NEB--推荐双酶切系统
- 1、下载文档前请自行甄别文档内容的完整性,平台不提供额外的编辑、内容补充、找答案等附加服务。
- 2、"仅部分预览"的文档,不可在线预览部分如存在完整性等问题,可反馈申请退款(可完整预览的文档不适用该条件!)。
- 3、如文档侵犯您的权益,请联系客服反馈,我们会尽快为您处理(人工客服工作时间:9:00-18:30)。
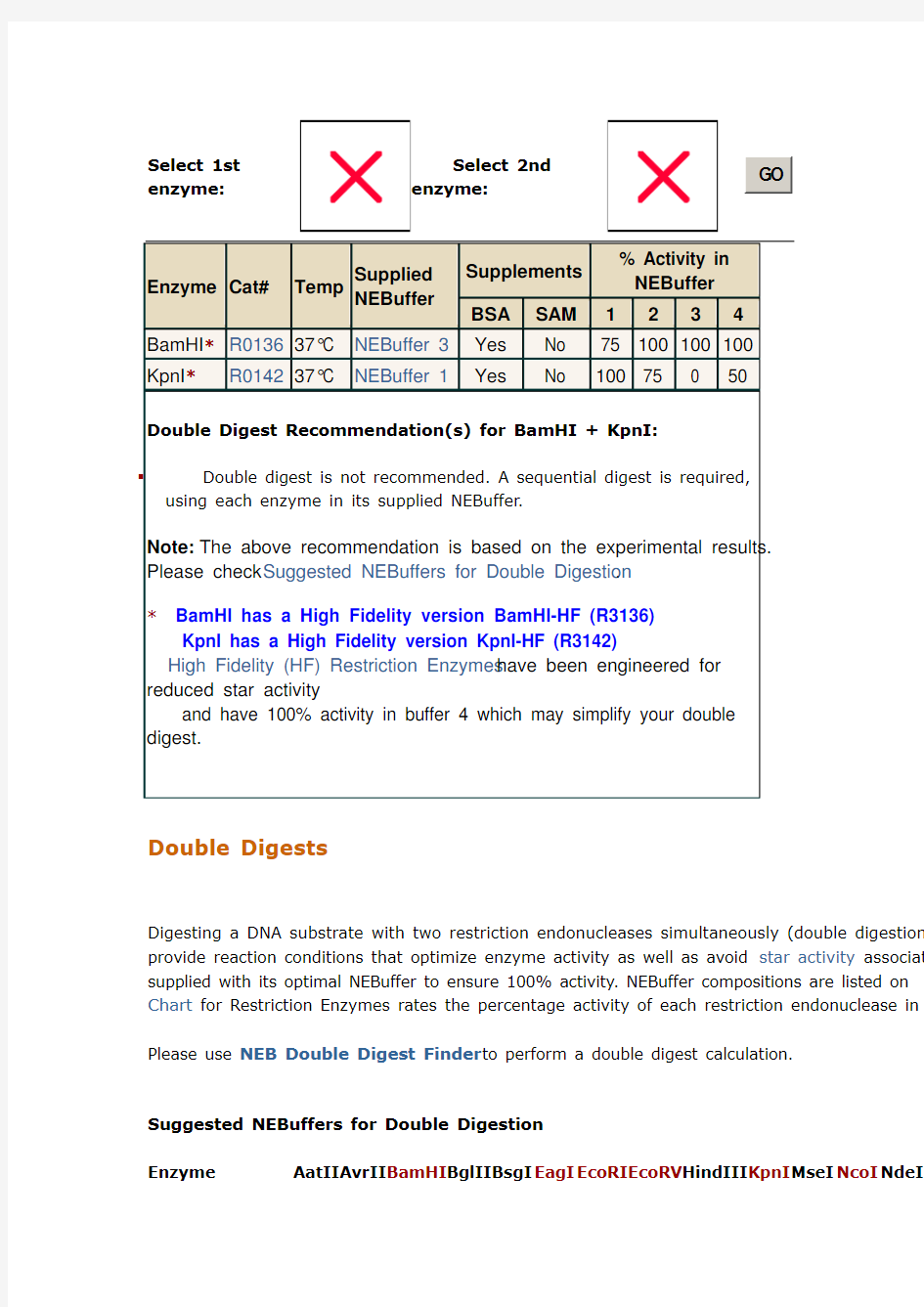
Select 1st enzyme:
Select 2nd
enzyme:
G O
Enzyme Cat#Temp Supplied
NEBuffer
Supplements
% Activity in
NEBuffer
BSA SAM1234
BamHI*R013637°C NEBuffer 3Yes No 75 100 100 100
KpnI*R014237°C NEBuffer 1Yes No 100 75 0 50
Double Digest Recommendation(s) for BamHI + KpnI:
▪Double digest is not recommended. A sequential digest is required,
using each enzyme in its supplied NEBuffer.
Note: The above recommendation is based on the experimental results.
Please check Suggested NEBuffers for Double Digestion.
* BamHI has a High Fidelity version BamHI-HF (R3136)
KpnI has a High Fidelity version KpnI-HF (R3142)
High Fidelity (HF) Restriction Enzymes have been engineered for
reduced star activity
and have 100% activity in buffer 4 which may simplify your double
digest.
Double Digests
Digesting a DNA substrate with two restriction endonucleases simultaneously (double digestion) provide reaction conditions that optimize enzyme activity as well as avoid star activity associate supplied with its optimal NEBuffer to ensure 100% activity. NEBuffer compositions are listed on b Chart for Restriction Enzymes rates the percentage activity of each restriction endonuclease in t Please use NEB Double Digest Finder to perform a double digest calculation.
Suggested NEBuffers for Double Digestion
Enzyme AatII A vrII B amHI B glII B sgI EagI EcoRI E coRV H indIII K pnI M seI NcoI NdeI N
NEBuffer 4 4 3 3 4 3 EcoRI 3 2 1 4 3 4
AatII 4 -- 4 seq seq 4 seq seq 4 4 4 4 4 4 AvrII 4 4 -- 3 2 4 3 EcoRI 2 2 1 4 4 4 BamHI 3 seq 3 -- 3 3 3 EcoRI 3 seq seq 3 3 3 BglII 3 seq 2 3 -- 3 3 EcoRI 3 2 2 2 3 3 BsgI 4 4 4 3 3 -- seq seq 4 2 seq 4 4 4 EagI 3 seq 3 3 3 seq -- EcoRI 3 seq seq 3 3 3 EcoRI EcoRI seq EcoRI EcoRI EcoRI seq EcoRI -- EcoRI seq 1 EcoRI E coRI E coRI EcoRV 3 4 2 3 3 4 3 EcoRI -- 2 2 2 3 2 HindIII 2 4 2 seq 2 2 seq seq 2 -- 2 2 2 2 KpnI 1 4 1 seq 2 seq seq 1 2 2 -- 1 1 1 MseI 4 4 4 3 2 4 3 EcoRI 2 2 1 -- 4 4 NcoI 3 4 4 3 3 4 3 EcoRI 3 2 1 4 -- 4 NdeI 4 4 4 3 3 4 3 EcoRI 2 2 1 4 4 -- NheI 2 4 2 seq 2 4 seq 1 2 2 1 2 2 4 NotI 3 seq 3 3 3 3 3 EcoRI 3 2 2 2 3 3 PstI 3 4 3 3 3 3 3 EcoRI 3 2 1 3 3 3 PvuI 3 seq 2 3 3 3 3 EcoRI 3 2 2 3 3 3 SacI 1 4 1 seq 2 4 seq 1 2 2 1 4 1 4 SacII 4 4 4 seq 2 4 seq EcoRI 2 2 4 4 4 4 SalI 3 seq 3 3 3 3 3 EcoRI 3 seq seq 3 3 3 SmaI 4 4 4 seq seq 4 seq seq 4 4 seq 4 4 4 SpeI 4 4 4 seq 2 4 seq EcoRI 2 2 1 4 4 4 SphI 2 4 2 3 2 4 3 EcoRI 2 2 1 2 2 2 XbaI 4 4 4 3 2 4 3 seq 2 2 2 4 4 4 XhoI 4 4 4 3 3 4 3 EcoRI 3 2 1 4 4 4 XmaI 4 4 4 seq 2 4 seq seq 4 seq 4 4 4 4
Note: Enzymes in Dark Red are available in High Fidelity (HF) Format. HF Enzymes have been en 4 which may simplify your double digest. Click for more HF information.
Setting up a Double Digestion
•Choose an NEBuffer that results in the most activity for both enzymes. If star activity is a using one of our High Fidelity (HF) enzymes.
•If BSA is required for either enzyme, add it to the double digest reaction (BSA does not inh enzyme).
•Set up reaction according to recommended conditions. The final concentration of glycero should be less than 5% to minimize the possibility of star activity. For example, in a 50 µl re amount of enzyme added should not exceed 5 µl.
•Incubate at recommended temperature. Overnight double digests should be avoide possibility of star activity.
