一个简单实用的遗传算法c程序完整版
遗传算法程序

遗传算法程序遗传算法程序(一):说明: fga.m 为遗传算法的主程序; 采用二进制Gray编码,采用基于轮盘赌法的非线性排名选择, 均匀交叉,变异操作,而且还引入了倒位操作!function[BestPop,Trace]=fga(FUN,LB,UB,eranum,popsize,pCross,pMutati on,pInversion,options) % [BestPop,Trace]=fmaxga(FUN,LB,UB,eranum,popsize,pcross,pmu tation) % Finds a maximum ofa function of several variables.% fmaxga solves problems of the form:% max F(X) subject to: LB <= X <= UB% BestPop - 最优的群体即为最优的染色体群% Trace - 最佳染色体所对应的目标函数值% FUN - 目标函数% LB - 自变量下限% UB - 自变量上限% eranum - 种群的代数,取100--1000(默认200)% popsize - 每一代种群的规模;此可取50--200(默认100)% pcross - 交叉概率,一般取0.5--0.85之间较好(默认0.8)% pmutation - 初始变异概率,一般取0.05-0.2之间较好(默认0.1) % pInversion - 倒位概率,一般取0.05-0.3之间较好(默认0.2) % options - 1*2矩阵,options(1)=0二进制编码(默认0),option(1)~=0十进制编%码,option(2)设定求解精度(默认1e-4)%% ------------------------------------------------------------------------T1=clock;if nargin<3, error('FMAXGA requires at least three input arguments'); endif nargin==3, eranum=200;popsize=100;pCross=0.8;pMutation=0.1;pInversio n=0.15;options=[0 1e -4];endif nargin==4, popsize=100;pCross=0.8;pMutation=0.1;pInversion=0.15;option s=[0 1e-4];endif nargin==5, pCross=0.8;pMutation=0.1;pInversion=0.15;options=[0 1e-4];endif nargin==6, pMutation=0.1;pInversion=0.15;options=[0 1e-4];endif nargin==7, pInversion=0.15;options=[0 1e-4];endif find((LB-UB)>0)error('数据输入错误,请重新输入(LB<ub):');< bdsfid="91" p=""></ub):');<>ends=sprintf('程序运行需要约%.4f 秒钟时间,请稍等......',(eranum*popsize/1000));disp(s);global m n NewPop children1 children2 VarNumbounds=[LB;UB]';bits=[];VarNum=size(bounds,1);precision=options(2);%由求解精度确定二进制编码长度bits=ceil(log2((bounds(:,2)-bounds(:,1))' ./ precision));%由设定精度划分区间[Pop]=InitPopGray(popsize,bits);%初始化种群[m,n]=size(Pop);NewPop=zeros(m,n);children1=zeros(1,n);children2=zeros(1,n);pm0=pMutation;BestPop=zeros(eranum,n);%分配初始解空间BestPop,TraceTrace=zeros(eranum,length(bits)+1);i=1;while i<=eranumfor j=1:mvalue(j)=feval(FUN(1,:),(b2f(Pop(j,:),bounds,bits)));%计算适应度end[MaxValue,Index]=max(value);BestPop(i,:)=Pop(Index,:);Trace(i,1)=MaxValue;Trace(i,(2:length(bits)+1))=b2f(BestPop(i,:),bounds,bits);[selectpop]=NonlinearRankSelect(FUN,Pop,bounds,bits);%非线性排名选择[CrossOverPop]=CrossOver(selectpop,pCross,round(unidrnd (eranum-i)/eranum));%采用多点交叉和均匀交叉,且逐步增大均匀交叉的概率%round(unidrnd(eranum-i)/eranum)[MutationPop]=Mutation(CrossOverPop,pMutation,VarNum );%变异[InversionPop]=Inversion(MutationPop,pInversion);%倒位Pop=InversionPop;%更新pMutation=pm0+(i^4)*(pCross/3-pm0)/(eranum^4);%随着种群向前进化,逐步增大变异率至1/2交叉率p(i)=pMutation;i=i+1;endt=1:eranum;plot(t,Trace(:,1)');title('函数优化的遗传算法');xlabel('进化世代数(eranum)');ylabel('每一代最优适应度(maxfitness)');[MaxFval,I]=max(Trace(:,1));X=Trace(I,(2:length(bits)+1));hold on; plot(I,MaxFval,'*');text(I+5,MaxFval,['FMAX=' num2str(MaxFval)]);str1=sprintf ('进化到 %d 代 ,自变量为 %s 时,得本次求解的最优值%f\n对应染色体是:%s',I,num2str(X),MaxFval,num2str(BestPop(I,:))); disp(str1);%figure(2);plot(t,p);%绘制变异值增大过程T2=clock;elapsed_time=T2-T1;if elapsed_time(6)<0elapsed_time(6)=elapsed_time(6)+60;elapsed_time(5)=elapsed_time(5)-1;endif elapsed_time(5)<0elapsed_time(5)=elapsed_time(5)+60;elapsed_time(4)=elaps ed_time(4)-1;end %像这种程序当然不考虑运行上小时啦str2=sprintf('程序运行耗时 %d 小时 %d 分钟 %.4f 秒',elapsed_time(4),elapsed_time(5),elapsed_time(6));disp(str2);%初始化种群%采用二进制Gray编码,其目的是为了克服二进制编码的Hamming悬崖缺点 function [initpop]=InitPopGray(popsize,bits) len=sum(bits);initpop=zeros(popsize,len);%The whole zero encodingindividualfor i=2:popsize-1pop=round(rand(1,len));pop=mod(([0 pop]+[pop 0]),2);%i=1时,b(1)=a(1);i>1时,b(i)=mod(a(i-1)+a(i),2)%其中原二进制串:a(1)a(2)...a(n),Gray串:b(1)b(2)...b(n)initpop(i,:)=pop(1:end-1);endinitpop(popsize,:)=ones(1,len);%The whole one encoding individual%解码function [fval] = b2f(bval,bounds,bits)% fval - 表征各变量的十进制数% bval - 表征各变量的二进制编码串% bounds - 各变量的取值范围% bits - 各变量的二进制编码长度scale=(bounds(:,2)-bounds(:,1))'./(2.^bits-1); %The range of the variables numV=size(bounds,1);cs=[0 cumsum(bits)];for i=1:numVa=bval((cs(i)+1):cs(i+1));fval(i)=sum(2.^(size(a,2)-1:-1:0).*a)*scale(i)+bounds(i,1);end%选择操作%采用基于轮盘赌法的非线性排名选择%各个体成员按适应值从大到小分配选择概率:%P(i)=(q/1-(1-q)^n)*(1-q)^i, 其中P(0)>P(1)>...>P(n), sum(P(i))=1function[selectpop]=NonlinearRankSelect(FUN,pop,bounds,bits)global m nselectpop=zeros(m,n);fit=zeros(m,1);for i=1:mfit(i)=feval(FUN(1,:),(b2f(pop(i,:),bounds,bits)));%以函数值为适应值做排名依据endselectprob=fit/sum(fit);%计算各个体相对适应度(0,1)q=max(selectprob);%选择最优的概率x=zeros(m,2);x(:,1)=[m:-1:1]';[y x(:,2)]=sort(selectprob);r=q/(1-(1-q)^m);%标准分布基值newfit(x(:,2))=r*(1-q).^(x(:,1)-1);%生成选择概率newfit=cumsum(newfit);%计算各选择概率之和rNums=sort(rand(m,1));fitIn=1;newIn=1;while newIn<=mif rNums(newIn)<newfit(fitin)< bdsfid="195" p=""></newfit(fitin)<>selectpop(newIn,:)=pop(fitIn,:);newIn=newIn+1;elsefitIn=fitIn+1;endend%交叉操作function [NewPop]=CrossOver(OldPop,pCross,opts)%OldPop为父代种群,pcross为交叉概率global m n NewPopr=rand(1,m);y1=find(r<pcross);< bdsfid="208" p=""></pcross);<>y2=find(r>=pCross);len=length(y1);if len>2&mod(len,2)==1%如果用来进行交叉的染色体的条数为奇数,将其调整为偶数 y2(length(y2)+1)=y1(len);endif length(y1)>=2for i=0:2:length(y1)-2if opts==0[NewPop(y1(i+1),:),NewPop(y1(i+2),:)]=EqualCrossOver(Old Pop(y1(i+1),:),OldPop(y1(i+2),:)) ;else[NewPop(y1(i+1),:),NewPop(y1(i+2),:)]=MultiPointCross(Old Pop(y1(i+1),:),OldPop(y1(i+2),:) );endendendNewPop(y2,:)=OldPop(y2,:);%采用均匀交叉function[children1,children2]=EqualCrossOver(parent1,parent2) global n children1 children2hidecode=round(rand(1,n));%随机生成掩码crossposition=find(hidecode==1);holdposition=find(hidecode==0);children1(crossposition)=parent1(crossposition);%掩码为1,父1为子1提供基因children1(holdposition)=parent2(holdposition);%掩码为0,父2为子1提供基因children2(crossposition)=parent2(crossposition);%掩码为1,父2为子2提供基因children2(holdposition)=parent1(holdposition);%掩码为0,父1为子2提供基因%采用多点交叉,交叉点数由变量数决定function[Children1,Children2]=MultiPointCross(Parent1,Parent2) global n Children1 Children2 VarNumChildren1=Parent1;Children2=Parent2;Points=sort(unidrnd(n,1,2*VarNum));for i=1:VarNumChildren1(Points(2*i-1):Points(2*i))=Parent2(Points(2*i-1):Points(2*i));Children2(Points(2*i-1):Points(2*i))=Parent1(Points(2*i-1):Points(2*i));end%变异操作function [NewPop]=Mutation(OldPop,pMutation,VarNum) global m n NewPopposition=find(r<=pMutation);len=length(position);if len>=1for i=1:lenk=unidrnd(n,1,VarNum); %设置变异点数,一般设置1点for j=1:length(k)if OldPop(position(i),k(j))==1OldPop(position(i),k(j))=0;elseOldPop(position(i),k(j))=1;EndendendendNewPop=OldPop;%倒位操作function [NewPop]=Inversion(OldPop,pInversion)global m n NewPopNewPop=OldPop;r=rand(1,m);PopIn=find(r<=pInversion);len=length(PopIn);if len>=1for i=1:lend=sort(unidrnd(n,1,2));if d(1)~=1&d(2)~=nNewPop(PopIn(i),1:d(1)-1)=OldPop(PopIn(i),1:d(1)-1);NewPop(PopIn(i),d(1):d(2))=OldPop(PopIn(i),d(2):-1:d(1)); NewPop(PopIn(i),d(2)+1:n)=OldPop(PopIn(i),d(2)+1:n);Endendend遗传算法程序(二):function youhuafunD=code;N=50; % Tunablemaxgen=50; % Tunablecrossrate=0.5; %Tunablemuterate=0.08; %Tunablegeneration=1;num = length(D);fatherrand=randint(num,N,3);score = zeros(maxgen,N);while generation<=maxgenind=randperm(N-2)+2; % 随机配对交叉A=fatherrand(:,ind(1:(N-2)/2));B=fatherrand(:,ind((N-2)/2+1:end));% 多点交叉rnd=rand(num,(N-2)/2);ind=rnd tmp=A(ind);A(ind)=B(ind);B(ind)=tmp;% % 两点交叉% for kk=1:(N-2)/2% rndtmp=randint(1,1,num)+1;% tmp=A(1:rndtmp,kk);% A(1:rndtmp,kk)=B(1:rndtmp,kk);% B(1:rndtmp,kk)=tmp;% endfatherrand=[fatherrand(:,1:2),A,B];% 变异rnd=rand(num,N);ind=rnd [m,n]=size(ind);tmp=randint(m,n,2)+1;tmp(:,1:2)=0;fatherrand=tmp+fatherrand;fatherrand=mod(fatherrand,3);% fatherrand(ind)=tmp;%评价、选择scoreN=scorefun(fatherrand,D);% 求得N个个体的评价函数score(generation,:)=scoreN;[scoreSort,scoreind]=sort(scoreN);sumscore=cumsum(scoreSort);sumscore=sumscore./sumscore(end);childind(1:2)=scoreind(end-1:end);for k=3:Ntmprnd=rand;tmpind=tmprnd difind=[0,diff(tmpind)];if ~any(difind)difind(1)=1;endchildind(k)=scoreind(logical(difind));endfatherrand=fatherrand(:,childind);generation=generation+1;end% scoremaxV=max(score,[],2);minV=11*300-maxV;plot(minV,'*');title('各代的目标函数值');F4=D(:,4);FF4=F4-fatherrand(:,1);FF4=max(FF4,1);D(:,5)=FF4;save DData Dfunction D=codeload youhua.mat% properties F2 and F3F1=A(:,1);F2=A(:,2);F3=A(:,3);if (max(F2)>1450)||(min(F2)<=900)error('DATA property F2 exceed it''s range (900,1450]') end % get group property F1 of data, according to F2 value F4=zeros(size(F1));for ite=11:-1:1index=find(F2<=900+ite*50);F4(index)=ite;endD=[F1,F2,F3,F4];function ScoreN=scorefun(fatherrand,D)F3=D(:,3);F4=D(:,4);N=size(fatherrand,2);FF4=F4*ones(1,N);FF4rnd=FF4-fatherrand;FF4rnd=max(FF4rnd,1);ScoreN=ones(1,N)*300*11;% 这里有待优化for k=1:NFF4k=FF4rnd(:,k);for ite=1:11F0index=find(FF4k==ite);if ~isempty(F0index)tmpMat=F3(F0index);tmpSco=sum(tmpMat);ScoreBin(ite)=mod(tmpSco,300);endendScorek(k)=sum(ScoreBin);ScoreN=ScoreN-Scorek;遗传算法程序(三):%IAGAfunction best=gaclearMAX_gen=200; %最大迭代步数best.max_f=0; %当前最大的适应度STOP_f=14.5; %停止循环的适应度RANGE=[0 255]; %初始取值范围[0 255] SPEEDUP_INTER=5; %进入加速迭代的间隔advance_k=0; %优化的次数popus=init; %初始化for gen=1:MAX_genfitness=fit(popus,RANGE); %求适应度f=fitness.f;picked=choose(popus,fitness); %选择popus=intercross(popus,picked); %杂交popus=aberrance(popus,picked); %变异if max(f)>best.max_fadvance_k=advance_k+1;x_better(advance_k)=fitness.x;best.max_f=max(f);best.popus=popus;best.x=fitness.x;endif mod(advance_k,SPEEDUP_INTER)==0RANGE=minmax(x_better);RANGEadvance=0;endreturn;function popus=init%初始化M=50;%种群个体数目N=30;%编码长度popus=round(rand(M,N));return;function fitness=fit(popus,RANGE)%求适应度[M,N]=size(popus);fitness=zeros(M,1);%适应度f=zeros(M,1);%函数值A=RANGE(1);B=RANGE(2);%初始取值范围[0 255] for m=1:M x=0;for n=1:Nx=x+popus(m,n)*(2^(n-1));endx=x*((B-A)/(2^N))+A;for k=1:5f(m,1)=f(m,1)-(k*sin((k+1)*x+k));endendf_std=(f-min(f))./(max(f)-min(f));%函数值标准化fitness.f=f;fitness.f_std=f_std;fitness.x=x; return;function picked=choose(popus,fitness)%选择f=fitness.f;f_std=fitness.f_std;[M,N]=size(popus);choose_N=3; %选择choose_N对双亲picked=zeros(choose_N,2); %记录选择好的双亲p=zeros(M,1); %选择概率d_order=zeros(M,1);%把父代个体按适应度从大到小排序f_t=sort(f,'descend');%将适应度按降序排列for k=1:Mx=find(f==f_t(k));%降序排列的个体序号d_order(k)=x(1);endfor m=1:Mpopus_t(m,:)=popus(d_order(m),:);endpopus=popus_t;f=f_t;p=f_std./sum(f_std); %选择概率c_p=cumsum(p)'; %累积概率for cn=1:choose_Npicked(cn,1)=roulette(c_p); %轮盘赌picked(cn,2)=roulette(c_p); %轮盘赌popus=intercross(popus,picked(cn,:));%杂交endpopus=aberrance(popus,picked);%变异return;function popus=intercross(popus,picked) %杂交[M_p,N_p]=size(picked);[M,N]=size(popus);for cn=1:M_pp(1)=ceil(rand*N);%生成杂交位置p(2)=ceil(rand*N);p=sort(p);t=popus(picked(cn,1),p(1):p(2));popus(picked(cn,1),p(1):p(2))=popus(picked(cn,2),p(1):p(2)); popus(picked(cn,2),p(1):p(2))=t;endreturn;function popus=aberrance(popus,picked) %变异P_a=0.05;%变异概率[M,N]=size(popus);[M_p,N_p]=size(picked)U=rand(1,2);for kp=1:M_pif U(2)>=P_a %如果大于变异概率,就不变异continue;endif U(1)>=0.5a=picked(kp,1);elsea=picked(kp,2);endp(1)=ceil(rand*N);%生成变异位置p(2)=ceil(rand*N);if popus(a,p(1))==1%0 1变换popus(a,p(1))=0;elsepopus(a,p(1))=1;endif popus(a,p(2))==1popus(a,p(2))=0;elsepopus(a,p(2))=1;endendreturn;function picked=roulette(c_p) %轮盘赌[M,N]=size(c_p);M=max([M N]);U=rand; if U<c_p(1)< bdsfid="491" p=""></c_p(1)<>picked=1;return;endfor m=1:(M-1)if U>c_p(m) & U<c_p(m+1)< bdsfid="497" p=""></c_p(m+1)<>picked=m+1;break;endend全方位的两点杂交、两点变异的改进的加速遗传算法(IAGA)遗传算法优化pid参数matlab程序chap5_4m%GA(Generic Algorithm) program to optimize Parameters of PID clear all;clear all;global rin yout timefG=100;Size=30;CodeL=10;MinX(1)=zeros(1);MaxX(1)=20*ones(1);MinX(2)=zeros(1);MaxX(2)=1.0*ones(1);MinX(3)=zeros(1);MaxX(3)=1.0*ones(1);E=round(rand(Size,3*CodeL));%Initian Code! BsJ=0;for kg=1:1:Gtime(kg)=kg;for s=1:1:Sizem=E(s,:);y1=0;y2=0;y3=0;m1=m(1:1:CodeL);for i=1:1:CodeLy1=y1+m1(i)*2^(i-1);endKpid(s,1)=(MaxX(1)-MinX(1))*y1/1023+MinX(1); m2=m(CodeL+1:1:2*CodeL);for i=1:1:CodeLy2=y2+m2(i)*2^(i-1);endKpid(s,2)=(MaxX(2)-MinX(2))*y2/1023+MinX(2); m3=m(2*CodeL+1:1:3*CodeL);for i=1:1:CodeLy3=y3+m3(i)*2^(i-1);endKpid(s,3)=(MaxX(3)-MinX(3))*y3/1023+MinX(3); %*******Step 1:Evaluate Best J*******Kpidi=Kpid(s,:);[Kpidi,BsJ]=chap5_3f(Kpidi,BsJ);BsJi(s)=BsJ;end[OderJi,IndexJi]=sort(BsJi);BestJ(kg)=OderJi(1);BJ=BestJ(kg);Ji=BsJi+1e-10;fi=1./Ji;%Cm=max(Ji);%fi=Cm-Ji; %Avoiding deviding zero[Oderfi,Indexfi]=sort(fi);%Arranging fi small to bigger%Bestfi=Oderfi(Size); %Let Bestfi=max(fi)%BestS=Kpid(Indexfi(Size),:); %Let BestS=E(m),m is the Indexfi belong to %max(fi)Bestfi=Oderfi(Size);%Let Bestfi=max(fi)BestS=E(Indexfi(Size),:);%Let BestS=E(m),m is the Indexfi belong to max(fi)kgBJBestS;%****Step 2:Select and Reproduct Operation***fi_sum=sum(fi);fi_Size=(Oderfi/fi_sum)*Size;fi_S=floor(fi_Size); %Selecting Bigger fi valuekk=1;for i=1:1:Sizefor j=1:1:fi_S(i) %Select and ReproduceTempE(kk,:)=E(Indexfi(i),:);kk=kk+1; %kk is used to reproduceendend%**********Step 3:Crossover Operation******pc=0.06;n=ceil(20*rand);for i=1:2:(Size-1)temp=rand;if pc>tempfor j=n:1:20TempE(i,j)=E(i+1,j);TempE(i+1,j)=E(i,j);endendendTempE(Size,:)=BestS;E=TempE;%***************Step 4: Mutation Operation************** %pm=0.001;pm=0.001-[1:1:Size]*(0.001)/Size;%Bigger fi,smaller pm%pm=0.0; %No mutation%pm=0.1; %Big mutationfor i=1:1:Sizefor j=1:1:3*CodeLtemp=rand;if pm>temp %Mutation Conditionif TempE(i,j)==0TempE(i,j)=1;elseTempE(i,j)=0;Endendendend%Guarantee TempE(Size,:)belong to the best individualTempE(Size,:)=BestS;E=TempE;%*************************************************** endBestfiBestSKpidiBest_J=BestJ(G)figure(1);plot(time,BestJ);xlabel('Time');ylabel('Best J');figure(2);plot(timef,rin,'r',timef,yout,'b');xlabel('Time(s)');ylabel('ran,yout');chap5_3f.mfunction [Kpidi,BsJ]=pid_gaf(Kpidi,BsJ) global rin yout timefts=0.001;sys=tf(400,[1,50,0]);dsys=c2d(sys,ts,'z');[num,den]=tfdata(dsys,'v');rin=1.0;u_1=0.0;u_2=0.0;y_1=0.0;y_2=0.0;x=[0,0,0]'; B=0;error_1=0;tu=1;s=0;P=100;for k=1:1:Ptimef(k)=k*ts;r(k)=rin;u(k)=Kpidi(1)*x(1)+Kpidi(2)*x(2)+Kpidi(3)*x(3); if u(k)>=10u(k)=10;endif u(k)<=-10u(k)=-10;endyout(k)=-den(2)*y_1-den(3)*y_2+num(2)*u_1+num(3)*u_2; error(k)=r(k)-yout(k);%-------------------Return of PID parameters---------------- u_2=u_1;u_1=u(k);y_2=y_1;y_1=yout(k);x(1)=error(k); % Calculating Px(2)=(error(k)-error_1)/ts; % Dx(3)=x(3)+error(k)*ts; % Ierror_2=error_1;error_1=error(k);if s==0if yout(k)>0.95&yout(k)<1.05tu=timef(k);s=1;endendendfor i=1:1:PJi(i)=0.999*abs(error(i))+0.01*u(i)^2*0.1; B=B+Ji(i);f i>1erry(i)=yout(i)-yout(i-1);if erry(i)<0B=B+100*abs(erry(i));EndEndendBsj=B+0.2*tu*10。
(完整版)遗传算法c语言代码

}
}
}
//拷贝种群
for(i=0;i<num;i++)
{
grouptemp[i].adapt=group[i].adapt;
grouptemp[i].p=group[i].p;
for(j=0;j<cities;j++)
grouptemp[i].city[j]=group[i].city[j];
{
group[i].p=1-(double)group[i].adapt/(double)biggestsum;
biggestp+=group[i].p;
}
for(i=0;i<num;i++)
group[i].p=group[i].p/biggestp;
//求最佳路劲
bestsolution=0;
for(i=0;i<num;i++)
printf("\n******************是否想再一次计算(y or n)***********************\n");
fflush(stdin);
scanf("%c",&choice);
}while(choice=='y');
return 0;
}
遗传算法代码
#include<stdio.h>
#include<string.h>
#include<stdlib.h>
#include<math.h>
#include<time.h>
#define cities 10 //城市的个数
一个简单实用的遗传算法c程序

一个简单实用的遗传算法c程序(转载)c++ 2009-07-28 23:09:03 阅读418 评论0 字号:大中小这是一个非常简单的遗传算法源代码,是由Denis Cormier (North Carolina State University)开发的,Sita S.Raghavan (University of North Carolina at Charlotte)修正。
代码保证尽可能少,实际上也不必查错。
对一特定的应用修正此代码,用户只需改变常数的定义并且定义“评价函数”即可。
注意代码的设计是求最大值,其中的目标函数只能取正值;且函数值和个体的适应值之间没有区别。
该系统使用比率选择、精华模型、单点杂交和均匀变异。
如果用Gaussian变异替换均匀变异,可能得到更好的效果。
代码没有任何图形,甚至也没有屏幕输出,主要是保证在平台之间的高可移植性。
读者可以从,目录coe/evol 中的文件prog.c中获得。
要求输入的文件应该命名为‘gadata.txt’;系统产生的输出文件为‘galog.txt’。
输入的文件由几行组成:数目对应于变量数。
且每一行提供次序——对应于变量的上下界。
如第一行为第一个变量提供上下界,第二行为第二个变量提供上下界,等等。
/**************************************************************************//* This is a simple genetic algorithm implementation where the *//* evaluation function takes positive values only and the *//* fitness of an individual is the same as the value of the *//* objective function *//**************************************************************************/#include <stdio.h>#include <stdlib.h>#include <math.h>/* Change any of these parameters to match your needs */#define POPSIZE 50 /* population size */#define MAXGENS 1000 /* max. number of generations */#define NVARS 3 /* no. of problem variables */#define PXOVER 0.8 /* probability of crossover */#define PMUTATION 0.15 /* probability of mutation */#define TRUE 1#define FALSE 0int generation; /* current generation no. */int cur_best; /* best individual */FILE *galog; /* an output file */struct genotype /* genotype (GT), a member of the population */{double gene[NVARS]; /* a string of variables一个变量字符串*/ double fitness; /* GT's fitness适应度*/double upper[NVARS]; /* GT's variables upper bound 变量的上限*/ double lower[NVARS]; /* GT's variables lower bound变量的下限*/ double rfitness; /* relative fitness 相对适应度*/double cfitness; /* cumulative fitness 累计适应度*/};struct genotype population[POPSIZE+1]; /* population */struct genotype newpopulation[POPSIZE+1]; /* new population; *//* replaces the *//* old generation */ /* Declaration of procedures used by this genetic algorithm */void initialize(void);double randval(double, double);void evaluate(void);void keep_the_best(void);void elitist(void);void select(void);void crossover(void);void Xover(int,int);void swap(double *, double *);void mutate(void);void report(void);/***************************************************************//* Initialization function: Initializes the values of genes *//* within the variables bounds. It also initializes (to zero) *//* all fitness values for each member of the population. It *//* reads upper and lower bounds of each variable from the */ /* input file `gadata.txt'. It randomly generates values *//* between these bounds for each gene of each genotype in the *//* population. The format of the input file `gadata.txt' is *//* var1_lower_bound var1_upper bound */ /* var2_lower_bound var2_upper bound ... */ /***************************************************************/void initialize(void){FILE *infile;int i, j;double lbound, ubound;if ((infile = fopen("gadata.txt","r"))==NULL){fprintf(galog,"\nCannot open input file!\n");exit(1);}/* initialize variables within the bounds */for (i = 0; i < NVARS; i++){fscanf(infile, "%lf",&lbound);fscanf(infile, "%lf",&ubound);for (j = 0; j < POPSIZE; j++){population[j].fitness = 0;population[j].rfitness = 0;population[j].cfitness = 0;population[j].lower[i] = lbound;population[j].upper[i]= ubound;population[j].gene[i] = randval(population[j].lower[i],population[j].upper[i]);}}fclose(infile);}/***********************************************************//* Random value generator: Generates a value within bounds */ /***********************************************************/double randval(double low, double high){double val;val = ((double)(rand()%1000)/1000.0)*(high - low) + low;return(val);}/*************************************************************//* Evaluation function: This takes a user defined function. *//* Each time this is changed, the code has to be recompiled. */ /* The current function is: x[1]^2-x[1]*x[2]+x[3] *//*************************************************************/void evaluate(void){int mem;int i;double x[NVARS+1];for (mem = 0; mem < POPSIZE; mem++){for (i = 0; i < NVARS; i++)x[i+1] = population[mem].gene[i];population[mem].fitness = (x[1]*x[1]) - (x[1]*x[2]) + x[3];}}/***************************************************************//* Keep_the_best function: This function keeps track of the */ /* best member of the population. Note that the last entry in */ /* the array Population holds a copy of the best individual *//***************************************************************/void keep_the_best(){int mem;int i;cur_best = 0; /* stores the index of the best individual */for (mem = 0; mem < POPSIZE; mem++){if (population[mem].fitness > population[POPSIZE].fitness){cur_best = mem;population[POPSIZE].fitness = population[mem].fitness;}}/* once the best member in the population is found, copy the genes */ for (i = 0; i < NVARS; i++)population[POPSIZE].gene[i] = population[cur_best].gene[i]; }/****************************************************************//* Elitist function: The best member of the previous generation *//* is stored as the last in the array. If the best member of *//* the current generation is worse then the best member of the *//* previous generation, the latter one would replace the worst *//* member of the current population */ /****************************************************************/void elitist(){int i;double best, worst; /* best and worst fitness values */int best_mem, worst_mem; /* indexes of the best and worst member */ best = population[0].fitness;worst = population[0].fitness;for (i = 0; i < POPSIZE - 1; ++i){if(population[i].fitness > population[i+1].fitness){if (population[i].fitness >= best){best = population[i].fitness;best_mem = i;}if (population[i+1].fitness <= worst){worst = population[i+1].fitness;worst_mem = i + 1;}}else{if (population[i].fitness <= worst){worst = population[i].fitness;worst_mem = i;}if (population[i+1].fitness >= best){best = population[i+1].fitness;best_mem = i + 1;}}}/* if best individual from the new population is better than */ /* the best individual from the previous population, then *//* copy the best from the new population; else replace the *//* worst individual from the current population with the *//* best one from the previous generation */if (best >= population[POPSIZE].fitness){for (i = 0; i < NVARS; i++)population[POPSIZE].gene[i] = population[best_mem].gene[i];population[POPSIZE].fitness = population[best_mem].fitness;}else{for (i = 0; i < NVARS; i++)population[worst_mem].gene[i] = population[POPSIZE].gene[i];population[worst_mem].fitness = population[POPSIZE].fitness;}}/**************************************************************//* Selection function: Standard proportional selection for *//* maximization problems incorporating elitist model - makes *//* sure that the best member survives *//**************************************************************/void select(void){int mem, i, j, k;double sum = 0;double p;/* find total fitness of the population */for (mem = 0; mem < POPSIZE; mem++){sum += population[mem].fitness;}/* calculate relative fitness */for (mem = 0; mem < POPSIZE; mem++){population[mem].rfitness = population[mem].fitness/sum;}population[0].cfitness = population[0].rfitness;/* calculate cumulative fitness */for (mem = 1; mem < POPSIZE; mem++){population[mem].cfitness = population[mem-1].cfitness +population[mem].rfitness;}/* finally select survivors using cumulative fitness. */for (i = 0; i < POPSIZE; i++){p = rand()%1000/1000.0;if (p < population[0].cfitness)newpopulation[i] = population[0];else{for (j = 0; j < POPSIZE;j++)if (p >= population[j].cfitness &&p<population[j+1].cfitness)newpopulation[i] = population[j+1];}}/* once a new population is created, copy it back */for (i = 0; i < POPSIZE; i++)population[i] = newpopulation[i];}/***************************************************************//* Crossover selection: selects two parents that take part in *//* the crossover. Implements a single point crossover */ /***************************************************************/void crossover(void){int i, mem, one;int first = 0; /* count of the number of members chosen */ double x;for (mem = 0; mem < POPSIZE; ++mem){x = rand()%1000/1000.0;if (x < PXOVER){++first;if (first % 2 == 0)Xover(one, mem);elseone = mem;}}}/**************************************************************//* Crossover: performs crossover of the two selected parents. *//**************************************************************/void Xover(int one, int two){int i;int point; /* crossover point *//* select crossover point */if(NVARS > 1){if(NVARS == 2)point = 1;elsepoint = (rand() % (NVARS - 1)) + 1;for (i = 0; i < point; i++)swap(&population[one].gene[i], &population[two].gene[i]);}}/*************************************************************//* Swap: A swap procedure that helps in swapping 2 variables */ /*************************************************************/void swap(double *x, double *y){double temp;temp = *x;*x = *y;*y = temp;}/**************************************************************//* Mutation: Random uniform mutation. A variable selected for *//* mutation is replaced by a random value between lower and */ /* upper bounds of this variable */ /**************************************************************/void mutate(void){int i, j;double lbound, hbound;double x;for (i = 0; i < POPSIZE; i++)for (j = 0; j < NVARS; j++){x = rand()%1000/1000.0;if (x < PMUTATION){/* find the bounds on the variable to be mutated */lbound = population[i].lower[j];hbound = population[i].upper[j];population[i].gene[j] = randval(lbound, hbound);}}}/***************************************************************//* Report function: Reports progress of the simulation. Data *//* dumped into the output file are separated by commas */ /***************************************************************/void report(void){int i;double best_val; /* best population fitness */double avg; /* avg population fitness */double stddev; /* std. deviation of population fitness */ double sum_square; /* sum of square for std. calc */ double square_sum; /* square of sum for std. calc */ double sum; /* total population fitness */sum = 0.0;sum_square = 0.0;for (i = 0; i < POPSIZE; i++){sum += population[i].fitness;sum_square += population[i].fitness * population[i].fitness;}avg = sum/(double)POPSIZE;square_sum = avg * avg * POPSIZE;stddev = sqrt((sum_square - square_sum)/(POPSIZE - 1));best_val = population[POPSIZE].fitness;fprintf(galog, "\n%5d, %6.3f, %6.3f, %6.3f \n\n", generation,best_val, avg, stddev); }/**************************************************************//* Main function: Each generation involves selecting the best *//* members, performing crossover & mutation and then */ /* evaluating the resulting population, until the terminating *//* condition is satisfied */ /**************************************************************/void main(void){int i;if ((galog = fopen("galog.txt","w"))==NULL){exit(1);}generation = 0;fprintf(galog, "\n generation best average standard \n"); fprintf(galog, " number value fitness deviation \n"); initialize();evaluate();keep_the_best();while(generation<MAXGENS){generation++;select();crossover();mutate();report();evaluate();elitist();}fprintf(galog,"\n\n Simulation completed\n");fprintf(galog,"\n Best member: \n");for (i = 0; i < NVARS; i++){fprintf (galog,"\n var(%d) = %3.3f",i,population[POPSIZE].gene[i]);}fprintf(galog,"\n\n Best fitness = %3.3f",population[POPSIZE].fitness);fclose(galog);printf("Success\n");}/***************************************************************/链接库文件?这个简单一般的第三方库文件有2种提供方式1.lib静态库,这样必须在工程设置里面添加。
遗传算法的C语言程序案例
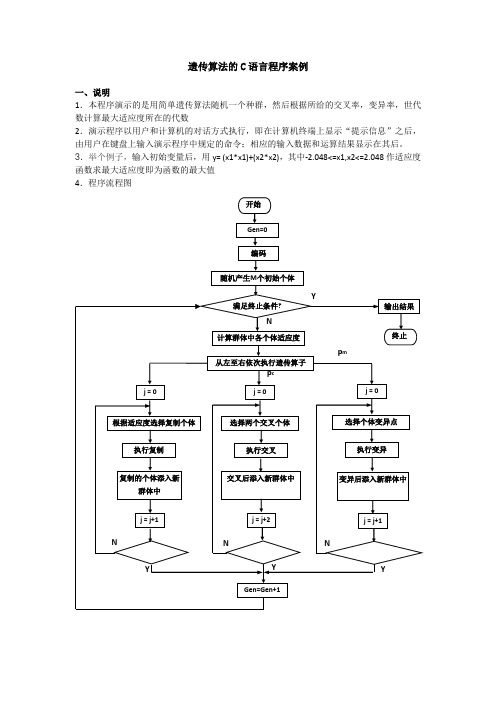
遗传算法的C语言程序案例一、说明1.本程序演示的是用简单遗传算法随机一个种群,然后根据所给的交叉率,变异率,世代数计算最大适应度所在的代数2.演示程序以用户和计算机的对话方式执行,即在计算机终端上显示“提示信息”之后,由用户在键盘上输入演示程序中规定的命令;相应的输入数据和运算结果显示在其后。
3.举个例子,输入初始变量后,用y= (x1*x1)+(x2*x2),其中-2.048<=x1,x2<=2.048作适应度函数求最大适应度即为函数的最大值4.程序流程图5.类型定义int popsize; //种群大小int maxgeneration; //最大世代数double pc; //交叉率double pm; //变异率struct individual{char chrom[chromlength+1];double value;double fitness; //适应度};int generation; //世代数int best_index;int worst_index;struct individual bestindividual; //最佳个体struct individual worstindividual; //最差个体struct individual currentbest;struct individual population[POPSIZE];3.函数声明void generateinitialpopulation();void generatenextpopulation();void evaluatepopulation();long decodechromosome(char *,int,int);void calculateobjectvalue();void calculatefitnessvalue();void findbestandworstindividual();void performevolution();void selectoperator();void crossoveroperator();void mutationoperator();void input();void outputtextreport();6.程序的各函数的简单算法说明如下:(1).void generateinitialpopulation ()和void input ()初始化种群和遗传算法参数。
用C 实现遗传算法

/*本程序试用遗传算法来解决Rosenbrock函数的全局最大值计算问题:max f(x1,x2)=100(x1^2-x2^2)^2+(1-x1)^2s.t.-2.048≤xi≤2.048(i=1,2)*/#include<iostream>#include<time.h>#include<stdlib.h>#include<cmath>using namespace std;const int M=8,T=2;//M群体大小,pc交叉概率,pm变异概率,T终止代数const double pc=0.6,pm=0.01;struct population//定义群体结构{int x[20];double x1,x2;double fit;double sumfit;}p[M];void initial(population*);//初始化函数void evaluefitness(population*);//计算适应度void select(population*);//选择复制函数void crossover(population*);//交叉函数void mutation(population*);//变异函数void decoding(population*);//解码函数void print(population*);//显示函数int main()//遗传算法主函数{int gen=0;initial(&p[0]);//随机获得初始解cout<<"initialed!"<<endl;print(&p[0]);decoding(&p[0]);//先解码cout<<"decoded!"<<endl;print(&p[0]);evaluefitness(&p[0]);//计算适应值与累计适值cout<<"evalued!"<<endl;print(&p[0]);while(gen<=T){cout<<"gen="<<gen+1<<endl;select(&p[0]);decoding(&p[0]);evaluefitness(&p[0]);cout<<"selected!"<<endl;print(&p[0]);crossover(&p[0]);decoding(&p[0]);evaluefitness(&p[0]);cout<<"crossovered!"<<endl;print(&p[0]);mutation(&p[0]);decoding(&p[0]);evaluefitness(&p[0]);cout<<"mutated!"<<endl;print(&p[0]);gen++;}decoding(&p[0]);evaluefitness(&p[0]);cout<<"最后得出的满意解为:"<<endl;for(int i=0;i<M;i++)cout<<"x1:"<<p[i].x1<<"x2:"<<p[i].x2<<"值:"<<p[i].fit<<endl;return0;}/*****************************初始化函数*****************************/int rand01()//用于随机取0或1的函数{int r;float q;q=rand()/(RAND_MAX+0.0);if(q<0.5)r=0;else r=1;return r;}void initial(population*t)//群体初始化函数{int j;population*po;srand(time(0));for(po=t;po<t+M;po++)for(j=0;j<20;j++)(*po).x[j]=rand01();}/*************************计算适应值函数*********************************/ void evaluefitness(population*t)//计算适应值函数{double f,x1,x2,temp=0.0;population*po,*po2;for(po=t;po<t+M;po++){x1=(*po).x1;x2=(*po).x2;f=100.0*(x1*x1-x2*x2)*(x1*x1-x2*x2)+(1.0-x1)*(1.0-x1);(*po).fit=f;}for(po=t;po<t+M;po++)//计算累计适应值{for(po2=t;po2<=po;po2++)temp=temp+(*po2).fit;(*po).sumfit=temp;temp=0.0;}}/**************************选择复制函数********************************/ double randab(double a,double b)//在区间(a,b)内生成一个随机数{double c,r;c=b-a;r=a+c*rand()/(RAND_MAX+1.0);return r;}void select(population*t)//选择算子函数{int i=0;population pt[M],*po;double s;srand(time(0));while(i<M){s=randab((*t).sumfit,(*(t+M-1)).sumfit);for(po=t;po<t+M;po++){if((*po).sumfit>=s){pt[i]=(*po);break;}else continue;}i++;}for(i=0;i<M;i++)//将复制后数据pt[M]转入p[M] for(int j=0;j<20;j++)p[i].x[j]=pt[i].x[j];}/***************************交叉函数*******************************/ void crossover(population*t)//交叉算子函数{population*po;double q;//用于存一个0到1的随机数int d,e,tem[20];//d存放从1到19的一个随机整数,用来确定交叉的位置//e存放从0到M的一个随机且与当前P[i]中i不同的整数,用来确定交叉的对象srand(time(0));for(po=t;po<t+M/2;po++){q=rand()/(RAND_MAX+0.0);if(q<pc){for(int j=0;j<M;j++)//运算M次,避免产生群体中某个体与自己交叉的情况{e=rand()%M;//随机确定交叉对象if(t+e!=po)break;}//不能重复d=1+rand()%19;//随机确定交叉位置for(int i=d;i<20;i++){tem[i]=(*po).x[i];(*po).x[i]=(*(t+e)).x[i];(*(t+e)).x[i]=tem[i];}}else continue;}}/***************************变异函数*******************************/void mutation(population*t)//变异算子函数{double q;//q用来存放针对每一个基因座产生的[0,1]的随机数population*po;srand(time(0));for(po=t;po<t+M;po++){int i=0;while(i<20){q=rand()/(RAND_MAX+0.0);if(q<pm)(*po).x[i]=1-(*po).x[i];i++;}}}/***************************解码函数*******************************/void decoding(population*t)//解码函数{population*po;int temp,s1=0,s2=0;float m,n,dit;n=ldexp(1,10);//n=2^10dit=4.096/(n-1);//dit=(U(max)-U(min))/n-1for(po=t;po<t+M;po++){for(int i=0;i<10;i++){temp=ldexp((*po).x[i],i);s1=s1+temp;}m=-2.048+s1*dit;(*po).x1=m;s1=0;for(int j=10;j<20;j++){temp=ldexp((*po).x[j],j-10);s2=s2+temp;}m=-2.048+s2*dit;(*po).x2=m;s2=0;}}/*******************************显示函数**************************************/ void print(population*t){population*po;for(po=t;po<t+M;po++){for(int j=0;j<20;j++){cout<<(*po).x[j];if((j+1)%5==0)cout<<"";}cout<<endl;cout<<(*po).x1<<""<<(*po).x2<<""<<(*po).fit<<""<<(*po).sumfit<<endl;}}。
遗传算法C

#include<stdlib.h>#include<stdIO.h>#include<time.h>#include<math.h>#define POPSIZE500//种群大小#define chromlength8//染色体长度int popsize;//种群大小int maxgeneration;//最大世代数double pc=0.0;//交叉率double pm=0.0;//变异率struct individual//定义染色体个体结构体{int chrom[chromlength];//定义染色体二进制表达形式,edit by ppme 将char转为intdouble value;//染色体的值double fitness;//染色体的适应值};int generation;//当前执行的世代数int best_index;//最好的染色体索引序号int worst_index;//最差的染色体索引序号struct individual bestindividual;//最佳染色体个体struct individual worstindividual;//最差染色体个体struct individual currentbest;//当前最好的染色体个体currentbest struct individual population[POPSIZE];//种群数组//函数声明void generateinitialpopulation();//ok-初始化当代种群void generatenextpopulation();//??产生下一代种群void evaluatepopulation();//评价种群void calculateobjectfitness();//计算种群适应度//long decodechromosome(char*,int,int);//染色体解码double decodechromosome(int,int);//染色体解码void findbestandworstindividual();//寻找最好的和最坏的染色体个体void performevolution();//进行演变进化void selectoperator();//选择操作void crossoveroperator();//交换操作void mutationoperator();//变异操作void input();//输入接口void outputtextreport();//输出文字报告void main()//主函数{int i;srand((unsigned)time(NULL));//强制类型转化,以当前时间戳定义随机数种子printf("本程序为求函数y=x*x的最大值\n");generation=0;//初始化generation当前执行的代input();//初始化种群大小、交叉率、变异率/*edit by ppme*///调试用。
遗传算法的c语言程序
一需求分析1.本程序演示的是用简单遗传算法随机一个种群,然后根据所给的交叉率,变异率,世代数计算最大适应度所在的代数2.演示程序以用户和计算机的对话方式执行,即在计算机终端上显示“提示信息”之后,由用户在键盘上输入演示程序中规定的命令;相应的输入数据和运算结果显示在其后。
3.测试数据输入初始变量后用y=100*(x1*x1-x2)*(x1*x2-x2)+(1-x1)*(1-x1)其中-2.048<=x1,x2<=2.048作适应度函数求最大适应度即为函数的最大值二概要设计1.程序流程图2.类型定义int popsize; //种群大小int maxgeneration; //最大世代数double pc; //交叉率double pm; //变异率struct individualchar chrom[chromlength+1];double value;double fitness; //适应度};int generation; //世代数int best_index;int worst_index;struct individual bestindividual; //最佳个体struct individual worstindividual; //最差个体struct individual currentbest;struct individual population[POPSIZE];3.函数声明void generateinitialpopulation();void generatenextpopulation();void evaluatepopulation();long decodechromosome(char *,int,int);void calculateobjectvalue();void calculatefitnessvalue();void findbestandworstindividual();void performevolution();void selectoperator();void crossoveroperator();void mutationoperator();void input();void outputtextreport();4.程序的各函数的简单算法说明如下:(1).void generateinitialpopulation ()和void input ()初始化种群和遗传算法参数。
遗传算法程序实现
遗传算法程序实现#include <stdlib.h>#include <stdio.h>#include <time.h>#include <math.h>#include "GA.h"unsigned seed=1;//产生一个不大于x的随机数double crandom(int x){seed++;srand((unsigned)(seed*time(NULL)));return(rand()%x);}//产生一个0~1的随机数double random(){seed++;srand((unsigned)(seed*time(NULL)));return (double)rand()/(double)RAND_MAX;}//产生(0.1,0.2,...,1)之间的一个随机数double crandom(void){double t;t=random();if((10*t-floor(10*t))>=0.5) return (floor(10*t)+1)/10;elsereturn(floor(10*t)/10);}//初始化种群void Initial_group(double group[M][N_para], double up_limit[N_para],double low_limit[N_para]){ double beta;int i,j;for (i=0;i<M;i++)for (j=0;j<N_para;j++){beta=crandom();group[i][j]=low_limit[j]+beta*(up_limit[j]-low_limit[j]);}}}//加载实时测试数据void Load_data(double realvalue[][4]){FILE *fp;float x;//changedif((fp=fopen("realtime\\Realdata.txt","r"))==NULL){printf("cannot open the file Realvalue.txt\n");exit(0);}for(int i=0;i<N_hit+N_Begin;i++){for(int j=0;j<4;j++)//修改了{fscanf(fp,"%f ",&x);if(i>=N_Begin)realvalue[i-N_Begin][j]=(double)x;//修改了}}fclose(fp);}//计算适应度值void Fit_calculate(double group[][N_para],double realvalue[][4],double fit_degree[],int n){int i;//double Mcar,m1,r1,r2,L,J1,J2,c1,c2,b;double mcar,m1,l1,J1,b,g,f1,eks,eksd,theta1,theta1d,h;double x[4],x_temp[4],k[4][4],lefA[4];double err,A11,A12,A21,A22,B1,B2,B3,B4,C11,C12,C21,C22,D2;g=9.8;h=0.005;for (i=0;i<n;i++){mcar=group[i][0];m1=group[i][1];l1=group[i][2];J1=group[i][3];b=group[i][4];f1=group[i][5];//printf("mcar=%6.3f,m1=%6.3f,l1=%6.3f,J1=%6.3f,b=%6.3,f1=%6.3\n,",mcar,m1, l1,J1,b,f1);err=0;for (int j=0;j<4;j++) //新加的{x[j]=realvalue[0][j];x_temp[j]=realvalue[0][j];}/*for (int j=0;j<4;j++)//修改了{x[j]=Initivalue[j];x_temp[j]=Initivalue[j];}*/for (int q=0;q<N_hit;q++){eks=x[0];eksd=x[1];theta1=x[2];theta1d=x[3];while(fabs(x[2])>(2*Pi)){printf("Error");printf(" q=%d",q);getchar();}A11=mcar+m1;A12=-m1*l1*cos(theta1);A22=m1*l1*l1+J1;A21=A12;C11=b;C12=m1*l1*sin(theta1)*theta1d;C21=0;C22=f1;D2=-m1*g*l1*sin(theta1);B1=eksd;B2=-C11*eksd-C12*theta1d;B3=theta1d;B4=-C22*theta1d-D2;/*%A=[1 0 0 0;% 0 A11 0 A12 ;% 0 0 1 0 ;% 0 A21 0 A22];%lefA = inv(A)*B;%A11x+A12y=B(2)%A21x+A22y=B(4)*/lefA[0]=B1;lefA[1]=(B2*A22-B4*A12)/(A22*A11-A21*A12);lefA[2]=B3;lefA[3]=(B2*A21-B4*A11)/(A12*A21-A22*A11);for (j=0;j<4;j++) //龙格-库塔法{k[j][0]=lefA[0];k[j][1]=lefA[1];k[j][2]=lefA[2];k[j][3]=lefA[3];for (int p=0;p<4;p++){if(j==0) x[p]=x_temp[p]+h/2*k[j][p];if(j==1) x[p]=x_temp[p]+h/2*k[j][p];if(j==2) x[p]=x_temp[p]+h*k[j][p];}}for(j=0;j<4;j++){x[j]=x_temp[j]+(k[0][j]+2*k[1][j]+2*k[2][j]+k[3][j])*h/6;x_temp[j]=x[j];}//getchar();//test uerr=err+pow((x[2]-realvalue[q][2]),2);//err=err+80*pow(x[1],2)+80*pow(x[3],2)+pow(u,2);}//end of for qfit_degree[i]=2+log(2/err)/log(200);///log(2);//9改成200了}}//种群排序void sort(double order_fit[],int index_fit[],int n){int i,j;int t;double temp;for (i=0;i<n;i++)index_fit[i]=i;for (j=1;j<=n-1;j++)for(i=1;i<=n-j;i++){if(order_fit[i]<order_fit[i-1]){temp=order_fit[i];order_fit[i]=order_fit[i-1];order_fit[i-1]=temp;t=index_fit[i];index_fit[i]=index_fit[i-1];index_fit[i-1]=t;}}}//反馈式突变void Feedback_mut(double group[][N_para],double up_limit[],double low_limit[],int index_fit[],double fit_opitimal,int nb){// printf("\nFeedback1 ok\n");double beta;int i,j,best,worse;int n=M;best=index_fit[M-1];for (i=0;i<n*0.9;i++){beta=crandom();worse=index_fit[i];if(nb>50||fit_opitimal<Sigma){if(nb>50) nb=0;for(j=0;j<N_para;j++)group[worse][j]=low_limit[j]+beta*(up_limit[j]-low_limit[j]);}elsefor(j=0;j<N_para;j++){group[worse][j]=(Nu+beta*Vi)*group[best][j];if (group[worse][j]<low_limit[j]||group[worse][j]>up_limit[j])group[worse][j]=group[best][j];}}//printf("\nFeedback 2 ok\n");}//选择操作void Slected(double cross[],double group[][N_para],double fit_degree[],double fit_sum){int j,i=0;double beta,slectvalue[M];beta=crandom();slectvalue[0]=fit_degree[0]/fit_sum;for(i=1;i<M;i++)slectvalue[i]=slectvalue[i-1]+fit_degree[i]/fit_sum;for(i=1;i<M;i++){if(slectvalue[i-1]<beta&&slectvalue[i]>beta)break;}for(j=0;j<N_para;j++)cross[j]=group[i][j];}//计算两个染色体的海明距离int haiming(double crossa[],double crossb[],int n){int num=0;int i;for (i=0;i<n;i++)if(crossa[i]!=crossb[i]) num++;return num;}//交叉操作void Cro_operation(double crossa[],double crossb[],double up_limit[],double low_limit[],int D_hm){int i,j,cro_point,d,zeta=0,sum=0;double cro_temp,beta;double x_temp[N_para];double y_temp[N_para];int rec[6]={0,0,0,0,0,0};if(D_hm==1){for(i=0;i<N_para;i++){if(crossa[i]!=crossb[i])break;}//cro_temp=crossa[i];//crossa[i]=crossb[i];//crossb[i]=cro_tempcro_point=i;}else{//zeta=(int)(crandom(4)+1);for(int ii=0;ii<N_para;ii++){for (int k=0;k<N_para;k++){x_temp[k]=0;y_temp[k]=0;}for(i=0;i<=ii;i++){x_temp[i]=crossa[i];y_temp[i]=crossb[i];}d=haiming(x_temp,y_temp,N_para);if(d>0&&d<D_hm){zeta++;rec[ii]=1;}//break;//>=改成<???}ii=(int)(crandom(zeta)+1);sum=0;for(i=0;sum<ii;i++){sum=sum+rec[i];cro_point=i;;}}//End of elsefor (j=0;j<cro_point;j++){cro_temp=crossa[j];crossa[j]=crossb[j];crossb[j]=cro_temp;}//cro_point--;//改了beta=crandom();cro_temp=(crossa[cro_point]<=crossb[cro_point])?crossa[cro_point]:crossb[cro_point];crossa[cro_point]=cro_temp+beta*fabs(crossa[cro_point]-crossb[cro_point]);beta=crandom();crossb[cro_point]=low_limit[cro_point]+beta*(up_limit[cro_point]-low_limit[cro_poi nt]);}//正交实验产生一个较优染色体void Cro_opiti(double crossa[N_para],double crossb[N_para],double cro_result[N_para],double realvalue[][4])//改了{double matrix[8][6]={//四因素两水平正交试验表1,1,1,1,1,1,1,1,1,2,2,2,1,2,2,1,1,2,1,2,2,2,2,1,2,1,2,1,2,1,2,1,2,2,1,2,2,2,1,1,2,2,2,2,1,2,1,1};int i,j;double fit_temp[8];double kx[N_para]={0,0,0,0,0,0};//changedouble ky[N_para]={0,0,0,0,0,0};//changefor(i=0;i<8;i++)for(j=0;j<N_para;j++){if(matrix[i][j]==1)matrix[i][j]=crossa[j];elsematrix[i][j]=crossb[j];}Fit_calculate(matrix,realvalue,fit_temp,8);for(i=0;i<8;i++)// 改了{for(j=0;j<N_para;j++){if(matrix[i][j]==crossa[j])kx[j]=kx[j]+fit_temp[i];if(matrix[i][j]==crossb[j])ky[j]=ky[j]+fit_temp[i];}}for(j=0;j<N_para;j++){if(kx[j]>ky[j])cro_result[j]=crossa[j];elsecro_result[j]=crossb[j];}}//变异操作void mut_operation(double cro_result[N_para],double up_limit[], double low_limit[]) {int mut_point1,mut_point2;double beta,x1,x2;do{mut_point1=(int)crandom(6);//改了13mut_point2=(int)crandom(6);//改了13}while(mut_point1==mut_point2);x1=(cro_result[mut_point1]-low_limit[mut_point1])/(up_limit[mut_point1]-low_limit [mut_point1]);x2=(cro_result[mut_point2]-low_limit[mut_point2])/(up_limit[mut_point2]-low_limit [mut_point2]);beta=crandom();if(beta<Pm){beta=crandom();cro_result[mut_point1]=(1-beta)*cro_result[mut_point1]+beta*low_limit[mut_point 1]+x2*(up_limit[mut_point1]-cro_result[mut_point1]);cro_result[mut_point2]=(1-beta)*cro_result[mut_point2]+beta*low_limit[mut_point 2]+x1*(up_limit[mut_point2]-cro_result[mut_point2]);}}//产生新一代种群void New_group(double group_temp[][N_para],double group[][N_para],double newfit_degree[],double fit_degree[]){int i,j;int temp;int index[M+Ncross];sort(newfit_degree,index,M+Ncross);for(i=0;i<M;i++){temp=index[Ncross+i];for(j=0;j<N_para;j++){group[i][j]=group_temp[temp][j];}fit_degree[i]=newfit_degree[Ncross+i];}}//种群最优个体,适应度值数据存盘void Data_record(double group[][N_para],double fit_degree[],double fit_population,int index_fit[]){FILE *fp;if((fp=fopen("Result\\Result.txt","a"))==NULL){printf("cannot open the fiel Result.txt\n");exit(0);}int nbest=index_fit[M-1];for(int i=0;i<N_para;i++)fprintf(fp,"%6.5f ",group[nbest][i]);printf("\npara=%6.5f %6.5f %6.5f %6.5f %6.5f %6.5f\n",group[nbest][0],group[n best][1],group[nbest][2],group[nbest][3],group[nbest][4],group[nbest][5]);//家的fprintf(fp,"%6.5f %6.5f\n",fit_degree[M-1],fit_population);fclose(fp);。
遗传算法求解函数极值C语言代码
#include "stdio.h"#include "stdlib.h"#include "conio.h"#include "math.h"#include "time.h"#define num_C 12 //个体的个数,前6位表示x1,后6位表示x2 #define N 100 //群体规模为100#define pc 0.9 //交叉概率为0.9#define pm 0.1 //变异概率为10%#define ps 0.6 //进行选择时保留的比例#define genmax 2000 //最大代数200int RandomInteger(int low,int high);void Initial_gen(struct unit group[N]);void Sort(struct unit group[N]);void Copy_unit(struct unit *p1,struct unit *p2);void Cross(struct unit *p3,struct unit *p4);void Varation(struct unit group[N],int i);void Evolution(struct unit group[N]);float Calculate_cost(struct unit *p);void Print_optimum(struct unit group[N],int k);/* 定义个体信息*/typedef struct unit{int path[num_C]; //每个个体的信息double cost; //个体代价值};struct unit group[N]; //种群变量groupint num_gen=0; //记录当前达到第几代int main(){int i,j;srand((int)time(NULL)); //初始化随机数发生器Initial_gen(group); //初始化种群Evolution(group); //进化:选择、交叉、变异getch();return 0;}/* 初始化种群*/void Initial_gen(struct unit group[N]){int i,j;struct unit *p;for(i=0;i<=N-1;i++) //初始化种群里的100个个体{p=&group[i]; //p指向种群的第i个个体for(j=0;j<12;j++){p->path[j]=RandomInteger(0,9); //end }Calculate_cost(p); //计算该种群的函数值}//end 初始化种群}/* 种群进化,进化代数由genmax决定*/void Evolution(struct unit group[N]){int i,j;int temp1,temp2,temp3,temp4,temp5;temp1=N*pc/2;temp2=N*(1-pc);temp3=N*(1-pc/2);temp4=N*(1-ps);temp5=N*ps;for(i=1;i<=genmax;i++){//选择for(j=0;j<=temp4-1;j++){ Copy_unit(&group[j],&group[j+temp5]); }//交叉for(j=0;j<=temp1-1;){Cross(&group[temp2+j],&group[temp3+j]);j+=2;}//变异Varation(group,i);}Sort(group);Print_optimum(group,i-1); //输出当代(第i-1代)种群}/* 交叉*/void Cross(struct unit *p3,struct unit *p4){int i,j,cross_point;int son1[num_C],son2[num_C];for(i=0;i<=num_C-1;i++) //初始化son1、son2{son1[i]=-1;son2[i]=-1;}cross_point=RandomInteger(1,num_C-1); //交叉位随机生成//交叉,生成子代//子代1//子代1前半部分直接从父代复制for(i=0;i<=cross_point-1;i++) son1[i]=p3->path[i];for(i=cross_point;i<=num_C-1;i++)for(j=0;j<=num_C-1;j++) //补全p1{son1[i]=p4->path[j];}//end 子代1//子代2//子代1后半部分直接从父代复制for(i=cross_point;i<=num_C-1;i++) son2[i]=p4->path[i];for(i=0;i<=cross_point-1;i++){for(j=0;j<=num_C-1;j++) //补全p1{son2[i]=p3->path[j];}}//end 子代2//end 交叉for(i=0;i<=num_C-1;i++){p3->path[i]=son1[i];p4->path[i]=son2[i];}Calculate_cost(p3); //计算子代p1的函数值Calculate_cost(p4); //计算子代p2的函数值}/* 变异*/void Varation(struct unit group[N],int flag_v){int flag,i,j,k,temp;struct unit *p;flag=RandomInteger(1,100);//在进化后期,增大变异概率if((!flag>(flag_v>100))?(5*100*pm):(100*pm)){i=RandomInteger(0,N-1); //确定发生变异的个体j=RandomInteger(0,num_C-1); //确定发生变异的位k=RandomInteger(0,num_C-1);p=&group[i]; //变异temp=p->path[j];p->path[j]=p->path[k];p->path[k]=temp;Calculate_cost(p); //重新计算变异后的函数值}}/* 将种群中个体按函数值从小到大排序*/void Sort(struct unit group[N]){int i,j;struct unit temp,*p1,*p2;for(j=1;j<=N-1;j++) //排序总共需进行N-1轮{for(i=1;i<=N-1;i++){p1=&group[i-1];p2=&group[i];if(p1->cost>p2->cost) //值大的往后排{Copy_unit(p1,&temp);Copy_unit(p2,p1);Copy_unit(&temp,p2);}}//end 一轮排序}//end 排序}/* 计算某个个体的函数值*/float Calculate_cost(struct unit *p){double x1,x2;x1=0;if(p->path[0]>5){x1=p->path[1]+p->path[2]*0.1+p->path[3]*0.01+p->path[4]*0.001+p->path[5]*0.0001;}else if(p->path[0]<6){x1=0-(p->path[1]+p->path[2]*0.1+p->path[3]*0.01+p->path[4]*0.001+p->path[5]*0.0001);}x2=0;if(p->path[6]>5){}else if(p->path[6]<6){x2=0-(p->path[7]+p->path[8]*0.1+p->path[9]*0.01+p->path[10]*0.001+p->path[11]*0.0001);}p->cost=20+x1*x1+x2*x2-10*(cos(2*3.14*x1)+cos(2*3.14*x2));return(p->cost);}/* 复制种群中的p1到p2中*/void Copy_unit(struct unit *p1,struct unit *p2){int i;for(i=0;i<=num_C-1;i++)p2->path[i]=p1->path[i];p2->cost=p1->cost;}/* 生成一个介于两整型数之间的随机整数*/int RandomInteger(int low,int high){int k;double d;k=rand();k=(k!=RAND_MAX)?k:(k-1); //RAND_MAX是VC中可表示的最大整型数d=(double)k/((double)(RAND_MAX));k=(int)(d*(high-low+1));return (low+k);}/* 输出当代种群中的最优个体*/void Print_optimum(struct unit group[N],int k){struct unit *p;double x1,x2;x1=x2=0;p=&group[0];if(p->path[0]>5){x1=p->path[1]+p->path[2]*0.1+p->path[3]*0.01+p->path[4]*0.001+p->path[5]*0.0001;}else if(p->path[0]<6){}x2=0;if(p->path[6]>5){x2=p->path[7]+p->path[8]*0.1+p->path[9]*0.01+p->path[10]*0.001+p->path[11]*0.0001;}else if(p->path[6]<6){x2=0-(p->path[7]+p->path[8]*0.1+p->path[9]*0.01+p->path[10]*0.001+p->path[11]*0.0001);}printf(" 当x1=%f x2=%f\n",x1,x2);printf(" 函数最小值为:%f \n",p->cost);}。
C语言版遗传算法
//distance[i][j]代表i城市与j城市的距离
/*
const static DISTANCE distance[][_CITY_AMOUNT]
={
0, 1, 4, 6, 8, 1, 3, 7, 2, 9, 7, 3, 4, 5, 8, 9, 2, 8, 2, 8,
{
cout << "第" << i+1 << "代" << std::endl;
CUnit.fnDispProbability();
CUnit.fnDispHistoryMin();
}
}
CUnit.fnDispHistoryMin();
int fnEvalOne(T &Gene); //测试某一个基因的适应度
void fnDispProbability(); //显示每个个体的权值
void fnDispHistoryMin();
private:
bool fnGeneAberranceOne(const int &i, const int &j);//变异某个基因
//#define _XCHG_GENE_AMOUNT_WHEN_MIX 2 //每次杂交所交换的碱基数量
#define _TIMES 50 //定义进化次数
#define _DISP_INTERVAL 5 //每隔多少次显示基因中的最高适应度
#define _CONTAINER std::vector<int> //定义个体基因容器类型
#define _CONTAINER_P std::vector<int> //定义适应度容器类型
- 1、下载文档前请自行甄别文档内容的完整性,平台不提供额外的编辑、内容补充、找答案等附加服务。
- 2、"仅部分预览"的文档,不可在线预览部分如存在完整性等问题,可反馈申请退款(可完整预览的文档不适用该条件!)。
- 3、如文档侵犯您的权益,请联系客服反馈,我们会尽快为您处理(人工客服工作时间:9:00-18:30)。
一个简单实用的遗传算法c程序HEN system office room 【HEN16H-HENS2AHENS8Q8-HENH1688】一个简单实用的遗传算法c程序(转载) 2009-07-28 23:09:03 阅读418 评论0 字号:大中小这是一个非常简单的遗传算法源代码,是由Denis Cormier (North Carolina State University)开发的,Sita (University of North Carolina at Charlotte)修正。
代码保证尽可能少,实际上也不必查错。
对一特定的应用修正此代码,用户只需改变常数的定义并且定义“评价函数”即可。
注意代码的设计是求最大值,其中的目标函数只能取正值;且函数值和个体的适应值之间没有区别。
该系统使用比率选择、精华模型、单点杂交和均匀变异。
如果用Gaussian变异替换均匀变异,可能得到更好的效果。
代码没有任何图形,甚至也没有屏幕输出,主要是保证在平台之间的高可移植性。
读者可以从,目录 coe/evol中的文件中获得。
要求输入的文件应该命名为‘’;系统产生的输出文件为‘’。
输入的文件由几行组成:数目对应于变量数。
且每一行提供次序——对应于变量的上下界。
如第一行为第一个变量提供上下界,第二行为第二个变量提供上下界,等等。
/**************************************************************************//* This is a simple genetic algorithm implementation where the *//* evaluation function takes positive values only and the *//* fitness of an individual is the same as the value of the *//* objective function *//**************************************************************************/#include <>#include <>#include <>/* Change any of these parameters to match your needs */#define POPSIZE 50 /* population size */#define MAXGENS 1000 /* max. number of generations */#define NVARS 3 /* no. of problem variables */#define PXOVER /* probability of crossover */#define PMUTATION /* probability of mutation */#define TRUE 1#define FALSE 0int generation; /* current generation no. */int cur_best; /* best individual */FILE *galog; /* an output file */struct genotype /* genotype (GT), a member of the population */{double gene[NVARS]; /* a string of variables一个变量字符串 */ double fitness; /* GT's fitness适应度 */double upper[NVARS]; /* GT's variables upper bound 变量的上限*/ double lower[NVARS]; /* GT's variables lower bound变量的下限 */ double rfitness; /* relative fitness 相对适应度*/double cfitness; /* cumulative fitness 累计适应度*/};struct genotype population[POPSIZE+1]; /* population */struct genotype newpopulation[POPSIZE+1]; /* new population; *//* replaces the *//* old generation *//* Declaration of procedures used by this genetic algorithm */ void initialize(void);double randval(double, double);void evaluate(void);void keep_the_best(void);void elitist(void);void select(void);void crossover(void);void Xover(int,int);void swap(double *, double *);void mutate(void);void report(void);/***************************************************************/ /* Initialization function: Initializes the values of genes */ /* within the variables bounds. It also initializes (to zero) */ /* all fitness values for each member of the population. It */ /* reads upper and lower bounds of each variable from the */ /* input file `'. It randomly generates values *//* between these bounds for each gene of each genotype in the */ /* population. The format of the input file `' is *//* var1_lower_bound var1_upper bound */ /* var2_lower_bound var2_upper bound ... */ /***************************************************************/ void initialize(void){FILE *infile;int i, j;double lbound, ubound;if ((infile = fopen("","r"))==NULL){fprintf(galog,"\nCannot open input file!\n");exit(1);}/* initialize variables within the bounds */for (i = 0; i < NVARS; i++){fscanf(infile, "%lf",&lbound);fscanf(infile, "%lf",&ubound);for (j = 0; j < POPSIZE; j++){population[j].fitness = 0;population[j].rfitness = 0;population[j].cfitness = 0;population[j].lower[i] = lbound;population[j].upper[i]= ubound;population[j].gene[i] = randval(population[j].lower[i], population[j].upper[i]);}}fclose(infile);}/***********************************************************//* Random value generator: Generates a value within bounds *//***********************************************************/ double randval(double low, double high){double val;val = ((double)(rand()%1000)/*(high - low) + low;return(val);}/*************************************************************//* Evaluation function: This takes a user defined function. *//* Each time this is changed, the code has to be recompiled. */ /* The current function is: x[1]^2-x[1]*x[2]+x[3] */ /*************************************************************/ void evaluate(void){int mem;int i;double x[NVARS+1];for (mem = 0; mem < POPSIZE; mem++){for (i = 0; i < NVARS; i++)x[i+1] = population[mem].gene[i];population[mem].fitness = (x[1]*x[1]) - (x[1]*x[2]) + x[3]; }}/***************************************************************/ /* Keep_the_best function: This function keeps track of the */ /* best member of the population. Note that the last entry in */ /* the array Population holds a copy of the best individual */ /***************************************************************/ void keep_the_best(){int mem;int i;cur_best = 0; /* stores the index of the best individual */for (mem = 0; mem < POPSIZE; mem++){if (population[mem].fitness > population[POPSIZE].fitness){cur_best = mem;population[POPSIZE].fitness = population[mem].fitness;}}/* once the best member in the population is found, copy the genes */ for (i = 0; i < NVARS; i++)population[POPSIZE].gene[i] = population[cur_best].gene[i];}/****************************************************************//* Elitist function: The best member of the previous generation *//* is stored as the last in the array. If the best member of *//* the current generation is worse then the best member of the *//* previous generation, the latter one would replace the worst *//* member of the current population */ /****************************************************************/ void elitist(){int i;double best, worst; /* best and worst fitness values */ int best_mem, worst_mem; /* indexes of the best and worst member */ best = population[0].fitness;worst = population[0].fitness;for (i = 0; i < POPSIZE - 1; ++i){if(population[i].fitness > population[i+1].fitness){if (population[i].fitness >= best){best = population[i].fitness;best_mem = i;}if (population[i+1].fitness <= worst){worst = population[i+1].fitness;worst_mem = i + 1;}}else{if (population[i].fitness <= worst){worst = population[i].fitness;worst_mem = i;}if (population[i+1].fitness >= best){best = population[i+1].fitness;best_mem = i + 1;}}}/* if best individual from the new population is better than */ /* the best individual from the previous population, then */ /* copy the best from the new population; else replace the */ /* worst individual from the current population with the *//* best one from the previous generation */if (best >= population[POPSIZE].fitness){for (i = 0; i < NVARS; i++)population[POPSIZE].gene[i] = population[best_mem].gene[i]; population[POPSIZE].fitness = population[best_mem].fitness;}else{for (i = 0; i < NVARS; i++)population[worst_mem].gene[i] = population[POPSIZE].gene[i]; population[worst_mem].fitness = population[POPSIZE].fitness;}}/**************************************************************//* Selection function: Standard proportional selection for *//* maximization problems incorporating elitist model - makes *//* sure that the best member survives *//**************************************************************/ void select(void){int mem, i, j, k;double sum = 0;double p;/* find total fitness of the population */for (mem = 0; mem < POPSIZE; mem++){sum += population[mem].fitness;}/* calculate relative fitness */for (mem = 0; mem < POPSIZE; mem++){population[mem].rfitness = population[mem].fitness/sum;}population[0].cfitness = population[0].rfitness;/* calculate cumulative fitness */for (mem = 1; mem < POPSIZE; mem++){population[mem].cfitness = population[mem-1].cfitness + population[mem].rfitness;}/* finally select survivors using cumulative fitness. */for (i = 0; i < POPSIZE; i++){p = rand()%1000/;if (p < population[0].cfitness)newpopulation[i] = population[0];else{for (j = 0; j < POPSIZE;j++)if (p >= population[j].cfitness &&p<population[j+1].cfitness)newpopulation[i] = population[j+1];}}/* once a new population is created, copy it back */for (i = 0; i < POPSIZE; i++)population[i] = newpopulation[i];}/***************************************************************/ /* Crossover selection: selects two parents that take part in */ /* the crossover. Implements a single point crossover */ /***************************************************************/void crossover(void){int i, mem, one;int first = 0; /* count of the number of members chosen */ double x;for (mem = 0; mem < POPSIZE; ++mem){x = rand()%1000/;if (x < PXOVER){++first;if (first % 2 == 0)Xover(one, mem);elseone = mem;}}}/**************************************************************/ /* Crossover: performs crossover of the two selected parents. */ /**************************************************************/void Xover(int one, int two){int i;int point; /* crossover point *//* select crossover point */if(NVARS > 1){if(NVARS == 2)point = 1;elsepoint = (rand() % (NVARS - 1)) + 1;for (i = 0; i < point; i++)swap(&population[one].gene[i], &population[two].gene[i]); }}/*************************************************************/ /* Swap: A swap procedure that helps in swapping 2 variables */ /*************************************************************/ void swap(double *x, double *y){double temp;temp = *x;*x = *y;*y = temp;}/**************************************************************//* Mutation: Random uniform mutation. A variable selected for *//* mutation is replaced by a random value between lower and *//* upper bounds of this variable *//**************************************************************/void mutate(void){int i, j;double lbound, hbound;double x;for (i = 0; i < POPSIZE; i++)for (j = 0; j < NVARS; j++){x = rand()%1000/;if (x < PMUTATION){/* find the bounds on the variable to be mutated */lbound = population[i].lower[j];hbound = population[i].upper[j];population[i].gene[j] = randval(lbound, hbound);}}}/***************************************************************//* Report function: Reports progress of the simulation. Data *//* dumped into the output file are separated by commas *//***************************************************************/void report(void){int i;double best_val; /* best population fitness */double avg; /* avg population fitness */double stddev; /* std. deviation of population fitness */ double sum_square; /* sum of square for std. calc */double square_sum; /* square of sum for std. calc */double sum; /* total population fitness */sum = ;sum_square = ;for (i = 0; i < POPSIZE; i++){sum += population[i].fitness;sum_square += population[i].fitness * population[i].fitness; }avg = sum/(double)POPSIZE;square_sum = avg * avg * POPSIZE;stddev = sqrt((sum_square - square_sum)/(POPSIZE - 1));best_val = population[POPSIZE].fitness;fprintf(galog, "\n%5d, %, %, % \n\n", generation,best_val, avg, stddev);}/**************************************************************/ /* Main function: Each generation involves selecting the best */ /* members, performing crossover & mutation and then */ /* evaluating the resulting population, until the terminating */ /* condition is satisfied */ /**************************************************************/ void main(void){int i;if ((galog = fopen("","w"))==NULL){exit(1);}generation = 0;fprintf(galog, "\n generation best average standard \n"); fprintf(galog, " number value fitness deviation \n"); initialize();evaluate();keep_the_best();while(generation<MAXGENS){generation++;select();crossover();mutate();report();evaluate();elitist();}fprintf(galog,"\n\n Simulation completed\n");fprintf(galog,"\n Best member: \n");for (i = 0; i < NVARS; i++){fprintf (galog,"\n var(%d) = %",i,population[POPSIZE].gene[i]);}fprintf(galog,"\n\n Best fitness = %",population[POPSIZE].fitness);fclose(galog);printf("Success\n");}/***************************************************************/链接库文件?这个简单一般的第三方库文件有2种提供方式静态库,这样必须在工程设置里面添加。
