promega 胶回收完整说明书
胶回收QC操作标准

琼脂糖凝胶DNA回收试剂盒(离心柱型)QC标准操作流程1. 实验试剂和耗材1.2 实验试剂1.3 耗材2. 实验操作流程胶回收试剂盒的检测须进行两个实验验证,第一个实验为GelRed染色法,第二个实验为EB染色法。
2.1 GelRed染色法2.1.1 1.5%琼脂糖凝胶的制备(1)准确称取1.5g琼脂糖置于250ml三角锥形瓶中,添加125ml 1×TAE溶液。
(2)轻微混匀后将其置于微波炉中,高火加热6min至溶液沸腾,取出锥形瓶放置室温2min。
(3)添加1×TAE溶液至110ml,再次于微波炉中高火加热3min至溶液沸腾。
(4)室温放置10min后将溶液倒入插有梳子的胶槽内,待胶块凝固后使用。
2.1.2 胶回收实验(1)取6个2ml离心管,依次编号为1-6号,使用分析天平准确称取空离心管的重量。
(2)用干净锋利的手术刀片切下不含核酸染料的1.5%琼脂糖凝胶胶块,放入已称重的2ml离心管中,称取胶块与离心管总重,计算凝胶块的重量。
要求胶块重量不超过200mg 为宜。
(3)添加溶胶液Binding Solution B,要求1、2号管中每100mg琼脂糖凝胶加入200ul Binding Solution B,3、4号管中每100mg琼脂糖凝胶加入300ul Binding Solution B,5、6号管中每100mg琼脂糖凝胶加入400ul Binding Solution B.(4)每管中添加50ul DL2502,混匀后置于60℃水浴5min,期间间断混合,直至凝胶块完全融化。
【注意事项】观察各管内溶胶液溶解胶块的快慢程度。
(5)将上述混合液转移至套放于2ml收集管的GenClean柱中,室温放置2min,10000rpm 室温离心1min,取出GenClean 柱,并倒掉收集管中废液。
(6)将GenClean 柱重新放回收集管中,加入500ul Wash Solution,于12000rpm,室温离心1min,倒掉收集管中废液。
胶回收 PPT

➢ 丙烯酰胺电泳凝胶产物回收的方法之一。
12
2.1.5 DEAE纤维素膜纸片法
➢ 将DEAE纤维素膜裁成小条活化处理。电泳后在 目的条带前切一刀,将比条带略宽的DEAE 纤维 素膜插入切口,不留气泡,继续电泳,条带上的 DNA被膜片截留,取出膜片冲洗后转移到离心管 中加缓冲液65℃保温洗脱,直到膜上DNA被完 全洗脱,将溶液用酚氯仿抽提沉淀。
2
向吸附柱CA2中 加入μL漂洗液PW
12000rpm, 离心30~60sec
3
弃废液,将吸附 柱重新放回收集管 中。重复操作1,2,3
注:1.漂洗液PW使用前先检查是否已加入无水乙醇。 2.如果回收的DNA是用于盐敏感的实验,例如平末端连接实验或
直接测序,建议PW加入后静置2~5min再离心。
25
4.1 实验流程
4.1.6 去除漂洗液
1
2
将吸附柱CA2放回 收集管中
12000rpm, 离心2min
3
将吸附柱CA2置于 室温放置数分钟,
晾干
注:1.尽量除尽漂洗液,以防止残留的漂洗液中的乙醇影响下一步的 实验(如酶切、PCR等)。
26
4.1 实验流程
4.1.7 收集DNA
1
2
将吸附柱CA2放到 一个干净离心管中
胶回收
目录
一 胶回收基础知识 二 方法及应用 三 试剂耗材与设备 四 操作流程 五 常见问题解答
目录
一 胶回收基础知识 二 方法及应用 三 试剂耗材与设备 四 操作流程 五 常见问题解答
1.胶回收的概念 2.胶回收的原理
1.1 胶回收概念
➢ 从电泳后的凝胶中回收目的DNA的过程,其目的 是将目标DNA重新利用。
PROMEGA逆转录试剂盒说明书

Reverse Transcription System
INSTRUCTIONS FOR USE OF PRODUCT A3500.
1. Description..........................................................................................................1 2. Product Components and Storage Conditions ............................................2 3. Reverse Transcription Protocol.......................................................................2
Printed in USA. Revised 3/09 Part# TB099 Page 1
2.
Product Components and Storage Conditions
Size 100 reactions Cat. # A3500
Product Reverse Transcription System
3.A. Reverse Transcri magnesium concentration may be optimized for any given sequence to achieve better yields. **Final concentration of reaction components: 5mM MgCl2; 1X Reverse Transcription Buffer (10mM Tris-HCl [pH 9.0 at 25°C]; 50mM KCl; 0.1% Triton® X-100); 1mM each dNTP; 1u/μl Recombinant RNasin® Ribonuclease Inhibitor; 15u/μg AMV Reverse Transcriptase (High Conc.); 0.5μg Oligo(dT)15 Primer or Random Primers per microgram RNA; 50ng/μl 1.2kb Kanamycin Positive Control RNA, poly(A)+ mRNA or total RNA.
胶回收方法全攻略

资料范本本资料为word版本,可以直接编辑和打印,感谢您的下载胶回收方法全攻略地点:__________________时间:__________________说明:本资料适用于约定双方经过谈判,协商而共同承认,共同遵守的责任与义务,仅供参考,文档可直接下载或修改,不需要的部分可直接删除,使用时请详细阅读内容胶回收方法全攻略说到电泳凝胶的片断回收,想来在实验室久已的“老手”们都会不屑一顾。
确实,一般在各个和分子生物学沾到一点边儿的实验室里,从琼脂糖凝胶中回收DNA,仅仅是一种简单不过的常规实验操作。
虽说早期的凝胶电泳片断回收没有1—2个小时是搞不定的,每个实验室也有自己的独门秘诀,可后来各大品牌纷纷推出了了多种不同用途,不同价钱的快速凝胶回收试剂盒,一下子将胶回收的时间缩短到10多分钟,操作也大大简化,胶回收就变成“小菜一碟”了。
然而,从bbs逛上一圈下来,发现实际上还是有不少人为胶回收这种看似简单的操作而烦恼。
到底是什么导致了实验新手,甚至是老手在这个已经发展近40年的常规性实验中卡壳?由于胶回收的质量和数量直接影响后继的一系列实验——比如酶切连接、转化筛选、测序或者PCR 扩增、标记乃至显微注射等等,为大家搜罗各种产品和方法的优缺点,注意事项,做一个胶回收全攻略篇。
一、胶回收的关键参数胶回收的质量直接影响后继实验的成功与否。
要想做好胶回收,无论是自己亲力亲为还是借助目前五花八门的胶回收试剂,最基本的评定标准无外乎这么几个:质量( 回收产物的纯度和浓度),回收效率,操作方便(速度),柱子的载量等等。
回收产物质量:回收产物的质量主要指纯度。
常规电泳过程中,普通级别的琼脂糖自带的一些性状不明的多糖,会连同DNA一起从凝胶中抽提出来,会强烈抑制后继的连接、酶切、或者标记、扩增等实验。
从凝胶中回收DNA片断,产物的纯度自然是我们考虑的第一要务。
对于大片断DNA回收,质量还包括了产物的完整与否,如果机械剪切力使得回收产物大小不一致,后果决不是你希望碰见的。
pcr切胶回收的步骤

pcr切胶回收的步骤
PCR切胶回收呀,这可是个很有趣的小实验呢。
先说说准备工作吧。
你得有已经跑过PCR并且经过琼脂糖凝胶电泳的胶块哦。
然后呢,把要用的工具都准备好,像干净的刀片啦,还有专门的切胶回收试剂盒。
开始切胶啦。
在紫外灯下看胶块的时候,就像寻宝一样呢。
找到你要的那条DNA 条带,然后用刀片小心翼翼地把它周围的胶切下来。
可别切太大块啦,不然会有好多杂质的。
切的时候就像在给小宝贝做精细的手术一样,要很小心哦。
切好胶之后,就按照试剂盒的说明来操作啦。
一般是把切下来的胶放到一个离心管里,然后加入一些溶液,让胶融化。
这时候你就看着胶慢慢变成液体,感觉就像魔法一样呢。
接着呢,要把融化后的液体加到吸附柱里。
这个吸附柱就像一个小卫士,专门把DNA吸附住,而那些杂质就被留在外面啦。
把液体加进去之后,离心一下,就像坐小过山车一样,让液体在离心力的作用下快速通过吸附柱。
然后呢,要清洗一下吸附柱。
这一步就像是给小卫士洗个澡,把它身上可能残留的杂质都洗干净。
再离心一下,把清洗的液体甩掉。
最后就是把我们想要的DNA从吸附柱上洗脱下来啦。
加入洗脱液,再离心,这个时候我们的DNA就乖乖地跑到洗脱液里啦。
就像把小宝贝从保护它的小房子里接出来一样。
这样,PCR切胶回收就完成啦。
虽然过程有点小复杂,但是只要按照步骤来,就可以得到我们想要的纯净的DNA啦。
每次做这个实验都感觉像是在和小小的DNA分子玩一场有趣的游戏呢。
promega-tm600-甘油三酯检测试剂盒说明书

G9711, G9712 and G9713中文说明书适用产品目录号J3160 和 J31612020 版 CTM600原英文技术手册TM600Triglyceride-Glo™ Assay普洛麦格(北京)生物技术有限公司Promega (Beijing) Biotech Co., Ltd 地址:北京市东城区北三环东路36号环球贸易中心B座907-909电话:************网址:技术支持电话:800 810 8133(座机拨打),400 810 8133(手机拨打)技术支持邮箱:*************************CTM6002020制作1Triglyceride-Glo™ Assay所有技术文献的英文原版均可在/ protocols获得。
请访问该网址以确定您使用的说明书是否为最新版本。
如果您在使用该试剂盒时有任何问题,请与Promega 北京技术服务部联系。
电子邮箱:*************************1. 产品描述 (2)2. 产品组分和储存条件 (4)3. 甘油三酯检测 (5)3. A. 用户需提供的材料 (5)3. B. 试剂制备 (5)3. C. 检测操作步骤 (6)4. 实验示例 (8)4. A. 肝细胞和小鼠肝组织中甘油三酯检测 (8)4. B. 癌细胞和人肝脏微组织的脂肪变性 (11)4. C. 脂肪细胞分化:染色法与甘油三酯定量法的比较 (12)5. 附录 (13)5. A. 信号稳定性 (13)5. B. 甘油三酯回收率 (13)5. C. 甘油三酯特异性 (15)5. D. 温度和试剂兼容性 (15)5. E. 微孔板和检测仪器 (15)5. F. 多重检测和归一化 (15)6. 参考文献 (16)7. 相关产品 (17)普洛麦格(北京)生物技术有限公司Promega (Beijing) Biotech Co., Ltd 地址:北京市东城区北三环东路36号环球贸易中心B座907-909电话:************网址:技术支持电话:800 810 8133(座机拨打),400 810 8133(手机拨打)技术支持邮箱:*************************CTM6002020制作21. 产品描述Triglyceride-Glo™ Assay(a)为检测培养细胞裂解物和其他生物样品(如细胞培养基、血清和组织匀浆)中的甘油三酯提供了一种发光检测方法。
PCR 或酶切产物凝胶回收试剂盒说明书
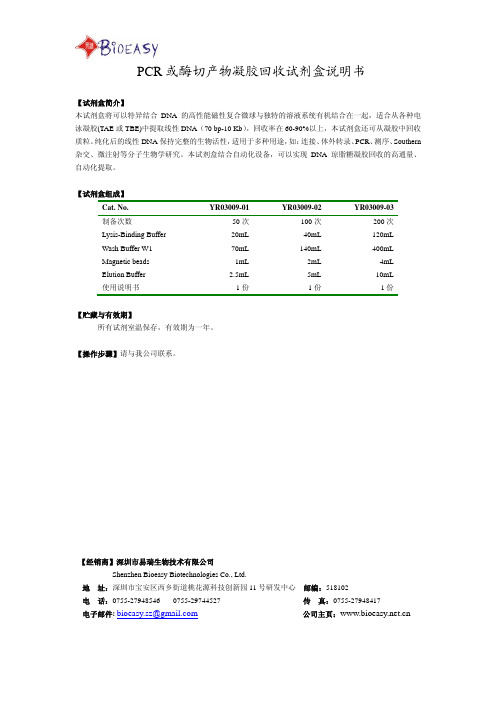
PCR或酶切产物凝胶回收试剂盒说明书
【试剂盒简介】
本试剂盒将可以特异结合DNA的高性能磁性复合微球与独特的溶液系统有机结合在一起,适合从各种电泳凝胶(TAE或TBE)中提取线性DNA(70 bp-10 Kb),回收率在60-90%以上,本试剂盒还可从凝胶中回收质粒。
纯化后的线性DNA保持完整的生物活性,适用于多种用途,如:连接、体外转录、PCR、测序、Southern 杂交、微注射等分子生物学研究。
本试剂盒结合自动化设备,可以实现DNA琼脂糖凝胶回收的高通量、自动化提取。
【试剂盒组成】
Cat. No. YR03009-01YR03009-02YR03009-03
制备次数 50次100次200次
Lysis-Binding Buffer 20mL40mL120mL
Wash Buffer W1 70mL140mL400mL
Magnetic beads 1mL2mL4mL
Elution Buffer 2.5mL5mL10mL
使用说明书1份1份1份
【贮藏与有效期】
所有试剂室温保存,有效期为一年。
【操作步骤】请与我公司联系。
【经销商】深圳市易瑞生物技术有限公司
Shenzhen Bioeasy Biotechnologies Co., Ltd.
地址:深圳市宝安区西乡街道桃花源科技创新园11号研发中心邮编:518102
电话:**************************传真:*************
电子邮件:********************公司主页:。
Omega胶回收试剂盒操作

Introduction . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 2 Benefits . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 2 Binding Capacity . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 2 Kit Contents . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 3 Materials Supplied By User . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 3 Gel Extraction Protocol . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 4 Optional Vacuum Protocol . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 5 Trouble Shooting Guide . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 6 Short Protocol For Experienced Users . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 7
- 1、下载文档前请自行甄别文档内容的完整性,平台不提供额外的编辑、内容补充、找答案等附加服务。
- 2、"仅部分预览"的文档,不可在线预览部分如存在完整性等问题,可反馈申请退款(可完整预览的文档不适用该条件!)。
- 3、如文档侵犯您的权益,请联系客服反馈,我们会尽快为您处理(人工客服工作时间:9:00-18:30)。
Promega Corporation ·2800 Woods Hollow Road ·Madison, WI 53711-5399 USA Toll Free in USA 800-356-9526·Phone 608-274-4330 ·Fax 608-277-2516 · Printed in USA.Part# TB308Revised 11/09Page 11.Description ..........................................................................................................12.Product Components and Storage Conditions ............................................33.General Considerations ....................................................................................44.Gel Slice and PCR Product Preparation .. (4)A.Preparing the Membrane Wash Solution (4)B.Dissolving the Gel Slice (5)C. Processing PCR Amplification Products (6)5.DNA Purification (6)A.DNA Purification by Centrifugation (6)B.DNA Purification by Vacuum............................................................................76.Troubleshooting .................................................................................................97.References .........................................................................................................118.Appendix .. (11)position of Buffers and Solutions (11)B. Related Products (12)1.DescriptionThe Wizard ®SV Gel and PCR Clean-Up System is designed to extract and purify DNA fragments of 100bp to 10kb from standard or low-melt agarose gels in either Tris acetate (TAE) or Tris borate (TBE), or to purify PCR products directly from a PCR amplification. Up to 95% recovery is achieved depending upon the DNA fragment size (see Table 1). PCR products are commonly purified to remove excess nucleotides and primers. This membrane-based system, which can bind up to 40μg DNA, allows recovery of isolated DNA fragments or PCR products in as little as 20 minutes, depending on the number of samples processed and the protocol used. The purified DNA can be used for automated fluorescent DNA sequencing, cloning, labeling, restriction enzyme digestion or in vitro transcription/translation without further manipulation.®Clean-Up System All technical literature is available on the Internet at /tbs Please visit the web site to verify that you are using the most current version of this Technical Bulletin. Please contact Promega Technical Services if you have questions on useof this system. E-mail techserv@.The Wizard ®SV Gel and PCR Clean-Up System is based on the ability of DNA to bind to silica membranes in the presence of chaotropic salts. After electrophoresis to separate the DNA fragments, the band(s) of interest is excised and dissolved in the presence of guanidine isothiocyanate (Membrane Binding Solution).Alternatively, after amplification, an aliquot of the PCR is added to theMembrane Binding Solution and directly purified. The system allows a choice of methods for isolation of DNA from the dissolved agarose gel slice or PCRamplification. DNA can be isolated using microcentrifugation to force thedissolved gel slice or PCR product through the membrane while simultaneously binding the DNA on the surface of the silica (Section 5.A). After washing the isolated DNA fragment or PCR product, the DNA is eluted in water. Another option is pulling the dissolved gel or PCR product through the SV Minicolumn and washing the DNA fragment using vacuum pressure (Section 5.B). TheVacuum Adapters allow the use of a vacuum manifold (e.g., Vac-Man ®Laboratory Vacuum Manifold, 20-sample capacity [Cat.# A7231], or Vac-Man ®boratory Vacuum Manifold, 2-sample capacity [Cat.# A7660]). The Vacuum Adapters (Cat. # A1331) are only supplied with Cat.# A9280, Wizard ®SV Gel and PCR Clean-Up System, 10 preps, but may be purchased separately.The Wizard ®SV Gel and PCR Clean-Up System can be used with linear DNA fragments, supercoiled plasmid DNA, or single-stranded linear or circular DNA.Expected yields with single-stranded DNA are lower than for double-stranded DNA.Table 1. Percent Recovery Versus Double-Stranded DNA Fragment Size. PCRproducts (55–1,000bp), linearized pGEM ®-3Zf(+) plasmid (3,199bp), or Lambda Hin d III fragments (9,416bp and 23,130bp) were purified in triplicate from a 1% agarose gelslice in 1X TAE buffer and quantified by ethidium bromide staining.Promega Corporation ·2800 Woods Hollow Road ·Madison, WI 53711-5399 USA Toll Free in USA 800-356-9526·Phone 608-274-4330 ·Fax 608-277-2516 · Part# TB308Printed in USA.Revised 11/09Page 22.Product Components and Storage ConditionsProduct Size Cat. # Wizard®SV Gel and PCR Clean-Up System10 preps A9280For Laboratory Use. Each system contains sufficient reagents for 10 purifications.Includes:•4ml Membrane Binding Solution•3ml Membrane Wash Solution (concentrated)• 1.25ml Nuclease-Free Water•10Wizard®SV Minicolumns•10Collection Tubes (2ml)•5Vacuum AdaptersProduct Size Cat. # Wizard®SV Gel and PCR Clean-Up System50 preps A9281For Laboratory Use. Each system contains sufficient reagents for 50 purifications.Includes:•20ml Membrane Binding Solution•15ml Membrane Wash Solution (concentrated)• 3.75ml Nuclease-Free Water•50Wizard®SV Minicolumns•50Collection Tubes (2ml)Product Size Cat. # Wizard®SV Gel and PCR Clean-Up System250 preps A9282For Laboratory Use. Each system contains sufficient reagents for 250purifications.Includes:•100ml Membrane Binding Solution•75ml Membrane Wash Solution (concentrated)•13ml Nuclease-Free Water•250Wizard®SV Minicolumns•250Collection Tubes (2ml)Product Size Cat. # Wizard®SV Gel and PCR Clean-Up System1000 preps A9285For Laboratory Use. Each system contains sufficient reagents for 4 × 250purifications. Includes:• 4 × 100ml Membrane Binding Solution• 4 × 75ml Membrane Wash Solution (concentrated)• 4 × 13ml Nuclease-Free Water• 4 × 250Wizard®SV Minicolumns• 4 × 250Collection Tubes (2ml)Storage Conditions:Store all components at room temperature (22–25°C). No refrigeration is required. Keep Membrane Binding Solution protected fromlight. See expiration date on product label.Promega Corporation·2800 Woods Hollow Road ·Madison, WI 53711-5399 USA Toll Free in USA 800-356-9526·Phone 608-274-4330 ·Fax 608-277-2516 · Printed in USA.Part# TB308 Revised 11/09Page 3Promega Corporation ·2800 Woods Hollow Road ·Madison, WI 53711-5399 USA Toll Free in USA 800-356-9526·Phone 608-274-4330 ·Fax 608-277-2516 · Part# TB308Printed in USA.Revised 11/09Page 43.General ConsiderationsAgarose, a linear polymer extracted from seaweed, is commonly used forelectrophoretic separation of nucleic acids. Standard agarose melts at 87–89°C and solidifies at 36–39°C. In low-melt agarose, hydroxyethyl groups have beenintroduced into the polysaccharide chain, resulting in an agarose that both melts and solidifies at much lower temperatures (65°C and 24–28°C, respectively). Low-melt agarose is often used for applications that require recovery of intact DNA fragments from the gel after electrophoresis. The Wizard ®SV Gel and PCR Clean-Up System can be used to recover DNA from either standard or low-melt agarose gels with no changes to the protocol or differences in recovery (Section 5).Standard safety apparel should be worn, especially when handling ethidiumbromide-stained agarose gels. This includes gloves and a UV-blocking face shield to protect the eyes and face from UV light. When excising the gel band, work quickly to minimize personal exposure to UV light and to minimize nicking of the DNA (1–4).The Wizard ®SV Gel and PCR Clean-Up System is compatible with PCR products generated using a variety of amplification enzymes, buffers or PCR-enhancing additives. Mineral oil does not interfere with purification.4.Gel Slice and PCR Product PreparationMaterials to Be Supplied by the User(Solution compositions are provided in Section 8.A.)•1.5ml microcentrifuge tubes •ethanol (95%)•Vacuum Adapters (Cat.# A1331; only for vacuum purification)•agarose gel (standard or low-melt; only for gel purification)•1X TAE or TBE electrophoresis buffer (only for gel purification)•50–65°C heating block (only for gel purification)4.A.Preparing the Membrane Wash SolutionAdd the indicated volume of 95% ethanol to the Membrane Wash Solution prior to beginning the procedure (see Table 2). Mark the bottle label to record that this addition was made. Tightly close the bottle cap after each use to preventevaporation.Table 2. Volume of 95% Ethanol to Add to Membrane Wash Solution for Each System Size.Promega Corporation ·2800 Woods Hollow Road ·Madison, WI 53711-5399 USA Toll Free in USA 800-356-9526·Phone 608-274-4330 ·Fax 608-277-2516 · Printed in USA.Part# TB308Revised 11/09Page 54.B.Dissolving the Gel Slice1.Load and run the gel using an established protocol. DNA can be extractedfrom standard or low-melt agarose gels run with either TAE or TBE buffer.2.Weigh a 1.5ml microcentrifuge tube for each DNA fragment to be isolatedand record the weight.3.Visualize and photograph the DNA using a long-wavelength UV lamp andan intercalating dye such as ethidium bromide. To reduce nicking, irradiatethe gel for the absolute minimum time possible (1–4). Excise the DNAfragment of interest in a minimal volume of agarose using a clean scalpelor razor blade. Transfer the gel slice to the weighed microcentrifuge tubeand record the weight. Subtract the weight of the empty tube from the totalweight to obtain the weight of the gel slice (see Notes 1–3 below).Note: The gel slice may be stored at 4°C or at –20°C for up to one week in atightly closed tube under nuclease-free conditions before purification.4.Add Membrane Binding Solution at a ratio of 10μl of solution per 10mg ofagarose gel slice.5.Vortex the mixture (see Note 4) and incubate at 50–65°C for 10 minutes oruntil the gel slice is completely dissolved. Vortex the tube every few minutesto increase the rate of agarose gel melting. Centrifuge the tube briefly atroom temperature to ensure the contents are at the bottom of the tube. Oncethe agarose gel is melted, the gel will not resolidify at room temperature.6.To purify the DNA using a microcentrifuge, proceed to Section V.A. Topurify the DNA using a vacuum manifold, proceed to Section V.B.Notes:1.Recovery from 1% high-melting-point agarose is comparable to that from1–2% low-melting-point agarose. High-melting-point agarose concentrationsof up to 3% have been tested. Gel slices with higher agarose concentrations(2–3%) may require a longer time to melt completely than a 1% agarose gelslice and may show reduced yields.2.The maximum capacity of the column is 350mg of gel mass dissolved in350μl of Membrane Binding Solution per column pass. For gel slices >350mg,continue to pass additional sample through the SV Minicolumn until all ofthe sample has been processed. The maximal amount of agarose that can beprocessed through a single column is approximately 3.5g (10 × 350mg) total.3.The maximum binding capacity of the column is approximately 40μg percolumn, and as little as 10ng has been successfully purified.4.DNA fragments that are larger than 5kb should be mixed gently to preventshearing. Do not vortex if DNA fragment is larger than 5kb; mix byinversion.4.C.Processing PCR Amplification Products1.Amplify target of choice using standard amplification conditions.2.Add an equal volume of Membrane Binding Solution to the PCRamplification (see Notes 1–4 below).3.To purify the DNA using a microcentrifuge, proceed to Section 5.A.To purify the DNA using a vacuum manifold, proceed to Section 5.B.Notes:1.The maximal capacity of a single SV Minicolumn is approximately 1mlof PCR amplification added to 1ml Membrane Binding Solution (2mltotal). For PCR volumes >350μl, continue to pass the sample through thecolumn until all of the sample has been processed.2.The maximum binding capacity is approximately 40μg per column, andas little as 10ng has been successfully purified.3.Mineral oil does not interfere with purification.4.For amplification reactions that do not produce a single product orwhere amplification has been inefficient and there is highly visibleprimer dimer, gel purification of the band of interest is recommended.Alternatively, an 80% ethanol wash solution can be substituted for thesupplied Membrane Wash Solution to reduce primer-dimer carryover.5.DNA PurificationPrepare the gel slice or PCR product as described in Section 4. Use either thecentrifugation procedure (Section 5.A) or the vacuum procedure (Section 5.B)to recover the DNA from the dissolved gel slice or PCR amplification. Afterthe procedure is completed, the DNA may be used in downstreamapplications.5.A.DNA Purification by Centrifugation1.Place one SV Minicolumn in a Collection Tube for each dissolved gelslice or PCR amplification.2.Transfer the dissolved gel mixture or prepared PCR product to the SVMinicolumn assembly and incubate for 1 minute at room temperature.3.Centrifuge the SV Minicolumn assembly in a microcentrifuge at 16,000 × g(14,000rpm) for 1 minute. Remove the SV Minicolumn from the SpinColumn assembly and discard the liquid in the Collection Tube. Returnthe SV Minicolumn to the Collection Tube.Note: Failure to spin at 16,000 × g(14,000rpm) can result in reducedyield.Promega Corporation·2800 Woods Hollow Road ·Madison, WI 53711-5399 USA Toll Free in USA 800-356-9526·Phone 608-274-4330 ·Fax 608-277-2516 · Part# TB308Printed in USA. Page 6Revised 11/09Promega Corporation ·2800 Woods Hollow Road ·Madison, WI 53711-5399 USA Toll Free in USA 800-356-9526·Phone 608-274-4330 ·Fax 608-277-2516 · Printed in USA.Part# TB308Revised 11/09Page 74.Wash the column by adding 700μl of Membrane Wash Solution, previouslydiluted with 95% ethanol (see Section 4.A), to the SV Minicolumn.Centrifuge the SV Minicolumn assembly for 1 minute at 16,000 × g(14,000rpm). Empty the Collection Tube as before and place the SVMinicolumn back in the Collection Tube. Repeat the wash with 500μl ofMembrane Wash Solution and centrifuge the SV Minicolumn assembly for5 minutes at 16,000 × g .5.Remove the SV Minicolumn assembly from the centrifuge, being careful notto wet the bottom of the column with the flowthrough. Empty the CollectionTube and recentrifuge the column assembly for 1 minute with the micro-centrifuge lid open (or off) to allow evaporation of any residual ethanol.6.Carefully transfer the SV Minicolumn to a clean 1.5ml microcentrifuge tube.Apply 50μl of Nuclease-Free Water directly to the center of the columnwithout touching the membrane with the pipette tip. Incubate at roomtemperature for 1 minute. Centrifuge for 1 minute at 16,000 × g (14,000rpm).7.Discard the SV Minicolumn and store the microcentrifuge tube containingthe eluted DNA at 4°C or –20°C.Note:The volume of the eluted DNA will be approximately 42–47μl. If theDNA needs to be further concentrated, perform an ethanol precipitation.Alternatively, the DNA may be eluted in as little as 15μl of Nuclease-FreeWater without significant reduction in yield. If using an elution volume of15μl, verify that the membrane is completely covered with Nuclease-FreeWater before centrifugation. Elution volumes less than 15μl are notrecommended (see Table 3).5.B.DNA Purification by Vacuum1.Attach one Vacuum Adapter with a Luer-Lok ®fitting to one port of themanifold (e.g., Vac-Man ®or Vac-Man ®Jr. Laboratory Vacuum Manifold)for each dissolved gel slice or PCR amplification. Insert SV Minicolumninto the Vacuum Adapter until it fits snugly in place.2.Transfer the dissolved gel mixture or PCR amplification to the SVMinicolumn and incubate for 1 minute at room temperature. Apply avacuum to pull the liquid completely through the SV Minicolumn.Note:The minimum vacuum pressure is 15 inches of mercury. See the tablebelow for comparison of inches of Hg to other pressure measurements.Promega Corporation ·2800 Woods Hollow Road ·Madison, WI 53711-5399 USA Toll Free in USA 800-356-9526·Phone 608-274-4330 ·Fax 608-277-2516 · Part# TB308Printed in USA.Revised 11/09Page 8 3.Wash the column by adding 700μl Membrane Wash Solution previouslydiluted with 95% ethanol (see Section IV.A) to the SV Minicolumn. Make sureany droplets remaining on the sides of the SV Minicolumn from the last stepare washed away. Apply a vacuum to pull the liquid through the SVMinicolumn. Repeat this wash a second time with 500μl of Membrane WashSolution.4.Turn off the vacuum source and open an unused port to vent the manifold.Remove the SV Minicolumn from the vacuum manifold and transfer to aCollection Tube. Centrifuge the SV Minicolumn assembly for 5 minutes at16,000 × g (14,000rpm) to remove any remaining Membrane Wash Solution.5.Empty the Collection Tube and recentrifuge the column assembly for 1 minutewith the microcentrifuge lid open (or off) to allow evaporation of any residualethanol.6.Carefully transfer the SV Minicolumn to a clean 1.5ml microcentrifuge tube,being careful not to wet the bottom of the SV Minicolumn with theflowthrough. Apply 50μl of Nuclease-Free Water directly to the center of thecolumn without contacting the membrane. Incubate at room temperature for1 minute. Centrifuge for 1 minute at 16,000 × g (14,000rpm).7.Discard the SV Minicolumn and store the microcentrifuge tube containing the eluted DNA at 4°C or –20°C.Note:The volume of the eluted DNA will be approximately 42–47μl. If the DNAneeds to be further concentrated, perform an ethanol precipitation. Alternatively,the DNA may be eluted in as little as 15μl of Nuclease-Free Water without asignificant reduction in yield. If using an elution volume of 15μl, verify that themembrane is completely covered with Nuclease-Free Water before centrifugation.Elution volumes less than 15μl are not recommended (see Table 3).Table 3. Percent Recovery VersusElution Volume. A 700bp PCR productwas directly purified in triplicate andquantified by ethidium bromide staining.6.TroubleshootingFor questions not addressed here, please contact your local Promega Branch Office or Distributor. Contact information available at: . E-mail: techserv@Symptoms Causes and CommentsLow DNA yield Verify that an equal volume of MembraneBinding Solution was added to the gel slice orPCR (10μl per 10mg gel slice or 10μl PCR).Make certain that the gel slice is completelymelted before proceeding with the purification.Incubation at 50–65°C is necessary tocompletely melt the gel slice.If the amount of DNA purified is too small toquantitate by spectrophotometry, quantitate byagarose gel electrophoresis followed byethidium bromide or PicoGreen®staining.Be sure to centrifuge at 16,000 × g(14,000rpm).Verify that ethanol was added to the MembraneWash Solution (see Section IV.A) and repeat thepurification.Poor results with automated Too little DNA may have been used. Increase the fluorescent sequencing amount of DNA used in sequencing reactionsor concentrate the DNA by ethanol precipitation.Up to 7μl of the eluted DNA can be used perfluorescent sequencing reaction.Too much DNA can interfere with fluorescentsequencing. Use less eluted DNA or diluteDNA prior to sequencing.If TE was used for elution, ethanol precipitatethe DNA or repurify the DNA fragments andelute with Nuclease-Free Water.Excessive thymidine-dimer formation may haveoccurred during UV exposure. See references1–4 for a method to minimize thymidine-dimerformation of AT-rich templates.Poor restriction digestion Increase the amount of restriction enzymeand/or the length of incubation time. Digest atthe appropriate temperature and in the optimalbuffer for the restriction enzyme used.Ethanol or salt carryover into the eluted DNAmay have occurred. Ethanol precipitate theDNA or keep the DNA volume to 10% or less ofthe final reaction volume.Promega Corporation·2800 Woods Hollow Road ·Madison, WI 53711-5399 USA Toll Free in USA 800-356-9526·Phone 608-274-4330 ·Fax 608-277-2516 · Printed in USA.Part# TB308 Revised 11/09Page 96.Troubleshooting (continued)Symptoms Causes and CommentsDNA yields on gel look low Trace contaminants in eluted DNA can artificially compared to spectrophotometric inflate spectrophotometer readings. Use agarosereadings gel electrophoresis followed by ethidium bromideor PicoGreen®staining to determine DNA yields.Ethanol precipitate the DNA.Low A260/A230ratios Typically due to guanidine isothiocyanatecontamination. Low ratios do not necessarilyindicate that the DNA will function poorly indownstream applications. Ethanol precipitatethe DNA if low A260/A230ratio is a concern.Clogged spin basket Increase the length of the 50–65°C incubation toensure the gel slice is completely melted.Verify that an equal ratio of Membrane BindingSolution to gel slice mass is used (10μl per 10mg).A vacuum pressure of >15 inches of mercury isrequired to use the SV Minicolumn in thevacuum protocol. If the vacuum is insufficient,use the spin protocol.Purified DNA floats out of the Ethanol carryover. Be certain that the Membranewell when loaded on a gel Wash Solution is not carried over from the washsteps. If the column has been wet, empty theCollection Tube and recentrifuge the columnassembly for 1 minute. Centrifuge the SVMinicolumn for 5 minutes to remove residualMembrane Wash Solution. After washing,centrifuge the column assembly with themicrocentrifuge lid open or off (Section 5.A.,Step 5; Section 5.B., Step 5) to allow evaporationof any residual ethanol.Add 3X loading dye to the DNA sample beforeloading onto the gel.Purified DNA bands are not sharp DNA may be sheared. Mix the agarose gel slicegently with the Membrane Binding Solution.Nuclease contamination may be an issue.Autoclave the gel running buffer before use.Store the gel slice at 4°C or –20°C for no morethan 1 week under nuclease-free conditions.Low cloning efficiency May be due to guanidine isothiocyanatecontamination. Ethanol precipitate the DNA,washing the pellet with 70% ethanol to reducecontamination.Promega Corporation·2800 Woods Hollow Road ·Madison, WI 53711-5399 USA Toll Free in USA 800-356-9526·Phone 608-274-4330 ·Fax 608-277-2516 · Part# TB308Printed in USA. Page 10Revised 11/097.References1.Zimmermann, M., Veeck, J. and Wolf, K. (1998) Minimizing the exposure to UV light when extracting DNA from agarose gels. BioTechniques 25, 586.2.Hengen, P. (1997) Methods and reagents. Protecting vector DNA from UV light.Trends Biochem. Sci.22, 182–3.3.Grundemann, D. and Schomig, E. (1996) Protection of DNA during preparativeagarose gel electrophoresis against damage induced by ultraviolet light. BioTechniques 21, 898–903.4.Cariello, N.F. et al . (1988) DNA damage produced by ethidium bromide staining and exposure to ultraviolet light. Nucl. Acids Res.16, 4157.8.Appendix8.A.Composition of Buffers and SolutionsPromega Corporation ·2800 Woods Hollow Road ·Madison, WI 53711-5399 USA Toll Free in USA 800-356-9526·Phone 608-274-4330 ·Fax 608-277-2516 ·Printed in USA.Part# TB308Revised 11/09Page 11Membrane Wash Solution (after ethanol addition)10mM potassium acetate (pH 5.0)80%ethanol16.7μM EDTA (pH 8.0)To prepare this solution, add 95%ethanol to the supplied Membrane Wash Solution (concentrated) as described in Table 2 in Section IV.A.Membrane Binding Solution4.5M guanidine isothiocyanate 0.5M potassium acetate (pH5.0)1X TE buffer10mM Tris-HCl (pH 7.5)1mM EDTA (pH 8.0)1X TBE buffer 89mM Tris base 89mM boric acid 2mM EDTA (pH 8.0)1X TAE buffer 40mM Tris base5mM sodium acetate 1mM EDTA (pH 8.0)8.B.Related ProductsProductSize Cat. #Vacuum Adapters20 each A1331Wizard ®SV 96 PCR Clean-Up System 11 × 96 preps A93404 × 96 preps A93418 × 96 preps A9342100 × 96 preps A9345GoTaq ®Green Master Mix 1,2100 reactions M71221,000 reactions M7123GoTaq ®Colorless Master Mix 1,2100 reactions M71321,000 reactions M7133GoTaq ®DNA Polymerase 1,2100 units M3001500 units M30052,500 units M3008GoTaq ®HotStart Polymerase 2,3100 units*M5001GoTaq ®HotStart Green Master Mix 2,3100 reactions*M5122GoTaq ®HotStart Colorless Master Mix 2,3100 reactions*M5132Access RT-PCR System 120 reactions A1260100 reactions A1250500 reactions A1280AccessQuick™ RT-PCR System 120 reactions A1701100 reactions A1702500 reactions A1703Agarose, LE, Analytical Grade 1100g V3121500g V3125Ethidium Bromide Solution, Molecular Grade 110ml H5041TAE Buffer,10X 1,000ml V4271TBE Buffer, 10X 1,000ml V42511For Laboratory Use.2Different Cat.# may apply for customers in Europe. Visit /catalog/ forthe amplifcation product catalog numbers appropriate for your location.3For Research Use. Not for use in diagnostic procedures.*Additional sizes available.Promega Corporation ·2800 Woods Hollow Road ·Madison, WI 53711-5399 USA Toll Free in USA 800-356-9526·Phone 608-274-4330 ·Fax 608-277-2516 ·Part# TB308Printed in USA.Revised 11/09Page 12© 2002, 2004, 2005 and 2009 Promega Corporation. All Rights Reserved.GoTaq, Vac-Man and Wizard are registered trademarks of Promega Corporation. AccessQuick is a trademark of Promega Corporation.Luer-Lok is a registered trademark of Becton, Dickinson and Company. PicoGreen is a registered trademark of Molecular Probes, Inc.Products may be covered by pending or issued patents or may have certain limitations. Please visit our Web site for more information.All prices and specifications are subject to change without prior notice.Product claims are subject to change. Please contact Promega Technical Services or access the Promega online catalog for the most up-to-date information on Promega products.。
