。 Z-DNA可能也是天然DNA存在的二级结构形式,其 DNA可能也是天然DNA存在的二级结构形式, 可能也是天然DNA存在的二级结构形式 功能可能与基因突变、基因复制和表达调控有关。 功能可能与基因突变、基因复制和表达调控有关。 可能与基因突变
目录Biblioteka 四、核苷酸(nucleotide) 核苷酸(nucleotide)
核苷酸是核苷在戊糖羟基上的磷酸酯。 核苷酸是核苷在戊糖羟基上的磷酸酯。 是核苷在戊糖羟基上的磷酸酯 核糖核苷酸: 核糖核苷酸: AMP、GMP、CMP、UMP 、 、 、 脱氧核糖核苷酸: 脱氧核糖核苷酸: dAMP、dGMP、dCMP、dTMP 、 、 、
目录
有的核苷一磷酸能形成3ˊ,5ˊ-磷酸二酯键,构成环 , 有的核苷一磷酸能形成 -磷酸二酯键, 化核苷酸, 化核苷酸,如3ˊ,5ˊ-环腺苷酸(cyclic ademosine , -环腺苷酸( monophophate, cAMP)及3ˊ,5ˊ-环鸟苷酸 ) , - ( cyclic guanosine monophosphate,cGMP),是一些激 ) 素等信号分子的第二信使,在信号转导中起重要作用。 素等信号分子的第二信使,在信号转导中起重要作用。
目录
5ˊ-核苷酸还可在5ˊ的磷酸基上再连上一或
两个磷酸基形成核苷二磷酸或核苷三磷酸。 两个磷酸基形成核苷二磷酸或核苷三磷酸。
目录
多种核苷三磷酸,特别是腺苷三磷酸( ),在 多种核苷三磷酸,特别是腺苷三磷酸(ATP),在 腺苷三磷酸 ), 细胞的能量代谢中起重要作用, 细胞的能量代谢中起重要作用,ATP属于高能磷酸 属于高能磷酸 化合物,当它被水解为腺苷二磷酸时, 化合物,当它被水解为腺苷二磷酸时,能释放较多 的自由能,后者可被机体直接利用。 的自由能,后者可被机体直接利用。
labinvest2009126a
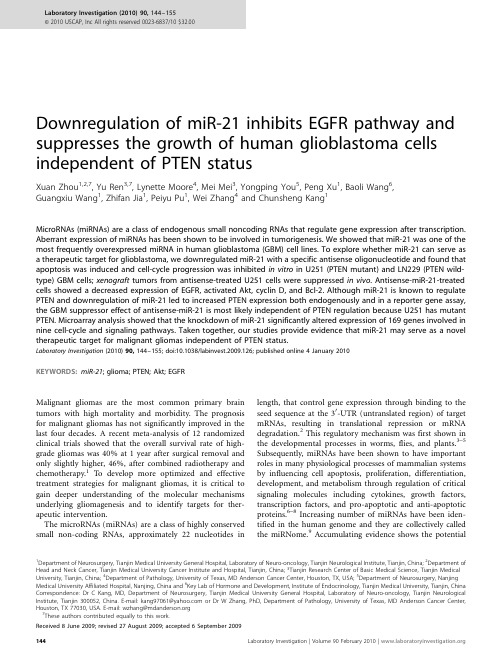
Downregulation of miR-21inhibits EGFR pathway and suppresses the growth of human glioblastoma cells independent of PTEN statusXuan Zhou 1,2,7,Yu Ren 3,7,Lynette Moore 4,Mei Mei 3,Yongping You 5,Peng Xu 1,Baoli Wang 6,Guangxiu Wang 1,Zhifan Jia 1,Peiyu Pu 1,Wei Zhang 4and Chunsheng Kang 1MicroRNAs (miRNAs)are a class of endogenous small noncoding RNAs that regulate gene expression after transcription.Aberrant expression of miRNAs has been shown to be involved in tumorigenesis.We showed that miR-21was one of the most frequently overexpressed miRNA in human glioblastoma (GBM)cell lines.To explore whether miR-21can serve as a therapeutic target for glioblastoma,we downregulated miR-21with a specific antisense oligonucleotide and found that apoptosis was induced and cell-cycle progression was inhibited in vitro in U251(PTEN mutant)and LN229(PTEN wild-type)GBM cells;xenograft tumors from antisense-treated U251cells were suppressed in vivo .Antisense-miR-21-treated cells showed a decreased expression of EGFR,activated Akt,cyclin D,and Bcl-2.Although miR-21is known to regulate PTEN and downregulation of miR-21led to increased PTEN expression both endogenously and in a reporter gene assay,the GBM suppressor effect of antisense-miR-21is most likely independent of PTEN regulation because U251has mutant PTEN.Microarray analysis showed that the knockdown of miR-21significantly altered expression of 169genes involved in nine cell-cycle and signaling pathways.Taken together,our studies provide evidence that miR-21may serve as a novel therapeutic target for malignant gliomas independent of PTEN status.Laboratory Investigation (2010)90,144–155;doi:10.1038/labinvest.2009.126;published online 4January 2010KEYWORDS:miR-21;glioma;PTEN;Akt;EGFRMalignant gliomas are the most common primary brain tumors with high mortality and morbidity.The prognosis for malignant gliomas has not significantly improved in the last four decades.A recent meta-analysis of 12randomized clinical trials showed that the overall survival rate of high-grade gliomas was 40%at 1year after surgical removal and only slightly higher,46%,after combined radiotherapy and chemotherapy.1To develop more optimized and effective treatment strategies for malignant gliomas,it is critical to gain deeper understanding of the molecular mechanisms underlying gliomagenesis and to identify targets for ther-apeutic intervention.The microRNAs (miRNAs)are a class of highly conserved small non-coding RNAs,approximately 22nucleotides inlength,that control gene expression through binding to the seed sequence at the 30-UTR (untranslated region)of target mRNAs,resulting in translational repression or mRNA degradation.2This regulatory mechanism was first shown in the developmental processes in worms,flies,and plants.3–5Subsequently,miRNAs have been shown to have important roles in many physiological processes of mammalian systems by influencing cell apoptosis,proliferation,differentiation,development,and metabolism through regulation of critical signaling molecules including cytokines,growth factors,transcription factors,and pro-apoptotic and anti-apoptotic proteins.6–8Increasing number of miRNAs have been iden-tified in the human genome and they are collectively called the miRNome.9Accumulating evidence shows the potentialReceived 8June 2009;revised 27August 2009;accepted 6September 20091Department of Neurosurgery,Tianjin Medical University General Hospital,Laboratory of Neuro-oncology,Tianjin Neurological Institute,Tianjin,China;2Department of Head and Neck Cancer,Tianjin Medical University Cancer Institute and Hospital,Tianjin,China;3Tianjin Research Center of Basic Medical Science,Tianjin Medical University,Tianjin,China;4Department of Pathology,University of Texas,MD Anderson Cancer Center,Houston,TX,USA;5Department of Neurosurgery,Nanjing Medical University Affiliated Hospital,Nanjing,China and 6Key Lab of Hormone and Development,Institute of Endocrinology,Tianjin Medical University,Tianjin,China Correspondence:Dr C Kang,MD,Department of Neurosurgery,Tianjin Medical University General Hospital,Laboratory of Neuro-oncology,Tianjin Neurological Institute,Tianjin 300052,China.E-mail:kang97061@ or Dr W Zhang,PhD,Department of Pathology,University of Texas,MD Anderson Cancer Center,Houston,TX 77030,USA.E-mail:wzhang@ 7These authors contributed equally to this work.Laboratory Investigation (2010)90,144–155&2010USCAP,Inc All rights reserved 0023-6837/10$32.00involvement of altered regulation of miRNAs in initiation and progression in a wide range of human cancers.Altered expression profiles of miRNAs are associated with genetic and epigenetic alterations including deletion,amplifica-tion,point mutation,and aberrant DNA methylation.10 The miRNA cluster miR-15a–16,including miR-15a and miR-16–1,is located near13q14,a region that is character-ized by a high frequency of deletion in chronic lymphocytic lymphoma.11The expression of miR-15a and miR-16–1were inversely correlated with the anti-apoptotic Bcl-2gene expression in chronic lymphocytic leukemia and both miR-NAs negatively regulated Bcl-2at the post-transcriptional level,resulting in the potentiation of the normal apoptotic response.12In a mouse model of lymphoma,increased expression of the miR-17–92cluster strongly accelerated lymphomagenesis and was the first functional evidence of an miRNA acting as a mammalian oncogene.13Another example of oncogenic miRNAs is miR-29.Enforced miR-29reduced the expression level of Mcl-1,an anti-apoptotic factor, to facilitate oncogenesis of cholangiocarcinoma.14Some miRNAs function as tumor suppressor genes.Let-7was reported to negatively regulate Ras and c-Myc expression after the let-7a-1precursor construct was transfected into human colon tumor cell lines and expression of miR-34a induced apoptosis in neuroblastoma cells.15–17Recent reports suggested that miR-21functions as an oncogene in human cancers.Ciafre`et al18profiled the expression of245miRNAs in10glioblastoma(GBM)cell lines and nine freshly resected GBM samples and observed that miR-21was overexpressed in human brain tumors.18 It was shown that when miR-21was suppressed,cell growth inhibition and caspase-dependent apoptosis were observed in A172,U87,LN229,and LN308cells.19Multiple target genes of miR-21have been reported.It has been shown that miR-21 modulates breast cancer cell anchorage-independent growth through suppressing TMP1expression.20In human color-ectal,breast cancer,and renal cell carcinoma,miR-21con-tributes to invasion and metastasis cell by inhibiting Pdcd4 mRNA at the post-transcription level.21–23A recent study showed that miR-21targets PTEN gene through a binding site on the30-UTR in hepatocellular carcinoma.24PTEN has been shown to be a critical tumor suppressor gene that is commonly inactivated in GBM by deletion,mutation, or attenuated expression.25Thus,increased expression of miR-21may contribute to the attenuated expression of PTEN in GBM.To further characterize the potential of miR-21as a target for treating GBM and to clarify the role of PTEN in the response,we used both in vitro and in vivo systems to study the effect of miR-21suppression in the PTEN wild-type (LN229)and PTEN mutant(U251)GBM cell lines.26 We found for the first time that downregulation of miR-21 inhibited the EGFR and Akt pathways and the anti-GBM effect was independent of PTEN status,suggesting that miR-21is a broader therapeutic target for GBM.MATERIALS AND METHODSCell Culture ConditionsThe human U251and LN229glioblastoma cell lines were purchased from the Institute of Biochemistry and Cell Biology,Chinese Academy of Science.Human glioblastoma cell lines,TJ861,TJ905,and TJ899,were established and characterized in Laboratory of Neuro-oncology,Tianjin Neurological Institute.27Human glioblastoma cell line,A172, and astrocytoma cell line,H4,were provided by Professor Jinhuan Wang(Tianjin First Central Hospital,China).All cell lines were maintained in Dulbecco’s modified Eagle’s medium(DMEM)(Invitrogen,Carlsbad,CA,USA)supple-mented with10%fetal bovine serum(Invitrogen),2mM glutamine(Sigma,St Louis,MO,USA),100units of peni-cillin/ml(Sigma),and100m g of streptomycin/ml(Sigma), at371C with5%CO2.Oligonucleotides and Cell TransfectionThe20-O-methyl(20-OMe-)oligonucleotides were chemically synthesized by SBS Genetech(Beijing,China).The20-O-Methyl oligos were composed entirely of20-O-methyl bases with the following sequences:scramble sequence50-AAGGC AAGCUGACCCUGAAGU-30and20-O-Me-miR-2150-GUCA ACAUCAGUCUGAUAAGCUA-30.19Oligonucleotides(50nm/l) were transfected into U251cells at70%confluence using Oligofectamine according to the manufacturer’s instructions (Invitrogen).MicroRNA Array and Reverse Transcription(RT)-Real-Time PCRTotal RNA was extracted from GBM cells(U251,TJ866, TJ905,TJ899,and A172)and human astrocytoma cell line H4using Trizol reagent(Invitrogen)for miRNA profile examination.Two separate total RNA samples of anti-sense-miR-21-treated U251cell and control U251cell were prepared for mRNA expression profiling.The miRNA microarray was obtained from CapitalBio Corporation (Beijing,China).The individual oligonucleotide probes were printed in triplicate on chemically modified glass slides in a21Â21 spot configuration of each subarray for miRNA microarray. All of the oligonucleotide probes were printed in triplicate on one microarray,and each of the four subarrays contained16 controls(Zip5,Zip13,Zip15,Zip21,Zip23,Zip25,Y2,Y3, U6,New-U2-R,tRNA-R,hsa-let-7a,hsa-let-7b,hsa-let-7c, 50%dimethyl sulphoxide(DMSO),and Hex).Total RNA(FirstChoice Total RNA,Ambion,Austin,TX, USA)from normal brain was used as a control.The miRNAs were enriched from total RNA using a mirVana miRNA Isolation Kit(Ambion)and labeled with mirVana Array La-beling Kit(Ambion).Labeled miRNAs were used for hy-bridization on each miRNA microarray containing509 probes in triplicate at421C overnight.A double-channel laser scanner(LuxScan10K/A,CapitalBio)was used to scan the arrays and quantified using image analysis software Downregulation of miR-21suppresses glioblastomaX Zhou et al(LuxScan3.0,CapitalBio).Raw data were normalized to the result of normal brain RNA and analyzed using the sig-nificance analysis of microarrays(SAM,version2.1,http:// /B tibs/SAM,Stanford University,CA, USA)software.For transfected cells,reverse transcription(RT)reaction was conducted with the mirVana t qRT-PCR miRNA detec-tion kit(Ambion).The real-time PCR was carried out with the mirVana qRT-PCR miRNA detection kit(Ambion). Amplification reaction was performed using MJ-real-time PCR(Bio-Rad,Hercules,CA,USA)and the protocol was performed for40cycles at951C for3min,951C for15s,and 601C for30s.Both RT and PCR primer were purchased from Ambion.5S RNA was used for normalization.Relative quantification was conducted using amplification efficiencies derived from cDNA standard curves and obtained relative gene expression.Data were shown as fold change(2ÀDD Ct) and analyzed initially using Opticon Monitor Analysis Software V2.02software(MJ Research,Waltham,MA,USA). Microarray AnalysisTwo separate total RNA samples were extracted from control and miR-21knocked down U251cells using Trizol reagent (Invitrogen).Samples were then sent to Beijing CapitalBio Corporation for processing.A total of500ng of total RNA was processed for use on the microarray by using the Affymetrix GeneChip30IVT Express Kit(Affymetrix,Santa Clara,CA, USA)according to the manufacturer’s recommended proto-cols.In brief,a first-stranded cDNA was synthesized from the total RNA sample using Superscript II Reverse Transcriptase (Invitrogen)and an oligo d(T)primer was linked with a T7 RNA polymerase binding site sequence.The amplified,labeled cRNA was produced using T7RNA polymerase and biotiny-lated nucleotides.After removal of free nucleotides,cRNA yield was measured by UV260absorbance with Nanodrop spectrophotometer(Gene,Carlsbad,CA,USA).The labeled cRNA was incubate with the5Âarray fragmentation buffer at 941C for35min for fragmentation reaction and combined with hybridization internal positive control(ACTB,GAPD, LDHA,and RPS9)and positive control(50-Hex–GTCAC ATGCGATGGATCGAGCTCCTTTATCATCGTTCCCACCTTA ATGCA-30)for DNA immobilization.After the signal am-plified according to the standard protocols,arrays were scanned with the Affymetrix Model3000scanner and data were analyzed using GSEABase software(http://www. /).To identify differentially expressed genes,sig-nal values were transformed to logarithm base2and z-scores were calculated.A significant P-value was assigned to each z-score by z-test.Genes with P-value of r0.01were considered differentially expressed.Statistical analysis was conducted using MATLAB7.0(The MathWorks,Inc.).Gene Ontology and KEGG Pathway AnalysisGenes with P-value of o0.01were selected as differentially expressed genes and used for further analysis.Expression Analysis Systematic Explorer(EASE)was used to analyze gene ontology and KEGG pathways.28Overrepresentation of genes are present if a larger fraction of genes within that pathway is differentially expressed compared with all genes in the genome.An EASE score of r0.05was used as a cutoff. MiR-21Detection by In Situ HybridizationUsing antisense locked nucleic acid(LNA)-modified oligo-nucleotides probe,in situ hybridization was performed with In situ hybridization kit(Boster,Wuhan,China).LNA/DNA oligos contained locked nucleic acids at eight consecutive centrally located bases(indicated by the underline)and had the following sequences:LNA-miR-2150-TCAACATCAGT CTGATAAGCTA-30.At72h after transfection,U251cells were fixed with freshly prepared4%paraformaldehyde (containing0.1%DEPC).The in situ hybridization detection of miR-21in U251GBM cells were conducted according to the protocol of the manufactures.The fixed U251cells were incubated with20m l LNA-miR-21hybridization solution at 421C for16h,and Cy3-avidin was used to label miR-21at a concentration of0.5mg/ml.Nuclei were counterstained with DAPI karyotyping kit(Genmed,Boston,MA,USA)and visualized using FluoView Confocal Laser Scanning Micro-scopes-FV1000(Olympus,Tokyo,Japan)and analyzed using IPP5.1(Olympus).Cell Proliferation AssayU251and LN229cells were seeded into96-well plates at4000 cells per well.After transfection as described previously,20m l of MTT(5g/l)was added into each well at each day of consecutive6days after treatment and the cells were incubated for additional4h,and the supernatant was then discarded.Finally,200m l of DMSO was added to each well to dissolve the precipitate.Optical density(OD)was measured at the wavelength of570nm.The data are presented as the mean±s.d.,which are derived from triplicate samples of at least three independent experiments.Cell-Cycle AnalysisFor cell-cycle analysis using FCM(flow cytometry),trans-fected and control cells in the log phase of growth were harvested,washed with PBS,fixed with90%ethanol over-night at41C,and then incubated with RNase at371C for 30min.Nuclei of cells were stained with propidium iodide for additional30min.A total of104nuclei were examined in a FACS Calibur flow-cytometer(Becton Dickinson,Franklin Lakes,NJ,USA).Samples were analyzed using flow cytometry for FL-2area and DNA histograms were analyzed using Modifit software.Experiments were performed in triplicate. Results are presented as percentage of cell in phase. Apoptosis Assays Using Annexin Stainingand TUNEL MethodParental and transfected cells in the log phase of growth were harvested and collected by centrifugation for5min at500gDownregulation of miR-21suppresses glioblastoma X Zhou et aland cells were then resuspended at a density of1Â106cells/ml. For the Annexin V assay,the annexin V-Cy3-labeled Apoptosis Detection Kit(Abcam,Cambridge,MA,USA)was used.The apoptotic cells were detected and quantified using FACSCalibur(Becton Dickinson,San Jose,CA,USA).The data obtained were analyzed using CellQuest software.The apoptotic cell death in the tumor specimens of mouse models from the in vivo study was examined by TUNEL method using an in situ cell death kit(Roche,Indianapolis,IN, USA).For detecting miRNA-21expression and apoptotic cells simultaneously,in situ hybridization was performed as de-scribed above,and then apoptosis was detected using TUNEL method.Positive cells were visualized using fluorescence mi-croscopy.The reaction mixture was incubated without enzyme in a control coverslip to detect nonspecific staining.Nuclei were counterstained with DAPI karyotyping kit(Genmed)and visualized using FV-1000laser scanning confocal biological microscopes and analyzed using IPP5.1(Olympus). Western Blot AnalysisParental and transfected cells were thrice washed with pre-chilled phosphate-buffered saline(PBS).The cells were then solubilized in1%Nonidet P-40lysis buffer(20mM Tris,pH 8.0,137mM NaCl,1%Nonidet P-40,10%glycerol,1mM CaCl2,1mM MgCl2,1mM phenylmethylsulfonyl fluoride, 1mM sodium fluoride,1mM sodium orthovanadate,and a protease inhibitor mixture).A total of40m g lysates were subjected to SDS-PAGE on8%SDS-acrylamide gel.Separate proteins were transferred to PVDF membranes(Millipore, Bedford,MA,USA)and incubated with primary antibodies against Akt-2(Santa Cruz,Santa Cruz,CA,USA),29pAKT (for Ser473,Santa Cruz),PTEN,EGFR,STAT3,and Bcl-2 (Zhongshan Bio Corp.,Beijing,China)followed by incu-bation with an HRP-conjugated secondary antibody(Zymed, San Francisco,CA,USA).The specific protein was detected using a SuperSignal protein detection kit(Pierce,Rockford, IL,USA).The membrane was stripped and re-probed with a primary antibody against b-actin(Santa Cruz).DNA Constructs and Luciferase AssayLuciferase activity assay-related constructs were made by ligating PTEN30-UTR fragments(approximately500bp) containing the predicted binding sites into luciferase reporter vector(pGL3).PCR was performed to amplify a fragment containing the miR-21target sequence using human DNA as the template. The primers used for PTEN30-UTR were50-ACTCTAGA GTCGACACCACTGACTCTGATC-30and50-ACTCTAGACA TGACACAGCTACACAACC-30.The product was digested using Xba I enzyme and afterward ligated with Xba I-treated pGL3-control vector containing the SV40promoter.The ligated product was transformed into E.coli JM109and colony PCRs were used to screen for the clones harboring the forwardly oriented insert using the antisense primer of PTEN and a sense primer of the vector(50-AGGAGTTGTGTT TGTGGACG-30).The desired construct was subsequently sequenced by Invitrogen.For luciferase reporter experiments,the pGL3-PTEN-3-UTR construct,which contains the putative binding site for miR-21of the3-UTR of the PTEN,was amplified by PCR from human genomic DNA and inserted into the pGL3 control vector(Promega,Madison,WI,USA),using the Xba I site immediately downstream from the stop codon of luci-ferase in the reporter gene vector.U251,A172,and H4cells were plated(2Â106cells per well)in six-well plates.After transfection,the cells were split into96-well plates in duplicate and harvested for luciferase assays24h later using a luciferase assay kit(Promega).U251Xenograft Tumor AssayFemale immune-deficient nude mice(BALB/C-nu),5weeks old,were purchased from the animal center of the Cancer Institute of Chinese Academy of Medical Science,and were bred at Compare Medicine Center,Tianjin Medical University.All experimental procedures were performed according to Tianjin Medical University policies.In all,four mice were injected subcutaneously with1Â106 of U251GBM cells,in a volume of50m l of PBS pre-mixed with equal volume of matrigel matrix(Becton Dickinson). Mice were monitored daily and three out of four mice formed tumors subcutaneously.When the tumor size reached approximately5mm in length,the tumors were surgically removed,cut into pieces of1–2mm3and re-seeded into the left inguinal region of30mice.When the diameter of the subcutaneous tumor reached7mm the mice were divided into three groups(10mice per group)randomly:U251 control group,U251scramble oligonucleotide(ODN)-trea-ted group,and U251As-miR-21-treated group.A mixture of20m l oligofectamine and ODN(50nmol/l)mixture was injected into the xenograft tumor model in a multi-site injection manner.Mice in the U251control group received 10m l of PBS only.A second administration was conducted on day3.The tumor volume was measured with a caliper every3days,using the formula volume¼lengthÂwidth2/2. At the end of the22-day observation period,the mice bearing xenograft tumors were killed and the tumor tissues were removed for formalin fixation and preparation of paraffin-embedded sections.Immunofluorescence and Immunohistochemistry StainingThe paraffin-embedded tissue sections were used for examination of PTEN,EGFR,p-AKT,AKT-2,Ki67,cyclin D1,Bcl-2expression,and HE staining.For immuno-fluorescence staining,transfected cells were seeded on cov-erslips and fixed with4%paraformaldehyde(PFA,Sigma), treated with3%H2O2for10min and incubated with the antibodies described above overnight at41C.FITC-or TRITC-labeled secondary antibody(1:200dilutions)was added for2h at371C.DAPI reagent was used to stain the Downregulation of miR-21suppresses glioblastomaX Zhou et alU251cell nuclei and the cells was visualized using FV-1000 laser scanning confocal microscopes and analyzed using IPP5.1(Olympus).For immunohistochemistry study,sec-tions were incubated with primary antibodies(1:200dilu-tions)overnight at41C,followed by biotin-labeled secondary antibody(1:100dilutions)for1h at room temperature. Sections were then incubated with ABC-peroxidase and DAB (diaminobenzedine),counterstained with hematoxylin,and visualized using light microscope.Statistical AnalysisData were expressed as means±s.e.Statistics was determined using ANOVA,w2test,or Student’s t-test using SPSS11.0 (Windows).Statistical significance was determined as P o0.05(*)or P o0.01(**).RESULTSMiR-21was Overexpressed in Human Glioblastoma Cell LinesWe first profiled miRNA expression in five glioblastoma cell lines,one astrocytoma cell line,and one normal brain tissue. Our analysis showed that8of the435human miRNAs (1.84%)were overexpressed with a greater than twofold increase and18of453(3.68%)showed a greater than twofold reduction in all glioma cell lines(Figure1a and b).Among the increased miRNAs,miR-21showed the most significant increase relative to normal brain tissue(7.0-fold).Other increased miRNAs include miR-221,miR-222,and miR-23a. The miR-21expression results from microarrays were con-firmed with quantitative RT-PCR assays(Figure1c).MiR-21 was chosen for further study because it was the most pro-minently overexpressed miRNA in glioma.Antisense-miR-21Suppressed U251Glioma Cell Proliferation and Induced ApoptosisTo evaluate the significance of miR-21overexpression in glioma cells,we used a loss-of-function antisense approach. An As-miR-21oligonucleotide(ODN)was used to knock down miR-21expression in U251and LN229cells.RT-real-time PCR results determined that the relative expression level of miR-21in As-miR-21ODN-treated U251cell was6.25% (P o0.01)and12.5%for LN229cells(P o0.01)compared with their control cells,respectively(Figure2a).In addition, LNA-based in situ hybridization showed that transfection of a scrambled ODN had no effect on miR-21expression. In contrast,the cy3red fluorescence signal in As-miR-21-transfected U251cells was lower(Figure2b).Thesedata TJ899TJ95U251A172H4hsa-miR-128ahsa-miR-128bhsa-miR-132hsa-miR-125ahsa-miR-323hsa-miR-219hsa-miR-1hsa-miR-223hsa-miR-126hsa-miR-329hsa-miR-495hsa-miR-330hsa-miR-137hsa-miR-95hsa-miR-497hsa-miR-127hsa-miR-368hsa-miR-335hsa-miR-338hsa-miR-451hsa-miR-342hsa-miR-124ahsa-miR-451hsa-miR-124hsa-miR-495hsa-miR-223hsa-miR-329hsa-miR-126hsa-miR-219hsa-miR-1hsa-miR-330hsa-miR-342hsa-miR-323hsa-miR-127hsa-miR-128hsa-miR-128hsa-miR-132hsa-miR-132hsa-miR-95hsa-miR-23bhsa-miR-137hsa-miR-23ahsa-miR-222hsa-miR-221hsa-miR-106hsa-miR-15bhsa-miR-21RelativeFoldTJ899TJ905TJ866A172H4NormalU2511.21.00.80.60.40.2Log2 (Expression in Glioma and Glioblastoma cell line/Ambion normal brain)-10-8-6-4-202468hsa-miR-108ahsa-miR-130ahsa-miR-17-5phsa-miR-22hsa-let-7ahsa-miR-106bhsa-miR-16hsa-miR-24hsa-miR-27ahsa-miR-221hsa-miR-222hsa-miR-15bhsa-miR-23bhsa-miR-21hsa-miR-23aFigure1miR-21is overexpressed in glioma cells.(a)Cluster analysis of miRNA expression profile across human glioblastoma cell lines.Glioma cell lines were presented in columns,miRNAs in rows.(b)The26most upregulated(right)and downregulated(left)miRNAs in six human glioma and globlastoma cell lines with respect to Ambion normal brain total RNA.The relative expression of miRNAs level is present in log2transformed for each glioblastoma cell line.(c)RT-real-time PCR shows miR21overexpression in glioma cell lines.U6snRNA was used as a loading control.Downregulation of miR-21suppresses glioblastomaX Zhou et alsuggested that As-miR-21can specifically inhibit the endogenous miR-21expression in U251and LN229cells. Cell viability was measured in ODN-transfected cells in up to4days after treatment.As-miR-21ODN-treated cells showed a significant decrease in viability compared with control ODN-treated cells or untreated cells(Figure2c).We found that the growth-inihibitory effect of decreased miR-21 reached maximum at3days after transfection.The lowest survival rate was57±16%for U251cell and49±9%for LN229cell.****Control200160120804001282563845122001601208040012825638451220016012080400128256384512200160120804001282563845122001601208040012825638451220016012080400128256384512U251LN229DNA content1201008060402012010080604020G/G1S G2/MControl Scramble As-miR-21G/G1S G2/MControl Scramble As-miR-21CellCycleDistribution(%)U251Control Scramble As-miR-212.62%3.91%252015105********U251LN229Control Scramble As-miR-21ApoptosisPercentage(%)252015105Control Scramble As-miR-21ApoptosisPercentage(%)5.30%FL1-H18.74%3.59%11.88%104103102101100104103102101100104103102101100104103102101100104103102101100104103102101100104103102101100104103102101100104103102101100104103102101100104103102101100104103102101100FL2-HFL2-HLN229CellNumberCellNumberScramble As-miR-21Control Scramble As-miR-2110080604020Day After T ransfection01234LN229CellSurvivalRate(%)10080604020Day After Transfection01234U251CellSurvivalRate(%)Control Scramble As-miR-21U251LN229Control Scramble As-miR-21miR-21relativeexpression(fold) 1.21.00.80.40.20.6Figure2The effect of miR-21knockdown on U251and LN229GBM cell proliferation.(a)miR-21expression was suppressed in U251and LN229cells using RT-real-time PCR.(b)In situ examination of miR-21expression in U251cells.Arrows highlight miR-21in situ expression in U251cells.Bar¼20m m.(c)MTT cell proliferation assay.miR-21knockdown in U251and LN229GBM inhibits cell proliferation.(d)Cell-cycle profiles after PI staining.miR-21 knockdown induced G1arrest in both U251and LN229GBM cells.(e)Graphical representation of the cell-cycle profiles in(d).As-miR-21and scramble ODN-transfected U251and LN229GBM cells were analyzed using FCM to determine cell-cycle status.w2Test was performed.**P o0.01.(f)Analysis of apoptosis using Annexin V staining.MiR-21knockdown led to U251and LN229GBM cell apoptosis.(g)Graphical representation of(f).As-miR-21and scramble ODN-transfected U251and LN229GBM cells were analyzed using FCM to determine cell apoptosis.One-way ANOVA was performed.**P o0.01.Downregulation of miR-21suppresses glioblastomaX Zhou et al。
第5章 DNA损伤和修复

Chapter 5 DNA damage and repair1.主要内容1)诱变2)DNA损伤3)DNA修复2.教学要求:1) 熟悉DNA损伤的原因、类型2)掌握DNA修复的方式5.1 Mutagenesis5.2 DNA damage5.3 DNA Repair5.1 Mutagenesis (诱变)• 5.1.1 The reasons of mutation• 5.1.2 Types of mutations• 5.1.3 Mutagens (诱变剂)• 5.1.4 mutagenesis (诱变)5.1 Mutagenesis (诱变)•Mutation (突变)= Permanent, heritable alterations in the base sequence of a DNA molecule(是DNA碱基序列水平上永久性的、可遗传的变化)5.1.1 The reasons of mutation•Spontaneous errors(自发性错误): in DNA replication or meiotic recombination •Mutagen (诱变物): A consequence of the damaging effects of physical or chemical mutagens on DNA•Mutagen= mutation causing agent•Essentially all mutagens are carcinogens(致癌物)•Most carcinogens are mutagens5.1.2 Types of mutations•Multisite(多位点):-Cause gross(严重的) chromosome abnormalities(畸形);-Involve large regions of DNA ;-Arise during meiosis(在减数分裂期发生)•Point mutations(点突变)-Involve only one or a few nucleotides;- Arise during DNA replication-Require two errors :1)An error during DNA replication;2)Failure to correct that errorTypes of point mutationsInversions: ACBDEFDuplications ABCDEEFDeletions: ABCD-FInsertions: ABCDSEFSubstitutions: ATCDEFTypes of point mutationsThe phenotypic effect of point mutation(点突变的表型效应)•Missense mutation(错义突变): 碱基序列的改变引起了产物氨基酸序列的改变。
3 Maize Centromere玉米着丝粒

Maize is the product of a fairly recent allotetraploidization event (prior to 4.8 million years ago; Swigonova et al., 2004), and has experienced massive genome expansion due to retrotransposon activity induced by that event. The high-quality maize genome sequence (Schnable et al., 2009) therefore provides a unique opportunity to study the interplay of retrotransposons and centromeric DNA, as well as the effects of large-scale chromosomal rearrangements that were required to reduce the initial allotetraploid chromosome number of 20 to today’s number of 10. Due to their size, the large A chromosomes are amenable to cytogenetic analysis—each chromosome has one functional centromere. Maize also has very well-studied B chromosomes and neocentromeres.
25
26
PLANT CENTROMERE BIOLOGY
大豆立枯丝核菌G蛋白

作物学报ACTA AGRONOMICA SINICA 2009, 35(2): 370−374/zwxb/ ISSN 0496-3490; CODEN TSHPA9E-mail: xbzw@本研究由教育部长江学者和创新团队发展计划项目(IRT0453)资助。
*通讯作者(Corresponding author):李仕贵, E-mail: lishigui_sc@; Tel: 028-******** 第一作者联系方式: E-mail: btma02@; Tel: 028-********Received(收稿日期): 2008-05-18; Accepted(接受日期): 2008-09-10.DOI: 10.3724/SP.J.1006.2009.00370大豆立枯丝核菌G蛋白β亚基基因的克隆与分析马炳田1,2曲广林1黄文娟1林瑜凡1李仕贵1,2,*1四川农业大学水稻研究所, 成都温江611130; 2四川农业大学 / 西南作物基因资源与遗传改良教育部重点实验室, 四川雅安625014 摘要: 由立枯丝核菌[Rhizoctonia solani Kühn, 有性世代:Thanatephorus cucumeris (Frank) Donk] 引起的大豆纹枯病(soybean sharp eyespot)是一种重要病害。
G蛋白β亚基(guanine nucleotide binding protein beta-subunit)作为重要的信号传导因子, 在植物病原菌致病分子机制中起着重要作用。
为了解G蛋白β亚基基因的结构与功能, 根据同源物种G蛋白β亚基相关序列设计引物, 利用PCR和RT-PCR技术克隆了大豆立枯丝核菌G蛋白β亚基的基因序列和开放阅读框(G-protein beta-subunit of soybean Rhizoctonia solani, 简写为gbsrs1, GenBank登录号为EU663628)。
02第二章 微生物发酵产酶 第三章 动植物细胞培养产酶

Amylase from Bacillus Protease from Bacillus Phosphatase from Bacillus Glucoamylase from Aspergillus …… Plant cell culture Animal cell culture
Few examples
30
进 位
成肽 转 位
31
合成终止
32
高效率的蛋白 质合成体系
33
蛋白质的折叠
蛋白质的空间结构如何形成? 功能与结构如何统一? 体外、体内的结构如何变化?
蛋白质分子设计及蛋白质工程的需要 越来越多的基因工程产物需要复性复活, 要求蛋白质折叠 的理论及技术的指导。基因工程重组蛋白类产物必须要形 成正确的折叠才能表现出功能和活性。
19
蛋白质合成的几个要素-遗传密码,nucleotide triplet codon
mRNA分子中,每三个相邻的核苷酸组成的三联体 代表某种氨基酸或其它信息,称为密码子或三联 密码。 四种核苷酸编成三联体可形成43个即64个密码子。 其中: 一个起始密码: AUG
三个终止密码: UAA
可产生一种组成型调节蛋白(regulatory protein) (一种变构蛋白),通过与效应物 (effector) (包括诱导物和辅阻遏物)的特异 结合而发生变构作用,从而改变它与操纵基因 的结合力。 调节基因常位于调控区的上游。
38
启动基因(promotor gene)(启动子):
有两个位点: (1)RNA聚合酶的结合位点 (2)cAMP-CAP的结合位点。 CAP:分解代谢产物基因活化蛋白(catabolite gene activator protein),又称环腺苷酸受体蛋白(cAMP receptor protein,CRP)。 只有cAMP-CRP复合物结合到启动子的位点上,RNA 聚合酶才能结合到其在启动子的位点上,酶的合成才 能开始。 P S DNA O R
投稿说明
references.KeywordsPlease provide 4 to 6 keywords which can be used for indexing purposes.TEXTText FormattingManuscripts should be submitted in Word.Use a normal, plain font (e.g., 10-point Times Roman) for text.Use italics for emphasis.Use the automatic page numbering function to number the pages.Do not use field functions.Use tab stops or other commands for indents, not the space bar.Use the table function, not spreadsheets, to make tables.Use the equation editor or MathType for equations.Note: If you use Word 2007, do not create the equations with the default equation editor but use the Microsoftequation editor or MathType instead.Save your file in doc format. Do not submit docx files.Word template (zip, 154 kB)Manuscripts with mathematical content can also be submitted in LaTeX.LaTeX macro package (zip, 182 kB)HeadingsPlease use no more than three levels of displayed headings.AbbreviationsAbbreviations should be defined at first mention and used consistently thereafter.FootnotesFootnotes can be used to give additional information, which may include the citation of a reference included in the reference list. They should not consist solely of a reference citation, and they should never include the bibliographic details of a reference. They should also not contain any figures or tables.Footnotes to the text are numbered consecutively; those to tables should be indicated by superscript lower-case letters (or asterisks for significance values and other statistical data). Footnotes to the title or the authors of the article are not given reference symbols.Always use footnotes instead of endnotes.AcknowledgmentsAcknowledgments of people, grants, funds, etc. should be placed in a separate section before the reference list. The names of funding organizations should be written in full.SCIENTIFIC STYLENomenclature: Insofar as possible, authors should use systematic names similar to those used by ChemicalAbstract Service or IUPAC.Genus and species names should be in italics.Sequence/nucleotide dataNew nucleotide data must be submitted and deposited in the DDBJ/EMBL/GenBank databases and an accession number obtained before the paper can be accepted for publication. Submission to any one of the three collaborating databanks is sufficient to ensure data entry in all. The accession number should be included in the manuscript e.g. as a footnote on the title page:'Note: Nucleotide sequence data reported are available in the DDBJ/EMBL/GenBank databases under the accession numbers(s)----'.If requested the database will withhold release of data until publication.The most convenient method for submitting sequence data is by World Wide Web:EMBL via Webin: /embl/Submission/webin.htmlGenBank via Bankit: /BankIt/DDBJ via Sakura: http://sakura.ddbj.nig.ac.jp/Alternatively, the stand-alone submission tool 'Sequin' is available from the EBI at /Services/Sequin and from NCBI at /Sequin/For special types of submissions (e.g. genomes, bulk submissions etc) additional submission systems are available from the above sites.Database Contact Information:EMBL:e-mail: datasubs@WWW URL: GenBank:e-mail: info@WWW URL: htpp://DDBJ:e-mail: ddbj@ddbj.nig.ac.jpWWW URL: http://www.ddbj.nig.ac.jp.Expression data"Current Genetics" requires microarray data to be repoted in accordance with the MIAME standards and this to be clearly documented in the Materials and Methods section. Please refer to . In addition, expression data should be deposited in a relevant repository such as:Gene expression omnibus: /geo/EBI Microarray Databases: /Databases/microarray.htmlCenter for Information Biology gene EXpression database: http://cibex.nig.ac.ip/Submission should be documented in the manuscript.REFERENCESCitationCite references in the text by name and year in parentheses. Some examples:Negotiation research spans many disciplines (Thompson 1990).This result was later contradicted by Becker and Seligman (1996).This effect has been widely studied (Abbott 1991; Barakat et al. 1995; Kelso and Smith 1998; Medvec et al.1993).Reference listThe list of references should only include works that are cited in the text and that have been published or accepted for publication. Personal communications and unpublished works should only be mentioned in the text. Do not use footnotes or endnotes as a substitute for a reference list.Reference list entries should be alphabetized by the last names of the first author of each work.Journal articleGamelin FX, Baquet G, Berthoin S, Thevenet D, Nourry C, Nottin S, Bosquet L (2009) Effect of high intensityintermittent training on heart rate variability in prepubescent children. Eur J Appl Physiol 105:731-738. doi:10.1007/s00421-008-0955-8Ideally, the names of all authors should be provided, but the usage of “et al” in long author lists will also beaccepted:Smith J, Jones M Jr, Houghton L et al (1999) Future of health insurance. N Engl J Med 965:325–329Article by DOISlifka MK, Whitton JL (2000) Clinical implications of dysregulated cytokine production. J Mol Med.doi:10.1007/s001090000086BookSouth J, Blass B (2001) The future of modern genomics. Blackwell, LondonBook chapterBrown B, Aaron M (2001) The politics of nature. In: Smith J (ed) The rise of modern genomics, 3rd edn. Wiley,New York, pp 230-257Online documentCartwright J (2007) Big stars have weather too. IOP Publishing PhysicsWeb. /articles/news/11/6/16/1. Accessed 26 June 2007DissertationTrent JW (1975) Experimental acute renal failure. Dissertation, University of CaliforniaAlways use the standard abbreviation of a journal’s name according to the ISSN List of Title Word Abbreviations, see /2-22661-LTWA-online.phpFor authors using EndNote, Springer provides an output style that supports the formatting of in-text citations and reference list.EndNote style (zip, 3 kB)TABLESAll tables are to be numbered using Arabic numerals.Tables should always be cited in text in consecutive numerical order.For each table, please supply a table caption (title) explaining the components of the table.Identify any previously published material by giving the original source in the form of a reference at the end of thetable caption.Footnotes to tables should be indicated by superscript lower-case letters (or asterisks for significance values andother statistical data) and included beneath the table body.ARTWORKFor the best quality final product, it is highly recommended that you submit all of your artwork – photographs, line drawings, etc. – in an electronic format. Your art will then be produced to the highest standards with the greatest accuracy to detail. The published work will directly reflect the quality of the artwork provided.Electronic Figure SubmissionSupply all figures electronically.Indicate what graphics program was used to create the artwork.For vector graphics, the preferred format is EPS; for halftones, please use TIFF format. MS Office files are alsoacceptable.Vector graphics containing fonts must have the fonts embedded in the files.Name your figure files with "Fig" and the figure number, e.g., Fig1.eps.Line ArtDefinition: Black and white graphic with no shading.Do not use faint lines and/or lettering and check that all lines and lettering within the figures are legible at final size.All lines should be at least 0.1 mm (0.3 pt) wide.Scanned line drawings and line drawings in bitmap format should have a minimum resolution of 1200 dpi.Vector graphics containing fonts must have the fonts embedded in the files.Halftone ArtDefinition: Photographs, drawings, or paintings with fine shading, etc.If any magnification is used in the photographs, indicate this by using scale bars withinthe figures themselves.Halftones should have a minimum resolution of 300 dpi.Combination ArtDefinition: a combination of halftone and line art, e.g., halftones containing line drawing, extensive lettering, color diagrams, etc.Combination artwork should have a minimum resolution of 600 dpi.Color ArtColor art is free of charge for online publication.If black and white will be shown in the print version, make sure that the main information will still be visible. Many colors are not distinguishable from one another when converted to black and white. A simple way to check this is to make a xerographic copy to see if the necessary distinctions between the different colors are still apparent.If the figures will be printed in black and white, do not refer to color in the captions.Color illustrations should be submitted as RGB (8 bits per channel).Figure LetteringTo add lettering, it is best to use Helvetica or Arial (sans serif fonts).Keep lettering consistently sized throughout your final-sized artwork, usually about 2–3 mm (8–12 pt).Variance of type size within an illustration should be minimal, e.g., do not use 8-pt type on an axis and 20-pt type for the axis label.Avoid effects such as shading, outline letters, etc.Do not include titles or captions within your illustrations.Figure NumberingAll figures are to be numbered using Arabic numerals.Figures should always be cited in text in consecutive numerical order.Figure parts should be denoted by lowercase letters (a, b, c, etc.).If an appendix appears in your article and it contains one or more figures, continue the consecutive numbering of the main text. Do not number the appendix figures, "A1, A2, A3, etc." Figures in online appendices (Electronic Supplementary Material) should, however, be numbered separately.Figure CaptionsEach figure should have a concise caption describing accurately what the figure depicts. Include the captions in the text file of the manuscript, not in the figure file.Figure captions begin with the term Fig. in bold type, followed by the figure number, also in bold type.No punctuation is to be included after the number, nor is any punctuation to be placed at the end of the caption.Identify all elements found in the figure in the figure caption; and use boxes, circles, etc., as coordinate points ingraphs.Identify previously published material by giving the original source in the form of a reference citation at the end of the figure caption.Figure Placement and SizeWhen preparing your figures, size figures to fit in the column width.For most journals the figures should be 39 mm, 84 mm, 129 mm, or 174 mm wide and not higher than 234 mm.For books and book-sized journals, the figures should be 80 mm or 122 mm wide and not higher than 198 mm.PermissionsIf you include figures that have already been published elsewhere, you must obtain permission from the copyright owner(s) for both the print and online format. Please be aware that some publishers do not grant electronic rights for free and that Springer will not be able to refund any costs that may have occurred to receive these permissions. In such cases, material from other sources should be used.AccessibilityIn order to give people of all abilities and disabilities access to the content of your figures, please make sure thatAll figures have descriptive captions (blind users could then use a text-to-speech software or a text-to-Braillehardware)Patterns are used instead of or in addition to colors for conveying information (color-blind users would then beable to distinguish the visual elements)Any figure lettering has a contrast ratio of at least 4.5:1ELECTRONIC SUPPLEMENTARY MATERIALSpringer accepts electronic multimedia files (animations, movies, audio, etc.) and other supplementary files to be published online along with an article or a book chapter. This feature can add dimension to the author's article, as certain information cannot be printed or is more convenient in electronic form.SubmissionSupply all supplementary material in standard file formats.Please include in each file the following information: article title, journal name, author names; affiliation and e-mail address of the corresponding author.To accommodate user downloads, please keep in mind that larger-sized files may require very long downloadtimes and that some users may experience other problems during downloading.Audio, Video, and AnimationsAlways use MPEG-1 (.mpg) format.Text and PresentationsSubmit your material in PDF format; .doc or .ppt files are not suitable for long-term viability.A collection of figures may also be combined in a PDF file.SpreadsheetsSpreadsheets should be converted to PDF if no interaction with the data is intended.If the readers should be encouraged to make their own calculations, spreadsheets should be submitted as .xlsfiles (MS Excel).Specialized FormatsSpecialized format such as .pdb (chemical), .wrl (VRML), .nb (Mathematica notebook), and .tex can also besupplied.Collecting Multiple FilesIt is possible to collect multiple files in a .zip or .gz file.NumberingIf supplying any supplementary material, the text must make specific mention of the material as a citation, similarto that of figures and tables.Refer to the supplementary files as “Online Resource”, e.g., "... as shown in the animation (Online Resource 3)",“... additional data are given in Online Resource 4”.Name the files consecutively, e.g. “ESM_3.mpg”, “ESM_4.pdf”.CaptionsFor each supplementary material, please supply a concise caption describing the content of the file.Processing of supplementary filesElectronic supplementary material will be published as received from the author without any conversion, editing,or reformatting.AccessibilityIn order to give people of all abilities and disabilities access to the content of your supplementary files, please make sure thatThe manuscript contains a descriptive caption for each supplementary materialVideo files do not contain anything that flashes more than three times per second (so that users prone to seizurescaused by such effects are not put at risk)AFTER ACCEPTANCEUpon acceptance of your article you will receive a link to the special Author Query Application at Springer’s web page where you can sign the Copyright Transfer Statement online and indicate whether you wish to order OpenChoice, offprints, or printing of figures in color.Once the Author Query Application has been completed, your article will be processed and you will receive the proofs.Open ChoiceIn addition to the normal publication process (whereby an article is submitted to the journal and access to that article is granted to customers who have purchased a subscription), Springer provides an alternative publishing option: Springer Open Choice. A Springer Open Choice article receives all the benefits of a regular subscription-based article, but in addition is made available publicly through Springer’s online platform SpringerLink. We regret that Springer Open Choice cannot be ordered for published articles.Springer Open ChoiceCopyright transferAuthors will be asked to transfer copyright of the article to the Publisher (or grant the Publisher exclusive publication and dissemination rights). This will ensure the widest possible protection and dissemination of information under copyright laws. Open Choice articles do not require transfer of copyright as the copyright remains with the author. In opting for open access, they agree to the Springer Open Choice Licence.OffprintsOffprints can be ordered by the corresponding author.Color illustrationsOnline publication of color illustrations is free of charge. For color in the print version, authors will be expected to make a contribution towards the extra costs.Proof readingThe purpose of the proof is to check for typesetting or conversion errors and the completeness and accuracy of the text, tables and figures. Substantial changes in content, e.g., new results, corrected values, title and authorship, are not allowed without the approval of the Editor.After online publication, further changes can only be made in the form of an Erratum, which will be hyperlinked to the article.Online First。
生物化学5-10章练习题英文版
⽣物化学5-10章练习题英⽂版Exercises of Chapter 5-10A1.Which can transport acyl-CoA into the Mitochondria? ( )A .apolipoprotein B. lipoprotein C. albumin D. citrate E. carnitine2. In which following conditions are ketone bodies synthesis greatly accelerated? ( )A .glycolysis is enhanced B. fat mobilization is enhanced C. fatty acid synthesis is enhancedD. malonyl CoA is reducedE. aerobic oxidation of glucose is enhanced3. How many moles of ATP are yielded when 1 mol glycerol is oxidized to CO2 and H2O ? ( )A .19 B. 16.5/18.5 C. 20/22 D.23 E. 174. which is the key enzyme in the regulation of FA synthesis? ( )A .fatty acid synthase B. acetyl-CoA carboxylase C. keto-acyl-ACP synthaseD.enoyl-ACP reductaseE. thioesterase5. NADPH in FA synthesis comes from ( ):A .aerobic oxidation of glucose B. glycolytic pathway C. gluconeogenesisD.pentose phosphate pathwayE. glucuronate pathway6. which is not concerned with the cholesterol biosynthesis?A .acetyl-CoA B. HMGCoA C. NADPH +H+ D.seualene E. glycerol7. the active form of choline in phospholipid biosynthesis is:A .ADP-choline B. UDP-choline C. CDP-choline D. GDP-choline E. CMP-choline8. which is not concerned with the glycerophospholipid biosynthesis?A .serine B. GTP C. ATP D. CTP E. choline9.which is specific function of apoAⅠ?A .activates LPL B. inhibits HL C. activates LCAT D. inhibits LCAT E. activates ACAT10.which is not required in β-Oxidation of Fatty AcidsA .CoASH B. NADP+ C. carnitine D. FAD E. NAD+11. which is the primary carrier of cholesterol in the blood of humans?A .CM B. HDL C.VLDL D. LDL E. IDL12. How many moles of ATP are yielded when 1 mol stearic acid (18 C) is oxidized to CO2 and H2O ? ( )A .146 B. 120 C. 165 D.182 E. 19913. which is an intermediate on the pathway for synthesis of ketone bodies and cholesterol?A .malonyl CoA B. acetoacetyl-CoA C. HMGCoA D. acetyl-CoA E. acetoacetate14. which is prosthetic group of acetyl-CoA carboxylase?A . NAD+ B. biotin C. NADP+ D. FAD E. FMN15. which is the key enzyme in the regulation of cholesterol synthesis? ( )A . HMGCoA synthase B. squalene monooxygenase C. HMGCoA reductaseD. squalene synthaseE. HMGCoA thiolase16.which is the correct sequence of 4 reactions in fatty acids oxidation?A . dehydrogenation, redegydrogenation, hydration, thiolysisB. thiolysis, dehydrogenation, redegydrogenation, hydrationC. dehydrogenation, hydration, redegydrogenation, thiolysisD. hydration, dehydrogenation, thiolysis, redegydrogenationE. dehydrogenation, thiolysis, redegydrogenation, hydration17. which is the major form that transports endogenous TAGs from the liver to the tissues?A .HDL B. LDL C.VLDL D.CM E. IDL18. which lipoprotein contains apoB100 mostly?A .HDL B. LDL C.VLDL D.CM E. CM remnants19. which lipoprotein contains apoB48 mostly?A .HDL B. LDL C.VLDL D.CM E. IDL20. which enzyme ‘s activator is apo CⅡ?A .LPL B. LCAT C.ACAT D.HL E. CETP21. the net result of the oxidation of 1 mol palmitic acid (16C) will be ( ) mol ATP?A .129 B. 96 C. 165 D.182 E. 10622.which is the major form that participates in the reverse cholesterol transport?A . IDL B. HDL C. LDL D. VLDL E. CM23.which is the rate –limiting enzyme in the TAG degradation?A .HSL B. LPL C.DAG lipase D.HL E. MAG lipase24.which hormone can accelerate fat mobilization?A .epinephrine B.glucagons C.TSH D.ACTH E. all above25.which is anti-lipolytic hormone ?A .epinephrine B.glucagons C.TSH D.ACTH E. prostaglandin E226.which is essential fatty acid?A . stearic acid B. oleic acid C.linoleic acid D. palmitic acid E. oxaloacetic acid27.which is the pathway that acetyl-CoA is transported from the mitochondria into the cytosol to allow FA synthesis to occur?A . TAC cycle B. malate-asparate shuttle C.glucose pyruvate cycleD. citrate pyruvate cycleE. α-glycerophosphate shuttle28. which is the key e nzyme in β-Oxidation of Fatty Acids? ( )A . carnitine acyl transferaseⅠ B. carnitine acyl transferaseⅡ C. acyl-CoA synthetaseD. carnitine-acylcaritine translocaseE. acetyl-CoA carboxylase29. the lack of ( ) means that ketone bodies are synthesized exckusively in the liver and must be used elsewhere.A . HMGCoA synthase B. succinyl CoA transsulfurase C. HMGCoA reductaseD. acetoacetyl-CoA thiolaseE. HMGCoA thiolase30. which agent is an inhibitor of electron transport at Complex Ⅲ?A .rotenone B. antimycin A C. piericidin A D. amobarbital E. DNP31. which agent is not an inhibitor of electron transport at Complex Ⅳ?A .carbon monoxide B. azide C. sulfureted hydrogen D. amobarbital E. cyanide32. which is not the energy-rich compound?A .CP B. ATP C. PEP D. 1,3-bisphoglycerate E. 2,3-bisphoglycerate33.the sequence of cytochrome in respiratory chain is :A. c----c1----b----aa3----O2B. c1---- c ----b----aa3----O2C. b ----c1---- c ----aa3----O2D. b ---- c ---- c1 ----aa3----O2E. c---- b ---- c1 ----aa3----O234. Which of the following state is nitrogen equilibrium?A. advanced cancer patientB. pregnant womenC. patient recovering from illnessD. people suffer form long-time hungerE. normal adult35. Which of the following state that nutrition sufficicent infant in?A.nitrogen balanceB.positive nitrogen balance/doc/4c28100b79563c1ec5da7190.html ative nitrotgen balanceD. nitrogen equilibriumE.none of the above36. Which of the following state is positive nitrogen balance?A.advanced cancerB. pregnant womenC.people suffer from feverD. people suffer form long-time hungerE.normal adult37. Which of the following is not one carbon unit?CH3; CH2; CO2; CH; CHO38. Which of the following is not produced by Tyr?norepinephrine;epinephrine;dopamine; Phe;melanin39. Which of the following is the main deamination style in muscle? Transamination;ornithine cycle;L-glutamate oxidation deamination;purine nucleotide cycle;γ-glutamyl cycle40. Which of the following is the storage and transportation style of ammonia?Glu; Tyr;GSH; Asn; Gln41. Which of the following is the main outlet of ammonia in mammalian? infiltrate to intestinaltract;synthesis of Gln;synthesis of urea in liver;synthesis of amino acid again; excrete out of the body in kidney42. Which of the following amino acid can not produce one carbon unit?Serine;Valine ;Tryptophan;Histidine ;Glycine43.Which of the following ammino acids take part in the synthesis of creatine? Glycine,mehionine,arginine; Methionine,ornithine,citrulline; Glycine,methionine,citrulline; Glycine,arginine,ornithine;Ornithine,citrulline, arginine44. Which of the following is the carrier of one carbon unit ?biotin; SAM; FH4; FH2; folic acid45. Which of the following is ketogenic amino acid only?Glycine; Leucine; Threonine; Isoleucine; Tyrosine46. Which of the following are the nitrogen source of urea ?carbomoyl phosphate and aspartate ;carbomoyl phosphate and ornithine;α-amino group andγ-amino group of ornithine;α-amino group of citrulline andα-amino group of arginine;γ-amino group of ornithine and glycine47. Which of the following amino acid can produce PAPS?taurine; methionine; cysteine; homocysteine; glutamate48. Which of the following is the main deamination style in the body?oxidation deamination; reduction deamination;direct deamination; transamination; union deamination49. Which of the following is the process that transport ammonia from muscle to liver? TAC cycle;ornithine cycle;alanine-glucose cycle;methionine cycle ; γ-glutamyl cycle 50. Which of the following vitamin that the coenzyme of amino acid decarboxylase contain?C; B2; B12;B6;B151. Which of the following is the direct donor of CH3?N5-CH3-FH4; SAM; N5,N10- CH2-FH4; N5,N10=CH-FH4; FAD52. Which of the following is ketogenic and glucogenic amino acid ?Ala; Phe; His; Pro; Leu53. Which of the following is the main source of ammonia in kidney?union deamination of amino acid; hydrolysis of Gln;hydrolysis of urea ; nonoxidative deamination of amino aicd;oxidation of amine54. In the synthesis of urea, one N come from NH3, which of the following amino acid provide another N for urea?Pro; Asp; Glu; Phe; Lys55. Which of the following is first synthesized in the process of de novo synthesis of purine nucleotide?GMP; AMP; IMP; ATP; GTP56. Which of the following is the end product of purine nucleotide catabolic metabolism?urea; creatine; creatinine; uric acid; β-alanine57. Which of the following substance directly link nucleotide synthesis and glucose metabolism?glucose; glucose-6-phosphate ; glucose-1-phosphate ; glucose-1,6-diphosphate; ribose-5-phosphate58. Which of the following is not the direct material of de nove synthesis of purine nucleotide ?glycine; aspartate ; glutamate ; CO2 ; one carbon unit59. Which of the following material could provide C4 and C5 for purine ring? alanine; glycine; glutamate ; aspartate ; glutamine60. Which of the following provide only one C atom for purine synthesis?CO2; glutamine; glycine; aspartate; formic acid61. Which of the following provide two N atoms for pyrimidine ring?glutamine and ammonia ; glutamine and glutamateglutamine and carbomoyl phosphate; aspartate and carbomoyl phosphate; aspartate and glutamate62.Which of the following is the common material for the synthesis of pyrimidine and purine nucleotide?fumarate; glutamine ; formic acid ; asparagine; glycine63. Which of the following amino acid totally participate in the synthesis of purine nucleotide?glycine ; aspartate; glutamate ; glutamine ; asparagines64.Which of the following provide C2 for purine ring?N5-CH3FH4; N10-CHO-FH4;N5,N10=CH-FH4; N5,N10-CH2-FH4; CO2 65. Which of the following is the energy source of brain for long-term starvation patient ?glucose; amino acid; glycerol; acetone body; glucogen66. Which of the following is not correct about chemical modification of key enzyme? the enzyme have two style: active and inactive ; the active style and inactive style can be interconverted by the other enzyme ; the enzyme that catalyze the interconversion can be controlled by the hormon; do not need energy; phosphorylation and dephosphorylation is the main style67. Which of the following is the main style of chemical modification?methylation and demethylation ; acetylation and deacetylation; phosphorylation and dephosphorylation; polymerization and depolymerization ; synthesis and decomposition of enzyme 68. Which of the following reaction take place in plasma?TAC ; oxidative phosphorylation ; pyruvate carboxylation; β-oxidation of fatty acid ; synthesis of fatty acid69. In the condition of starvation, which of the following is not correct? insulin secretion is enhanced ; glucagon secretion is enhanced;fat mobilization is enhanced ; synthesis of acetone body is enhanced ; glyconeogenesis is enhanced70. Which of the following is not correct about the substance metabolism in different tissues and organs ?liver is the hinge of the substance metabolism;glucose decomposition in heart is mainly aerobic oxidation ;the energy of brain usually come from glucose ;the energy of red cell usually come from glycolytic pathway;liver is the only organ that have the reaction of glyconeogenesis71. Which of the following is not correct about the allosteric regulation ?the allosteric enzyme usually has two subunits ;the allosteric effectors usually are small materials ;the allosteric effectors usually bind to the site out of the active center of the enzyme ; the end production of the pathway usually is the allosteric inhibitors of the enzyme that catalyze the initiation reaction of the pathway ; can be amplified72. Which of the following is not correct about the key enzyme?the key enzyme usually catalyze the first reaciton of the metabolic pathway ; the key enzyme has the highest activity; the key enzyme usually is the allosteric enzyme ; the key enzyme usually is regulated by hormone; the reaction that catalyzed by the key enzyme usually is the one-way reaction or equilibrium reaction 73. Which of the following enzyme do not belong to chemical modification enzyme ? phosphorylase; glycogen synthetase; pyruvate kinase; malate dehydrogenase ; phosphorylase b kinase74. Which of the following metabolic pathway take place in the mitochondira? glycolytic pathway; the synthesis of acetone body; pentose phosphate pathway ; the synthesis of the fatty acid ; the synthesis of the glycogen75. Which of the following is the material of glyconeogenesis for short time starvation? glycerol ; glycerol-3-phosphate; alanine; pyruvate; lactate76. In the oxidation decomposition of glucose ,fat and amino acid , which is the main compound that go into the TAC cycle? isocitrate ; pyruvate ; α-ketoglutarate ; α-keto acid ; acetyl CoA77. Which of the following is not the outlet of Acetyl CoA?go into the TAC cycle ; use to synthesize fat ; transition ot ketogenic amino acid ; transition ot acetone body; transition to cholesterol78. Which of the following is the rate-limiting enzyme of glyconeogenesis ?Acetyl CoA carboxylase ; 7-α-hydroxylase; HMGCoA reductase; pyruvate carboxylase; 6-phosphofructokinase-179. Which of the following does not participate in DNA replication?helicase; single strand binding protein; transciption factor; ligase ; primase 80. DNA replication is a ( ) process. conservative; destructive; non-conservative; semi- conservative, random dipersive81. he primary structure of DNA is the squence of its nucletide residues connected by ( )?3’,5’-phosphodiester bonds; hydrogen bonds; peptide bonds; salt bonds; disulfide bonds82. When the two daughter strands of DNA replicated , ( )one strand in the 5’ to 3’ and the other in the 3’ to 5’direction; the both strands are all in the 3’ to 5’ direction; the both strands are all in the 5’ to 3’ direction; the b oth strands are all conservative replicated; the DNAsynthesis of leading strand is continuous83.Which one can separate the parental DNA strands?DnaC; DNA-pol; topoisomerase; primase ; helicase84.Human telomerase RNA is one of the ( ).reverse transcriptase ; DNA-pol; RNA-pol; DNA ligase; DNA topoisomerase 85.Which of these enzyme is not concerned with DNA replication?DNA dependent DNA polymerase; DNA dependent RNA polymerase; RNA dependent DNA polymerase; topoisomerase; DNA ligase86.In the initiation of DNA replication, the proper order that following enzyme and protein acting is? 1, DNA-polⅢ 2,SSB 3,primase 4, helicase1,2,3,4; 4,2,3,1; 3,1,2,4; 1,4,2,3; 2,3,4,187.Which of the following could proofreading, DNA repairing, filling of gaps? DNA polα; DNA polγ ; DNA pol δ; DNA polⅠ; DNA polⅢ88.Which of the following have primase activity?DNA pol α; DNA polγ; DNA pol δ; DNA polⅠ; DNA polⅢ89.Which of the following serve as DNA replication in mitochondria ?DNA polα; DNA polγ; DNA pol δ; DNA polⅠ ; DNA polⅢ90.In DNA replication, what is the reason of Okazaki fragment generated?the speed of DNA replication is too fast ; bidirectional replication; come fromprimer; the direction of replication is different with that of unwinding; DNA be coiled and tied knots in replication 91.Which of the following is the product of the replication discontinuously? Klenow fragment; primer; leading strand ; lagging strand, Okazaki fragment92.Which of the following is a short RNA?Klenow fragment; primer ;leading strand; lagging strand; Okazaki fragment. 93.Which of the following could contain the length of eukaryotic chromosome? telomerase;primase;SSB;DNA ligase;topoiserase94.The template DNA is 5′-ATAGC-3′,and the daughter DNA is:5′-GCTAG-3′;5′-GCTAT-3′;5′-GCATA-3′;5′-GATAT-3′;5′-TATCG-3′. 95.Which of the following is the function of SSBunwingding;to identify the ori of replication;synthetic primer;to stabilize the single strand;filling the gapX:which are correlated with FA synthesis?A . acetyl-CoA carboxylase B.ACP C.biotin D. NADPH +H+ E. acyl-CoA synthetase2. which is essential fatty acid?A . oleic acid B. linolenic acid C.linoleic acid D. arachidonate E. palmitic acid3. which materials can not be transported from the mitochondria into the cytosol?A . acetyl-CoA B.pyruvate C. acyl-CoA D. oxaloacetic acid E. malate4. the function of cholesterol is:A . conversion into vitamin D B. conversion into bile acid C. as a biological membraneD. providing energyE. conversion into cholesterol hormones5. which material synthesis requires acetyl-CoA?A . fatty acid B.ketone bodies C. cholesterol D. lipoprotein E. sugar6. which does not exist in hepatic cells?A . succinyl CoA transsulfurase B. acetyl-CoA carboxylase C. HMGCoA reductaseD. acetoacetyl-CoA thiolaseE. acetoacetyl-CoA thiokinase7. utilization of ketone bodies in ( )A . heart B. brain C. kidney D. liver E. skeletal8.proton pumps are ( ) where protons flow from the matrix to the intermembrane space and a membrane potential is formed.A . ComplexⅠ B. ComplexⅡ C. Complex Ⅲ D. Complex Ⅳ E. ATP Synthase9. which agents are inhibitors of electron transport at ComplexⅠ?A .rotenone B. antimycin A C. piericidin A D. amobarbital E. BAL10. Which of the following are essential amino acids?phenylalanine;tyrosine;serine;threonine;leucine11. Which of the following substance need one carbon unit to synthesize?AMP; cholesterol; TMP; heme; fatty acid12. Which of the following are produced by Cysteine ?5-HT ; γ-GABA ; taurine ; GSH ; PAPS13. Which of the following are the source of ammonia?deamination of amino acid ;absorption of ammonia in digestive canal; secretory ammonia in kidney; decomposion of purine and pyrimidine nucleotide; decomposition of urea14. Which of the following α-keto acid can be directly converted to amino acid by transamination ?oxaloacetate;pyruvate; α-ketoglutarate;α-isobutyric acid ; hydroxyproline15. Which of the following are the main transportation style of ammonia between tissues?NH4CL; alanine; urea; glutamine; oxaloacetate16. Which of the following are one carbon unit?-CH=NH;-CHO;-CH2-;-CH3;CO217. Which of the following are glucogenic aminio acid?arginine;lysine;histidine;alanine; leucine18. Which of the following are ketogenic amino acid ?tyrosine; phenylalanine;ornithine;leucine ;lysine19. Which of the following are ketogenic and glucogenic amino acid?isoleucine; tyrosine; tryptophan; phenylalanine; serine20. Which of the following amino aicds take part in the synthesis of creatine ? glycine; arginine; methionine; cysteine; ornithine21. Which of the following substance can produce uric acid ?AMP ; UMP ; IMP ; TMP ; GMP22. Which of the following are the products of pyrimidine nucleotide decomposition? NH3; uric acid; CO2; β-alanine;β-aminoisobutyrate23.Which of the following provide C and N atoms for purine nucleotide ? glutamine; glycine; aspartate ; glutamate; alanine24. Which of the following are the materials for de novo synthesis of pyrimidine nucleotide?glutamine and aspartate; one carbon unit ; ribose phosphate; CO2 ; glycine 25. Which of the following are the compound that have the high-energy bound ? phosphoenolpyruvic acid ; Acetyl CoA; creatine phosphate ; AMP; 3-phosphoglycerate26. Which of the following are right about the allosteric enzyme ?all the reaction catalyzed by the allosteric enzyme is reversible ; the binding to the allosteric effectors is reversible ; all the enzyme have the regulatory subunit and catalytic subunit; the allosterism of the enzyme ofen accompany with the polymerization and depolymerization of the subunit; allosteric effectors can be the substrate or the production27. The central dogma of biological genetic information contain:DNA→DNA;DNA→RNA;RNA→DNA;RNA→R NA ;RNA→Potein 28.Which of the following could catalize the synthesis of phosphodiester bond? DNA-pol;topoismerase;telomerase;reverse transcriptase;DNA ligase.29。
投稿说明
references.KeywordsPlease provide 4 to 6 keywords which can be used for indexing purposes.TEXTText FormattingManuscripts should be submitted in Word.Use a normal, plain font (e.g., 10-point Times Roman) for text.Use italics for emphasis.Use the automatic page numbering function to number the pages.Do not use field functions.Use tab stops or other commands for indents, not the space bar.Use the table function, not spreadsheets, to make tables.Use the equation editor or MathType for equations.Note: If you use Word 2007, do not create the equations with the default equation editor but use the Microsoftequation editor or MathType instead.Save your file in doc format. Do not submit docx files.Word template (zip, 154 kB)Manuscripts with mathematical content can also be submitted in LaTeX.LaTeX macro package (zip, 182 kB)HeadingsPlease use no more than three levels of displayed headings.AbbreviationsAbbreviations should be defined at first mention and used consistently thereafter.FootnotesFootnotes can be used to give additional information, which may include the citation of a reference included in the reference list. They should not consist solely of a reference citation, and they should never include the bibliographic details of a reference. They should also not contain any figures or tables.Footnotes to the text are numbered consecutively; those to tables should be indicated by superscript lower-case letters (or asterisks for significance values and other statistical data). Footnotes to the title or the authors of the article are not given reference symbols.Always use footnotes instead of endnotes.AcknowledgmentsAcknowledgments of people, grants, funds, etc. should be placed in a separate section before the reference list. The names of funding organizations should be written in full.SCIENTIFIC STYLENomenclature: Insofar as possible, authors should use systematic names similar to those used by ChemicalAbstract Service or IUPAC.Genus and species names should be in italics.Sequence/nucleotide dataNew nucleotide data must be submitted and deposited in the DDBJ/EMBL/GenBank databases and an accession number obtained before the paper can be accepted for publication. Submission to any one of the three collaborating databanks is sufficient to ensure data entry in all. The accession number should be included in the manuscript e.g. as a footnote on the title page:'Note: Nucleotide sequence data reported are available in the DDBJ/EMBL/GenBank databases under the accession numbers(s)----'.If requested the database will withhold release of data until publication.The most convenient method for submitting sequence data is by World Wide Web:EMBL via Webin: /embl/Submission/webin.htmlGenBank via Bankit: /BankIt/DDBJ via Sakura: http://sakura.ddbj.nig.ac.jp/Alternatively, the stand-alone submission tool 'Sequin' is available from the EBI at /Services/Sequin and from NCBI at /Sequin/For special types of submissions (e.g. genomes, bulk submissions etc) additional submission systems are available from the above sites.Database Contact Information:EMBL:e-mail: datasubs@WWW URL: GenBank:e-mail: info@WWW URL: htpp://DDBJ:e-mail: ddbj@ddbj.nig.ac.jpWWW URL: http://www.ddbj.nig.ac.jp.Expression data"Current Genetics" requires microarray data to be repoted in accordance with the MIAME standards and this to be clearly documented in the Materials and Methods section. Please refer to . In addition, expression data should be deposited in a relevant repository such as:Gene expression omnibus: /geo/EBI Microarray Databases: /Databases/microarray.htmlCenter for Information Biology gene EXpression database: http://cibex.nig.ac.ip/Submission should be documented in the manuscript.REFERENCESCitationCite references in the text by name and year in parentheses. Some examples:Negotiation research spans many disciplines (Thompson 1990).This result was later contradicted by Becker and Seligman (1996).This effect has been widely studied (Abbott 1991; Barakat et al. 1995; Kelso and Smith 1998; Medvec et al.1993).Reference listThe list of references should only include works that are cited in the text and that have been published or accepted for publication. Personal communications and unpublished works should only be mentioned in the text. Do not use footnotes or endnotes as a substitute for a reference list.Reference list entries should be alphabetized by the last names of the first author of each work.Journal articleGamelin FX, Baquet G, Berthoin S, Thevenet D, Nourry C, Nottin S, Bosquet L (2009) Effect of high intensityintermittent training on heart rate variability in prepubescent children. Eur J Appl Physiol 105:731-738. doi:10.1007/s00421-008-0955-8Ideally, the names of all authors should be provided, but the usage of “et al” in long author lists will also beaccepted:Smith J, Jones M Jr, Houghton L et al (1999) Future of health insurance. N Engl J Med 965:325–329Article by DOISlifka MK, Whitton JL (2000) Clinical implications of dysregulated cytokine production. J Mol Med.doi:10.1007/s001090000086BookSouth J, Blass B (2001) The future of modern genomics. Blackwell, LondonBook chapterBrown B, Aaron M (2001) The politics of nature. In: Smith J (ed) The rise of modern genomics, 3rd edn. Wiley,New York, pp 230-257Online documentCartwright J (2007) Big stars have weather too. IOP Publishing PhysicsWeb. /articles/news/11/6/16/1. Accessed 26 June 2007DissertationTrent JW (1975) Experimental acute renal failure. Dissertation, University of CaliforniaAlways use the standard abbreviation of a journal’s name according to the ISSN List of Title Word Abbreviations, see /2-22661-LTWA-online.phpFor authors using EndNote, Springer provides an output style that supports the formatting of in-text citations and reference list.EndNote style (zip, 3 kB)TABLESAll tables are to be numbered using Arabic numerals.Tables should always be cited in text in consecutive numerical order.For each table, please supply a table caption (title) explaining the components of the table.Identify any previously published material by giving the original source in the form of a reference at the end of thetable caption.Footnotes to tables should be indicated by superscript lower-case letters (or asterisks for significance values andother statistical data) and included beneath the table body.ARTWORKFor the best quality final product, it is highly recommended that you submit all of your artwork – photographs, line drawings, etc. – in an electronic format. Your art will then be produced to the highest standards with the greatest accuracy to detail. The published work will directly reflect the quality of the artwork provided.Electronic Figure SubmissionSupply all figures electronically.Indicate what graphics program was used to create the artwork.For vector graphics, the preferred format is EPS; for halftones, please use TIFF format. MS Office files are alsoacceptable.Vector graphics containing fonts must have the fonts embedded in the files.Name your figure files with "Fig" and the figure number, e.g., Fig1.eps.Line ArtDefinition: Black and white graphic with no shading.Do not use faint lines and/or lettering and check that all lines and lettering within the figures are legible at final size.All lines should be at least 0.1 mm (0.3 pt) wide.Scanned line drawings and line drawings in bitmap format should have a minimum resolution of 1200 dpi.Vector graphics containing fonts must have the fonts embedded in the files.Halftone ArtDefinition: Photographs, drawings, or paintings with fine shading, etc.If any magnification is used in the photographs, indicate this by using scale bars withinthe figures themselves.Halftones should have a minimum resolution of 300 dpi.Combination ArtDefinition: a combination of halftone and line art, e.g., halftones containing line drawing, extensive lettering, color diagrams, etc.Combination artwork should have a minimum resolution of 600 dpi.Color ArtColor art is free of charge for online publication.If black and white will be shown in the print version, make sure that the main information will still be visible. Many colors are not distinguishable from one another when converted to black and white. A simple way to check this is to make a xerographic copy to see if the necessary distinctions between the different colors are still apparent.If the figures will be printed in black and white, do not refer to color in the captions.Color illustrations should be submitted as RGB (8 bits per channel).Figure LetteringTo add lettering, it is best to use Helvetica or Arial (sans serif fonts).Keep lettering consistently sized throughout your final-sized artwork, usually about 2–3 mm (8–12 pt).Variance of type size within an illustration should be minimal, e.g., do not use 8-pt type on an axis and 20-pt type for the axis label.Avoid effects such as shading, outline letters, etc.Do not include titles or captions within your illustrations.Figure NumberingAll figures are to be numbered using Arabic numerals.Figures should always be cited in text in consecutive numerical order.Figure parts should be denoted by lowercase letters (a, b, c, etc.).If an appendix appears in your article and it contains one or more figures, continue the consecutive numbering of the main text. Do not number the appendix figures, "A1, A2, A3, etc." Figures in online appendices (Electronic Supplementary Material) should, however, be numbered separately.Figure CaptionsEach figure should have a concise caption describing accurately what the figure depicts. Include the captions in the text file of the manuscript, not in the figure file.Figure captions begin with the term Fig. in bold type, followed by the figure number, also in bold type.No punctuation is to be included after the number, nor is any punctuation to be placed at the end of the caption.Identify all elements found in the figure in the figure caption; and use boxes, circles, etc., as coordinate points ingraphs.Identify previously published material by giving the original source in the form of a reference citation at the end of the figure caption.Figure Placement and SizeWhen preparing your figures, size figures to fit in the column width.For most journals the figures should be 39 mm, 84 mm, 129 mm, or 174 mm wide and not higher than 234 mm.For books and book-sized journals, the figures should be 80 mm or 122 mm wide and not higher than 198 mm.PermissionsIf you include figures that have already been published elsewhere, you must obtain permission from the copyright owner(s) for both the print and online format. Please be aware that some publishers do not grant electronic rights for free and that Springer will not be able to refund any costs that may have occurred to receive these permissions. In such cases, material from other sources should be used.AccessibilityIn order to give people of all abilities and disabilities access to the content of your figures, please make sure thatAll figures have descriptive captions (blind users could then use a text-to-speech software or a text-to-Braillehardware)Patterns are used instead of or in addition to colors for conveying information (color-blind users would then beable to distinguish the visual elements)Any figure lettering has a contrast ratio of at least 4.5:1ELECTRONIC SUPPLEMENTARY MATERIALSpringer accepts electronic multimedia files (animations, movies, audio, etc.) and other supplementary files to be published online along with an article or a book chapter. This feature can add dimension to the author's article, as certain information cannot be printed or is more convenient in electronic form.SubmissionSupply all supplementary material in standard file formats.Please include in each file the following information: article title, journal name, author names; affiliation and e-mail address of the corresponding author.To accommodate user downloads, please keep in mind that larger-sized files may require very long downloadtimes and that some users may experience other problems during downloading.Audio, Video, and AnimationsAlways use MPEG-1 (.mpg) format.Text and PresentationsSubmit your material in PDF format; .doc or .ppt files are not suitable for long-term viability.A collection of figures may also be combined in a PDF file.SpreadsheetsSpreadsheets should be converted to PDF if no interaction with the data is intended.If the readers should be encouraged to make their own calculations, spreadsheets should be submitted as .xlsfiles (MS Excel).Specialized FormatsSpecialized format such as .pdb (chemical), .wrl (VRML), .nb (Mathematica notebook), and .tex can also besupplied.Collecting Multiple FilesIt is possible to collect multiple files in a .zip or .gz file.NumberingIf supplying any supplementary material, the text must make specific mention of the material as a citation, similarto that of figures and tables.Refer to the supplementary files as “Online Resource”, e.g., "... as shown in the animation (Online Resource 3)",“... additional data are given in Online Resource 4”.Name the files consecutively, e.g. “ESM_3.mpg”, “ESM_4.pdf”.CaptionsFor each supplementary material, please supply a concise caption describing the content of the file.Processing of supplementary filesElectronic supplementary material will be published as received from the author without any conversion, editing,or reformatting.AccessibilityIn order to give people of all abilities and disabilities access to the content of your supplementary files, please make sure thatThe manuscript contains a descriptive caption for each supplementary materialVideo files do not contain anything that flashes more than three times per second (so that users prone to seizurescaused by such effects are not put at risk)AFTER ACCEPTANCEUpon acceptance of your article you will receive a link to the special Author Query Application at Springer’s web page where you can sign the Copyright Transfer Statement online and indicate whether you wish to order OpenChoice, offprints, or printing of figures in color.Once the Author Query Application has been completed, your article will be processed and you will receive the proofs.Open ChoiceIn addition to the normal publication process (whereby an article is submitted to the journal and access to that article is granted to customers who have purchased a subscription), Springer provides an alternative publishing option: Springer Open Choice. A Springer Open Choice article receives all the benefits of a regular subscription-based article, but in addition is made available publicly through Springer’s online platform SpringerLink. We regret that Springer Open Choice cannot be ordered for published articles.Springer Open ChoiceCopyright transferAuthors will be asked to transfer copyright of the article to the Publisher (or grant the Publisher exclusive publication and dissemination rights). This will ensure the widest possible protection and dissemination of information under copyright laws. Open Choice articles do not require transfer of copyright as the copyright remains with the author. In opting for open access, they agree to the Springer Open Choice Licence.OffprintsOffprints can be ordered by the corresponding author.Color illustrationsOnline publication of color illustrations is free of charge. For color in the print version, authors will be expected to make a contribution towards the extra costs.Proof readingThe purpose of the proof is to check for typesetting or conversion errors and the completeness and accuracy of the text, tables and figures. Substantial changes in content, e.g., new results, corrected values, title and authorship, are not allowed without the approval of the Editor.After online publication, further changes can only be made in the form of an Erratum, which will be hyperlinked to the article.Online First。
Nguyen_et_al-2009-Journal_of_Cellular_and_Molecular_Medicine
IntroductionBoth physiological and pathological angiogenesis are regulated by adelicate balance between angiogenic growth factors and endogenousinhibitors [1]. Tumour growth and metastasis are promoted whenthis balance is shifted towards angiogenesis. Angiogenesis inhibitorscan be used to reverse this trend [2]. Endostatin, a 20-kD fragmentof the non-collagenous 1 domain of collagen XVIII-␣1, is a well-characterized endogenous angiogenesis inhibitor [1, 3, 4] and isknown to inhibit endothelial cell proliferation, and cell migration [5],and to induce apoptosis [6]. Recently, a mutant form of endostatin
with a proline to alanine substitution at position 125 was identifiedand characterized [7]. P125A-endostatin showed higher binding toendothelial cell surface and exhibited stronger inhibitory effects onboth cell proliferation and cell migration [7, 8]. Integrin ␣51and
