DNAMAN使用说明
序列分析软件DNAMAN 的使用方法简介

序列分析软件DNAMAN 的使用方法简介DNAMAN 是一种常用的核酸序列分析软件。
由于它功能强大,使用方便,已成为一种普遍使用的DNA 序列分析工具。
本文以DNAMAN 5.2.9 Demo version 为例,简单介绍其使用方法。
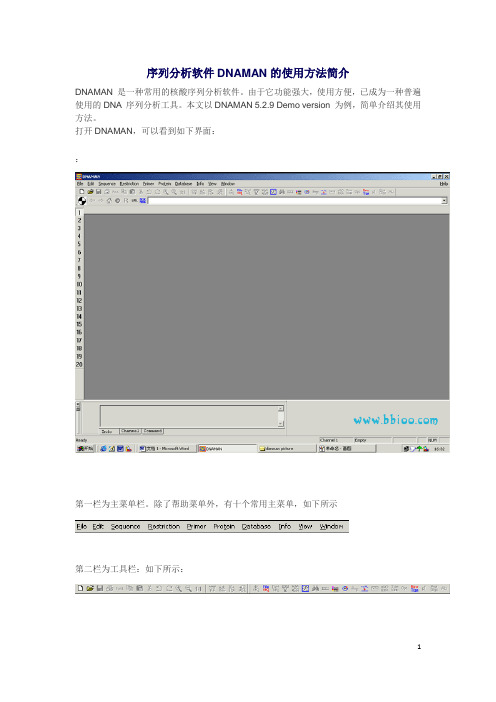
打开DNAMAN,可以看到如下界面:第一栏为主菜单栏。
除了帮助菜单外,有十个常用主菜单,第二栏为工具栏:第三栏为浏览器栏:在浏览器栏下方的工作区左侧,可见Channel 工具条,DNAMAN 提供20 个Channel,(如左所示:)点击Channel 工具条上相应的数字,即可击活相应的Channel。
每个Channel 可以装入一个序列。
将要分析的序列(DNA 序列或氨基酸序列)放入Channel 中可以节约存取序列时间,加快分析速度。
此版本DNAMAN 提供自动载入功能,用户只需激活某个Channel,然后打开一个序列文件,则打开的序列自动载入被激活的Channel 中。
本文以具体使用DNAMAN 的过程为例来说明如何使用DNAMAN 分析序列。
1.将待分析序列装入Channel(1)通过File Open 命令打开待分析序列文件,则打开的序列自动装入默认Channel。
(初始为channel1)可以通过激活不同的channel (例如:channel5)来改变序列装入的Channel。
(2)通过Sequence/Load Sequence 菜单的子菜单打开文件或将选定的部分序列装入Channel 。
通过Sequence/Current Sequence/Analysis Defination 命令打开一个对话框,通过此对话框可以设定序列的性质(DNA 或蛋白质),名称,要分析的片段等参数。
2.以不同形式显示序列通过Sequence//Display Sequence 命令打开对话框,如下图所示:根据不同的需要,可以选择显示不同的序列转换形式。
对话框选项说明如下:Sequence &Composition 显示序列和成分Reverse Complement Sequence 显示待分析序列的反向互补序列Reverse Sequence 显示待分析序列的反向序列Complement Sequence 显示待分析序列的互补序列Double Stranded Sequence 显示待分析序列的双链序列RNA Sequence 显示待分析序列的对应RNA 序列3.DNA 序列的限制性酶切位点分析将待分析的序列装入Channel,点击要分析的Channel,然后通过Restriction/Analysis 命令打开对话框,如下所示:参数说明如下:Results 分析结果显示其中包括:Show summary(显示概要) Show sites on sequence(在结果中显示酶切位点)Draw restriction map(显示限制性酶切图)Draw restriction pattern(显示限制性酶切模式图)Ignore enzymes with more than(忽略大于某设定值的酶切位点)Ignore enzymes with less than(忽略小于某设定值的酶切位点)Target DNA (目标DNA 特性)circular(环型DNA),dam/dcm methylation(dam/dcm 甲基化)all DNA in Sequence Channel(选择此项,在Sequence Channel 中的所有序列将被分析,如果选择了Draw restriction pattern,那么当所有的channel 中共有两条DNA 时,则只能选择两个酶分析,如果共有三个以上DNA 时,则只能用一个酶分析。
DNAman使用说明

查看文章DNAMAN使用说明书(中文)2008年04月16日星期三下午10:50DNAMAN 是一种常用的核酸序列分析软件。
由于它功能强大,使用方便,已成为一种普遍使用的DNA 序列分析工具。
本文以DNAMAN 5.2.9 Demo version 为例,简单介绍其使用方法。
打开DNAMAN,可以看到如下界面:第一栏为主菜单栏。
除了帮助菜单外,有十个常用主菜单,第二栏为工具栏:第三栏为浏览器栏:在浏览器栏下方的工作区左侧,可见Channel 工具条,DNAMAN 提供20 个Channel,(如左所示:)点击Channel 工具条上相应的数字,即可击活相应的Channel。
每个Channel 可以装入一个序列。
将要分析的序列(DNA 序列或氨基酸序列)放入Channel 中可以节约存取序列时间,加快分析速度。
此版本DNAMAN 提供自动载入功能,用户只需激活某个Channel,然后打开一个序列文件,则打开的序列自动载入被激活的Channel 中。
本文以具体使用DNAMAN 的过程为例来说明如何使用DNAMAN 分析序列。
1.将待分析序列装入Channel(1)通过File Open 命令打开待分析序列文件,则打开的序列自动装入默认Channel。
(初始为channel1)可以通过激活不同的channel (例如:channel5)来改变序列装入的Channel。
(2)通过Sequence/Load Sequence 菜单的子菜单打开文件或将选定的部分序列装入Channel 。
通过Sequence/Current Sequence/Analysis Defination 命令打开一个对话框,通过此对话框可以设定序列的性质(DNA 或蛋白质),名称,要分析的片段等参数。
2.以不同形式显示序列通过Sequence//Display Sequence 命令打开对话框,如下图所示:根据不同的需要,可以选择显示不同的序列转换形式。
dnaman使用方法

dnaman使用方法使用DNAMAN的方法DNAMAN是一款功能强大的生物信息学软件,广泛应用于DNA 和蛋白质序列分析、比对、编辑和可视化等领域。
本文将介绍DNAMAN的使用方法,帮助读者快速上手并熟练运用该软件。
一、安装和启动DNAMAN1. 下载软件安装包:在DNAMAN官方网站上下载适用于您的操作系统的安装包,并保存到本地。
2. 安装软件:双击安装包,按照提示完成软件的安装过程。
3. 启动软件:安装完成后,双击桌面上的DNAMAN图标,即可启动软件。
二、导入和编辑序列1. 导入序列:在DNAMAN的主界面,点击"文件"菜单,选择"导入序列",然后选择您要导入的序列文件(支持FASTA、GenBank 等格式)。
2. 编辑序列:在序列编辑界面,您可以对序列进行添加、删除、替换等操作。
点击"编辑"菜单,选择相应的编辑功能,然后在弹出的对话框中进行操作。
三、序列比对和分析1. 序列比对:点击"工具"菜单,选择"序列比对",然后选择比对算法和参数设置,点击"开始比对"按钮即可进行序列比对。
2. 序列分析:DNAMAN提供了丰富的序列分析工具,如ORF预测、限制酶切位点分析、引物设计等。
点击"工具"菜单,选择相应的分析工具,按照提示进行操作。
四、序列可视化和输出1. 序列可视化:DNAMAN提供了多种序列可视化方式,如线性图、环形图、比对图等。
点击"视图"菜单,选择相应的可视化方式,即可查看序列的结构和特征。
2. 输出结果:完成序列分析后,您可以将结果导出为文本文件或图像文件。
点击"文件"菜单,选择"导出结果",然后选择输出格式和保存路径,点击"保存"按钮即可导出结果。
五、保存和管理项目1. 保存项目:在使用DNAMAN进行序列分析时,建议您保存项目以便后续操作。
序列分析软件DNAMAN_的使用方法中文

4.DNA 序列比对分析 (Dot Matrix Comparision)
要比较两个序列,可以使用DNAMAN 提供的序列比对工具Dot Matrix Comparision (点矩阵比较)通过 Sequense/Dot matrix comparision 命令打开比对界面, 点击对比界面左上角的按钮,出现下列 对话框:
Annotations 是否显示注释 Comparision 比对参数, 其中Window 代表Window size(单位比对长度), Mismatch 代表Mismatch size(单位比对长度中许 可的错配值)要快速比对,需将此项设为0。 Both stran 代表Both strand(双链比对)选择此项, 是指用Sequence 2 中的序列的正链和负链分别和 Sequence 1 比较。 Sequence 2 正链与Sequence 1 比较结果用黑色点 表示,Sequence 2 负链比对结果用红色点表示。
饶志明
博士/教授/ 博士生导师
江南大学生物工程学院工业微生物中心 江南大学工业生物技术教育部重点实验室
E-mail: raozhm@
DNAMAN 是一种常用的核酸序列分析 软件。由于它功能强大,使用方便,已 成为一种普遍使用的DNA 序列分析工具。
打开DNAMAN,可以看到如下界面:
3.DNA 序列的限制性酶切位点分析
将待分析的序列装入Channel,点击要分析的 Channel,然后通过Restriction/Analysis 命令打开 对话框, 参数说明如下: Results 分析结果显示 其中包括: Show summary(显示概要) Show sites on sequence(在结果中显示酶切位点) Draw restriction map(显示限制性酶切图) Draw restriction pattern(显示限制性酶切模式图)
序列分析软件DNAMan
序列分析软件DNAMAN 的使用方法简介DNAMAN 是一种常用的核酸序列分析软件。
由于它功能强大,使用方便,已成为一种普遍使用的DNA 序列分析工具。
本文以DNAMAN 5.2.9 Demo version 为例,简单介绍其使用方法。
打开DNAMAN,可以看到如下界面::第一栏为主菜单栏。
除了帮助菜单外,有十个常用主菜单,如下所示第二栏为工具栏:如下所示:第三栏为浏览器栏:如下所示:在浏览器栏下方的工作区左侧,可见Channel 工具条,DNAMAN 提供20 个Channel,如左所示:点击Channel 工具条上相应的数字,即可击活相应的Channel。
每个Channel 可以装入一个序列。
将要分析的序列(DNA 序列或氨基酸序列)放入Channel 中可以节约存取序列时间,加快分析速度。
此版本DNAMAN 提供自动载入功能,用户只需激活某个Channel ,然后打开一个序列文件,则打开的序列自动载入被激活的Channel 中。
本文以具体使用DNAMAN 的过程为例来说明如何使用DNAMAN 分析序列。
1.将待分析序列装入Channel(1)通过File|Open 命令打开待分析序列文件,则打开的序列自动装入默认Channel。
(初始为channel1)可以通过激活不同的channel(例如:channel5)来改变序列装入的Channel。
(2)通过Sequence|Load Sequence 菜单的子菜单打开文件或将选定的部分序列装入Channel。
可以通过Sequence|Current Sequence|Analysis Defination 命令打开一个对话框,通过此对话框可以设定序列的性质(DNA 或蛋白质),名称,要分析的片段等参数。
2.以不同形式显示序列通过Sequence|Display Sequence 命令打开对话框,如下图所示:根据不同的需要,可以选择显示不同的序列转换形式。
序列分析软件DNAMAN的使用方法

DNAMAN 是一种常用的核酸序列分析 软件。由于它功能强大,使用方便,已 成为一种普遍使用的DNA 序列分析工具。
打开DNAMAN,可以看到如下界面:
第一栏为主菜单栏。除了帮助菜单外,有十个 常用主菜单, 第二栏为工具栏: 第三栏为浏览器栏: 在浏览器栏下方的工作 区左侧,可见Channel 工具条,DNAMAN 提 供20 个Channel,点击Channel 工具条上相 应的数字,即可击活相应的Channel。 每个Channel 可以装入一个序列。将要分析 的序列(DNA 序列或氨基酸序列)放入 Channel 中可以节约存取序列时间,加快分 析速度。
Annotations 是否显示注释 Comparision 比对参数, 其中Window 代表Window size(单位比对长度), Mismatch 代表Mismatch size(单位比对长度中许 可的错配值)要快速比对,需将此项设为0。 Both stran 代表Both strand(双链比对)选择此项, 是指用Sequence 2 中的序列的正链和负链分别和 Sequence 1 比较。 Sequence 2 正链与Sequence 1 比较结果用黑色点 表示,Sequence 2 负链比对结果用红色点表示。
ห้องสมุดไป่ตู้
选择所需的项目,然后按提示操作点击按扭,出现下 列对话框: 参数说明如下: Enzyme 代表(enzyme data file),点击旁边的下拉按钮, 出现两个默认选项,restrict.enz 和dnamane.enz, 如果添加过自制的酶列表,则附加显示自制酶列表文 件名。 其中restrict.enz 数据文件包含180 种限制酶, dnamane.enz 数据文件包含2524 种限制酶。 选择其中一个数据文件,相应的酶在左边的显示框中 列出(按酶名称字母表顺序),鼠标双击酶名称,则 对应的酶被选中,在右边空白框中列出。
DNAMAN中文使用说明
DNAMAN中文使用说明好不容易找到了一个中文说明,希望可以帮助初学使用DNAMAN的朋友,更快的进入状态,当然现在网上也有汉化版的软件了,但是这个说明还是可以起到很好的帮助作用,与大家分享!DNAMAN 是一种常用的核酸序列分析软件。
由于它功能强大,使用方便,已成为一种普遍使用的DNA 序列分析工具。
本文以DNAMAN 5.2.9 Demo version 为例,简单介绍其使用方法。
打开DNAMAN,可以看到如下界面:第一栏为主菜单栏。
除了帮助菜单外,有十个常用主菜单,第二栏为工具栏:第三栏为浏览器栏:在浏览器栏下方的工作区左侧,可见Channel 工具条,DNAMAN 提供20 个Channel,(如左所示:)点击Channel 工具条上相应的数字,即可击活相应的Channel。
每个Channel 可以装入一个序列。
将要分析的序列(DNA 序列或氨基酸序列)放入Channel 中可以节约存取序列时间,加快分析速度。
此版本DNAMAN 提供自动载入功能,用户只需激活某个Channel,然后打开一个序列文件,则打开的序列自动载入被激活的Channel 中。
本文以具体使用DNAMAN 的过程为例来说明如何使用DNAMAN 分析序列。
1.将待分析序列装入Channel(1)通过File Open 命令打开待分析序列文件,则打开的序列自动装入默认Channel。
(初始为channel1)可以通过激活不同的channel (例如:channel5)来改变序列装入的Channel。
(2)通过Sequence/Load Sequence 菜单的子菜单打开文件或将选定的部分序列装入Channel 。
通过Sequence/Current Sequence/Analysis Defination 命令打开一个对话框,通过此对话框可以设定序列的性质(DNA 或蛋白质),名称,要分析的片段等参数。
2.以不同形式显示序列通过Sequence//Display Sequence 命令打开对话框,如下图所示:根据不同的需要,可以选择显示不同的序列转换形式。
序列分析软件DNAMAN的使用方法中文演示文稿
序列分析软件DNAMAN的使用方法中文演示文稿第一部分:软件介绍1.DNAMAN是什么?-DNAMAN是一款用于DNA和蛋白质序列分析的软件。
-它提供多种功能,包括序列比对、引物设计、限制酶分析和进化树构建等。
2.DNAMAN的应用领域-DNAMAN广泛应用于生物学、生物技术和医药领域。
-它可以帮助研究人员进行序列分析、设计实验方案和解读实验结果。
第二部分:基本序列比对1.创建新项目-打开DNAMAN软件,点击“新建”按钮创建新项目。
-输入项目名称和序列信息,保存并打开该项目。
2.导入序列-点击“导入”按钮,选择需要比对的序列文件,点击“确定”导入。
-系统会自动将序列导入到项目中。
3.序列比对-选择需要比对的序列,点击“比对”按钮进行序列比对。
-系统会自动比对序列并生成比对结果。
4.结果解读-比对结果以图形和文本形式展示。
-可以通过选择不同的比对算法和调整参数来优化比对结果。
第三部分:引物设计1.创建新项目-打开DNAMAN软件,点击“新建”按钮创建新项目。
-输入项目名称和序列信息,保存并打开该项目。
2.导入标记序列-点击“导入”按钮,选择需要设计引物的标记序列文件,点击“确定”导入。
-系统会自动将标记序列导入到项目中。
3.引物设计-点击“引物设计”按钮,选择设计引物的参数和算法。
-系统会根据所选参数和算法自动生成引物设计结果。
4.结果解读-引物设计结果以图形和文本形式展示。
-可以通过选择不同的参数和算法来优化引物设计结果。
第四部分:限制酶分析1.创建新项目-打开DNAMAN软件,点击“新建”按钮创建新项目。
-输入项目名称和序列信息,保存并打开该项目。
2.导入限制酶序列-点击“导入”按钮,选择需要分析的限制酶序列文件,点击“确定”导入。
-系统会自动将限制酶序列导入到项目中。
3.限制酶分析-点击“限制酶分析”按钮,选择分析参数和算法。
-系统会根据所选参数和算法自动进行限制酶分析。
4.结果解读-限制酶分析结果以图形和文本形式展示。
DNAMAN的使用方法
DNAMAN的使用方法DNAMAN是一种快速、准确、高通量的DNA检测工具,可以用于各种生物学研究和临床应用中。
DNAMAN的使用非常简单,但有一些关键步骤和注意事项需要注意。
以下是DNAMAN的详细使用方法,以帮助您充分利用这个工具。
第一步:准备样本第二步:DNA提取DNA提取是DNAMAN使用的关键步骤,它有助于从样本中纯化和分离出DNA。
您可以使用商用的DNA提取试剂盒,按照说明书中的步骤进行操作。
确保遵循所有操作步骤,并注意避免任何与其他样本的交叉污染。
第三步:DNA定量第四步:稀释DNA根据DNAMAN的说明书,使用PBS或其他适当的缓冲溶液将DNA稀释到所需的浓度。
确保在实验室条件下进行稀释,并遵循操作规范,以避免任何误差。
第五步:添加DNA至DNAMAN芯片将稀释后的DNA样品添加到DNAMAN芯片中的指定位置。
根据芯片的标记,确保每个样品正确放置,并避免任何交叉污染。
如果需要,可以使用多通道移液器进行操作,以提高效率。
第六步:启动DNAMAN分析在DNAMAN分析仪上启动相应的分析程序。
根据仪器的说明书,选择适当的参数和设置。
确保校准仪器,以获得准确的结果。
第七步:数据收集和解读注意事项:1.在操作DNAMAN之前,确保您已经熟悉仪器的说明书和操作流程。
遵循所有操作规程,以确保准确性和结果的可靠性。
2.在使用DNAMAN之前,请确保仪器已经进行了校准和维护,并且仪器状态良好。
如有必要,请进行常规性能检查,并在需要时进行任何维护和修理。
3.在操作中避免交叉污染。
使用带有盖子的离心管和一次性移液器尖,避免任何样品之间的接触。
4.尽量减少操作时间,以避免DNA的降解。
在整个实验过程中,尽量保持操作环境的清洁和无尘。
5.定期评估和验证DNAMAN的性能。
与其他方法进行对比试验,以确保DNAMAN的结果的准确性和可靠性。
总结:DNAMAN是一种非常有用的DNA检测工具,可以在各种生物学研究和临床应用中得到应用。
DNAman使用方法
7.画质粒模式图
参数说明如下: Name : 要添加的酶切位点 的名称(例如HindIII) Position:位置(以碱基数表 示) 点击 Add Element选项,出 现如下对话框:
• Type :要素类型(共有三种类型, 鼠标点击即可切换) • (c)olor/Pattern: 填充色(共有 16种颜色供选择) • Name: 要素名称 • Start /End/Size:要素起点/终点/粗 细度 • 点击Add Text选项,出现如下对 话框:
6.PCR引物设计
点击 按钮,出现下列对话框: 选择选项,点击 按钮,出现:
7.画质粒模式图
• 我们常常要用到各种质粒图,无论是制作幻灯片,还是发 表文章,常常需要质粒图。DNAMAN 提供强大的绘质粒 图功能,能满足我们的需要。通过Restriction/Draw map 命令打开质粒绘图界面:
DNAMAN 使用方法
• 1.将待分析序列装入Channel
• • • • • •
2.以不同形式显示序列 3.DNA序列的限制性酶切位点分析 4.DNA序列比对分析 5.序列同源性分析 6. PCR引物设计 7.质粒模式图绘制
DNAMAN 使用方法
• DNAMAN是一种常用的核酸序列分析软件。由于它功能 强大,使用方便,已成为普遍使用的DNA序列分析工具。 以DNAMAN 5.2.9 Demo version为例,简单介绍其使用 方法。打开DNAMAN,可以看到如下界面:
• 输入要保存酶列表的文件名,点击 按钮即可保存。 自制酶列表可以方便分析特定的酶切位点。 • • Cutter 酶切识别序列长度;End 酶切产生的末端,其中 包括,Blunt(平头末端),5’Overhang(5’突出粘性末 端),3’Overhang(3’突出粘性末端),系统根据cutter和 end的设定情况,在左边酶列表中显示符合条件的酶。最后, 点击 按钮执行操作。
- 1、下载文档前请自行甄别文档内容的完整性,平台不提供额外的编辑、内容补充、找答案等附加服务。
- 2、"仅部分预览"的文档,不可在线预览部分如存在完整性等问题,可反馈申请退款(可完整预览的文档不适用该条件!)。
- 3、如文档侵犯您的权益,请联系客服反馈,我们会尽快为您处理(人工客服工作时间:9:00-18:30)。
DNAMAN for WindowsDNAMAN uses advanced features of Microsoft Windows and offers versatile visual tool analysis.Sequence Manipulation生物秀-专心做生物w w w .b b i o o .c o mSequence inputDNAMAN provides 20 sequence channels to keep active sequences in memory. These channels simplify multiple functional analyses and substantially increase the your works. A panel window shows the current working sequence. You can edit th directly in the panel.Information exchangeDNAMAN accepts sequence files in GenBank, GCG, CUSTAL, FASTA, PIR and GDE form export multiple sequences in GCG, CUSTAL, PIR and GDE format.DNAMAN provides a word processor for sequence editing. With this word processo incorporate charts, images, graphics from any other Windows software into your DNAMAN works as an object linking and embedding (OLE) server to exchange the i restriction maps with other software. With OLE, you can incorporate your restr other Windows applications, and directly modify the maps within the applicatio You can directly access the Internet with DNAMAN through the Netscape browser.the browser into a document of the DNAMAN word processor, you can directly exc information between your documents and the Internet.生物秀-专心做生物w w w .b b i o o .c o mEditingEditing a text file with DNAMAN is as easy as working with your favorite word the word processor of DNAMAN, you can edit original sequence files, and analys well.Sequence composition and conversionDNAMAN reports the composition and molecular weight of a sequence. It performs of a sequence to its reverse, complementary, reverse complementary, double str sequences.生物秀-专心做生物w w w .b b i o o .c o mBLAST documents/Internet accessIn addition to accessing the Internet through Web browser, DNAMAN prepares a f document formats with a query sequence for directly accessing BLAST E-mail Ser document formats are currently available: Blastn, Blastx, Tblastx, Blastp and Examples:Sequence SearchSearch for nucleotide sequencesWith DNAMAN, you can search for nucleotide sequences from one or both str DNA sequence. Gaps and ambiguous nucleotides are allowed in the query seq can also provide multiple query sequences for searching.生物秀-专心做生物w w w .b b i o o .c o mDNAMAN instantly reports the searching results in graphics. Colors and ar indicate different sequence groups and sites. You can magnify any region fragment and display the regional sequence of by selection.The versatility of query sequences and graphical presentations make the s function useful in many other areas, such as presenting restriction sites and gene structures.Search for consensus sequences With DNAMAN, you can search for DNA or protein consensus sequences from b or six reading frames of DNA sequences. DNAMAN provides a database of DNA protein consensus sequences. The database is expandable and editable. You custom consensus sequence databases.Search for open reading frames生物秀-专心做生物w w w .b b i o o .c o mYou can search for open reading frames from six reading frames of a DNA s The searching result is shown in a text table. DNAMAN can also display th a graphic window.Search for repeat sequencesYou can search for direct repeat and reverse repeat sequences from both stands o DNA sequence. You can also search for reverse complementary repeat sequences tha may form hairpin/stem-loop structures.Search for amino acid sequencesYou can search for an amino acid sequence and its variations from the six frames of a DNA sequence. DNAMAN allows ambiguous amino acids as well as mismatches in a query sequence.Restriction AnalysisRestriction site analysisYou can search any restriction site on a DNA sequence. DNAMAN supplies twrestriction enzyme files; the restrict.enz file with 180 most frequently used restriction enzymes, and the dnamanre.enz file with 1364 enzyme records. You can also create your own enzyme files. All the enzyme files are editable and DNAMAN provides custom restriction enzyme filters on cutter, ends, freque methylation sensitivity. Users can define the DNA molecule as a linear or type.DNAMAN reports the restriction analysis results in an easy-to-read table.cutting sites are shown in alphabetical order of enzymes, and in site pos as well. The non-cutting enzymes are also listed. In addition, DNAMAN dis enzyme sites on the top of the DNA sequence.Restriction mapDNAMAN provides easy-to-use tools to produce publication-quality restrict These tools can be used to draw linear or circular restriction maps with DNA sequence information. You can also draw maps for other projects, such strategy diagrams, gene structural maps, etc.生物秀-专心做生物w w w .b b i o o .c o mDNAMAN accompanies with a high quality drawing program, LBDraw. Working w you can easily make sophisticated diagrams with many restriction maps. Restriction pattern illustrationDNAMAN predicts the patterns of restriction enzyme digested DNA fragments electrophoresis. You can perform single enzyme digestion on multiple sequ single or multiple digestion on a single sequence. DNAMAN shows the infor restriction fragments on their sizes and ends when you click on these fraElectronic cloningDNA cloning is a time consuming and expensive process. DNAMAN provides ea tools to design a cloning strategy and performs evaluation analyses on ta sequences. This feature could improve the efficiency of your cloning worklaboratory.DNAMAN mimics basic steps of the actual cloning process:l Restriction analysis on the original vector and insert sequences l Selection of vector and insert fragments from restriction pattern l Verification of the end compatibility of DNA fragments l Modification of fragment ends if necessary l Insertion of linkers if necessary lProducing the final clone sequenceConstructing restriction mapsDNAMAN can help you reconstruct a restriction map in the absence of DNA s You must provide all fragment sizes in single and double digestion. DNAMA the possible restriction map(s) from the information of restriction fragmSilent mutation analysisSilent mutation analysis allows you to design a desired mutation site on sequence. This mutation will result in the modification of restriction pr without changing the coding amino acid sequence. This function searches f potential mutation positions to create or destroy restriction enzyme siteDirected mismatch analysisDirected mismatch analysis allows you to create or remove restriction sit sequence or its mutants (variants) by incorporating a single or double mi site near the mutation. Using this function you can create or destroy a r site in order to distinguish the wild type allele and a common mutant allSequence AssemblySequence assembly method生物秀-专心做生物w w w .b b i o o .c o mDNAMAN uses fast alignment algorithms to assemble quickly and accurately a lar overlapping sequences. DNAMAN can automatically adjust the orientation of each remove vector sequences as well. You can set sensitivity parameters to control ambiguous sequences in contigs.Sequence assembly editorDNAMAN displays sequence assembly project in three windows:lGraphic window provides an overview of the assembly construction. You can edit t graphical presentation to produce high quality diagrams for publications.lName list window contains all assembled sequence names. You can change the seque by moving the sequence names in this window.lSequence window shows all original sequences and a consensus sequence. You can e of the original sequences to improve the result of sequence assembly.DNAMAN exports sequence the assembly result in a text window. You have op reporting the consensus sequence only, or all sequences including the con and original sequences.Sequence Homology AnalysesDot matrix plotWith DNAMAN, you can compare two DNA sequences or two protein sequences in a d There are two methods for comparison. Method 1 uses a fast algorithm and is su sequences. Method 2 performs progressive comparison to reduce the background o regions. The two methods yield different speed and accuracy in analysis. DNAMAN displays the actual sequences and their alignment on any selected regio 生物秀-专心做生物w w w .b b i o o .c o mwindow.Two sequence alignmentDNAMAN uses fast or optimal algorithms to align two DNA or protein sequences. to control the sensitivity of alignment. DNAMAN also allows you to select any sequences for alignment.For DNA sequence alignment, you have an option to use the minus strand for com protein sequence alignment, DNAMAN can report the amino acid similarity of two sequences in a text window. The amino acid similarity matrix is editable by us生物秀-专心做生物w w w .b b i o o .c o mMultiple Sequence AlignmentMultiple sequence alignment methodsFast alignmentDNAMAN uses Wilbur and Lipman algorithm for fast alignment. You can quickly al number of DNA or protein sequences. This algorithm is able to produce an accur sequence divergence is low. Optimal alignmentFeng-Doolittle and Thompson method (CLUSTAL) is used for optimal sequence alig input individual sequences and/or multiple alignment profiles. There are three alignment:l Full alignment to completely align all sequences.l Profile alignment to align two multiple alignment profiles without distur original alignments.lNew sequence on profile to align a new sequence with an existing multiple profile.You can optimize multiple alignment by selecting alignment criteria in the eas panel.Multiple sequence alignment editorDNAMAN provides a high performance alignment editor. A graphical view of the a you to quickly move to an interesting region. You can change the alignment lis and drop sequence names. You are also able to add or delete gap insertions, mowithin a gap and truncate aligned sequences.生物秀-专心做生物w w w .b b i o o .c o mYou can modify the appearance of multiple alignment sequences:ldisplaying identical residues in colors or blocks l displaying consensus sequence lchanging text fontMultiple alignment input and outputYou can directly input sequences or multiple alignment profiles for alignment following sources:l GenBank,l EMBL/Swiss Prot, l GCG/MSF, l CUSTAL, l FASTA, l NBRF/PIR lGDEThe multiple alignment editor can output an alignment in different formats:NBRF/PIR , and GDE . The multiple input and out put capacity of DNAMAN makes it compati major sequence analysis software.Phylogenetic treesDNAMAN calculates the homology matrix and establishes related distances betweesequences. Consequently, DNAMAN can output a distance matrix of multiple alignment, a phylogenetic trees or homology tree s. You can carry out bootstrapping tests for the c value of a phylogenetic tree.生物秀-专心做生物w w w .b b i o o .c o mRestriction analysisIf the sequences in multiple alignment editor are DNA, you can perform a restr on these sequences. This analysis is useful in restriction site comparison of sequences.Hydrophobic / hydrophilic profilesIf the sequences in multiple alignment editor are protein, you can plot the hy hydrophilic profile of all sequences for comparison.Protein secondary structure predictionDNAMAN predicts the secondary structure of multiple protein sequences using th 生物秀-专心做生物w w w .b b i o o .c o mdeveloped by King and Sternberg.Primer AnalysisPrimer designThe function of primer design includes not only primer filtration by Tm, but a and restriction analyses on the primers. DNAMAN can help you to find optimal p satisfy your requirements.lDNAMAN allows you to set numerous control criteria for optimal primer fil the regions of target DNA, size of PCR products, primer characteristics, conditions and primer configurations.lYou can carry out a restriction analysis on the primers in order to selec without restriction site(s).l You can discard the primers that are easy to anneal to secondary sites of using mispriming analysis.Melting temperature predictionDNAMAN calculates and reports the thermodynamic Tm, hybridization Tm, and GC+A hybridization. You can also have the Tm information on the hybridization of RNA . These Tms can be used for PCR primers as well as hybridization probes.Complementarity of primersPrimer complementarity may affect the performance of PCR primers or hybridizat DNAMAN analyzes the following three kinds of primer complementarity.lSelf-complementaryDNAMAN searches for the most possible self-complementary configuration of the lowest free energy.l Complementarity with target DNADNAMAN searches for the complementary sequences between the primer and bo target DNA.lTwo primer complementarity生物秀-专心做生物w w w .b b i o o .c o mDNAMAN searches for complementary sequences between two primers. It repor continuous and discontinuous complementary sequences.Mispriming analysisWith mispriming analysis you can search for all possible annealing sites of aDNA sequence. DNAMAN allows you to set up Score matrix : perfect match, mismatch and G Position weight matrix , Gap penalty and Cut-off score . This analysis can eliminate PC that are easy to anneal to secondary sites.Silent mutation primersSilent mutation analysis allows you to design a desired mutation site on a DNA mutation will result in the modification of restriction property without chang amino acid sequence. This function searches for potential mutation positions t destroy restriction enzyme sites. You can use this function to design primers silent muation on target DNA sequence.Directed mismatch primersDirected mismatch analysis allows you to create or remove restriction sites on or its mutants (variants) by incorporating mismatch at a site near the mutatio function you can design PCR primers to create or destroy a restriction site in distinguish the wild type allele and a common mutant allele.Translation and Protein AnalysisTranslationDNAMAN deduces protein sequences from three reading frames of a DNA sequence a results with many options. By setting the number of amino acid per one line, y the layout of the translation file.DNAMAN allows you to select any region from a sequence file, and perform trans In the report file DNAMAN shows both translated and untranslated regions.Genetic code tableDNAMAN provides seven genetic code tables with the options of adding new code editing any existing one. You can select any genetic code table for translatio 生物秀-专心做生物w w w .b b i o o .c o manalysis.Reading frame overviewDNAMAN presents a graphical overview on six reading frames of a DNA sequence. feature to locate the ORFs on a large DNA sequence.Codon usage analysisDNAMAN provides a codon usage table for any reading frame of a DNA sequence. T indicates the number and frequency of each codon used in a reading frame.Amino acid compositionDNAMAN reports the amino acid composition, pI and molecular weight of a proteiProtein hydrophobic and hydrophilic profileDNAMAN shows protein hydrophobic and hydrophilic profiles in a graphic windowyou to predict hydrophobic clusters or antigen regions in a protein sequence. The gra profiles are editable to produce high quality illustrations for publications.Protein charge and pI analysisDNAMAN calculates protein charge at a given pH. It shows also the predicted is of the protein. In addition, DNAMAN can deduce the suitable buffer pH for a deProtein secondary structure predictionDNAMAN predicts the secondary structure of a protein sequence using the DSC me by King and Sternberg.生物秀-专心做生物w w w .b b i o o .c o mReverse translationDNAMAN provides the reverse translation of a protein sequence. It reports the translated DNA sequence with ambiguous nucleotides at variant positions. This used to degenerate primers from peptide sequences.Database ManagementOligo databaseYou may have a large number of oligo nucleotides for experiments of sequencing etc....DNAMAN provides an oligo database manager that can help you to effectiv use these oligoes.When you create oligo databases for different projects. DNAMAN allows you to a for the security of the database.l Any oligo database is expandable and all records are editable.lYou can provide information for each record in seven fields: name, source GC content, melting temperature and sequence.l The name, source, memo, length can be used as sorting keys.l You can import a large number of oligo records from a text file, or expor a text file.DNA and protein databaseDNAMAN database manager is used to collect and store DNA and protein sequences or delete a DNA or protein database and set security to protect the database.DNAMAN has an easy-to-use database manager. All databases are expandable and a editable. You can directly import records from text files, GCG and GenBank fil information related to any record is shown in seven fields: sequence name, def keywords, source, reference, memo, coding region. The first four fields can be keys.Searching in DNA or protein databaselDNAMAN can search for homology sequences of a target DNA by scanning all database. The algorithm for the comparison is fast alignment method. l DNAMAN searches for a nucleotide sequence from both strands of all record database.lDNAMAN searches for a peptide sequence from all records of a protein data the six reading frames of all records in a DNA database.System Requirementsl Intel Pentium 90 MHz or higher l Windows 95/98/NT l At least 16MB memorylA hard drive with at least 6MB of available disk space生物秀-专心做生物w w w .b b i o o .c o mlA 1.44 floppy drive (for installation)DNAMAN for MacintoshDNAMAN uses advanced features of MacOS and offers versatile visual tools for s analysis.Features of DNAMANSequence ManipulationSequence inputDNAMAN provides 20 sequence channels to keep active sequences in memory. sequence channels simplify multiple functional analyses and substantially the efficiency of your works. A panel window shows the current working se can edit the sequence directly in the panel.Information exchangeDNAMAN accepts sequence files in GenBank, GCG, CUSTAL, FASTA, PIR and GDE can export multiple sequences in GCG, CUSTAL, PIR and GDE format. You can exchange information on the Internet by copying and pasting seque between DNAMAN and your favorite web browser..EditingEditing a text file with DNAMAN is as easy as working with your favorite processor. With the word processor of DNAMAN, you can edit original seque and analysis results as well.Sequence composition and conversionDNAMAN reports the composition and molecular weight of a sequence. It per conversion of a sequence to its reverse, complementary, reverse complemen double strand, and RNA sequences.生物秀-专心做生物w w w .b b i o o .c o mBLAST documents/Internet accessIn addition to accessing the Internet through Web browser, DNAMAN prepare BLAST document formats with a query sequence for directly accessing BLASTServer. Five BLAST document formats are currently available: Blastn, Blastx, Tblastx, Blastp and Tblastn . Examples:Sequence SearchSearch for nucleotide sequencesWith DNAMAN, you can search for nucleotide sequences from one or both str DNA sequence. Gaps and ambiguous nucleotides are allowed in the query seq can also provide multiple query sequences for searching.生物秀-专心做生物w w w .b b i o o .c o mDNAMAN instantly reports the searching results in graphics. Colors and ar indicate different sequence groups and sites. You can magnify any region fragment and display the regional sequence of by selection.The versatility of query sequences and graphical presentations make the s function useful in many other areas, such as presenting restriction sites and gene structures.Search for consensus sequencesWith DNAMAN, you can search for DNA or protein consensus sequences from b or six reading frames of DNA sequences. DNAMAN provides a database of DNA protein consensus sequences. The database is expandable and editable. You custom consensus sequence databases.Search for open reading framesYou can search for open reading frames from six reading frames of a DNA s The searching result is shown in a text table. DNAMAN can also display th a graphic window.Search for repeat sequencesYou can search for direct repeat and reverse repeat sequences from both stands o DNA sequence. You can also search for reverse complementary repeat sequences tha may form hairpin/stem-loop structures.Search for amino acid sequencesYou can search for an amino acid sequence and its variations from the six frames of a DNA sequence. DNAMAN allows ambiguous amino acids as well as mismatches in a query sequence.生物秀-专心做生物w w w .b b i o o .c o mRestriction AnalysisRestriction site analysisYou can search any restriction site on a DNA sequence. DNAMAN supplies twrestriction enzyme files; the restrict.enz file with 180 most frequently used restriction enzymes, and the dnamanre.enz file with 1364 enzyme records. You can also create your own enzyme files. All the enzyme files are editable andDNAMAN provides custom restriction enzyme filters on cutter, ends, freque methylation sensitivity. Users can define the DNA molecule as a linear or type.DNAMAN reports the restriction analysis results in an easy-to-read table.cutting sites are shown in alphabetical order of enzymes, and in site pos as well. The non-cutting enzymes are also listed. In addition, DNAMAN dis enzyme sites on the top of the DNA sequence.Restriction mapDNAMAN provides easy-to-use tools to produce publication-quality restrict These tools can be used to draw linear or circular restriction maps with DNA sequence information. You can also draw maps for other projects, such strategy diagrams, gene structural maps, etc.生物秀-专心做生物w w w .b b i o o .c o mRestriction pattern illustrationDNAMAN predicts the patterns of restriction enzyme digested DNA fragments electrophoresis. You can perform single enzyme digestion on multiple sequ single or multiple digestion on a single sequence. DNAMAN shows the infor restriction fragments on their sizes and ends when you click on these fraElectronic cloningDNA cloning is a time consuming and expensive process. DNAMAN provides ea tools to design a cloning strategy and performs evaluation analyses on ta生物秀-专心做生物w w w .b b i o o .c o msequences. This feature could improve the efficiency of your cloning work laboratory.DNAMAN mimics basic steps of the actual cloning process:l Restriction analysis on the original vector and insert sequences l Selection of vector and insert fragments from restriction pattern l Verification of the end compatibility of DNA fragments l Modification of fragment ends if necessary l Insertion of linkers if necessary lProducing the final clone sequenceConstructing restriction mapsDNAMAN can help you reconstruct a restriction map in the absence of DNA s You must provide all fragment sizes in single and double digestion. DNAMA the possible restriction map(s) from the information of restriction fragmSilent mutation analysisSilent mutation analysis allows you to design a desired mutation site on sequence. This mutation will result in the modification of restriction pr without changing the coding amino acid sequence. This function searches f potential mutation positions to create or destroy restriction enzyme siteDirected mismatch analysisDirected mismatch analysis allows you to create or remove restriction sit sequence or its mutants (variants) by incorporating a single or double mi site near the mutation. Using this function you can create or destroy a r site in order to distinguish the wild type allele and a common mutant allSequence AssemblySequence assembly method生物秀-专心做生物w w w .b b i o o .c o mDNAMAN uses fast alignment algorithms to assemble quickly and accurately number of overlapping sequences. DNAMAN can automatically adjust the orie each fragment and remove vector sequences as well. You can set sensitivit parameters to control gaps and ambiguous sequences in contigs.Sequence assembly editorDNAMAN displays sequence assembly project in three windows:lGraphic window provides an overview of the assembly construction. You can e this graphical presentation to produce high quality diagrams for publName list window contains all assembled sequence names. You can change the sequence order by moving the sequence names in this window.lSequence window shows all original sequences and a consensus sequence. You edit any of the original sequences to improve the result of sequenceDNAMAN exports sequence the assembly result in a text window. You ha of reporting the consensus sequence only, or all sequences including consensus sequence and original sequences.Sequence Homology AnalysesDot matrix plotWith DNAMAN, you can compare two DNA sequences or two protein sequences i matrix plot. There are two methods for comparison. Method 1 uses a fast a and is suitable for long sequences. Method 2 performs progressive compari reduce the background of low homology regions. The two methods yield diff生物秀-专心做生物w w w .b b i o o .c o mand accuracy in analysis.DNAMAN displays the actual sequences and their alignment on any selected sequence window.Two sequence alignmentDNAMAN uses fast or optimal algorithms to align two DNA or protein sequen have options to control the sensitivity of alignment. DNAMAN also allows select any region of target sequences for alignment.For DNA sequence alignment, you have an option to use the minus strand fo comparison. For protein sequence alignment, DNAMAN can report the amino a similarity of two protein sequences in a text window. The amino acid simi matrix is editable by users.生物秀-专心做生物w w w .b b i o o .c o mMultiple Sequence AlignmentMultiple sequence alignment methodsFast alignmentDNAMAN uses Wilbur and Lipman algorithm for fast alignment. You can quick large number of DNA or protein sequences. This algorithm is able to produ accurate alignment if sequence divergence is low.Optimal alignmentFeng-Doolittle and Thompson method (CLUSTAL) is used for optimal sequence You can input individual sequences and/or multiple alignment profiles. Th three types of alignment:l Full alignment to completely align all sequences.l Profile alignment to align two multiple alignment profiles without d the original alignments.lNew sequence on profile to align a new sequence with an existing mul alignment profile.You can optimize multiple alignment by selecting alignment criteria in th use control panel.Multiple sequence alignment editorDNAMAN provides a high performance alignment editor. A graphical view of alignment allows you to quickly move to an interesting region. You can ch alignment list order by drag and drop sequence names. You are also able t delete gap insertions, move a fragment within a gap and truncate aligned生物秀-专心做生物w w w .b b i o o .c o mYou can modify the appearance of multiple alignment sequences:l displaying identical residues in colors or blocks l displaying consensus sequence l changing text fontMultiple alignment input and outputYou can directly input sequences or multiple alignment profiles for align the following sources:l GenBank,l EMBL/Swiss Prot, l GCG/MSF, l CUSTAL, l FASTA, l NBRF/PIR lGDEThe multiple alignment editor can output an alignment in different formatCUSTAL, NBRF/PIR , and GDE . The multiple input and out put capacity of DNAMAN mak it compatible with major sequence analysis software.Phylogenetic treesDNAMAN calculates the homology matrix and establishes related distances bpairs of sequences. Consequently, DNAMAN can output a distance matrix of multipl alignment, and draw phylogenetic trees or homology tree s. You can carry out bootstrappingtests for the confidence value of a phylogenetic tree.生物秀-专心做生物w w w .b b i o o .c o m。
