组蛋白乙酰化 去乙酰化 综述
表观遗传的调控机制

表观遗传的调控机制摘要: 表观遗传是非DNA 序列遗传信息的传递, 它不涉及基因序列的改变, 不符合孟德尔式的遗传方式。
表观遗传学研究的是生物可遗传的染色质修饰。
目前,其主要研究内容包括DNA 甲基化、翻译后组蛋白修饰、组蛋白组成变化。
其中DNA 甲基化是一种重要的表观遗传修饰方式, 是调节基因组功能的重要手段。
组蛋白修饰作为表观传中重要的调控机制之一, 在包括基因表达调控等多种生物学过程中起着重要作用。
组蛋白甲基转移酶和组蛋白去甲基化酶共同参与形成和维持不同的组蛋白甲基化状态, 继而通过多种分子参与对组蛋白甲基化修饰的识别而引起下游过程的发生。
组蛋白乙酰化和去乙酰化修饰也是调控表观遗传机制之一。
最近人们还发现非编码的RNA也参与了表观遗传调控。
关键词:表观遗传,DNA甲基化,组蛋白修饰,RNA调控。
一 DNA甲基化调控表观遗传经典遗传学认为,生命的遗传信息储存在 DNA的碱基序列上,几乎所有的生命活动都受基因调控。
但是,作为开放的复杂系统,生命活动从来就不是由一种因素就能完全决定的。
随着科学的发展,人们发现一些 DNA 或染色体水平的修饰也会造成基因表达模式的改变。
这种通过有丝分裂或减数分裂来传递非DNA 序列遗传信息的现象称为表观遗传(epigenetic inheritance)。
由于它不涉及基因序列的改变,不符合孟德尔式的遗传方式,因此它是一种全新的遗传机制。
表观遗传修饰有许多,其中 DNA 甲基化是基因组DNA 的一种最重要的表观遗传修饰方式,是调节基因组功能的重要手段。
在植物中,DNA 甲基化参与细胞的许多生物学过程,在植物生长发育及进化过程中起着重要的调节作用。
1 植物DNA胞嘧啶甲基转移酶植物DNA的甲基化是在 DNA 甲基转移酶(DNAMethyltransferase,DMT)的作用下,将 S- 腺苷甲硫氨酸上的甲基基团转移到 DNA 分子的胞嘧啶碱基上。
在植物细胞中广泛存在的有三类结构和功能上不同的胞嘧啶甲基转移酶[1,2]。
乙酰化和去乙酰化修饰

乙酰化和去乙酰化修饰
乙酰化修饰通常发生在蛋白质的赖氨酸残基上,通过将乙酰基添加到赖氨酸上来改变蛋白质的电荷、构象和亲疏水性质。
这可以影响蛋白质的互作、局部结构和整体折叠状态。
乙酰化修饰也可以发生在DNA上,通过将乙酰基添加到组蛋白蛋白质上来影响DNA的紧密度和可读性,从而影响基因表达。
去乙酰化修饰则是通过去除蛋白质、DNA或RNA上的乙酰基来调节它们的功能。
这种修饰方式常常由去乙酰化酶催化,是维持细胞内化学平衡的重要机制。
例如,去乙酰化修饰可以改变某些DNA结合蛋白质与DNA的亲和力,从而影响染色质的结构和转录调控。
总之,乙酰化和去乙酰化修饰是细胞内非常重要的化学修饰方式,通过调节蛋白质、DNA或RNA的功能和稳定性,维持着细胞内的正常生理活动。
- 1 -。
(细胞生物学专业优秀论文)组蛋白乙酰化修饰调控果蝇热休克基因表达和寿..

摘要衰老是一个普遍的生物学现象,衰老控制着生物寿命的长短,主要受遗传因子和环境因素所影响。
了解衰老的分子机制,对于延缓衰老、保持生命活力具有重要的意义。
热休克蛋白(HSP)作为高度保守的“分子伴侣”,在细胞内广泛地参与许多复杂的功能活动,可以抵制衰老过程中一些有害蛋白的发生。
其基因的表达调控是一种特殊的真核基因表达模式,包括基础水平和诱导水平的表达。
由组蛋白乙酰转移酶(HAT)和组蛋白去乙酰化酶(HDAC)催化的乙酰化反应在真核基因的表达调控中起着重要作用,这两种酶通过对核心组蛋白进行可逆修饰来调节核心组蛋白的乙酰化水平,从而调控转录的起始与延伸。
组蛋白去乙酰化酶抑制剂(HDI)可以通过抑制HDAC活性提高组蛋白乙酰化水平,是研究乙酰化修饰在真核基因表达调控中的作用的有用工具。
本论文一方面采用HDItrichostatinA<TSA)和丁酸钠(BuA)喂食果蝇,改变果蝇体内组蛋白乙酰化水平,系统地研究组蛋白乙酰化修饰、HSP的表达以及寿命调控三者之间的关系。
结果发现hsp基因在长寿果蝇中具有较高的基础表达、较快的热激诱导反应速度以及较强的高温抵抗性。
同时,不同的hsp基因在果蝇衰老过程中的作用不尽相同,hsp22的作用最为重要,hsp70次之,而hsp26的表达几乎与寿命无关。
使用HDITSA和BuA喂食果蝇可以延长其寿命,但不同的HDI的作用机制不尽相同,同一种HDI对不同寿命品系的果蝇的延长程度也不尽相同。
TSA的处理有一种时间依赖性,更长时间的TSA处理对寿命是有利的;而BuA的处理却与此不同,过长时间的处理反而加速衰老。
同样的去乙酰化酶抑制剂,同一剂量处理,在不同果蝇品系种的作用不同,它们对短寿果蝇寿命的延长程度更为明显。
另外,HDI处理还促进果蝇衰老过程中hsp基因的基础表达和诱导表达,但是随着衰老的进行,这种促进作用逐渐减弱。
同样在不同寿命的果蝇品系中,其提高hsp基因表达的程度也不一样。
26-第10章 基因组表观遗传-组蛋白乙酰化

人类 基因 组组 蛋白 修饰 作图
上图图示为Chr.21染色质的部分作图结果,下面为染色质免疫沉淀抗体: H3K4me1, H3K4me2, H3K4me3; H3K9me1, H3K9me2, H3K9me3; H3K27me1, H3K27me1, H3K27me3; H3K36me1, H3K36me3; H3K79me1,H3K79me2, H3K79me3; H4K20me1, H4K20me3; H3R2me2 (as), H2A+H4R3me2, H2BK5me1, H2A.Z, Pol II, CTCF
HAT功能域有四个保守的基序(A,B,C和D)。 染色质又被去乙酰化。
组蛋白去乙酰化酶家族
根据功能与DNA序列相似性,将组 蛋白去乙酰化酶分为四大类: HDACI (histone deacetylase 1): HDAC1, HDAC2, HDAC3和 HDAC8. HDAC1, HDAC2和 HDAC8主要分布于细胞核, HDAC3主要分布于细胞质; HDACII (histone deacetylase 2): HDAC4, HDAC5, HDA6C, HDAC7, HDAC8,HDAC9 和HDAC10, 类同于HDACI. HDACII可在细胞核和细胞 质之间穿梭分布. HDACIII(histone deacetylase 3): SIRT1, SIRT2, SIRT3, SIRT4, SIRT5, SIRT6 和 SIRT7. HDACIV (histone deacetylase 4): HDAC11, 非典型去乙酰化酶.
-
组蛋白乙酰化酶
根据在细胞中的分布特点, 传统上将HA T 分为两大类。 A 型:位于细胞核,通过 核小体组蛋白的乙酰化调 控基因表达,可识别并乙 酰化组蛋白赖氨酸, Gcn5 p300/CBP和TAF11 250属 于此类,含bromodomain 结构域。B型:位于细胞 质,在组装为核小体前将 组蛋白乙酰化。B型HA T 缺少bromodomain结构域, 其功能是将新生的核心组 蛋白乙酰化。这些乙酰化 组蛋白一旦进入核内渗入
组蛋白去乙酰化技术在表观遗传学研究中的应用

组蛋白去乙酰化技术在表观遗传学研究中的
应用
随着科学技术的不断发展,表观遗传学作为一门新兴的生物学科学日渐受到科学研究人员的关注。
在表观遗传学领域中,组蛋白去乙酰化技术被广泛用于研究基因组结构和功能,其在表观遗传学研究中的应用也越来越广泛。
表观遗传学是研究遗传信息以外的遗传信息传递的一门学科,主要是研究如何通过化学修饰来调节基因表达,以及在通过遗传方式传递的特征外,如何通过表观遗传途径传递表型特征。
而组蛋白是在核酸糖苷酸骨架上重要的蛋白质,在细胞核中占有重要的地位。
乙酰化是一种常见的组蛋白修饰形式,可以影响基因的调控和表达。
因此,组蛋白去乙酰化技术成为了研究表观遗传学的重要手段之一。
通过组蛋白去乙酰化技术,研究人员可以探索组蛋白乙酰化和去乙酰化对基因表达的影响,了解细胞信号传导通路等生理生化过程的调控机制,并深入了解越来越多的疾病发生发展机理。
在研究中,组蛋白去乙酰化技术可以起到去乙酰化的作用,从而在细胞内的DNA中恢复原有的生化信息,帮助研究人员更好地理解DNA乙酰化和去乙酰化的机制。
组蛋白去乙酰化技术的应用已经从初期的对基础生命科学领域,进一步拓展到药物研发和治疗方面。
例如,在药物研发中,组蛋
白去乙酰化技术可以较好地检测药物的毒性及其可能的副作用,
为研发安全高效的药物提供了新的突破口。
此外,组蛋白去乙酰
化技术在产前诊断和妇科疾病等领域也有广泛的应用。
总之,组蛋白去乙酰化技术在表观遗传学研究中的应用及其发
展前景是非常广阔的。
未来,在不断发展的技术和工具的支持下,组蛋白去乙酰化技术将在表观遗传学的研究中扮演更加重要的角色。
组蛋白去乙酰化酶的研究进展

组蛋白去乙酰化酶的研究进展组蛋白去乙酰化酶(HDACs)是一类重要的酶类蛋白质,在细胞核内对染色质修饰起着重要的作用,调节基因的表达。
在过去的几十年中,对HDACs的研究取得了重大进展,深化了人们对这些蛋白质的认知和理解。
一、HDACs的分类HDACs主要分为四类:I、II、III和IV类。
其中I、II类又分为A、B、C、D子型。
这四类HDACs在受体结构及功能上都有所不同。
从结构上来看,HDACs主要由三个结构域组成,包括一个催化结构域、一个辅酶A结合结构域和一个维持结构稳定的结构域。
二、HDACs的生物学功能HDACs主要通过催化组蛋白的去乙酰化反应来调节基因的表达。
在生物体内,组蛋白去乙酰化是一个非常关键的生物过程,能够帮助细胞调节基因转录的水平,从而影响细胞的生长、分化、凋亡和代谢等生物过程。
HDACs通过去掉组蛋白的乙酰基,使其变得更加紧密结合,从而抑制相应的基因表达。
三、HDACs在疾病治疗上的应用近年来,越来越多的研究表明,在很多重要的疾病中,包括癌症、炎症和神经系统疾病等,HDACs扮演着非常重要的角色。
特别是在抗癌治疗方面,HDACs已经成为了一个非常重要的领域。
很多针对HDACs的抗癌药物,如美罗华(Merck)、法诺司汀(Novartis)等,已经被广泛使用,并取得了很好的治疗效果。
四、HDACs与DNA甲基化的关系HDACs在细胞内还与DNA甲基化过程密切相关。
DNA甲基化是一种常见的表观修饰形式,在基因表达、DNA复制和细胞分化等生物过程中起着非常重要的作用。
通过调节HDACs的表达和活性,可以进一步影响细胞内DNA甲基化修饰,从而调节基因的表达。
总结起来,HDACs是一类非常重要的酶类蛋白质,对于细胞核内染色质修饰和基因转录调节起着重要的作用。
在细胞内,通过调节HDACs的表达和活性,可以进一步影响基因的表达和细胞的生物过程,具有广泛的研究和应用前景。
组蛋白去乙酰化酶(HDACs)的研究进展

组蛋白去乙酰化酶(HDACs)的研究进展【摘要】在肿瘤的表观遗传学研究中,组蛋白的乙酰化修饰对肿瘤的发生发展起重要作用。
正常细胞体一旦出现核内组蛋白乙酰化与去乙酰化的失衡,即会导致正常的细胞周期与细胞代谢行为的改变而诱发肿瘤。
组蛋白去乙酰化酶(histone deacetylases,HDACs)催化组蛋白的去乙酰化,维系组蛋白乙酰化与去乙酰化的平衡状态,与癌相关基因转录表达、细胞增殖分化及细胞凋亡等诸多过程密切相关。
从组蛋白去乙酰化酶HDACs的结构分类及其与肿瘤发生发展关系两方面对HDACs做一综述。
【关键词】组蛋白去乙酰化酶(HDACs);肿瘤;表观遗传学Abstract:The modification for histone acetylation is of great importance for formulation and development oftumors in the epigenetic study of tumors. The disequilibrium of histone acetylation and deacetylation may cause some changes of cell cycle and cell metabolism. Histone deacetylases (HDACs) catalyze the deacetylation of histones,and maintain the equilibrium between histone acetylation and deacetylation as well. They are related to many regulation processes containing transcription of oncogene,cell cycle,apoptosis and so on. The structure classification of HDACs and the relationship between the HDACs and the formation and advancement of tumor were reviewed in this paper.Key words:histone feacetylases (HDACs); tumor; epigenetics肿瘤的发生是一个复杂的病理过程,受多重因素的影响,包括个体遗传因素、环境因素、物理化学因素、分子生物学因素等等。
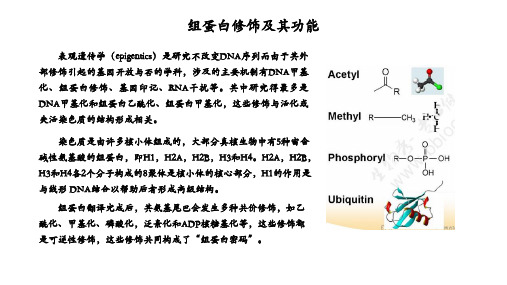
组蛋白修饰及其功能(乙酰化,甲基化,磷酸化等)-于凯
组蛋白密码学说的完善: 1. 更好地开发新药。研究组蛋白密码对药物开发具有战略
意义,多种组蛋白修饰酶已成为相关疾病治疗的靶目标。比如,组蛋白去乙酰酶
(HDACs)抑制剂已应用于临床治疗多种肿瘤; 2. 深入探讨遗传调控和表观遗传调控相互作用的网络与不同生物学表型之间的关系;
3. 在控制真核基因选择性表达的网络体系内进一步深入理解染色质结构、调控序列以
②组蛋白的N末端尾巴可与参与维持染色质高级结构的多种蛋白质相互作用,更加稳定了核
小体的结构。而组蛋白乙酰化却减弱了上述作用,阻碍了核小体装配成规则的高级结构(如 螺线管);
③组蛋白乙酰基转移酶(HAT)对相关转录因子或活化因子进行乙酰化修饰以调节基因的表
达。如 CBP/P300对P53的乙酰化可增强其特异性 DNA结合能力、转录激活能力,并延长其 半衰期。
组蛋白乙酰化调节转录的机制
组蛋白乙酰化引起染色质结构改变及基因转录活性变化的至少包括以下几个方面: ①组蛋白尾部赖氨酸残基的乙酰化能够使组蛋白携带正电荷量减少,降低其与带负电荷的 DNA链的亲和性,导致局部 DNA与组蛋白八聚体解开缠绕,从而促使参与转录调控的各种 蛋白因子与DNA特异序列结合,进而发挥转录调控作用;
及调控蛋白之间交互作用的内在机制; 4. 建立基因表达的调控网络数据库及其分析系统。总之,随着越来越多组蛋白核心结
构区域和修饰方式的确定,组蛋白密码在基因调控过程中的作用会越来越明确。
局部乙酰化举例
当DNA与核小体尚未解开缠绕时,转录激活因子如糖皮质激素受体可以和DNA上相应 的反应元件(GRE)结合。当结合至GRE之后,糖皮质激素募集共激活因子如CBP到染色
体上的靶转录基因区。此时,共激活因子利用HAT活性使得结合在DNA启动子区域的核心
- 1、下载文档前请自行甄别文档内容的完整性,平台不提供额外的编辑、内容补充、找答案等附加服务。
- 2、"仅部分预览"的文档,不可在线预览部分如存在完整性等问题,可反馈申请退款(可完整预览的文档不适用该条件!)。
- 3、如文档侵犯您的权益,请联系客服反馈,我们会尽快为您处理(人工客服工作时间:9:00-18:30)。
the genetics and biochemistry of chromatin remodeling complexes: large, multisubunit catalytic entities perform the work of histone modification that leads either to transcriptional activation or repression of target genes. Here, promoter selectivity for sequence-specific DNA binding proteins must guide the assembly of these big chromatin-modifying machines,yet the genetic regulatory elements must also be able to respond rapidly to changing transcriptional requirements. Active investigation of chromatin remodeling continues in many laboratories, from the level of sequence-specific modification of specific histones to the level of multiprotein complex assembly.A particular protein motif called a “bromodomain” has been noticed in many of the proteins that compose the chromatin modifying machinery. It was first identified in 1992 as a 61 – 63amino acid signature (14). Although it lacked a known function at the time, it has subsequently been identified in transcription factors, co-activators and other proteins that are important in transcription or chromatin remodeling and its boundaries have been expanded to about 110 amino acids. The number of such proteins was about forty at last report (15,16)and several important additions to the family have been made since then. The first described bromodomain protein, yeast Gcn5 (17), was shown to be necessary for amino acid metabolism and was characterized as a transcriptional co-activator (18). It provides a histone acetylation (19) component of the ADA (Adapter) and SAGA (Spt-Ada-Gcn5acetyltransferase) transcription complexes (20), which is fundamental and essential for viability (21). Gcn5 is also structurally related to the mammalian proteins CBP, p300 and Hat1 (22). In mammals, CBP and p300 also have intrinsic HAT activity (23,24) and interact with many important transcription factors as co-activators of transcription. Virtually all of the nuclear histone acetyltransferases (HATs) contain bromodomains (16), but not all bromodomain proteins are HATs. For example, other classes of bromodomain proteins include MLL, a putative transcription factor (25,26) that interacts with the SWI/SNF chromatin remodeling complex (27); Spt7, an acidic transcriptional activator and component of the SAGA complex (28); and a helicase superfamily that includes Snf2, Rsc1/Rsc2 andSth1, components of the SWI/SNF (29) and RSC complexes (30); Brg1, which binds RB(31,32); and brahma , which also contacts RB, is related to Swi2/Snf2 (33,34) and hashomeotic functions in Drosophila (35–37). The role of bromodomains in transcriptioncomplexes has been controversial because their deletion has widely different consequences:in yeast, bromodomain deletion of Spt7 has no phenotype, of Snf2 causes slow growth, butdeletion of Sth1, Rsc1 and Rsc2 causes lethality (16). Much of the apparent significance ofbromodomain proteins lies in their either having intrinsic HAT activity, or being associatedwith promoter-bound complexes that contain HAT or histone deacetylase (HDAC) activity.Bromodomain proteins are thereby potentially important players in the transcriptionalcontrol of a wide variety of eukaryotic genes, including those that control growth.The bromodomain proteins that interact with RB highlight an important duality intranscriptional control: the need also to turn promoters off. In particular, the transcriptionalcontrol of E2F-regulated mammalian cell cycle genes is essential for proper progressionthrough each stage of the cell cycle. Whereas transcriptional activation of one set of genes isnecessary to enter a stage of the cell cycle, repression of certain other genes associated withthe previous stage is necessary to exit from that stage. RB (and its family members p107 andp130) bind to E2F proteins and block their transcription activation function (38,39).Recent evidence has revealed that in addition to this direct repression, RB also recruits ahistone deacetylase (40,41), as do p107 and p130 (42), through cooperation with mammalianbrahma and other proteins in the SWI/SNF complex (31,32). Coordinated transcriptionalactivation and repression of the key E2F-regulated mammalian cell cycle genes cyclin E ,cyclin A and cdc2 permit proper transitions between G1 and S phases, and S and G 2 phases(43). This dual nature of chromatin remodeling complexes was first suspected in yeast,NIH-PA Author ManuscriptNIH-PA Author ManuscriptNIH-PA Author Manuscriptwhere SWI/SNF complexes, initially associated with transcriptional activation (44), were later linked to repression as well: more genes are activated than repressed by SWI/SNF mutations (45). It now appears that SWI/SNF function may establish a widely applicable paradigm in chromatin remodeling complexes, whereby transcriptionally active euchromatin can be converted to inactive heterochromatin and vice versa in part through the exchange of HAT and HDAC enzymes in the complex (46). This model has been refined lately with the observation in yeast that several inducible genes active during interphase can recruit HAT activity independently of SWI/SNF, whereas mitotic genes require SWI/SNF to recruit HAT activity (47). This observation emphasizes the importance of coordinated complex formation for proper transit of the cell cycle.A central development in the field of bromodomain-containing proteins came with a report of Zhou and colleagues (48), who used nuclear Overhauser enhancements to solve the solution structure of the bromodomain of p/CAF in association with N-acetylated lysine.The highly conserved structure of bromodomain proteins suggests a hypothesis that many of them will bind N-acetyl-lysine in histones, however by no means will this necessarily be true for all. The presence of bromodomains in many proteins that are known independently to possess HAT activity strongly supports the Zhou hypothesis. A looser notion that this motif is present in proteins that are involved in chromatin modification and transcription regulation is the best guide to their classification at the moment. The future discovery of bromodomains in proteins that are uninvolved in chromatin restructuring will be a test of the utility of such a classification.In this special issue, several authors have been invited to contribute their perspectives on the developing field of bromodomain proteins and associated chromatin-modifying activities.Major questions that they address continue to provoke the development of the field, and include:A.What are the number and type of histone modifications, including phosphorylation,acetylation, methylation, ADP-ribosylation and ubiquitylation, that could regulatethe recruitment of different classes of chromatin-modifying enzymes and mightthese represent a kind of combinatorial “histone code”? How do modifications ofbromodomain-containing proteins reciprocally affect histone modificationactivities?B.Should bromodomain-containing proteins be thought of as a kind of bridge orplatform that recruits diverse enzymatic activities, such as HATs, HDACs, kinasesor helicases, to chromatin? Why are these activities present in some bromodomain-containing proteins as independently-folding domains of a single polypeptide chainand in other cases as separate proteins? Does the weak affinity constant for a singlebromodomain binding to N-acetyl-lysine (~0.1 mM) imply that bromodomains canfunction only in multiprotein complexes with multiple interaction sites?C.Do different bromodomains have different functions, including those that arepresent more than once in a single protein? For example, double bromodomains,such as those in TAF II 250 might provide mutual cooperativity for protein bindingto chromatin or might interfere with binding instead; or they might conferdifferential promoter specificity.D.Why are some bromodomains essential for enzymatic function or cell viabilitywhereas deletion of others has no apparent phenotype? Does this behavior reflectredundancy within bromodomain-containing complexes, so that for example SWI/SNF activities on some promoters can partially substitute for HAT-containingcomplexes such as SAGA?NIH-PA Author ManuscriptNIH-PA Author ManuscriptNIH-PA Author ManuscriptE.What is the significance of the time order of recruitment of SWI/SNF activities andHAT activities to certain promoters? Why does SWI/SNF recruitment of HATactivity impact yeast transcriptional activation during late mitosis (47), whereasmany inducible promoters recruit HATs independently of SWI/SNF earlier in thecell cycle, and how widespread is this behavior in eukaryotes?AcknowledgmentsThe author’s work is supported by grant CA75107 from NIH.References1. Bradbury EM. Reversible histone modifications and the chromosome cell cycle. Bioessays 1992;14:9–16. [PubMed: 1312335]2. Wolffe AP, Pruss D. Targeting chromatin disruption: Transcription regulators that acetylate histones. Cell 1996;84:817–819. [PubMed: 8601304]3. Roth SY, Allis CD. Histone acetylation and chromatin assembly: a single escort, multiple dances?Cell 1996;87:5–8. [PubMed: 8858142]4. Grunstein M. Histone acetylation in chromatin structure and transcription. Nature 1997;389:349–352. [PubMed: 9311776]5. Tyler JK, Kadonaga JT. The “dark side” of chromatin remodeling: repressive effects on transcription. Cell 1999;99:443–446. [PubMed: 10589670]6. Cheung P, Allis CD, Sassone-Corsi P. Signaling to chromatin through histone modifications. Cell 2000;103:263–271. [PubMed: 11057899]7. Orphanides G, Reinberg D. RNA polymerase II elongation through chromatin. Nature 2000;407:471–475. [PubMed: 11028991]8. Cheung P, Tanner KG, Cheung WL, Sassone-Corsi P, Denu JM, Allis CD. Synergistic coupling of histone H3 phosphorylation and acetylation in response to epidermal growth factor stimulation. Mol Cell 2000;5:905–915. [PubMed: 10911985]9. Lo WS, Trievel RC, Rojas JR, Duggan L, Hsu JY, Allis CD, Marmorstein R, Berger SL.Phosphorylation of serine 10 in histone H3 is functionally linked in vitro and in vivo to Gcn5-mediated acetylation at lysine 14. Mol Cell 2000;5:917–926. [PubMed: 10911986]10. Mahadevan LC, Willis AC, Barratt MJ. Rapid histone H3 phosphorylation in response to growthfactors, phorbol esters, okadaic acid and protein synthesis inhibitors. Cell 1991;65:775–783.[PubMed: 2040014]11. Sassone-Corsi P, Mizzen CA, Cheung P, Crosio C, Monaco L, Jacquot S, Hanauer A, Allis CD.Requirement of Rsk-2 for epidermal growth factor-activated phosphorylation of histone H3.Science 1999;285:886–891. [PubMed: 10436156]12. Chadee DN, Hendzel MJ, Tylipski CP, Allis CD, Bazett-Jones DP, Wright JA, Davie JR. IncreasedSer-10 phosphorylation of histone H3 in mitogen-stimulated and oncogene-transformed mousefibroblasts. J Biol Chem 1999;274:24914–24920. [PubMed: 10455166]13. Wei Y, Yu L, Bowen J, Gorovsky MA, Allis CD. Phosphorylation of histone H3 is required forproper chromosome condensation and segregation. Cell 1999;97:99–109. [PubMed: 10199406]14. Haynes SR, Dollard C, Winston F, Beck S, Trowsdale J, Dawid IB. The bromodomain: aconserved sequence found in human, Drosophila and yeast proteins. Nucl Acids Res1992;20:2603. [PubMed: 1350857]15. Jeanmougin F, Wurtz JM, Le Douarin B, Chambon P, Losson R. The bromodomain revisited.Trends Biochem Sci 1997;22:151–153. [PubMed: 9175470]16. Winston F, Allis CD. The bromodomain: a chromatin-targeting module? Nature Struct Biol1999;6:601–604. [PubMed: 10404206]17. Hinnebusch AG, Fink GR. Positive regulation in the general amino acid control of Saccharomycescerevisiae . Proc Nat’l Acad Sci USA 1983;80:5374–5378.18. Guarente L. Transcriptional coactivators in yeast and beyond. Trends Biochem Sci 1995;20:517–521. [PubMed: 8571454]NIH-PA Author ManuscriptNIH-PA Author ManuscriptNIH-PA Author Manuscript19. Struhl K. Histone acetylation and transcriptional regulatory mechanisms. Genes Dev 1998;12:599–606. [PubMed: 9499396]20. Wang L, Liu L, Berger SL. Critical residues for histone acetylation by Gcn5, functioning in Ada and SAGA complexes, are also required for transcriptional function in vivo . Genes Dev 1998;12:640–653. [PubMed: 9499400]21. Kuo MH, Brownell JE, Sobel RE, Ranalli TA, Cook RG, Edmondson DG, Roth SY, Allis CD.Transcription-linked acetylation by Gcn5p of histone H3 and H4 at specific lysines. Nature 1996;383:269–272. [PubMed: 8805705]22. Dutnall RN, Tafrov ST, Sternglanz R, Ramakrishnan V. Structure of the histone acetyltransferase Hat1: A paradigm for the GCN5-related N-acetyltransferase superfamily. Cell 1998;94:427–438.[PubMed: 9727486]23. Bannister AJ, Kouzarides T. The CBP co-activator is a histone acetyltransferase. Nature 1996;384:641–643. [PubMed: 8967953]24. Ogryzko VV, Schiltz OL, Russanova V, Howard BH, Nakatani Y. The transcriptional co-activators p300 and CBP are histone acetyltransferases. Cell 1996;87:953–959. [PubMed: 8945521]25. Gu Y, Nakamura T, Alder H, Prasad R, Canaani O, Cimino G, Croce CM, Canaani E. The t(4;11)chromosomal translocation of human acute leukemias fuses the ALL-1 gene, related to Drosophila trithorax , to the AF-4 gene. Cell 1992;71:701–708. [PubMed: 1423625]26. Tkachuk DC, Kohler S, Cleary ML. Involvement of a homolog of Drosophila trithorax by 11q23chromosomal translocations in acute leukemias. Cell 1992;71:691–700. [PubMed: 1423624]27. Rozenblatt-Rosen O, Rozovskaia T, Burakov D, Sedkov Y, Tillib S, Blechman J, Nakamura T,Croce CM, Mazo A, Canaani E. The C-terminal SET domains of ALL-1 and TRITHORAX interact with the INI1 and SNR1 proteins, components of the SWI/SNF complex. Proc Nat’l Acad Sci USA 1998;95:4152–4157.28. Gansheroff LJ, Dollard C, Tan P, Winston F. The Saccharomyces cerevisiae SPT7 gene encodes a very acidic protein important for transcription in vivo . Genetics 1995;139:523–536. [PubMed:7713415]29. Peterson CL, Tamkun JW. The SWI-SNF complex: a chromatin remodeling machine? Trends Biochem Sci 1995;20:143–146. [PubMed: 7770913]30. Cairns BR, Lorch Y, Li Y, Zhang M, Lacomis L, Erdjument-Bromage H, Tempst P, Du J, LaurentB, Kornberg RD. RSC, an essential, abundant chromatin-remodeling complex. Cell1996;87:1249–1260. [PubMed: 8980231]31. Trouche D, Le Chalony C, Muchardt C, Yaniv M, Kouzarides T. RB and hbrm cooperate torepress the activation functions of E2F1. Proc Nat’l Acad Sci USA 1997;94:11268–11273.32. Muchardt C, Yaniv M. The mammalian SWI/SNF complex and the control of cell growth. SemCell Dev Biol 1999;10:189–195.33. Tamkun JW, Deuring R, Scott MP, Kissenger M, Pattatucci AM, Kaufman TC, Kennison JA.brahma - a regulator of Drosophila homeotic genes structurally related to the yeast transcriptional activator SWI2/SNF2. Cell 1992;68:561–572. [PubMed: 1346755]34. Pazin MJ, Kadonaga JT. SWI2/SNF2 and related proteins: ATP-driven motors that disrupt protein-DNA interactions? Cell 1997;88:737–740. [PubMed: 9118215]35. Kennison JA, Tamkun JW. Dosage-dependent modifiers of Polycomb and Antennapedia mutationsin Drosophila . Proc Nat’l Acad Sci USA 1988;85:8136–8140.36. Elfring LK, Deuring R, McCallum CM, Peterson CL, Tamkun JW. Identification andcharacterization of Drosophila relatives of the yeast transcriptional activator SNF2/SWI2. Mol Cell Biol 1994;14:2225–2234. [PubMed: 7908117]37. Elfring LK, Daniel C, Papoulas O, Deuring R, Sarte M, Moseley S, Beek SJ, Waldrip WR,Daubresse G, DePace A, Kennison JA, Tamkun JW. Genetic analysis of brahma: the Drosophila homolog of the yeast chromatin remodeling factor SWI2/SNF2. Genetics 1998;148:251–265.[PubMed: 9475737]38. Dyson N. The regulation of E2F by pRB-family proteins. Genes Dev 1998;12:2245–2262.[PubMed: 9694791]39. Nevins JR. Toward an understanding of the functional complexity of the E2F and retinoblastomafamilies. Cell Growth Diff 1998;9:585–593. [PubMed: 9716176]NIH-PA Author ManuscriptNIH-PA Author ManuscriptNIH-PA Author Manuscript40. Brehm A, Miska EA, McCance DJ, Reid JL, Bannister AJ, Kouzarides T. Retinoblastoma protein recruits histone deacetylase to repress transcription. Nature 1998;391:597–601. [PubMed:9468139]41. Magnaghi-Jaulin L, Groisman R, Naguibneva I, Robin P, Lorain S, Le Villain JP, Troalen F,Trouche D, Harel-Bellan A. Retinoblastoma protein represses transcription by recruiting a histone deacetylase. Nature 1998;391:601–605. [PubMed: 9468140]42. Ferreira R, Magnaghi-Jaulin L, Robin P, Harel-Bellan A, Trouche D. The three members of the pocket proteins family share the ability to repress E2F activity through recruitment of a histone deacetylase. Proc Nat’l Acad Sci USA 1998;95:10493–10498.43. Zhang HS, Gavin M, Dahiya A, Postigo AA, Ma D, Luo RX, Harbour JW, Dean DC. Exit from G1and S phase of the cell cycle is regulated by repressor complexes containing HDAC-Rb-hSWI/SNF and Rb-hSWI/SNF. Cell 2000;101:79–89. [PubMed: 10778858]44. Burns LG, Peterson CL. The yeast SWI-SNF complex facilitates binding of a transcriptional activator to nucleosomal sites in vivo . Mol Cell Biol 1997;17:4811–4819. [PubMed: 9234737]45. Holstege FC, Jennings EG, Wyrick JJ, Lee TI, Hengartner CJ, Green MR, Golub TR, Lander ES,Young RA. Dissecting the regulatory circuitry of a eukaryotic genome. Cell 1998;95:717–728.[PubMed: 9845373]46. Heinzel T, Lavinsky RM, Mullen TM, Soderstrom M, Laherty CD, Torchia J, Yang WM, Brard G,Ngo SD, Davie JR, Seto E, Eisenman RN, Rose DW, Glass CK, Rosenfield MG. A complex containing N-CoR, mSin3 and histone deacetylase mediates transcriptional repression. Nature 1997;387:43–48. [PubMed: 9139820]47. Krebs JE, Fry CJ, Samuels ML, Peterson CL. Global role for chromatin remodeling enzymes in mitotic gene expression. Cell 2000;102:587–598. [PubMed: 11007477]48. Dhalluin C, Carlson JE, Zeng L, He C, Aggarwal AK, Zhou MM. Structure and ligand of a histone acetyltransferase bromodomain. Nature 1999;399:491–496. [PubMed: 10365964]NIH-PA Author Manuscript NIH-PA Author Manuscript NIH-PA Author Manuscript。
