Cap 139N dihydrostreptomycinand streptomycin三
OXOID抗生素敏感检测纸片

抗生素英文名全称中文名全称名称缩写浓度产品编号 AAmikacin阿米卡星(丁胺卡那霉素) AK30μg CT0107B Amoxycillin阿莫西林(羟氨苄青霉素)AML2μg CT0060B Amoxycillin阿莫西林(羟氨苄青霉素)AML 10μg CT0161B Amoxycillin阿莫西林(羟氨苄青霉素)AML 25μg CT0061BAMC3μg CT0538B Amoxycillin/clavulanic acid阿莫西林/克拉维酸(棒酸)(2:1)(澳格门汀)AMC30μg CT0223B Amoxycillin/clavulanic acid阿莫西林/克拉维酸(棒酸)(2:1)(澳格门汀)Ampicillin氨苄西林AMP2μg CT0002B Ampicillin氨苄西林AMP10μg CT0003B Ampicillin氨苄西林AMP 25μg CT0004B Ampicillin/sulbactam 1:1氨苄西林/舒巴坦(1:1)SAM20μg CT0520B Ampicillin/sulbactam 2:1氨苄西林/舒巴坦(2 :1)SAM30μg CT1653B Apramycin阿泊拉霉素(安普霉素)APR15μg CT0545B Azithromycin阿齐霉素(阿奇霉素)AZM15μg CT0906B Aztreonam安曲南ATM 30μg CT0264B BBacitracin杆菌肽B10 units CT0005B CCarbenicillin羧苄西林(羧苄青霉素) CAR100μg CT0006B Cefaclor头孢克洛(头孢克罗)CEC30μg CT0149B Cefadroxil头孢羟氨苄CFR 30μg CT0453B Cefamandole头孢孟多MA 30μg CT0108B Cefepime头孢吡肟(马斯平)FEP 30μg CT0771B Cefixime头孢克肟(世福素)CFM 5μg CT0653B Cefoperazone头孢哌酮(先锋必)CFP30μg CT0193B Cefoperazone头孢哌酮(先锋必)CFP75μg CT0249B Cefoperazone/sulbactam 2:1头孢哌酮/舒巴坦(2 :1)(舒普深)SCF105μg CT1727B Cefotaxime头孢噻肟(凯福隆)CTX5μg CT0407B Cefotaxime头孢噻肟(凯福隆) CTX30μg CT0166B Cefotetan头孢替坦CTT30μg CT0665B Cefoxitin头孢西丁(美福仙)FOX30μg CT0119B Cefpirome头孢匹罗CPO30μg CT1412B Cefpodoxime头孢泊肟CPD10μg CT1612B Cefprozil头孢丙烯CPR30μg CT1647B Cefsulodin头孢磺胺(达克舒林)CFS30μg CT0263B Ceftazidime头孢他啶CAZ10μg CT1629B Ceftazidime头孢他啶CAZ30μg CT0412B Ceftibuten头孢布烯CFT30μg CT1662B Ceftiofur头孢噻呋EFT30μg CT1751B Ceftizoxime头孢唑肟(安保速灵)ZOX30μg CT0477B Ceftriaxone头孢曲松(头孢三嗪)CRO 5μg CT1743B Ceftriaxone头孢曲松(头孢三嗪)CRO 30μg CT0417B Cefuroxime sodium头孢呋新钠CXM5μg CT0406B Cefuroxime sodium头孢呋新钠CXM30μg CT0127BCephalexin头孢氨苄 (头孢力新,先锋IV)CL30μg CT0007B Cephalothin头孢噻吩(头孢菌素,先锋I )KF30μg CT0010B Cephazolin头孢唑啉(先锋V)KZ30μg CT0011B Cephradine头孢拉定(先锋VI)CE30μg CT0063B Chloramphenicol氯霉素C10μg CT0012B Chloramphenicol氯霉素C30μg CT0013B Chloramphenicol氯霉素C50μg CT0014B Cinoxacin西诺沙星CIN100μg CT0162B Ciprofloxacin环丙沙星(悉复欢)CIP1μg CT0623B Ciprofloxacin环丙沙星(悉复欢)CIP5μg CT0425B Ciprofloxacin环丙沙星(悉复欢)CIP10μg CT1615B Clarithromycin克拉霉素CLR2μg CT1599B Clarithromycin克拉霉素CLR 5μg CT1623B Clarithromycin克拉霉素CLR15μg CT0693B Clindamycin克林霉素(氯林可霉素,氯洁霉素)DA2μg CT0064B Clindamycin克林霉素(氯林可霉素,氯洁霉素)DA10μg CT0015B Cloxacillin氯唑西林(邻氯青霉素)OB5μg CT0016B Colistin sulphate多粘菌素E (硫酸粘杆菌素)CT10μg CT0017B Colistin sulphate多粘菌素E (硫酸粘杆菌素)CT25μg CT0065B Colistin sulphate多粘菌素E (硫酸粘杆菌素)CT50μg CT0664B Compound sulphonamides磺胺复合物S3300μg CT0059B DDoxycycline强力霉素DO30μg CT0018B EEnrofloxacin恩诺沙星ENR5μg CT0639B Ertapenem厄他培南ETP10μg CT1761B Erythromycin红霉素E5μg CT0066B Erythromycin红霉素E10μg CT0019B Erythromycin红霉素E15μg CT0020B Erythromycin红霉素E30μg CT0021B FFlorfenicol氟苯尼考FFC30μg CT1754B Fluconazole氟康唑FCA25μg CT1806B Flumequine氟甲喹UB30μg CT0666B Fosfomycin磷霉素FOS50μg CT0183B Fosfomycin/trometamol磷霉素/氨丁三醇(复安欣)FOT200μg CT0758B Framycetin新霉素B FY100μg CT0071B Fusidic acid褐霉素(夫西地酸)FD5μg CT0493B Fusidic acid褐霉素(夫西地酸)FD10μg CT0023B Fusidic acid褐霉素(夫西地酸)FD50μg CT1617B GGentamicin庆大霉素CN10μg CT0024B Gentamicin庆大霉素CN30μg CT0072B Gentamicin庆大霉素CN120μg CT0794B Gentamicin庆大霉素CN200μg CT0695B IImipenem亚胺培南(配能)IPM10μg CT0455BKKanamycin卡那霉素K5μg CT0025B Kanamycin卡那霉素K30μg CT0026B LLatamoxef拉氧头孢MOX30μg CT0302B Levofloxacin左氧氟沙星(可乐必妥)LEV1μg CT1586B Levofloxacin左氧氟沙星(可乐必妥)LEV5μg CT1587B Lincomycin林可霉素(洁霉素)MY2μg CT0027B Lincomycin林可霉素(洁霉素)MY10μg CT0123B Lincomycin林可霉素(洁霉素)MY15μg CT0028B Lincomycin/neomycin林可霉素(洁霉素)/新霉素LN75μg CT1757B Lincomycin/spectinomycin林可霉素/壮观霉素LS109μg CT1758B Linezolid利奈唑胺LZD10μg CT1649B Linezolid利奈唑胺LZD30μg CT1650B Lomefloxacin洛美沙星LOM10μg CT1661B MMecillinam美西林MEL10μg CT0096B Mecillinam美西林MEL25μg CT0091B Meropenem美罗培南(美平)MEM10μg CT0774B Metronidazole甲硝唑(灭滴灵)MTZ5μg CT0067B Metronidazole甲硝唑(灭滴灵)MTZ 50μg CT0466B Mezlocillin美洛西林MEZ30μg CT0174B Mezlocillin美洛西林MEZ75μg CT0192B Minocycline米诺环素(二甲胺四环素)MH30μg CT0030B Moxalactam拉氧头孢MOX30μg CT0302B Moxifloxacin莫西沙星MXF1μg CT1683B Moxifloxacin莫西沙星MXF5μg CT1633B Mupirocin莫匹罗星MUP5μg CT0522B Mupirocin莫匹罗星MUP20μg CT1826B Mupirocin莫匹罗星MUP200μg CT0523B NNalidixic acid萘啶酸NA30μg CT0031B Neomycin新霉素N10μg CT0032B Neomycin新霉素N30μg CT0033B Netilmicin奈替米星(乙基西梭霉素)NET10μg CT0424B Netilmicin奈替米星(乙基西梭霉素)NET30μg CT0225B Nitrofurantoin呋喃妥因(呋喃妥英)F50μg CT0069B Nitrofurantoin呋喃妥因(呋喃妥英)F100μg CT0034B Nitrofurantoin呋喃妥因(呋喃妥英)F200μg CT0035B Nitrofurantoin呋喃妥因(呋喃妥英)F300μg CT0036B Norfloxacin诺氟沙星(氟哌酸)NOR2μg CT0687B Norfloxacin诺氟沙星(氟哌酸)NOR5μg CT0668B Norfloxacin诺氟沙星(氟哌酸)NOR10μg CT0434B Novobiocin新生霉素NV5μg CT0037B Novobiocin新生霉素NV30μg CT0038B Nystatin制霉菌素NS100units CT0073B OOfloxacin氧氟沙星(泰利必妥)OFX5μg CT0446B Oleandomycin竹桃霉素OL15μg CT0039B Oxacillin苯唑西林OX1μg CT0159B Oxacillin苯唑西林OX5μg CT0040B Oxolinic acid奥索利酸(恶喹酸)OA2μg CT0181B Oxytetracycline土霉素 (氧四环素,地霉素)OT30μg CT0041B PPefloxacin培氟沙星(甲氟哌酸)PEF5μg CT0661B Penicillin G青霉素G P1unit CT0152B Penicillin G青霉素G P 1.5unit CT0042B Penicillin G青霉素G P2units CT0088B Penicillin G青霉素G P5units CT0124B Penicllin G青霉素G P10units CT0043B Penicillin/novobiocin青霉素/新生霉素PNV40CT1755B Pipemidic acid吡哌酸PIP20μg CT0180B Piperacillin哌拉西林(氧哌嗪青霉素)PRL30μg CT1619B Piperacillin哌拉西林(氧哌嗪青霉素)PRL75μg CT0261B Piperacillin哌拉西林(氧哌嗪青霉素)PRL100μg CT0199B Piperacillin/tazobactam哌拉西林/他唑巴坦 (特治星)TZP36μg CT1616B Piperacillin/tazobactam哌拉西林/他唑巴坦(特治星)TZP40μg CT1628B Piperacillin/tazobactam哌拉西林/他唑巴坦(特治星)TZP85μg CT0720B Piperacillin/tazobactam哌拉西林/他唑巴坦(特治星)TZP110μg CT0725B Pirlimycin吡利霉素PIR2μg CT1668B Polymyxin B多粘菌素B PB300units CT0044B QQuinupristin/dalfopristin喹奴普汀/达福普汀QD15μg CT1644B RRifampicin利福平RD2μg CT0078B Rifampicin利福平RD5μg CT0207B Rifampicin利福平RD30μg CT0104B SSpectinomycin大观霉素(壮观霉素)SH10μg CT0046B Spectinomycin大观霉素(壮观霉素)SH25μg CT0411B Spectinomycin大观霉素(壮观霉素)SH100μg CT0823B Spiramycin螺旋霉素SP100μg CT0232B Streptomycin链霉素S10μg CT0047B Streptomycin链霉素S25μg CT0048B Sulbactam/ampicillin 1 : 1舒巴坦/氨苄西林SAM20μg CT0520B Sulbactam/ampicillin 1 : 2舒巴坦/氨苄西林(优立新)SAM30μg CT1653B Sulphafurazole磺胺异恶唑SF300μg CT0075B Sulphamethoxazole磺胺甲基异恶唑(新诺明)RL 25μg CT0051B Sulphamethoxazole磺胺甲基异恶唑(新诺明)RL100μg CT0074BSulphamethoxazole/trimethoprim 19 : 1磺胺甲基异恶唑(新诺明)/甲氧苄氨嘧啶SXT25μg CT0052BSulphonamides compound磺胺复合物S3300μg CT0059B TTeicoplanin替考拉宁(壁霉素)TEC30μg CT0647BTelithromycin 泰利霉素TEL 15μg CT1714B Tetracycline 四环素TE 10μg CT0053B Tetracycline 四环素TE 30μg CT0054B Ticarcillin替卡西林(羧噻吩青霉素)TIC 75μg CT0167B Ticarcillin/clavulanic acid 7.5 : 1替卡西林/克拉维酸(7.5:1)TIM 85μg CT0449B Tigecycline 替加环素TGC 15μg CT1841B Tilmicosin 替米考星TIL 15μg CT1756B Tobramycin 妥布霉素(托普霉素)TOB 10μg CT0056B Tobramycin 妥布霉素(托普霉素)TOB 30μg CT1618B Trimethoprim 甲氧苄氨嘧啶W 1.25μg CT0057B Trimethoprim 甲氧苄氨嘧啶W 2.5μg CT0070B Trimethoprim甲氧苄氨嘧啶W 5μg CT0076B Trimethoprim/sulphamethoxazole 1:19甲氧苄氨嘧啶/磺胺甲基异恶唑(复方新诺明)SXT 25μg CT0052BVVancomycin 万古霉素(稳可信)VA 5μg CT0188B Vancomycin 万古霉素(稳可信)VA 30μg CT0058B Voriconazole优立康唑VOR 1μg CT1807B注释:美国临床实验室标准化研究所 英国抗生素化疗协会 上海桥星贸易有限公司联系人:蒋经理电话:182********座机:021-********地址:上海市杨浦区杭州路193弄83号瑞典抗生素委员会 SRGA: Swedish Reference Group forAntibiotics 法国微生物学会SFM: Soci ét é Fran aise de MicrobiologieBSAC: British Society forAntimicrobial Chemotherapy 药敏纸片统一规格:5 ×50片/盒药敏纸片统一价格:232.00/盒CLSI: Clinical and Laboratory Standards Institute DIN: Deutsches Institut f ür Normung德国标准化学会CLSI DIN BSAC SRGA SFM√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√√究所。
氨基糖苷类抗生素定义和概述

抑制细菌蛋白质合成
1. 起始阶段:抑制70S始动复合物形成
2. 肽链延伸阶段:造成mRNA密码错 译,合成无功能的蛋白质
3. 终止阶段:阻止肽链的脱落和核糖体 循环
aminoglycosdes﹒antibacterial mechanisms
氨基糖苷类﹒抗菌机制
aminoglycosides
OH II
NH2
R6
O
OH
O
I NHR4
R5
OH
NHR3
Structure of aminoglycosides
Common Properties of Aminoglycosides 氨基糖苷类的共性
➢抗菌作用及机制 ➢细菌耐药性 ➢体内过程 ➢临床应用 ➢不良反应
aminoglycosides﹒antibacterial effects
➢离子吸附作用 ➢插入异常膜蛋白
aminoglycosdes﹒antibacterial mechanisms
氨基糖苷类﹒抗菌机制
抑制细菌蛋白质合成全过程 干扰细菌胞浆膜的通透性
aminoglycosides﹒resistance
氨基糖苷类﹒耐药性
耐药机制:
➢钝化酶 modifying enzyme
氨基糖苷类抗生素定义和 作用机制
aminoglycosides
天然来源
链霉素 streptomycin 妥布霉素 tobramycin 卡那霉素 kanamycin 大观霉素spectinomycin 新霉素 neomycin
庆大霉素 gentamicin
小诺霉素micronomicin 阿司米星 astromicin 西索米星 sisomicin
硫酸链霉素

硫酸链霉素[典][基]Streptomycin Sulfate (Strepolin)字体[大][中][小]链霉素系由土壤中放线菌属灰色链丝菌所产生的抗生素,其硫酸盐供药用。
【性状】白色或类白色粉末,无臭、味微苦,有引湿性,易溶于水,不溶于乙醇或氯仿,干燥状态下稳定,密闭避光保存4年以上效价不变,水溶液在室温pH3 ~7条件下可保存2~4周,遇酸、碱、氧化剂均易破坏失效。
【药效学】药物进入机体组织后,通过病原微生物的细胞膜,与病原微生物核糖体30s亚单位的特殊受体蛋白相结合,干扰信使核糖核酸与30s亚单位间起始复合物的形成,使DNA发生错误,亦即使应配对的氨基酸受到非配对氨基酸的竞争性对抗,导致无功能蛋白质的合成,使病原微生物多聚核糖体分裂,从而使合成蛋白质遭到最终破坏,造成病原微生物的死亡。
【药动学】肌内注射给药吸收良好。
广布于机体器官组织,但极少透过血脑屏障进入脑组织,药物主要存在于细胞外液,可进入胆汁,胸、腹水、结核性脓肿、干酪样组织;尿液中药物浓度较高,并可透过胎盘进入胎儿组织。
蛋白结合率仅为20%~30%,肌注给药1g,0.5~1.5 h血药浓度达高峰值25~50μg/ml,半衰期2.5~3h,24h内给药量的80%~98%以原型从尿中排出,约有1%从胆汁排出,亦有极少量从唾液和乳汁排出。
【临床应用】本品对革兰阳性菌与革兰阴性菌均有效,但对链球菌、肺炎球菌等革兰阳性菌的作用不及青霉素G,对立克次体,原虫、病毒、真菌无效;对结核杆菌及多种革兰阴性菌效果较好,如大肠杆菌、布氏杆菌、产气杆菌、流感杆菌、痢疾杆菌、鼠疫杆菌等。
临床主要用于治疗结核杆菌所致的肺结核、骨结核、淋巴结核、肠结核、结核性脑膜炎等及敏感菌株所致的脑膜炎、肺炎、败血症、脓胸、胸膜炎、腹膜炎等,还可用于敏感菌株所致的泌尿系、呼吸系感染。
【用法与用量】肌注给药:成人每次0.5~1g,每日1~2次;小儿每公斤体重每日15~25mg,分1~2次给,鞘内注射:成人每次50~100mg;小儿每公斤体重每次1mg,总量不超过20mg,用灭菌0.9%氯化钠注射液稀释至5mg/ml 使用,注入量不得超过引流的脑脊液量。
动物组织中氨基糖苷类药物残留量的测定高效液相色谱一质谱质谱法

动物组织中氨基糖苷类药物残留量的测定高效液相色谱一质谱/质谱法1 原理试样中氨基糖苷类药物残留,采用磷酸盐缓冲液提取,经过C18固相萃取柱净化,浓缩后,使用七氟丁酸作为离子对试剂,高效液相色谱一质谱/质谱测定,外标法定量。
2试剂和材料2. 1 甲醇:液相色谱级。
2. 2 冰乙酸:液相色谱级。
2. 3 甲酸:液相色谱级。
2. 4 七氟丁酸:纯度≥99%。
2. 5 浓盐酸。
2.6 氢氧化钠。
2. 7 三氯乙酸:纯度≥99%。
2. 8 乙二胺四乙酸二钠(Na2 EDTA):纯度≥99%。
2. 9 磷酸二氢钾。
2.10 七氟丁酸溶液(HFBA) :100 mmol/L,准确量取6. 5 mL七氟丁酸((2. 4),用水稀释至500 mL(4℃避光可保存6个月)。
2.11 七氟丁酸溶液:20 mmol/L准确量取100 mmol/L七氟丁酸溶液50 mL(2.10),用水稀释至250 mL(4℃避光可保存6个月)。
2.12 磷酸盐缓冲液(含0.4 mmol/L EDTA和2%三氯乙酸溶液):准确称取磷酸二氢钾(2. 9)1. 36 g,用980 mL水溶解,用1. 0 mol/L的盐酸调pH到4. 0,分别加人Na2EDTA(2.8)0. 15 g和三氯乙酸(2.7)20 g,溶解混匀并定容至1 000 mL(4℃避光可保存1个月)。
2.13 甲酸:0.1%(体积分数),准确吸取1. 0 mL甲酸(2. 3)于1 000 mL容量瓶中,用水稀释至刻度,混匀。
2.14 壮观霉素、潮霉素B、双氢链霉素、链霉素、丁胺卡那霉素、卡那霉素、安普霉素、妥布霉素、庆大霉素、新霉素标准品:纯度范围92. 0%~99%。
2.15 10种氨基糖苷类药物标准贮备液:分别准确称取适量的每种氨基糖苷类药物标准品(2.14),用水溶解,配制成浓度为100 μg/mL的标准贮备溶液(4 ℃避光可保存6个月)。
2. 16 l0种氨基糖苷类药物混合标准中间溶液:分别准确量取壮观霉素、双氢链霉素、链霉素、丁胺卡那霉素、卡那霉素、妥布霉素、庆大霉素标准贮备溶液(2.15)各1.0 mL,新霉素、潮霉素B、安普霉素标准贮备溶液((2.15)各5. 0 mL,于25 mL容量瓶中,用水定容至刻度,配制成壮观霉素、双氢链霉素、链霉素、丁胺卡那霉素、卡那霉素、妥布霉素和庆大霉素浓度为 4.0μg/mL,新霉素、潮霉素B和安普霉素浓度为20. 0 μg/mL的混合标准中溶液(4℃避光可保存1个月)。
2.04.09.-.Contagious Bovine Pleuropneumonia 牛传染性胸膜肺炎(2014)

NB: Version adopted by the World Assembly of Delegates of the OIE in May 2014C H A P T E R2.4.9.C O N T A G I O U S B O V I N E P L E U R O P N E U M O N I ASUMMARYContagious bovine pleuropneumonia (CBPP) is a disease of ruminants (Bos and Bubalus genuses)caused by Mycoplasma mycoides subsp. mycoides SC (Mmm SC; SC = small colony). It ismanifested by anorexia, fever and respiratory signs such as dyspnoea, polypnoea, cough and nasaldischarges in bovines. Diagnosis requires the isolation of the aetiological agent. The main problemsfor control or eradication are the frequent occurrence of subacute or subclinical infections, thepersistence of chronic carriers after the clinical phase and the lack of extensive vaccine coverage.Identification of the agent: Samples to be taken from live animals are nasal swabs and/orbroncho-alveolar washings or pleural fluid obtained by puncture. Samples to be taken at necropsyare lung lesions, lymph nodes, pleural fluid and synovial fluid from those animals with arthritis.For cultivation of the pathogen, the tissues are ground in buffered solution and inoculated intoselective broth and solid media with antibiotics or other inhibitors to prevent the growth of cell-walled bacteria. The growth of Mmm can take up to 10 days, depending upon the type of sampleand the mycoplasma titre.In broth, growth is visible as a homogeneous cloudiness which forms swirls when shaken; on agar,small colonies develop, 1 mm in diameter, with the classical ‘fried-egg’ appearance. Thebiochemical characteristics of Mmm are the following: sensitivity to digitonin, reduction oftetrazolium salts, fermentation of glucose, absence of arginine hydrolysis, and the absence of orvery slight phosphatase and proteolytic activities. Special media have been described that arerecommended for these tests. Diagnosis is confirmed by immunological tests, such as the growthinhibition and immunofluorescence tests (both use hyperimmune sera). The polymerase chainreaction is a rapid, specific, sensitive and easy-to-use test.Serological tests: For diagnosis, the modified Campbell & Turner complement fixation testremains a prescribed test for international trade. The competitive enzyme-linked immunosorbentassay is also designated as an prescribed test for international trade. An immunoblotting test hasundergone evaluation and is highly specific and sensitive.Requirements for vaccines: The attenuated strains T1/44 and T1sr are now recommended forvaccine production. The minimal required titre is 107 mycoplasmas per vaccine dose, but highertitres of at least 108 are recommended.A. INTRODUCTIONContagious bovine pleuropneumonia (CBPP) is an infectious and contagious respiratory disease of Bovidae caused by Mycoplasma mycoides subsp. mycoides “small colony” (Mmm SC) with a major impact on livestock production and a potential for rapid spread. As a result, CBPP-infected countries are excluded from international trade of live animals.Mmm SC is a mycoplasma, i.e. a wall-less bacteria (mollicute), belonging to the so-called “mycoides cluster” that groups five mycoplasma species that are ruminant pathogens (Manso-Silván et al., 2009). These five mycoplasmas share phenotypic and genotypic characteristics that cause cross-reactions in conventional diagnostic techniques. The closest relative to Mmm SC is M. mycoides susbp capri (Mmc), which is usually found in goats.In natural conditions, Mmm SC affects only the ruminants of the Bos genus, i.e. mainly bovine and zebu cattle but also the yak (Bos grunniens) and water buffaloes (Bubalus bubalis) (Santini et al., 1992). Mmm SC has beenisolated from sheep and goats in Africa, in Portugal and in India (Srivastava et al., 2000). Among wild animals, one single case has been reported in American buffaloes (Bison bison) and none in African buffaloes (Syncerus caffer) or other wild ruminants. Small ruminants and wild animals do not play a role in the epidemiology of the disease, and CBPP is not a zoonotic agent.The incubation period for naturally infected animals can range from 3 weeks to 6 months. The clinical manifestations in cattle range from hyperacute through acute, subacute and chronic forms.CBPP is manifested by anorexia, fever and respiratory signs, such as dyspnoea, polypnoea, cough and nasal discharges during the acute stage of the disease when the causative agent can spread rapidly; in the chronic stage there may be long-term persistence of the agent. Typical lesions include a unilateral pneumonia associated with pleurisy. During the chronic stage of the disease, clinical signs wane and infected animals are more difficult to identify. In these cases, lungs may contain typical encapsulated lesions called sequestra. These ‘silent’ carriers may be infectious and thus responsible for unnoticed persistence of the infection in a herd; they play an important role in the maintenance and in the epidemiology of the disease.CBPP has been unequivocally identified in Europe since the 18th century and it gained a world-wide distribution during the second half of the 19th century through cattle trade. CBPP was eradicated from many countries at the beginning of the 20th century, mostly through stamping-out strategies (UK, USA) or by vaccination campaigns followed by stamping-out strategies (Australia). Today, CBPP remains enzootic in many Sub-Saharan African countries, while in Europe the last CBPP cases were observed in Portugal in 1999. The situation in some Asian countries is unclear.In the field, CBPP might be confused with other diseases causing respiratory problems such as pasteurellosis or other mycoplasmosis. The absence of confirmatory diagnosis may lead to antibiotic treatments being used in the case of CBPP outbreaks.There is no known risk of human infection with Mmm. Biocontainment measures should be determined by risk analysis as described in Chapter 1.1.3a Standard for managing biorisk in the veterinary laboratory and animal facilities. In CBPP-free areas, it is advisable to manipulate Mmm SC in biosafety level (BSL) 2 laboratories, while BSL1 would be sufficient in enzootic zones. Good laboratory practices should be used for all procedures.B. DIAGNOSTIC TECHNIQUESClinical diagnosis of CBPP is unreliable as initial signs may be slight or non-existent and may be indistinguishable from any severe pneumonia. Therefore, CBPP should be investigated by pathological, microbiological, molecular or serological diagnostic methods. As the pathological lesions of CBPP are distinctive, and pathognomonic, abattoir surveillance for CBPP involving lung examination is a practical method for disease monitoring.It is recommended to isolate and identify the causative organism in order to confirm an outbreak. Table 1 lists the laboratory methods used for the diagnosis of CBPP.Table 1. Laboratory methods currently used for diagnosis of CBPP and their purposeMethodPurposePopulationfreedomfrominfectionIndividual animalfreedom frominfection prior tomovementContributiontoeradicationpoliciesConfirmationof clinicalcasesPrevalenceof infection –surveillanceImmune status inindividual animalsor populationspost-vaccination* Agent detection and identification1In-vitro cultureisolation (followedby speciesidentification tests)+++ – – +++ – –Direct moleculartest (PCR)– – – ++ – – 1 A combination of agent identification methods applied on the same clinical sample is recommended.Detection of immune response2CFT +++ +++ +++ ++ +++ – Immunoblotting ++ ++ ++ ++ +++ – C-ELISA +++ +++ +++ +++ +++ – *NB: at present, there is no test described in the table that allows evaluation of the immune statusof an animal after vaccination, with the current T1 strains.Key: +++ = recommended method; ++ = suitable method; + = may be used in some situations, but cost, reliability, or otherfactors severely limits its application; – = not appropriate for this purpose.Although not all of the tests listed as category +++ or ++ have undergone formal validation, their routine nature and the fact that they have been used widely without dubious results, makes them acceptable.PCR = polymerase chain reaction; CFT = complement fixation test;C-ELISA = competitive enzyme-linked immunosorbent assay.1. Identification of the agent1.1. SamplesA key to isolation success lies in collecting good quality samples. Mmm SC can be isolated fromsamples taken either from live animals or at necropsy. Samples taken from live animals are: nasalswabs or nasal discharges, broncho-alveolar lavage or transtracheal washing and pleural fluidcollected aseptically by puncture made in the lower part of the thoracic cavity between the seventh andeighth ribs. Samples taken at necropsy are: lungs with lesions, pleural fluid (‘lymph’), lymph nodes ofthe broncho-pulmonary tract, and synovial fluid from those animals with arthritis. The lung samplesshould be collected from lesions at the interface between diseased and normal tissue.When collecting nasal swab samples, a transport medium should be used to protect the mycoplasmasand prevent proliferation of cell-walled bacteria (heart-infusion broth without peptone and glucose, 10%yeast extract, 20% serum, 0.3% agar, 500 International Units [IU]/ml penicillin, thallium acetate0.2 g/litre).After collection all samples must be kept refrigerated at 4°C and sent to the laboratory within 24 hours.For longer periods they should be frozen at or below –20°C.1.2. In-vitro cultureThe presence of the pathogen varies greatly with the stage of development of the lesions, and anegative result is not conclusive, particularly if the animal was treated with an antibiotic.Mmm SC needs selective media to grow but it is not considered a fastidious mycoplasma. There areseveral media compositions used in different reference laboratories but, essentially, they shouldcontain: a basic medium such as heart-infusion broth or PPLO broth (pleuropneumonia-likeorganisms), 1–2.5% yeast extract, 10–20% inactivated horse serum, 0.1% glucose, 1% triptose, and0.0024% DNA. To avoid growth of other bacteria, the media can also contain an antibiotic of thepenicillin family (for example,500 IU/ml penicillin G) as mycoplasmas are naturally resistant. The mediashould be used both as broth and solid. All culture media prepared should be subjected to qualitycontrol and must support growth of low passage Mycoplasma spp. from small inocula. The referencestrain should be cultured in parallel with the suspicious samples to ensure that the tests are performedcorrectly.After grinding with broth, the tissue samples are diluted tenfold to minimise contaminating bacteria andare inoculated into five tubes of broth and onto five solid media plates. Alternatively, to avoidcontaminating bacteria and to reduce the number of tubes and plates per sample, inoculate thesupernatant of the ground sample through a 25-mm filter with 0.45 pore size. The pleural fluid can beinoculated directly without prior dilution or filtration as, when infected, it is almost a pure culture ofmycoplasma. To ensure the best conditions for mycoplasma growth, the tubes and Petri dishes areincubated at 37°C in a 5% CO2 atmosphere and should be inspected daily for up to 10 days. After thistime if there is no growth, the sample is considered negative. Positive samples in liquid medium show ahomogeneous cloudiness, usually within 2–4 days, frequently with a silky, fragile filament called a2 One of the listed serological tests is sufficient.‘comet’. During the following days a uniform opacity develops which forms swirls when shaken. On agar media, the colonies are small (1 mm in diameter) and have the classical appearance of ‘fried eggs’ with a dense centre. At this stage, biochemical tests, the indirect fluorescent antibody (IFA) test or polymerase chain reaction (PCR) can be performed to identify the colonies.1.3. Biochemical and immunological identification testsBiochemical tests were used routinely in the past but have now been superseded by other tests, namely the PCR. Biochemical tests alone do not allow identification of a precise Mycoplasma species because of overlapping of the few phenotypic traits that can be evaluated. Therefore, molecular tests such as PCR are recommended for identification of isolates. The biochemical methods are given below for historic reasons.Following subculture, antibiotics should be omitted from the medium to check if the isolate is a mycoplasma or an L-form of a bacterium that will regain its original form in the medium without inhibitors. Once this test is done, the organism can be identified using biochemical tests (Freundt et al., 1979).Mmm SC is sensitive to digitonin (like all members of the order Mycoplasmatales), does not produce ‘film and spots’, ferments glucose, reduces tetrazolium salts (aerobically or anaerobically), does not hydrolyse arginine, has no phosphatase activity, and has no or weak proteolytic properties.For these tests, special media have been developed that include the same basic ingredients (heart-infusion broth or Bacto PPLO broth, horse serum, 25% yeast extract solution, 0.2% DNA solution), to which is added 1% of a 50% glucose solution for glucose hydrolysis, 4% of a 38% arginine HCl solution for arginine hydrolysis, and 1% of a 2% triphenyl tetrazolium chloride solution for tetrazolium reduction, as well as a pH indicator (e.g. phenol red). (NOTE: a pH indicator should not be added to a medium containing triphenyl tetrazolium chloride.) For demonstration of proteolysis, growth is carried out on casein agar and/or coagulated serum agar.Once the biochemical characteristics have been checked, one of the following immunological tests can be performed to confirm the identification: disk growth inhibition test (DGIT) (Freundt et al., 1979), fluorescent antibody test (FAT), IFA, agar gel immunodiffusion test (AGID) (Provost, 1972), or dot immunobinding on a membrane filter (MF-dot) test (Brocchi et al., 1993).At present, few laboratories use immunochemical tests on a routine basis for detection and identification of Mmm SC because of the development of PCR-based tests that are more specific, sensitive, rapid and easy to perform and standardise.1.4. Molecular identification and typing methods — polymerase chain reaction (PCR)-based testsSee Chapter 1.1.5 Principles and methods of validation of diagnostic assays for infectious diseases for further details of implementation and validation of PCR-based tests.PCR has become the method of choice for the rapid and specific identification of Mmm SC when the organism is isolated from a clinical sample. To avoid cross-contamination and carryover-contamination, strict separation of laboratory rooms, used for PCR preparation and handling of reactions, is needed.Various authors have developed a PCR system for Mmm SC identification and there is no preferred one, though the more sensitive, nested PCR should be avoided because of the higher risk of PCR product carryover, resulting in false positives. As the DNA target is not from a sample, where the number of cells vary and PCR inhibitors can be present, but from an isolate the sensitivity is not a critical point. Primers complementary to DNA regions CAP-21, lppA gene and 16S rRNA gene of the genome of Mmm SC have been designed by different authors and used in PCR systems, followed by restriction endonuclease analysis of the amplified product (amplicon) (PCR-REA) or by a second amplification (nested-PCR) (Table 2).Table 2. Conventional PCR systems most often used for Mmm SC identificationTarget DNA region* Amplicom(bp)RestrictionenzymeHydrolysesproducts (bp)Specificity ReferenceCAP-21 (position: 181628) 574 Asn I 379,178220, 178, 153Mmm SCMmcBashiruddinet al.,1994Target DNA region* Amplicom(bp)RestrictionenzymeHydrolysesproducts (bp)Specificity Reference(position: 443115) 275 Ase I 232,43Mmm SC Dedieuet al., 1994**Gene lppA (nested)717 – Mmm SC Miserezet al., 1997***(position:20061)503*Mmm SC PG1 sequence Gene Bank accession number: NC005364**Primers sequence: MSC1: ATA-CTT-CTG-TTC-TAG-TAA-TAT-G; MSC2: CTG-ATT-ATG-ATG-ACA-GTG-GTC-A***Primers sequence: SC3NEST1-L: ACA-AAA-AGA-AGA-TAT-GGT-GTT-GG and SC3NEST1-R: ATC-AGG-TTT-ATC-CAT-TGG-TTG-G; SC3VII: ATT-AGG-ATT-AGC-TGG-TGG-AGG-AAC and SC3IV-S: TCT-GGG-TTA-TTC-GAA-CCA-TTA-TAs an example of one PCR system that is used in routine for Mmm SC identification, the PCR-REA procedure adapted from Bashiruddin et al.,1994, is provided below. A single step PCR has also been described by Miles et al. (2006).1.4.1. DNA extractionAny accepted method for DNA extraction would be appropriate. A simple and effective methodis to select a single colony, resuspend it in 100 µl of PCR-grade water, boil for 15 minutes tolyse the cells and release the DNA, and centrifuge at 1500 g for 1 minute. The supernatant withthe DNA will be used in the PCR reaction after diluting 1/10 in PCR-grade water. Alternatively500 µl of a 4-day broth culture, from a single colony, is centrifuged at 1500 g for 5 minutes, thepellet is resuspended in 100 µl of PCR-grade water and boiled and centrifuged as above. Afterdiluting 1/10, 1 µl of the supernatant is used in the PCR.1.4.2. PCR amplificationi) Preparation of the master mixSynthetic oligonucleotides should be dissolved in TE buffer (Tris-EDTA [ethylene diaminetetra-acetic acid]) to a 100 µM concentration. This stock solution is stable at –20°C for atleast 4 years. A working solution is prepared from the stock solution by a 1/2 dilution, toobtain a final concentration of 50 µM. In a final volume of 25 µl the PCR reaction shouldcontain the following:Concentration Volume in one reaction(total volume 25 µl) Final concentration in thereactionH2O PCR-grade– 16µl – Reaction buffer without MgCl210 × 2.5 µl 1×MgCl225 mM 3 µl 3 mMdNTPs 10 mM 1 µl 400 µM Primer MM450* 50 µM 0.5 µl 1 µMPrimer MM451** 50 µM 0.5 µl 1 µMTaq pol. 5U/µl 0.5 µl 2.5 U template DNA 1 µl*Sequence: 5’-GTA-TTT-TCC-TTT-CTA-ATT-TG-3’**Sequence: 5’-AAA-TCA-AAT-TAA-TAA-GTT-TG-3’ii) AmplificationconditionsOne µl of template DNA of tested sample, or positive control (DNA from Mmm SC-typestrain PG1) or negative control (water) is added to the mix. The amplification is performedunder the following conditions: 30 cycles of 94°C for 1 minute, 50°C for 1 minute, 72°C for2 minutes; and hold at 4°C indefinitely.iii) Detection of amplified productsAfter amplification, the reactions should be open in a separate room where the PCRproducts are detected by 1.5% agarose gel electrophoresis for 2 hours at 90 V. Positivereactions and the positive control, and not the negative control, should present anamplicom of 574 bp visible under UV light after staining with a suitable fluorescent dye.1.4.3. Restriction endonuclease analysis (REA)An enzymatic restriction of the PCR product is carried out in 10 µl reaction volume containing:2 µl of PCR reaction, 5 U Asn I (2 µl), 1× buffer, at 37°C for 1 hour. The result is analysed is a2% agarose gel electrophoresis for 1 hour at 100 V and the products visualised as above.Identification is based on the sizes of the restriction products as presented in Table 2.NB: Direct detection and identification of Mmm SC by PCR in clinical samples has not yet beenfully evaluated and so the conventional PCR may be inadequate because of the presence ofPCR inhibitors, fewer microorganisms in the sample and the presence of other bacteria, with aconcomitant reduction in sensitivity and specificity. These problems should be avoided throughthe use of real-time PCR assays for Mmm SC detection, as fluorescence resulting from specificgenomic amplification is detected and measured as the amplicon is being synthesised, and a 2–3 log increase in sensitivity, compared with conventional PCR, is obtained (Lorenzon et al.,2008). Using samples such as pleural fluid, the PCR can be performed after boiling the sampleand centrifuging to recover DNA in the supernatant. For lung fragments, the PCR is appliedafter DNA extraction procedures using an extraction kit or a phenol/chlorophorm extraction.1.4.4. Mmm SC strain typingAn enzymatic restriction of the PCR product is carried out in 10 µl reaction volume containing:2 µl of PCR reaction, 5 U Asn I (2 µl), 1× buffer, at 37°C for 1 hour. The result is analysed is a2% agarose gel electrophoresis for 1 hour at 100 V and the products visualised as above.2. Serological testsSerological tests for CBPP are valid at the herd level only because false positive or false negative results may occur in individual animals. Tests on single animals can be misleading, either because the animal is in the early stage of disease, which may last for several months, before specific antibodies are produced, or it may be in the chronic stage of the disease when very few animals are seropositive.False-positive results can occur (2%), of which an important cause is serological cross-reactions with other mycoplasmas, particularly other members of the M. mycoides cluster. The validity of the results has to be confirmed by post-mortem and bacteriological examination, and serological tests on blood taken at the time of slaughter.The complement fixation test (CFT) and enzyme-linked immunosorbent assays (ELISAs) are recommended for screening and eradication programmes. The highly specific immunoblotting test is useful as a confirmatory test but is not fit for mass screening.2.1. Complement fixation (recommended for determining disease free status and a prescribed testfor international trade)The modified Campbell & Turner CFT remains the recommended procedure and it is widely used in all countries where infection occurs (Provost et al., 1987). The CFT is most conveniently carried out in a microtitre format and has been harmonised within countries of the EU (European Commission, 2001).With a sensitivity of 63.8% and a specificity of 98% (Bellini et al., 1998), the CFT can detect nearly all sick animals with acute lesions, but a rather smaller proportion of animals in the early stages of the disease or of animals with chronic lesions.2.1.1. ReagentsFor CFT validation and accreditation, the quality control and standardisation of all the reagentsis a critical issue as well as the pipettes and tips that are used to dispense them. To facilitatethe accreditation procedure (ISO/IEC317025) appropriate antigen and sera controls can beobtained from OIE Reference Laboratories for CBPP.The features of the following reagents, should be taken into account, as they have an impact inthe final result.i) Ultra-pure sterilised water. The quality of the water is critical for the development of thereaction.ii) Positive reference sera: for harmonisation purposes a positive bovine reference standard serum (PRS) is available from the OIE Reference Laboratories in Portugal and Italy, andshould be used in the diagnostic laboratory for routine use and antigen titration. The PRShas been obtained from a naturally CBPP-infected bovine, and is negative againstBrucella, Mycobacterium paratuberculosis, Chlamydia, Coxiella burnetii, Leptospira,bovine viral diarrhoea virus, respiratory syncytial virus, infectious bovine rhinotracheitisvirus, adenovirus, bovine herpes virus 4, foot and mouth disease viruses, bovine leukosisvirus, and parainfluenza 3 virus, and should also be negative for adventitious viruses. ThePRS presents 100% haemolysis in a dilution of 1/160 and 50% haemolysis in a dilution of1/320. The PRS should be stored at –20°C in aliquots, this would prevent repetitivefreeze–thawing, which causes deterioration of the serum.iii) Negative reference sera: the negative control serum (NRS), also available from OIE Reference Laboratories in Portugal and in Italy, is a serum from a healthy bovine from abovine spongiform encephalopathy (BSE)-free source. It is CBPP free and should benegative against the above microorganisms. The NRS should be stored in aliquots at–20°C.iv) Antigen: the test must use an antigen that has been prepared from a suspension of a selected Mmm strain essayed previously and presenting the five specific antigenic bandsof 110, 98, 95, 62/60 and 48 kDa. The antigen is previously checkerboard titrated andused at a dose of 2 complement fixing units (CFU) and should be standardised to give50% fixation, at a dilution of 1/320 of PRS. It must be stored at 4°C ± 3°C and should notbe frozen. It is produced, standardised and delivered by OIE Reference Laboratories. Theuse of an antigen standardised against the OIE reference sera promotes internationalharmonisation of diagnostic testing.v) Buffer: veronal buffered saline (VB), pH 7.2 ± 1, is the standard diluent for the CFT. The VB can be prepared from tablets commercially available or it may be prepared from astock solution of sodium chloride (42.5 g), barbituric acid (2.875 g), sodium diethylbarbiturate (1.875 g), magnesium chloride (MgCl2 × 6 H2O – 0.501 g), and calciumchloride (CaCl2 × 6 H2O – 0.18 g) in 1 litre of distilled water. A concentrated stock solutionis used diluted 1/5 in double-distilled water, before use.vi) Haemolysin(amboceptor): the haemolysin is a hyperimmune rabbit serum to SRBC (sheep red blood cells). The quantity used is 6 haemolytic doses read at 50% end-point(HD50 [50% haemolysing dose]). The SRBC are obtained by aseptic puncture of thejugular vein. They can be preserved in Alsever’s solution or in sodium citrate. They areused in a 6% suspension. Haemolysin is also available commercially.vii) Haemolytic system(HS): HS is prepared by diluting haemolysin in VB to give a dose of12 HD50. An equal volume of 6% SRBC antibody suspension is added, and the system issensitised in a water bath at 37°C for 30 minutes with periodic shaking.viii) Complement(C’): C’ is obtained from normal guinea-pig serum. It is freeze-dried and reconstituted with double-distilled water. It must be kept at –20°C after reconstitution. Acommercially produced complement can be used, according to the manufacturer’sinstructions. It is titrated by making a close dilution series in VB containing an appropriatequantity of the antigen to be used in the test. After incubation at 37°C for 30 minutes, anappropriate quantity of sensitised SRBC is added to each dilution. The titration is readafter incubation for a further 30 minutes. The highest dilution giving complete haemolysisof the SRBC equals 1 C’ unit, from which can be calculated the dilution required for2.5 units in 25 µl.3 ISO/IEC: International Organisation for Standardisation/International Electrotechnical Commission2.1.2. Test procedure (using microplates)The most critical factors for CFT performance are the control of the reagents and that it is carried out by trained and qualified personnel. Temperature and incubation times should also be carefully controlled. The entire procedure should be performed in a temperature-controlled room at 21°C ± 3°C.Contaminated or haemolysed sera should not be tested, as this will interfere with the reaction.Undiluted test sera samples and appropriate working standards should be inactivated for30 minutes in water bath at 56°C ± 2°C. This will destroy the complement of the sera andreduce or eliminate the anti-complementary activity (ACA).Usually, only one serum dilution (1/10 in VB) is tested routinely but serial dilutions must be done for the positive control (PRS). Using standard 96-well microtitre plates with round (U) bottoms, the technique is performed as follows:i) Dispense 25 µl of test serum inactivated and diluted 1/10 in VB in the wells of the first andsecond rows. The first row is an anti-complementary control for each serum.ii) Volumes of 25 µl of antigen at a dose of 2 CFU are added to each well except the anti-complementary controls, to which 25 µl of VB should be added to compensate for lack ofantigen.iii) Add 25 µl of C’ at a dose of 2.5 units. Shake gently and incubate at 37°C for 30 minutes with periodic (at least twice during the incubation period) or continuous shaking.iv) Add 25 µl of HS. Shake gently and incubate at 37°C for 30 minutes. Microplates must be shaken twice during the incubation period.v) It is necessary to set up the following controls:a) C’: 0.5 units, 1 unit and 2.5 units.b) HS: 75 µl of VB + 25 µl of HS.c) Antigen: 25 µl of 2 CFU of antigen + 25 µl of C’ at 2.5 units + 25 µl of HS = 25 µl ofVB.These controls, along with the positive control serum (PRS) and the negative controlserum (NRS), should always be used in each microplate or in a series of microplateswhere the same batches of reagents are used.vi) Interpretation of resultsAfter centrifugation of the microplates at 125 g for 5 minutes, the analysis is carried outbased on the percentage of complement fixation observed.Positive result: 100% fixation at 1/10;Doubtful results: 25, 50 or 75% fixation at 1/10.vii) Validation of resultsExpected results for the plate controls are as follows:a) PRS: expected titre.b) NRS: 100% haemolysisc) Anticomplementary control of serum samples: 100% haemolysisd) Antigen control: 100% haemolysise) Complement units’ control: 100% haemolysis for 2.5 unitsf) HS control in absence of complement: 0% haemolysisIt is recommended that any CFT result, even partial (25, 50 or 75%), at a serum dilution of 1/10 be confirmed by additional investigations, such as immunoblotting or post-mortem examination and bacteriological tests according to contingency.。
抗生素可以诱导细菌形成生物膜
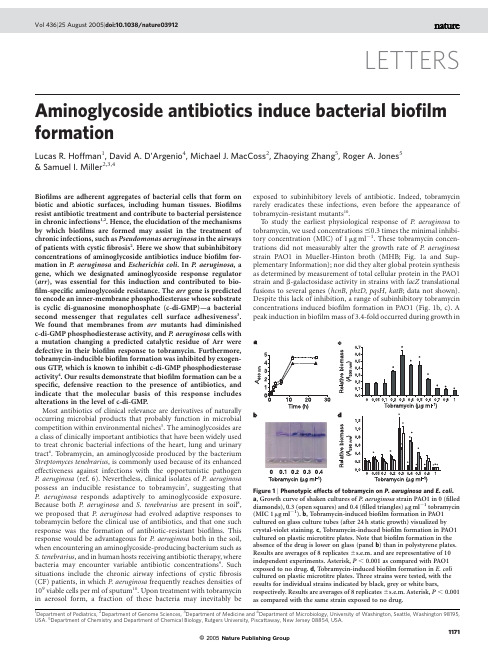
Aminoglycoside antibiotics induce bacterial biofilm formationLucas R.Hoffman1,David A.D’Argenio4,Michael J.MacCoss2,Zhaoying Zhang5,Roger A.Jones5&Samuel ler2,3,4Biofilms are adherent aggregates of bacterial cells that form on biotic and abiotic surfaces,including human tissues.Biofilms resist antibiotic treatment and contribute to bacterial persistence in chronic infections1,2.Hence,the elucidation of the mechanisms by which biofilms are formed may assist in the treatment of chronic infections,such as Pseudomonas aeruginosa in the airways of patients with cysticfibrosis2.Here we show that subinhibitory concentrations of aminoglycoside antibiotics induce biofilm for-mation in P.aeruginosa and Escherichia coli.In P.aeruginosa,a gene,which we designated aminoglycoside response regulator (arr),was essential for this induction and contributed to bio-film-specific aminoglycoside resistance.The arr gene is predicted to encode an inner-membrane phosphodiesterase whose substrate is cyclic di-guanosine monophosphate(c-di-GMP)—a bacterial second messenger that regulates cell surface adhesiveness3. We found that membranes from arr mutants had diminished c-di-GMP phosphodiesterase activity,and P.aeruginosa cells with a mutation changing a predicted catalytic residue of Arr were defective in their biofilm response to tobramycin.Furthermore, tobramycin-inducible biofilm formation was inhibited by exogen-ous GTP,which is known to inhibit c-di-GMP phosphodiesterase activity4.Our results demonstrate that biofilm formation can be a specific,defensive reaction to the presence of antibiotics,and indicate that the molecular basis of this response includes alterations in the level of c-di-GMP.Most antibiotics of clinical relevance are derivatives of naturally occurring microbial products that probably function in microbial competition within environmental niches5.The aminoglycosides are a class of clinically important antibiotics that have been widely used to treat chronic bacterial infections of the heart,lung and urinary tract6.Tobramycin,an aminoglycoside produced by the bacterium Streptomyces tenebrarius,is commonly used because of its enhanced effectiveness against infections with the opportunistic pathogen P.aeruginosa(ref.6).Nevertheless,clinical isolates of P.aeruginosa possess an inducible resistance to tobramycin7,suggesting that P.aeruginosa responds adaptively to aminoglycoside exposure. Because both P.aeruginosa and S.tenebrarius are present in soil8, we proposed that P.aeruginosa had evolved adaptive responses to tobramycin before the clinical use of antibiotics,and that one such response was the formation of antibiotic-resistant biofilms.This response would be advantageous for P.aeruginosa both in the soil, when encountering an aminoglycoside-producing bacterium such as S.tenebrarius,and in human hosts receiving antibiotic therapy,where bacteria may encounter variable antibiotic concentrations9.Such situations include the chronic airway infections of cysticfibrosis (CF)patients,in which P.aeruginosa frequently reaches densities of 109viable cells per ml of sputum10.Upon treatment with tobramycin in aerosol form,a fraction of these bacteria may inevitably be exposed to subinhibitory levels of antibiotic.Indeed,tobramycin rarely eradicates these infections,even before the appearance of tobramycin-resistant mutants10.To study the earliest physiological response of P.aeruginosa to tobramycin,we used concentrations#0.3times the minimal inhibi-tory concentration(MIC)of1m g ml21.These tobramycin concen-trations did not measurably alter the growth rate of P.aeruginosa strain PAO1in Mueller-Hinton broth(MHB;Fig.1a and Sup-plementary Information);nor did they alter global protein synthesis as determined by measurement of total cellular protein in the PAO1 strain and b-galactosidase activity in strains with lacZ translational fusions to several genes(hcnB,phzD,pqsH,katB;data not shown). Despite this lack of inhibition,a range of subinhibitory tobramycin concentrations induced biofilm formation in PAO1(Fig.1b,c).A peak induction in biofilm mass of3.4-fold occurred during growth inLETTERS Figure1|Phenotypic effects of tobramycin on P.aeruginosa and E.coli. a,Growth curve of shaken cultures of P.aeruginosa strain PAO1in0(filled diamonds),0.3(open squares)and0.4(filled triangles)m g ml21tobramycin (MIC1m g ml21).b,Tobramycin-induced biofilm formation in PAO1 cultured on glass culture tubes(after24h static growth)visualized by crystal-violet staining.c,Tobramycin-induced biofilm formation in PAO1 cultured on plastic microtitre plates.Note that biofilm formation in the absence of the drug is lower on glass(panel b)than in polystyrene plates. Results are averages of8replicates^s.e.m.and are representative of10 independent experiments.Asterisk,P,0.001as compared with PAO1 exposed to no drug.d,Tobramycin-induced biofilm formation in E.coli cultured on plastic microtitre plates.Three strains were tested,with the results for individual strains indicated by black,grey or white bars, respectively.Results are averages of8replicates^s.e.m.Asterisk,P,0.001 as compared with the same strain exposed to no drug.1Department of Pediatrics,2Department of Genome Sciences,3Department of Medicine and4Department of Microbiology,University of Washington,Seattle,Washington98195, USA.5Department of Chemistry and Department of Chemical Biology,Rutgers University,Piscattaway,New Jersey08854,USA.1171©2005Nature Publishing Groupthe presence of tobramycin at0.3times the MIC(Fig.1c)—an increase in biomass similar to that associated with P.aeruginosa mutants with increased propensities for biofilm formation2,11.Tobra-mycin induced biofilm formation in three different growth media (M63salts supplemented with glucose and casamino acids,MHB, Luria broth),on several abiotic surfaces(borosilicate glass,poly-styrene,polypropylene,polycarbonate)and in13out of14 P.aeruginosa clinical isolates tested(Fig.1b,c and Supplementary Information and data not shown).For both Gram-negative and Gram-positive bacteria,subinhibi-tory antibiotic treatment can stimulate production of exopoly-saccharides12,13.Subinhibitory levels of the b-lactam antibiotic imipenem augment production of the P.aeruginosa exopoly-saccharide alginate,and leads to increased biofilm volume13.How-ever,P.aeruginosa algD mutants(which are defective in alginate production)and the parental strain PAO1demonstrated equivalent tobramycin-induced biofilm formation(data not shown).Further-more,the tobramycin-induced increase in biomass(Fig.1c)corre-sponded to an increase in biofilm colony-forming units(CFU) (Supplementary Information),indicating that it resulted primarily from an increase in cell number and not an increase in extracellular matrix.We assayed other antibiotics at a range of concentrations(relative to the MIC)equivalent to those used for tobramycin.Three additional aminoglycoside antibiotics induced biofilm formation, although tobramycin had the strongest effect.The maximum induc-tion by amikacin,streptomycin and gentamicin was75%,66%and 25%of that by tobramycin,respectively.In contrast,polymyxin B (a peptide antibiotic that is cationic like aminoglycosides and interacts with membranes)had no effect on biofilm formation;nor did the protein synthesis inhibitor chloramphenicol or the cell wall synthesis inhibitor carbenicillin(data not shown).Therefore,biofilm induction by tobramycin is unlikely to be solely due to non-specific protein synthesis inhibition,cell damage or interaction with cell membranes through positive charge;rather,it appears to be a specific response by P.aeruginosa to aminoglycosides.In order to test whether this response was present in other Gram-negative bacteria,we examined the enteric bacterium E.coli.Sub-inhibitory levels of tobramycin induced biofilm formation in three clinical isolates of E.coli from three patients with bacteraemia (Fig.1d).As with P.aeruginosa,this induction occurred in a range of tobramycin concentrations centred on0.3m g ml21(0.3£the MIC for E.coli).Such conservation suggests that tobramycin acti-vates a signalling pathway present in both P.aeruginosa and E.coli. Signalling by the second messenger c-di-GMP is a good candidate for such a conserved system as this dinucleotide regulates cell adhesiveness in a diverse range of bacteria3,14,15.The PAO1genome includes at least38genes that are predicted to encode a regulator of intracellular c-di-GMP levels.These proteins contain either a GGDEF domain(found in putative cyclases for c-di-GMP synthesis), an EAL domain(found in putative phosphodiesterases for c-di-GMP degradation),or both16.To identify which,if any,of these genes might be involved in signalling the presence of tobramycin,we screened the relevant transposon-insertion mutants of PAO1 (ref.17)for tobramycin-inducible biofilm formation.We reasoned that the inactivation of a gene involved in signalling the presence of tobramycin would result in a strain that showed reduced biofilm induction by tobramycin,but normal biofilm production in the absence of tobramycin,relative to wild-type cells.Many of the mutants had altered biofilm formation even in the absence of tobramycin(Supplementary Information),and these were not analysed further.Three strains,each with a different insertion mutation in the monocistronic open reading frame PA2818,were defective for tobramycin-induced biofilm formation(Fig.2a,b and Supplemen-tary Information).We designated this gene arr,for aminoglycoside response regulator.The arr gene is predicted to encode an inner-membrane protein with two transmembrane domains,a periplasmic domain that could transduce an environmental stimulus,and an EAL domain(Fig.3a).Database searches revealed a similar domain architecture in the gene products of many Gram-negative bacteria, including plementation with a plasmid expressing arr restored wild-type tobramycin-induced biofilm formation in an arr mutant(Fig.2a),indicating that Arr is necessary for this response. There was no complementation with a plasmid expressing a mutant arr gene encoding a protein in which the conserved glutamate residue of the EAL domain was replaced with an alanine(E297A)(Fig.2d). As this mutation has abolished the biological activity attributed to other EAL domain proteins18,this result suggested that Arr phos-phodiesterase activity is required for tobramycin-induced biofilm formation.Consistent with Arr having c-di-GMP phosphodiesterase activity, membranes from arr mutant cells were54%less active in degrading c-di-GMP than membranes from PAO1cells(Fig.3b);this is a surprisingfinding given that23of the38predicted PAO1c-di-GMP regulators are expected to be transmembrane proteins.Wild-type phosphodiesterase activity was restored to the arr mutant by a plasmid expressing arr(Fig.3b),but not by a plasmid expressing the gene encoding the E297A Arr mutant(data not shown).Further-more,tobramycin-inducible biofilm formation in wild-type cells was inhibited by exogenous GTP—a c-di-GMP phosphodiesterase inhibitor4(Supplementary Information).Based on these results,we propose that tobramycin,either directly or indirectly,enhances the phosphodiesterase activity of the Arr cytoplasmic EAL domain, leading to c-di-GMP inactivation and augmented biofilm formation (Fig.3a).Figure2|The role of P.aeruginosa arr in tobramycin induction of biofilm formation and biofilm antibiotic resistance.a,Microtitre plate biofilm assay with the parental PAO1strain,the arr mutant strain,and the arr mutant strain carrying a plasmid expressing the wild-type arr gene(pArr). Results are averages of8replicates^s.e.m.and are representative of three independent experiments.Asterisk,P,0.001as compared with PAO1 cultured in the same tobramycin concentration.b,Crystal-violet-stained biofilms of PAO1and the arr mutant strain grown for24h on glass coverslips spanning the air–liquid interface of standing cultures.The dark portion above the air–liquid interface biofilm in tobramycin-induced PAO1 is adherent pellicle.c,Microtitre plate biofilm antibiotic resistance assay in which biofilms of the indicated strains were challenged with tobramycin at the concentrations shown,and the survival of suspended biofilm cells determined.Results are the average of three experiments^s.e.m.Asterisk, P,0.001as compared with PAO1biofilms exposed to the same drug concentration.d,The indicated plasmids were introduced into the arr mutant strain.Fold induction of biofilm formation by0.3m g ml21 tobramycin for each transformant was determined(average^s.e.m.of6 replicates).Asterisk,P,0.001as compared with cells carrying the empty vector.Cells were grown in500m g ml21carbenicillin for plasmid maintenance.LETTERS NATURE|Vol436|25August2005 1172©2005Nature Publishing Group© 2005Nature Publishing GroupAlthough Arr promotes biofilm formation,expression of EAL-type regulators in other organisms has been linked to a reduction in biofilm formation 15;this is in apparent contradiction with our model (Fig.3a).However,inactivation of putative c-di-GMP regulatory proteins (with either GGDEF,EAL,or both domains)had remark-ably varied effects on basal levels of biofilm formation in PAO1(Supplementary Information).This indicates a complex relationship between biofilm formation and the expression of individual regula-tors,which could involve the localized regulation of discrete cyto-plasmic c-di-GMP pools.The Caulobacter crescentus GGDEF-type regulator PleD (ref.14)and the P.aeruginosa GGDEF/EAL-type regulator FimX are both localized to one cell pole 3,14,19.Similarly,the Arr periplasmic domain could mediate the formation of localized protein complexes.Multiple cell surface appendages mediate bacterial aggregation and facilitate biofilm formation.Flagella and pili are involved in biofilm formation 11,20and are regulatory targets of c-di-GMP (refs 14,19).It was possible that tobramycin and Arr were regulating biofilm formation by altering these surface appendages.Indeed,subinhibi-tory tobramycin concentrations inhibited PAO1flagellar motility in a dose-dependent manner,and inhibition was reduced in an arr mutant (Supplementary Information).Nevertheless,tobramycin did not affect type IV pili-mediated twitching motility,and PAO1fliC mutants (lacking flagella)and pilA mutants (lacking type IV pili)showed tobramycin-induced biofilm formation that was equivalentto wild-type levels (data not shown).The arr mutant was also motile in assays for twitching (data not shown)and flagellar swimming (Supplementary Information),and polymyxin B inhibited flagellar swimming (Supplementary Information)without any effect on biofilm formation (data not shown).Therefore,it is unlikely that tobramycin or Arr affect biofilm formation simply via these surface appendages.Because biofilms are associated with increased antibiotic resist-ance 1,2,we tested whether Arr had a role in biofilm-mediated antibiotic resistance as well as biofilm formation.In a peg biofilm assay that was recently proposed for clinical use 21,biofilms of arr -mutant cells were approximately 100-fold more susceptible to tobramycin killing than PAO1biofilms,and the wild-type phenotype was restored by expressing arr from a plasmid (Fig.2c).Decreased biofilm tobramycin-resistance of the arr mutant was also observed in a standard biofilm crystal violet staining assay (data not shown),and by measuring CFU in colony biofilms (Supplementary Information).In contrast,planktonic cultures of arr -mutant cells exhibited the same killing by tobramycin as cultures of wild-type cells (MIC of 1m g ml 21).This indicates that arr ,like ndvB (ref.1),is a genetic determinant of biofilm-mediated antibiotic resistance in P.aerugi-nosa .The contribution of Arr to biofilm tobramycin-resistance could be clinically relevant,as improvement of lung function in CF patients treated with tobramycin in aerosol form correlates with an approxi-mate 100-fold reduction in P.aeruginosa CFU (ref.10);this is equivalent to the difference in killing between wild-type and arr -mutant biofilms treated with tobramycin (Fig.2c).Another P.aeruginosa EAL-type regulator,PvrR,plays a role in the formation of small colony variants (SCVs)that are hyperadherent and antibiotic resistant 22.Mutation of pvrR increased the frequency of appearance of SCVs growing in the presence of high concen-trations of the aminoglycoside kanamycin 22.This raised the possi-bility that mutation of arr altered the frequency of such variants with a consequent change in tobramycin-induced biofilm formation.To test this,we isolated and examined resuspended cells from PAO1and arr -mutant biofilms grown with and without subinhibitory tobra-mycin.The frequency of SCVs was equivalent in each of the four cell populations (data not shown).In addition,we tested these resus-pended cells for their ability to form new biofilms in the absence of antibiotic,and for their tobramycin susceptibilities in both static and shaken planktonic cultures.None of these characteristics were altered,either by mutation of arr or by subinhibitory tobramycin treatment (Supplementary Information).Therefore,subinhibitory tobramycin does not enrich for relatively resistant or adherent genetic variants,and the arr mutant phenotype is not due to the altered frequency of such variants.Taken together,our results suggest that inhibiting the activity of EAL-type regulators such as Arr might be of therapeutic benefit early in P.aeruginosa chronic infections:particularly airway infections in which the aerosol form of tobramycin is widely used.It might also be beneficial in acute disease,as the mutation of pvrR decreased P.aeruginosa virulence in burned mice 23.However,clinical strain variability could complicate such therapeutic manipulation of c-di-GMP metabolism.The pvrR locus of strain PA14is contained on a genetic island that is absent in strain PAO1(ref.23).Similarly,a genomic microarray analysis suggested that arr was absent or divergent in some P.aeruginosa isolates 24.This variability notwith-standing,13of the 14CF isolates we tested demonstrated tobramy-cin-induced biofilm formation,while the one remaining isolate appeared to lack arr by polymerase chain reaction (PCR)analysis (data not shown).Therefore,the relevant characteristic conserved among these strains and with E.coli (Fig.1c,d)is antibiotic induction of biofilm formation,probably through c-di-GMP as a second messenger.This biofilm response could contribute to differ-ences in therapeutic outcome upon antibiotic treatment,such as in CF where not all patients respond positively to tobramycin aerosols 10,and the ability to manipulate c-di-GMP metabolism mayhaveFigure 3|The role of c-di-GMP in the biofilm response toaminoglycosides.a ,A model for aminoglycoside effects on P.aeruginosa .Aminoglycoside antibiotics such as tobramycin (indicated by the three-ringed structure)could act as first messengers that trigger changes —mediated either by binding directly to proteins such as Arr or indirectly through intermediary molecules —in the level of the second messenger c-di-GMP .The proposed domain organization of Arr is indicated,including the location of the carboxy-terminal EAL domain.b ,Effect on c-di-GMP concentration of washed cell membranes containing 0.4mg of protein from either the PAO1strain,the arr mutant strain,or the arr mutant strain with a plasmid expressing the wild-type arr gene after incubation with 10m M c-di-GMP .Results are averages of at least 3replicates ^s.d.,and are representative of four separate experiments.Asterisk,P ,0.001as compared with PAO1.NATURE |Vol 436|25August 2005LETTERS1173therapeutic value by increasing bacterial susceptibility to standard therapy.There is growing evidence that bacteria respond specifically and defensively to subinhibitory antibiotic concentrations5,25.The evi-dence presented here indicates that Gram-negative bacteria can respond to aminoglycosides by forming antibiotic-resistant bio-films—perhaps only one of many strategies used to counter antibiotic production by Gram-positive soil bacteria such as the Strepto-mycetes.In P.aeruginosa,this biofilm response requires Arr—a regulator that alters c-di-GMP levels(Fig.3a).Both in soil and within animals,c-di-GMP regulation could contribute to the diver-sity of possible outcomes for bacterial communities challenged with antibiotics.METHODSBacterial strains,chemicals and media.The PAO1strain was obtained from B.Iglewski through C.Manoil,and the transposon-insertion mutants from M.Jacobs17.E.coli isolates were from patients with bacteraemia at the Massachusetts General Hospital(archived by ler),and P.aeruginosa longitudinal clinical isolates were from young CF patients less than8yr old (provided by J.Burns26).PAO1mutants containing a transposon TN5-derived insertion element17in the gene PA2818that were analysed in detail included the 50022(PA2818::IS phoA/hah-Tc),17026(PA2818::IS lacZ/hah-Tc)and5339 (PA2818::IS lacZ/hah-Tc)mutants;these mutants are described more fully at /UWGC/pseudomonas/index.cfm. Mutants in other genes are described in the Supplementary Methods.All data shown for mutants in the PA2818gene are for PAO1mutant50022;results were confirmed with PAO1mutants17026and5339.Bacteria were grown at378C in MHB(Difco)unless otherwise indicated.Polymyxin B was purchased from Amersham Life Sciences.GTP,as well as tobramycin sulphate and all other antibiotics,were obtained from Sigma.Unlabelled and18O-labelled c-di-GMP was synthesized,purified and characterized as described27.Phenotypic assays.P.aeruginosa PAO1was inoculated in duplicate onto Mueller-Hinton agar(MHA)with0.3%agar(to characterize swimming motility)or beneath MHA with1.5%agar(to characterize twitching motility), each with and without antibiotics,and then incubated at378C for12h, essentially as described20.For use as an inoculum for MIC determination(by broth microdilution),killing assays,growth curves and biofilm assays,overnight cultures of bacteria grown in MHB were diluted approximately1:100with MHB (to a density of107CFU ml21).Antibiotics were added to this inoculum as indicated.Cell number was determined by measuring CFU(by plating serial dilutions of cultures onto LB agar)or by measuring the absorbance at600nm (A600nm)of suspended cells.Biofilm formation was routinely quantified by measuring either A570nm or A595nm(which gave equivalent results)of crystal-violet staining of adherent cells as described previously21.To assay the induction of biofilm formation,biofilms were grown for24h without shaking at378C either in glass culture tubes,on glass cover slips,or in wells of untreated96-well polystyrene microtitre plates(Nunc)using100m l of culture per well(8 duplicates per condition per experiment).To assay biofilm antibiotic resistance and to confirm biofilm induction,biofilms were grown in microtitre plates without antibiotics as above and with a lid containing96polystyrene pegs (Nunc)such that a peg was inserted into each well.The pegs with adherent biofilms were removed after24h,washed three times in water,and placed into a new microtitre plate with fresh MHB supplemented with antibiotics.After static incubation for24h at378C,the pegs were removed,washed three times with water,inserted into a96-well plate containing fresh MHB,centrifuged at1,811g for20min,and then sonicated at approximately258C for5min(using a Branson1510water bath;Branson)to remove and disperse adherent cells for CFU determination as described21.Consistent removal of the biofilm was confirmed by crystal-violet staining of the pegs.For microtitre biofilm assays, reported values are the mean of at least three replicates(with the error calculated as s.e.m.).Student’s two-tailed t-test was used to establish the significance of differences between two means.In parallel with each biofilm experiment,an MIC was determined for the planktonic cells in the culture used as the inoculum. For all of the P.aeruginosa strains derived from PAO1,and for the E.coli strains tested,the MIC of tobramycin was consistently1m g ml21,including for resuspended biofilm-grown P.aeruginosa.Cloning of arr and complementation experiments.A PCR fragment contain-ing the gene PA2818(nucleotides3,170,500–3,174,000in the P.aeruginosa PAO1 genome)was cloned into the Eco RI and Hind III sites of pUCP18.The resulting plasmid was introduced into PAO1and the PA2818(arr)50022mutant by electroporation and selection for carbenicillin resistance using standard methods.These transformants were found to retain the plasmid for over36h of growth in the absence of selection.Therefore,biofilm experiments with the strain carrying the resulting plasmid with the wild-type arr gene were performed in the absence of carbenicillin,except as indicated.Measurement of c-di-GMP.Washed cell membranes were prepared and assayed for c-di-GMP degrading activity essentially as described28.Overnight cultures of arr mutant and wild-type PAO1were diluted1:100in MHB and grown statically at378C for18h.The cells were collected by centrifugation,the pellet resus-pended in TME buffer(50mM Tris-HCl pH8,0.9mM EDTA,10mM MgCl2) and the cells disrupted by sonication on ice.Unbroken cells were removed by centrifuging at3,000r.p.m.for5min.Membranes were separated from the supernatant by centrifuging at14,000r.p.m.for30min.The resulting pellets of washed membranes were resuspended in TME buffer,and protein concentration determined by a modified Lowry assay(BioRad).Aliquots of membrane corresponding to the indicated amount of protein were resuspended in200m l TME buffer with10m M c-di-GMP.The mixture was incubated for10min at 308C,and the reaction halted by boiling at1008C for2min.Membranes were removed by centrifugation at14,000r.p.m.for30min.The supernatant was analysed for c-di-GMP content as described in the Supplementary Methods. Received23March;accepted9June2005.1.Mah,T.F.et al.A genetic basis for Pseudomonas aeruginosa biofilm antibioticresistance.Nature426,306–-310(2003).2.Whiteley,M.et al.Gene expression in Pseudomonas aeruginosa biofilms.Nature413,860–-864(2001).3.D’Argenio,D.A.&Miller,S.I.Cyclic di-GMP as a bacterial second messenger.Microbiol.150,2497–-2502(2004).4.Ross,P.et al.The cyclic diguanylic acid regulatory system of cellulosesynthesis in Acetobacter xylinum.Chemical synthesis and biological activity of cyclic nucleotide dimer,trimer,and phosphothioate derivatives.J.Biol.Chem.265,18933–-18943(1990).5.Goh,E.B.et al.Transcriptional modulation of bacterial gene expression bysubinhibitory concentrations of antibiotics.Proc.Natl A99,17025–-17030(2002).6.Goodman,L.S.&Gilman,A.Goodman and Gilman’s The Pharmacological Basisof Therapeutics(Macmillan,New York,1985).7.Barclay,M.L.et al.Adaptive resistance to tobramycin in Pseudomonasaeruginosa lung infection in cysticfibrosis.J.Antimicrob.Chemother.37,1155–-1164(1996).8.Bergey,D.H.&Holt,J.G.Bergey’s Manual of Systematic Bacteriology(Williams&Wilkins,Baltimore,1984).9.Mukhopadhyay,S.et al.The quantitative distribution of nebulized antibiotic inthe lung in cysticfibrosis.Respir.Med.88,203–-211(1994).10.Ramsey,B.W.et al.Intermittent administration of inhaled tobramycin inpatients with cysticfibrosis.Cystic Fibrosis Inhaled Tobramycin Study Group.N.Engl.J.Med.340,23–-30(1999).11.Chiang,P.&Burrows,L.L.Biofilm formation by hyperpiliated mutants ofPseudomonas aeruginosa.J.Bacteriol.185,2374–-2378(2003).12.Rachid,S.,Ohlsen,K.,Witte,W.,Hacker,J.&Ziebuhr,W.Effect ofsubinhibitory antibiotic concentrations on polysaccharide intercellular adhesin expression in biofilm-forming Staphylococcus epidermidis.Antimicrob.AgentsChemother.44,3357–-3363(2000).13.Bagge,N.et al.Pseudomonas aeruginosa biofilms exposed to imipenem exhibitchanges in global gene expression and beta-lactamase and alginate production.Antimicrob.Agents Chemother.48,1175–-1187(2004).14.Paul,R.et al.Cell cycle-dependent dynamic localization of a bacterial responseregulator with a novel di-guanylate cyclase output domain.Genes Dev.18,715–-727(2004).15.Simm,R.,Morr,M.,Kader,A.,Nimtz,M.&Romling,U.GGDEF and EALdomains inversely regulate cyclic di-GMP levels and transition from sessility to motility.Mol.Microbiol.53,1123–-1134(2004).16.Galperin,M.Y.,Nikolskaya,A.N.&Koonin,E.V.Novel domains of theprokaryotic two-component signal transduction systems.FEMS Microbiol.Lett.203,11–-21(2001).17.Jacobs,M.A.et prehensive transposon mutant library of Pseudomonasaeruginosa.Proc.Natl A100,14339–-14344(2003).18.Bobrov,A.G.,Kirillina,O.&Perry,R.D.The phosphodiesterase activity of theHmsP EAL domain is required for negative regulation of biofilm formation in Yersinia pestis.FEMS Microbiol.Lett.247,123–-130(2005).19.Huang,B.,Whitchurch,C.B.&Mattick,J.S.FimX,a multidomain proteinconnecting environmental signals to twitching motility in Pseudomonasaeruginosa.J.Bacteriol.185,7068–-7076(2003).20.O’Toole,G.A.&Kolter,R.Flagellar and twitching motility are necessary forPseudomonas aeruginosa biofilm development.Mol.Microbiol.30,295–-304(1998).21.Moskowitz,S.M.,Foster,J.M.,Emerson,J.&Burns,J.L.Clinically feasiblebiofilm susceptibility assay for isolates of Pseudomonas aeruginosa frompatients with cysticfibrosis.J.Clin.Microbiol.42,1915–-1922(2004).22.Drenkard,E.&Ausubel,F.M.Pseudomonas biofilm formation and antibioticLETTERS NATURE|Vol436|25August2005 1174©2005Nature Publishing Group。
氨基糖苷类抗生素简介及应用
26
耐药机制
1.细菌产生氨基糖苷类钝化酶,这是临床菌株对 本类药物产生耐药性的最主要原因
Acetylase,AC Adenylase,AD Phosphorylase, P 乙酰化酶 腺苷化酶 磷酸化酶
27
ACⅡ
R1 HC
AC
ACⅠ
ACⅢ
ACⅡ
NHR2 O O NH2
NH2 NH2 OH
鼠疫、兔热病------首选 结核病---------------联合其他 细菌性心内膜炎------常合用青霉素 布鲁菌病
41
庆大霉素
38
注意事项
肾功能减退患者应用时,需调整给药剂量或给药间期
根据血浓 或按照内生肌酐清除率的改变
依替米星维持剂量=患者的Ccr/正常的Ccr*常规的维持剂量 阿米卡星:
Ccr 50-90ml/min:每12h给予正常剂量(7.5mg/kg)的60%-90% Ccr 10-50ml/min:每24-48h给予正常剂量(7.5mg/kg)的20%-30%
9
分布
消除
主要内容
结构和分类
药代动力学特征 抗菌谱及抗菌特点 作用机制 耐药情况及耐药机制
临床应用
不良反应
几种氨基糖苷类抗生素的特点
10
抗菌谱
对需氧G-杆菌作用强大
埃希菌属、变形杆菌属、克雷伯菌属、肠杆菌属、志贺菌属、枸橼酸菌属等
大部分对铜绿假单胞菌有效,除外:链霉素、大观霉素、卡那
23
抗菌机制总结
SNAP原理(v201007)
抗体+ 有催化底物能力的酶
SNAP® : 双流向酶联免疫技术
原理 阴性奶 Negative Milk
• 奶样中的β-内酰胺抗生素与试管内
β-内酰胺抗生素
阳性奶
的酶标抗体结合
• 未结合的酶标抗体将与事先包被了 青霉素类似物的样品点结合
• 显色剂中的底物遇到已结合到样品
点上的酶标抗体后,被酶催化而显 色
青霉素类
氨苄西林 Ampicillin 氯唑西林 Cloxacillin 苯唑西林(苯唑青霉素) Oxacillin 头孢氨苄 Cefalexin
头孢类
头孢喹肟 Cefquinome 头孢噻呋 Ceftiofur
Page 8
牛乳中最高残留限量规定 (其他抗生素)
农业部公告第235号 二00二年十二月二十四日
样品点和 对照点
ቤተ መጻሕፍቲ ባይዱ
勾刺
•
酶底物液池 吸湿块 洗涤液池
SNAP 试剂盒中的反应过程
3.样品流动, 反应窗口深蓝色变成 白色 1.加热样品被加入样品孔( β内酰胺抗生素和酶标抗体反应 后)
®
2.未结合的酶标抗体和样
4.按下反应键
品点抗原结合 7.控制点显色 7.样品点显色
5.洗涤液
6.酶反应底物
100 90 80 70 60 50 40 30 20 10 0
98
63
40 33 26 25 20 15
B-内酰胺
中药
氨基(庆大)
酶制剂
四环素
磺胺
林可
激素
Page 14
抗生素的使用 风险控制及抗生素分级建议
级别
使用比例
药物
青霉素G Benzylpenicillin 头孢氨苄 Cefalexin 阿莫西林 Amoxicillin 氨苄西林 Ampicillin 头孢噻呋Ceftiofur 庆大霉素Gentamycin 林可霉素Lincomycin 四环素类总量 Total Tetracyclines 磺胺类总量 Sulfonamides 苯唑西林(苯唑青霉素)Oxacillin 磺胺二甲嘧啶Sulfadimidine/Sulfamethazine 四环素 Tetracycline 金霉素 Chlortetracycline/ 土霉素 Oxytetracycline 氯霉素Chloramphenicol 链霉素/双氢链霉素 Streptomycin/Dihydrostreptomycin 大观霉素 Spectinomycin 新霉素Neomycin 达氟沙星 Danofloxacin 恩诺沙星 Enrofloxacin 红霉素Erythromycin 甲砜霉素Thiamphenicol 泰乐菌素Tylosin
- 1、下载文档前请自行甄别文档内容的完整性,平台不提供额外的编辑、内容补充、找答案等附加服务。
- 2、"仅部分预览"的文档,不可在线预览部分如存在完整性等问题,可反馈申请退款(可完整预览的文档不适用该条件!)。
- 3、如文档侵犯您的权益,请联系客服反馈,我们会尽快为您处理(人工客服工作时间:9:00-18:30)。
Cap 139N dihydrostreptomycinand streptomycin三 摘要:本文主要介绍了Cap 139N dihydrostreptomycinand streptomycin 的主要内容。
Cap 139N s 10 Order to recall food animals (1) The Director may order any food animal farmer or food animal trader who has supplied the food animals in respect of which a suspension order is in force to withdraw immediately other food animals of the same batch already supplied and to retrieve, in the manner and to the extent reasonably possible, those animals already supplied.
(2) The food animals withdrawn or retrieved under subsection (1) shall be disposed of in such manner as the Director may direct.
Cap 139N s 11 Possession of prohibited chemicals, etc. (1) Subject to section 17(7), a food animal farmer who has in his possession or under his control any prohibited chemical commits an offence.
(2) Subject to section 17(8), a food animal farmer who has in his possession or under his control fodder containing or mixed with any prohibited chemical commits an offence. (3) Subject to section 17(7), a food animal trader who knowingly and wilfully has in his possession or under his control any prohibited chemical commits an offence.
(4) Subject to section 17(8), a food animal trader who knowingly and wilfully has in his possession or under his control fodder containing or mixed with any prohibited chemical commits an offence.
Cap 139N s 12 Supply of prohibited chemicals, etc. (1) Subject to section 17(7), a person commits an offence if he supplies or offers to supply any prohibited chemical to any person whom he knows or has reason to believe is a food animal farmer or food animal trader.
(2) A person commits an offence if he supplies or offers to supply fodder containing or mixed with any prohibited chemical to any person whom he knows or has reason to believe is a food animal farmer or food animal trader.
Cap 139N s 13 Information in respect of fodder (1) No person shall supply or offer to supply any fodder which contains or is mixed with any agricultural and veterinary chemical unless the information set out in subsection (2) is- (a) in the case of the fodder being contained in a package, legibly displayed, in both the English and the Chinese languages, in a conspicuous position on the package; and
(b) in any other case, supplied together with the fodder in both the English and the Chinese languages.(2) The information referred to in subsection (1) is-
(a) a list of all the agricultural and veterinary chemicals contained or mixed in the fodder and their respective amounts;
(b) the instructions for use of the fodder; (c) the withholding period, that is to say, the period of time between feeding the food animal with the fodder for the last time and slaughtering; and
(d) the name and address of the fodder supplier.(3) A person commits an offence if he, in purported compliance with the requirement to provide information imposed by subsection (1), provides any information that is incorrect, false or misleading in a material particular and he knows it to be incorrect, false or misleading in a material particular.
Cap 139N s 14 Order to suspend supply of fodder (1) Upon receipt of reports or information from the Public Analyst or other sources that-
(a) any fodder is suspected of containing- (i) prohibited chemicals; (ii) agricultural and veterinary chemicals at a level that is likely to endanger animals or the health of any person; or
(iii) any other substance that is likely to endanger animals or the health of any person; or(b) any fodder is suspected of being supplied without the information required to be furnished under section 13, or where the information is furnished, the information is incorrect, false, misleading or insufficient,the Director may make an order requiring any person who supplies the fodder to suspend the supply of such fodder forthwith for such period as may be reasonably necessary if he considers it in the public interest to do so.
(2) The senior veterinary officer, or any person acting under his direction, may destroy or order the forfeiture of any of the fodder in respect of which an order has been made under subsection (1).
