6Synthesis and Microtubule Binding of Fluorescent Paclitaxel
人类所有持家基因列表

所有的看家基因的列表,大家在设计引物时,可以直接采用这些基因的序列,本表来自于Trends in Gentics上一篇文献,内容全面,所有的看家基因均标注了基因银行号(Genbank),可以直接到pubme d中查询得出。
引物设计可以推荐用primer 5,也可以直接与我们联系设计,有关事宜,请点击:/service/meeting/200506/130599.html(引物设计与合成服务)。
List of housekeeping genes derived from the article "Human Housekeeping genes are compact," published in Trends in Genetics19, 362-365 (2003).Each gene name/description is followed by its geometric average expression level accordi ng to the data published by Su et al. Genes designated by asterisk are in popular use as reference in real-time PCR.*NM_001101 Homo sapiens actin, beta (ACTB), mRNA 6988*NM_000034 Homo sapiens aldolase A,fructose-bisphosphate (ALDOA), mRNA 3425*NM_002046 Homo sapiens glyceraldehyde-3-phosphate dehydrogenase (GAPD), mRNA 828 *NM_000291 Homo sapiens phosphoglycerate kinase 1 (PGK1), mRNA 2727*NM_005566 Homo sapiens lactate dehydrogenase A (LDHA), mRNA 2105*NM_002954 Homo sapiens ribosomal protein S27a (RPS27A), mRNA 4156*NM_000981 Homo sapiens ribosomal protein L19 (RPL19), mRNA 6997*NM_000975 Homo sapiens ribosomal protein L11 (RPL11), mRNA 6060*NM_007363 Homo sapiens non-POU domain containing, octamer-binding (NONO), mRNA 1708*NM_004309 Homo sapiens Rho GDP dissociation inhibitor (GDI) alpha (ARHGDIA), mRNA 1358*NM_000994 Homo sapiens ribosomal protein L32 (RPL32), mRNA 9523*NM_022551 Homo sapiens ribosomal protein S18 (RPS18), mRNA 11261*NM_007355 Homo sapiens heat shock 90kDa protein 1, beta (HSPCB), mRNA 4119NM_004515 Homo sapiens interleukin enhancer binding factor 2, 45kDa (ILF2), mRNA 1000NM_004651 Homo sapiens ubiquitin specific protease 11 (USP11), mRNA 1950NM_004888 Homo sapiens ATPase, H+ transporting, lysosomal 13kDa, V1 subunit G isof orm 1 (ATP6V1G1), mRNA 928NM_003334 Homo sapiens ubiquitin-activating enzyme E1 (A1S9T and BN75 temperature sensitivity complementing) (UBE1), transcript variant 1, mRNA 1351NM_001320 Homo sapiens casein kinase 2, beta polypeptide (CSNK2B), mRNA 1233 NM_003915 Homo sapiens copine I (CPNE1), transcript variant 3, mRNA 698NM_001250 Homo sapiens tumor necrosis factor receptor superfamily, member 5 (TNFRS F5), transcript variant 1, mRNA 524NM_001904 Homo sapiens catenin (cadherin-associated protein), beta 1, 88kDa (CTNNB 1), mRNA 1165NM_003753 Homo sapiens eukaryotic translation initiation factor 3, subunit 7 zeta, 66/ 67kDa (EIF3S7), mRNA 1363NM_004541 Homo sapiens NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 1, 7. 5kDa (NDUFA1), nuclear gene encoding mitochondrial protein, mRNA 2307NM_001654 Homo sapiens v-raf murine sarcoma 3611 viral oncogene homolog 1 (ARAF 1), mRNA 846NM_002967 Homo sapiens scaffold attachment factor B (SAFB), mRNA 545NM_001183 Homo sapiens ATPase, H+ transporting, lysosomal interacting protein 1 (ATP 6IP1), mRNA 1089NM_003526 Homo sapiens H2B histone family, member L (H2BFL), mRNA 2232NM_004718 Homo sapiens cytochrome c oxidase subunit VIIa polypeptide 2 like (COX7 A2L), nuclear gene encoding mitochondrial protein, mRNA 1013NM_004436 Homo sapiens endosulfine alpha (ENSA), mRNA 659NM_001207 Homo sapiens basic transcription factor 3 (BTF3), mRNA 3348NM_004907 Homo sapiens immediate early protein (ETR101), mRNA 1249NM_004889 Homo sapiens ATP synthase, H+ transporting, mitochondrial F0 complex, sub unit f, isoform 2 (ATP5J2), mRNA 1513NM_003769 Homo sapiens splicing factor, arginine/serine-rich 9 (SFRS9), mRNA 1207NM_003910 Homo sapiens maternal G10 transcript (G10), mRNA 696NM_000100 Homo sapiens cystatin B (stefin B) (CSTB), mRNA 1422NM_004785 Homo sapiens solute carrier family 9 (sodium/hydrogen exchanger), isoform 3 regulatory factor 2 (SLC9A3R2), mRNA 455NM_001120 Homo sapiens tetracycline transporter-like protein (TETRAN), mRNA 935NM_000182 Homo sapiens hydroxyacyl-Coenzyme A dehydrogenase/3-ketoacyl-Coenzyme A thiolase/enoyl-Coenzyme A hydratase (trifunctional protein), alpha subunit (HADHA),mRNA 609NM_003377 Homo sapiens vascular endothelial growth factor B (VEGFB), mRNA 649NM_003576 Homo sapiens serine/threonine kinase 24 (STE20 homolog, yeast) (STK24), mRNA 769NM_000918 Homo sapiens procollagen-proline, 2-oxoglutarate 4-dioxygenase (proline 4-h ydroxylase), beta polypeptide (protein disulfide isomerase; thyroid hormone binding prot ein p55) (P4HB), mRNA 2719NM_004584 Homo sapiens RAD9 homolog (S. pombe) (RAD9), mRNA 860NM_004952 Homo sapiens ephrin-A3 (EFNA3), mRNA 505NM_004308 Homo sapiens Rho GTPase activating protein 1 (ARHGAP1), mRNA 893NM_003190 Homo sapiens TAP binding protein (tapasin) (TAPBP), mRNA 1213NM_004640 Homo sapiens HLA-B associated transcript 1 (BAT1), transcript variant 1, m RNA 1022NM_001064 Homo sapiens transketolase (Wernicke-Korsakoff syndrome) (TKT), mRNA 1139NM_002117 Homo sapiens major histocompatibility complex, class I, C (HLA-C), mRNA 4696NM_004161 Homo sapiens RAB1A, member RAS oncogene family (RAB1A), mRNA 216 3NM_003339 Homo sapiens ubiquitin-conjugating enzyme E2D 2 (UBC4/5 homolog, yeast) (UBE2D2), mRNA 514NM_003969 Homo sapiens ubiquitin-conjugating enzyme E2M (UBC12 homolog, yeast)(UBE2M), mRNA 1197NM_000516 Homo sapiens GNAS complex locus (GNAS), transcript variant 1, mRNA 4358NM_002819 Homo sapiens polypyrimidine tract binding protein 1 (PTBP1), transcript vari ant 1, mRNA 1138NM_001001 Homo sapiens ribosomal protein L36a-like (RPL36AL), mRNA 1740NM_004649 Homo sapiens chromosome 21 open reading frame 33 (C21orf33), mRNA 585NM_000175 Homo sapiens glucose phosphate isomerase (GPI), mRNA 1633NM_001867 Homo sapiens cytochrome c oxidase subunit VIIc (COX7C), nuclear gene en coding mitochondrial protein, mRNA 3004NM_001967 Homo sapiens eukaryotic translation initiation factor 4A, isoform 2 (EIF4A2), mRNA 2935NM_001863 Homo sapiens cytochrome c oxidase subunit VIb (COX6B), nuclear gene en coding mitochondrial protein, mRNA 1719NM_001997 Homo sapiens Finkel-Biskis-Reilly murine sarcoma virus (FBR-MuSV) ubiqui tously expressed (fox derived); ribosomal protein S30 (FAU), mRNA 3898NM_002088 Homo sapiens glutamate receptor, ionotropic, kainate 5 (GRIK5), mRNA 535NM_001862 Homo sapiens cytochrome c oxidase subunit Vb (COX5B), nuclear gene enc oding mitochondrial protein, mRNA 1087NM_004255 Homo sapiens cytochrome c oxidase subunit Va (COX5A), nuclear gene enc oding mitochondrial protein, mRNA 853NM_001788 Homo sapiens CDC10 cell division cycle 10 homolog (S. cerevisiae) (CDC1 0), mRNA 1340NM_004781 Homo sapiens vesicle-associated membrane protein 3 (cellubrevin) (V AMP3), mRNA 684NM_003801 Homo sapiens GPAA1P anchor attachment protein 1 homolog (yeast) (GPAA 1), mRNA 776NM_004643 Homo sapiens poly(A) binding protein, nuclear 1 (PABPN1), mRNA 502NM_001537 Homo sapiens heat shock factor binding protein 1 (HSBP1), mRNA 874NM_003680 Homo sapiens tyrosyl-tRNA synthetase (YARS), mRNA 535NM_003345 Homo sapiens ubiquitin-conjugating enzyme E2I (UBC9 homolog, yeast) (UB E2I), mRNA 1283NM_002568 Homo sapiens poly(A) binding protein, cytoplasmic 1 (PABPC1), mRNA 319 9NM_001487 Homo sapiens GCN5 general control of amino-acid synthesis 5-like 1 (yeast) (GCN5L1), mRNA 313NM_001861 Homo sapiens cytochrome c oxidase subunit IV isoform 1 (COX4I1), nuclea r gene encoding mitochondrial protein, mRNA 2738NM_004890 Homo sapiens sperm associated antigen 7 (SPAG7), mRNA 618NM_002812 Homo sapiens proteasome (prosome, macropain) 26S subunit, non-ATPase, 8 (PSMD8), mRNA 942NM_004926 Homo sapiens zinc finger protein 36, C3H type-like 1 (ZFP36L1), mRNA 1168NM_002539 Homo sapiens ornithine decarboxylase 1 (ODC1), mRNA 1361NM_000979 Homo sapiens ribosomal protein L18 (RPL18), mRNA 4417NM_000977 Homo sapiens ribosomal protein L13 (RPL13), transcript variant 1, mRNA 6407NM_001015 Homo sapiens ribosomal protein S11 (RPS11), mRNA 7614NM_001760 Homo sapiens cyclin D3 (CCND3), mRNA 676NM_003973 Homo sapiens ribosomal protein L14 (RPL14), mRNA 3135NM_002815 Homo sapiens proteasome (prosome, macropain) 26S subunit, non-ATPase, 11 (PSMD11), mRNA 536NM_000367 Homo sapiens thiopurine S-methyltransferase (TPMT), mRNA 1574NM_000973 Homo sapiens ribosomal protein L8 (RPL8), transcript variant 1, mRNA 813 8NM_004689 Homo sapiens metastasis associated 1 (MTA1), mRNA 506NM_001848 Homo sapiens collagen, type VI, alpha 1 (COL6A1), mRNA 757NM_004068 Homo sapiens adaptor-related protein complex 2, mu 1 subunit (AP2M1), m RNA 2188NM_001687 Homo sapiens ATP synthase, H+ transporting, mitochondrial F1 complex, del ta subunit (ATP5D), mRNA 1167NM_004197 Homo sapiens serine/threonine kinase 19 (STK19), transcript variant 1, mRN A 574NM_001028 Homo sapiens ribosomal protein S25 (RPS25), mRNA 4683NM_001022 Homo sapiens ribosomal protein S19 (RPS19), mRNA 6683NM_004759 Homo sapiens mitogen-activated protein kinase-activated protein kinase 2 (M APKAPK2), transcript variant 1, mRNA 641NM_001623 Homo sapiens allograft inflammatory factor 1 (AIF1), transcript variant 3, m RNA 497NM_004894 Homo sapiens chromosome 14 open reading frame 2 (C14orf2), mRNA 704NM_002375 Homo sapiens microtubule-associated protein 4 (MAP4), transcript variant 1, mRNA 717NM_001013 Homo sapiens ribosomal protein S9 (RPS9), mRNA 6868NM_003779 Homo sapiens UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypepti de 3 (B4GALT3), mRNA 565NM_001296 Homo sapiens chemokine binding protein 2 (CCBP2), mRNA 394NM_001009 Homo sapiens ribosomal protein S5 (RPS5), mRNA 6739NM_003021 Homo sapiens small glutamine-rich tetratricopeptide repeat (TPR)-containing (SGT), mRNA 488NM_004285 Homo sapiens hexose-6-phosphate dehydrogenase (glucose 1-dehydrogenase) (H6PD), mRNA 646NM_004142 Homo sapiens matrix metalloproteinase-like 1 (MMPL1), mRNA 695NM_001950 Homo sapiens E2F transcription factor 4, p107/p130-binding (E2F4), mRNA 956NM_003815 Homo sapiens a disintegrin and metalloproteinase domain 15 (metargidin) (A DAM15), mRNA 771NM_001119 Homo sapiens adducin 1 (alpha) (ADD1), transcript variant 1, mRNA 135 6NM_001111 Homo sapiens adenosine deaminase, RNA-specific (ADAR), transcript variant ADAR-a, mRNA 1036NM_003466 Homo sapiens paired box gene 8 (PAX8), transcript variant PAX8A, mRNA 901NM_001155 Homo sapiens annexin A6 (ANXA6), transcript variant 1, mRNA 718NM_003465 Homo sapiens chitinase 1 (chitotriosidase) (CHIT1), mRNA 561NM_003186 Homo sapiens transgelin (TAGLN), mRNA 1209NM_000802 Homo sapiens folate receptor 1 (adult) (FOLR1), transcript variant 2, mRNA 514NM_004924 Homo sapiens actinin, alpha 4 (ACTN4), mRNA 1187NM_002931 Homo sapiens ring finger protein 1 (RING1), mRNA 576NM_000020 Homo sapiens activin A receptor type II-like 1 (ACVRL1), mRNA 849NM_001785 Homo sapiens cytidine deaminase (CDA), mRNA 391NM_004339 Homo sapiens pituitary tumor-transforming 1 interacting protein (PTTG1IP), mRNA 1200NM_003860 Homo sapiens Breakpoint cluster region protein, uterine leiomyoma, 1; barrie r to autointegration factor (BCRP1), mRNA 1303NM_000214 Homo sapiens jagged 1 (Alagille syndrome) (JAG1), mRNA 536NM_002167 Homo sapiens inhibitor of DNA binding 3, dominant negative helix-loop-helix protein (ID3), mRNA 1192NM_001664 Homo sapiens ras homolog gene family, member A (ARHA), mRNA 4050NM_003166 Homo sapiens sulfotransferase family, cytosolic, 1A, phenol-preferring, memb er 3 (SULT1A3), mRNA 461NM_001746 Homo sapiens calnexin (CANX), mRNA 1923NM_001662 Homo sapiens ADP-ribosylation factor 5 (ARF5), mRNA 724NM_001660 Homo sapiens ADP-ribosylation factor 4 (ARF4), mRNA 1014NM_001658 Homo sapiens ADP-ribosylation factor 1 (ARF1), mRNA 2195NM_003313 Homo sapiens tissue specific transplantation antigen P35B (TSTA3), mRNA 440NM_001494 Homo sapiens GDP dissociation inhibitor 2 (GDI2), mRNA 1352NM_003145 Homo sapiens signal sequence receptor, beta (translocon-associated protein be ta) (SSR2), mRNA 1265NM_001619 Homo sapiens adrenergic, beta, receptor kinase 1 (ADRBK1), mRNA 695NM_001420 Homo sapiens ELA V (embryonic lethal, abnormal vision, Drosophila)-like 3 (Hu antigen C) (ELA VL3), mRNA 1070NM_004930 Homo sapiens capping protein (actin filament) muscle Z-line, beta (CAPZB), mRNA 1183NM_004596 Homo sapiens small nuclear ribonucleoprotein polypeptide A (SNRPA), mRN A 734NM_004168 Homo sapiens succinate dehydrogenase complex, subunit A, flavoprotein (Fp)(SDHA), nuclear gene encoding mitochondrial protein, mRNA 1106NM_004156 Homo sapiens protein phosphatase 2 (formerly 2A), catalytic subunit, beta isoform (PPP2CB), mRNA 1195NM_004910 Homo sapiens phosphatidylinositol transfer protein, membrane-associated (P I TPNM), mRNA 869NM_004517 Homo sapiens integrin-linked kinase (ILK), mRNA 654NM_004494 Homo sapiens hepatoma-derived growth factor (high-mobility group protein 1 -like) (HDGF), mRNA 1385NM_004121 Homo sapiens gamma-glutamyltransferase-like activity 1 (GGTLA1), mRNA 412NM_004404 Homo sapiens neural precursor cell expressed, developmentally down-regulate d 5 (NEDD5), mRNA 1571NM_004394 Homo sapiens death-associated protein (DAP), mRNA 623NM_004383 Homo sapiens c-src tyrosine kinase (CSK), mRNA 899NM_004074 Homo sapiens cytochrome c oxidase subunit VIII (COX8), nuclear gene enc oding mitochondrial protein, mRNA 3188NM_004039 Homo sapiens annexin A2 (ANXA2), mRNA 2417NM_001053 Homo sapiens somatostatin receptor 5 (SSTR5), mRNA 423NM_001328 Homo sapiens C-terminal binding protein 1 (CTBP1), mRNA 798NM_001273 Homo sapiens chromodomain helicase DNA binding protein 4 (CHD4), mRN A 775NM_003430 Homo sapiens zinc finger protein 91 (HPF7, HTF10) (ZNF91), mRNA 716NM_003314 Homo sapiens tetratricopeptide repeat domain 1 (TTC1), mRNA 651NM_003217 Homo sapiens testis enhanced gene transcript (TEGT), mRNA 1766NM_003132 Homo sapiens spermidine synthase (SRM), mRNA 1093NM_000199 Homo sapiens N-sulfoglucosamine sulfohydrolase (sulfamidase) (SGSH), mRN A 565NM_002818 Homo sapiens proteasome (prosome, macropain) activator subunit 2 (PA28 be ta) (PSME2), mRNA 697NM_002733 Homo sapiens protein kinase, AMP-activated, gamma 1 non-catalytic subunit (PRKAG1), mRNA 497NM_002631 Homo sapiens phosphogluconate dehydrogenase (PGD), mRNA 1009NM_002574 Homo sapiens peroxiredoxin 1 (PRDX1), mRNA 2241NM_002512 Homo sapiens non-metastatic cells 2, protein (NM23B) expressed in (NME2), nuclear gene encoding mitochondrial protein, mRNA 2553NM_002455 Homo sapiens metaxin 1 (MTX1), mRNA 845NM_002444 Homo sapiens moesin (MSN), mRNA 1798NM_000529 Homo sapiens melanocortin 2 receptor (adrenocorticotropic hormone) (MC2R), mRNA 420NM_003573 Homo sapiens latent transforming growth factor beta binding protein 4 (LTB P4), mRNA 740NM_002315 Homo sapiens LIM domain only 1 (rhombotin 1) (LMO1), mRNA 469NM_000884 Homo sapiens IMP (inosine monophosphate) dehydrogenase 2 (IMPDH2), m RNA 1288NM_003641 Homo sapiens interferon induced transmembrane protein 1 (9-27) (IFITM1), mRNA 4898NM_000841 Homo sapiens glutamate receptor, metabotropic 4 (GRM4), mRNA 1369NM_002070 Homo sapiens guanine nucleotide binding protein (G protein), alpha inhibitin g activity polypeptide 2 (GNAI2), mRNA 1877NM_001493 Homo sapiens GDP dissociation inhibitor 1 (GDI1), mRNA 1387NM_002048 Homo sapiens growth arrest-specific 1 (GAS1), mRNA 940NM_002032 Homo sapiens ferritin, heavy polypeptide 1 (FTH1), mRNA 2616NM_001418 Homo sapiens eukaryotic translation initiation factor 4 gamma, 2 (EIF4G2), mRNA 3646NM_001350 Homo sapiens death-associated protein 6 (DAXX), mRNA 510NM_001843 Homo sapiens contactin 1 (CNTN1), mRNA 535NM_001728 Homo sapiens basigin (BSG), mRNA 1508NM_001667 Homo sapiens ADP-ribosylation factor-like 2 (ARL2), mRNA 965NM_001659 Homo sapiens ADP-ribosylation factor 3 (ARF3), mRNA 1327NM_003746 Homo sapiens dynein, cytoplasmic, light polypeptide 1 (DNCL1), mRNA 275 8NM_002127 Homo sapiens HLA-G histocompatibility antigen, class I, G (HLA-G), mRN A 1090NM_004712 Homo sapiens hepatocyte growth factor-regulated tyrosine kinase substrate (H GS), mRNA 505NM_003475 Homo sapiens chromosome 11 open reading frame 13 (C11orf13), mRNA 413NM_004046 Homo sapiens ATP synthase, H+ transporting, mitochondrial F1 complex, alp ha subunit, isoform 1, cardiac muscle (ATP5A1), mRNA 1348NM_001894 Homo sapiens casein kinase 1, epsilon (CSNK1E), transcript variant 2, mRN A 613NM_003795 Homo sapiens sorting nexin 3 (SNX3), transcript variant 1, mRNA 1426NM_001909 Homo sapiens cathepsin D (lysosomal aspartyl protease) (CTSD), mRNA 151 2NM_002792 Homo sapiens proteasome (prosome, macropain) subunit, alpha type, 7 (PSM A7), transcript variant 1, mRNA 728NM_002799 Homo sapiens proteasome (prosome, macropain) subunit, beta type, 7 (PSMB 7), mRNA 545NM_002300 Homo sapiens lactate dehydrogenase B (LDHB), mRNA 4144NM_004176 Homo sapiens sterol regulatory element binding transcription factor 1 (SREB F1), mRNA 632NM_002796 Homo sapiens proteasome (prosome, macropain) subunit, beta type, 4 (PSMB 4), mRNA 1229NM_002794 Homo sapiens proteasome (prosome, macropain) subunit, beta type, 2 (PSMB 2), mRNA 1304NM_002793 Homo sapiens proteasome (prosome, macropain) subunit, beta type, 1 (PSMB1), mRNA 2084NM_002473 Homo sapiens myosin, heavy polypeptide 9, non-muscle (MYH9), mRNA 138 1NM_001810 Homo sapiens centromere protein B, 80kDa (CENPB), mRNA 532NM_002624 Homo sapiens prefoldin 5 (PFDN5), transcript variant 1, mRNA 1718NM_004710 Homo sapiens synaptogyrin 2 (SYNGR2), mRNA 1414NM_001127 Homo sapiens adaptor-related protein complex 1, beta 1 subunit (AP1B1), t r a nscript variant 1, mRNA 873NM_002107 Homo sapiens H3 histone, family 3A (H3F3A), mRNA 9328NM_003899 Homo sapiens Rho guanine nucleotide exchange factor (GEF) 7 (ARHGEF7), transcript variant 1, mRNA 558NM_003406 Homo sapiens tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activati on protein, zeta polypeptide (YWHAZ), transcript variant 1, mRNA 2008NM_002419 Homo sapiens mitogen-activated protein kinase kinase kinase 11 (MAP3K11), mRNA 535NM_001130 Homo sapiens amino-terminal enhancer of split (AES), mRNA 2395NM_003379 Homo sapiens villin 2 (ezrin) (VIL2), mRNA 1356NM_002636 Homo sapiens PHD finger protein 1 (PHF1), transcript variant 1, mRNA 640NM_002622 Homo sapiens prefoldin 1 (PFDN1), mRNA 658NM_001823 Homo sapiens creatine kinase, brain (CKB), mRNA 1407NM_003405 Homo sapiens tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, eta polypeptide (YWHAH), mRNA 2195NM_002939 Homo sapiens ribonuclease/angiogenin inhibitor (RNH), mRNA 726NM_003562 Homo sapiens solute carrier family 25 (mitochondrial carrier; oxoglutarate c a r rier), member 11 (SLC25A11), mRNA 517NM_001916 Homo sapiens cytochrome c-1 (CYC1), mRNA 895NM_002823 Homo sapiens prothymosin, alpha (gene sequence 28) (PTMA), mRNA 372 3NM_003096 Homo sapiens small nuclear ribonucleoprotein polypeptide G (SNRPG), mRN A 687NM_003321 Homo sapiens Tu translation elongation factor, mitochondrial (TUFM), mRN A 1543NM_003404 Homo sapiens tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activati on protein, beta polypeptide (YWHAB), transcript variant 1, mRNA 1144NM_002946 Homo sapiens replication protein A2, 32kDa (RPA2), mRNA 537NM_004356 Homo sapiens CD81 antigen (target of antiproliferative antibody 1) (CD81), mRNA 3638NM_001743 Homo sapiens calmodulin 2 (phosphorylase kinase, delta) (CALM2), mRNA 4675NM_004231 Homo sapiens ATPase, H+ transporting, lysosomal 14kDa, V1 subunit F (AT P6V1F), mRNA 997NM_004893 Homo sapiens H2A histone family, member Y (H2AFY), transcript variant 2, mRNA 788NM_004146 Homo sapiens NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 7, 18k Da (NDUFB7), mRNA 967NM_002128 Homo sapiens high-mobility group box 1 (HMGB1), mRNA 1295NM_002002 Homo sapiens Fc fragment of IgE, low affinity II, receptor for (CD23A) (F CER2), mRNA 969NM_000858 Homo sapiens guanylate kinase 1 (GUK1), mRNA 1166NM_001469 Homo sapiens thyroid autoantigen 70kDa (Ku antigen) (G22P1), mRNA 168 4NM_003766 Homo sapiens beclin 1 (coiled-coil, myosin-like BCL2 interacting protein) (B ECN1), mRNA 948NM_003906 Homo sapiens MCM3 minichromosome maintenance deficient 3 (S. cerevisia e) associated protein (MCM3AP), mRNA 430NM_000757 Homo sapiens colony stimulating factor 1 (macrophage) (CSF1), mRNA 797NM_002149 Homo sapiens hippocalcin-like 1 (HPCAL1), transcript variant 1, mRNA 102 6NM_001694 Homo sapiens ATPase, H+ transporting, lysosomal 16kDa, V0 subunit c (AT P6V0C), mRNA 2105NM_004047 Homo sapiens ATPase, H+ transporting, lysosomal 21kDa, V0 subunit c'' (A TP6V0B), mRNA 1212NM_001696 Homo sapiens ATPase, H+ transporting, lysosomal 31kDa, V1 subunit E isof orm 1 (ATP6V1E1), mRNA 784NM_001865 Homo sapiens cytochrome c oxidase subunit VIIa polypeptide 2 (liver) (COX7A2), nuclear gene encoding mitochondrial protein, mRNA 1818NM_004373 Homo sapiens cytochrome c oxidase subunit VIa polypeptide 1 (COX6A1), nuclear gene encoding mitochondrial protein, mRNA 5529NM_000801 Homo sapiens FK506 binding protein 1A, 12kDa (FKBP1A), transcript varia nt 12B, mRNA 4273NM_000992 Homo sapiens ribosomal protein L29 (RPL29), mRNA 6060NM_000988 Homo sapiens ribosomal protein L27 (RPL27), mRNA 6101NM_001004 Homo sapiens ribosomal protein, large P2 (RPLP2), mRNA 5924NM_001003 Homo sapiens ribosomal protein, large, P1 (RPLP1), mRNA 10300NM_000405 Homo sapiens GM2 ganglioside activator protein (GM2A), mRNA 1250NM_000967 Homo sapiens ribosomal protein L3 (RPL3), mRNA 7416NM_001428 Homo sapiens enolase 1, (alpha) (ENO1), mRNA 3668NM_000999 Homo sapiens ribosomal protein L38 (RPL38), mRNA 8302NM_000997 Homo sapiens ribosomal protein L37 (RPL37), mRNA 6689NM_000995 Homo sapiens ribosomal protein L34 (RPL34), transcript variant 1, mRNA 5424NM_002948 Homo sapiens ribosomal protein L15 (RPL15), mRNA 5450NM_002952 Homo sapiens ribosomal protein S2 (RPS2), mRNA 8825NM_001026 Homo sapiens ribosomal protein S24 (RPS24), transcript variant 2, mRNA 5701NM_001020 Homo sapiens ribosomal protein S16 (RPS16), mRNA 7477NM_001018 Homo sapiens ribosomal protein S15 (RPS15), mRNA 6261NM_001017 Homo sapiens ribosomal protein S13 (RPS13), mRNA 5430NM_000969 Homo sapiens ribosomal protein L5 (RPL5), mRNA 4653NM_000985 Homo sapiens ribosomal protein L17 (RPL17), mRNA 4369NM_000937 Homo sapiens polymerase (RNA) II (DNA directed) polypeptide A, 220kDa (POLR2A), mRNA 753NM_001016 Homo sapiens ribosomal protein S12 (RPS12), mRNA 8265NM_002140 Homo sapiens heterogeneous nuclear ribonucleoprotein K (HNRPK), transcrip t variant 1, mRNA 2429NM_002138 Homo sapiens heterogeneous nuclear ribonucleoprotein D (AU-rich element R NA binding protein 1, 37kDa) (HNRPD), transcript variant 3, mRNA 1157NM_004499 Homo sapiens heterogeneous nuclear ribonucleoprotein A/B (HNRPAB), trans cript variant 2, mRNA 654NM_001014 Homo sapiens ribosomal protein S10 (RPS10), mRNA 8074NM_002383 Homo sapiens MYC-associated zinc finger protein (purine-binding transcriptio n factor) (MAZ), mRNA 2580NM_002467 Homo sapiens v-myc myelocytomatosis viral oncogene homolog (avian) (MY C), mRNA 537NM_001436 Homo sapiens fibrillarin (FBL), mRNA 1408NM_004069 Homo sapiens adaptor-related protein complex 2, sigma 1 subunit (AP2S1),transcript variant AP17, mRNA 979NM_001614 Homo sapiens actin, gamma 1 (ACTG1), mRNA 6560NM_002355 Homo sapiens mannose-6-phosphate receptor (cation dependent) (M6PR), mR NA 388NM_004597 Homo sapiens small nuclear ribonucleoprotein D2 polypeptide 16.5kDa (SNR PD2), mRNA 1136NM_002308 Homo sapiens lectin, galactoside-binding, soluble, 9 (galectin 9) (LGALS9), transcript variant short, mRNA 888NM_000398 Homo sapiens diaphorase (NADH) (cytochrome b-5 reductase) (DIA1), nucle ar gene encoding mitochondrial protein, transcript variant M, mRNA 2190NM_000754 Homo sapiens catechol-O-methyltransferase (COMT), transcript variant MB-C OMT, mRNA 845NM_002406 Homo sapiens mannosyl (alpha-1,3-)-glycoprotein beta-1,2-N-acetylglucosami nyltransferase (MGAT1), mRNA 893NM_003752 Homo sapiens eukaryotic translation initiation factor 3, subunit 8, 110kDa (E I F3S8), mRNA 1871NM_001355 Homo sapiens D-dopachrome tautomerase (DDT), mRNA 631NM_004960 Homo sapiens fusion, derived from t(12;16) malignant liposarcoma (FUS), m RNA 1019NM_004729 Homo sapiens Ac-like transposable element (ALTE), mRNA 963NM_004587 Homo sapiens ribosome binding protein 1 homolog 180kDa (dog) (RRBP1), mRNA 658NM_004552 Homo sapiens NADH dehydrogenase (ubiquinone) Fe-S protein 5, 15kDa (N ADH-coenzyme Q reductase) (NDUFS5), mRNA 936NM_004450 Homo sapiens enhancer of rudimentary homolog (Drosophila) (ERH), mRNA 1028NM_004048 Homo sapiens beta-2-microglobulin (B2M), mRNA 4992NM_000239 Homo sapiens lysozyme (renal amyloidosis) (L YZ), mRNA 796NM_000269 Homo sapiens non-metastatic cells 1, protein (NM23A) expressed in (NME1), mRNA 887NM_000431 Homo sapiens mevalonate kinase (mevalonic aciduria) (MVK), mRNA 753NM_001247 Homo sapiens ectonucleoside triphosphate diphosphohydrolase 6 (putative fun ction) (ENTPD6), mRNA 495NM_003365 Homo sapiens ubiquinol-cytochrome c reductase core protein I (UQCRC1), mRNA 1026NM_003329 Homo sapiens thioredoxin (TXN), mRNA 1002NM_001069 Homo sapiens tubulin, beta polypeptide (TUBB), mRNA 1013NM_000356 Homo sapiens Treacher Collins-Franceschetti syndrome 1 (TCOF1), mRNA 673NM_003134 Homo sapiens signal recognition particle 14kDa (homologous Alu RNA bindi ng protein) (SRP14), mRNA 2911NM_003131 Homo sapiens serum response factor (c-fos serum response element-binding transcription factor) (SRF), mRNA 664NM_000454 Homo sapiens superoxide dismutase 1, soluble (amyotrophic lateral sclerosis1 (adult)) (SOD1), mRNA 1739NM_003091 Homo sapiens small nuclear ribonucleoprotein polypeptides B and B1 (SNRP B), mRNA 1609NM_003089 Homo sapiens small nuclear ribonucleoprotein 70kDa polypeptide (RNP antig en) (SNRP70), mRNA 1672NM_003016 Homo sapiens splicing factor, arginine/serine-rich 2 (SFRS2), mRNA 1476NM_003952 Homo sapiens ribosomal protein S6 kinase, 70kDa, polypeptide 2 (RPS6KB 2), mRNA 476NM_002950 Homo sapiens ribophorin I (RPN1), mRNA 495NM_002743 Homo sapiens protein kinase C substrate 80K-H (PRKCSH), mRNA 691NM_002686 Homo sapiens phenylethanolamine N-methyltransferase (PNMT), mRNA 501NM_002654 Homo sapiens pyruvate kinase, muscle (PKM2), mRNA 2474NM_002648 Homo sapiens pim-1 oncogene (PIM1), mRNA 1052NM_002635 Homo sapiens solute carrier family 25 (mitochondrial carrier; phosphate c a r rier), member 3 (SLC25A3), nuclear gene encoding mitochondrial protein, transcript variant 1b, mRNA 2310NM_002494 Homo sapiens NADH dehydrogenase (ubiquinone) 1, subcomplex unknown, 1, 6kDa (NDUFC1), mRNA 2247NM_002488 Homo sapiens NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 2, 8k Da (NDUFA2), mRNA 689NM_002434 Homo sapiens N-methylpurine-DNA glycosylase (MPG), mRNA 1808NM_002415 Homo sapiens macrophage migration inhibitory factor (glycosylation-inhibiting factor) (MIF), mRNA 3774NM_002227 Homo sapiens Janus kinase 1 (a protein tyrosine kinase) (JAK1), mRNA 990NM_001536 Homo sapiens HMT1 hnRNP methyltransferase-like 2 (S. cerevisiae) (HRMT 1L2), mRNA 832NM_000183 Homo sapiens hydroxyacyl-Coenzyme A dehydrogenase/3-ketoacyl-Coenzyme A thiolase/enoyl-Coenzyme A hydratase (trifunctional protein), beta subunit (HADHB), mRNA 1087NM_002085 Homo sapiens glutathione peroxidase 4 (phospholipid hydroperoxidase) (GPX 4), mRNA 1279NM_001502 Homo sapiens glycoprotein 2 (zymogen granule membrane) (GP2), mRNA 572NM_002080 Homo sapiens glutamic-oxaloacetic transaminase 2, mitochondrial (aspartate aminotransferase 2) (GOT2), nuclear gene encoding mitochondrial protein, mRNA 100 0NM_001440 Homo sapiens exostoses (multiple)-like 3 (EXTL3), mRNA 592NM_003754 Homo sapiens eukaryotic translation initiation factor 3, subunit 5 epsilon, 47k Da (EIF3S5), mRNA 1585NM_003755 Homo sapiens eukaryotic translation initiation factor 3, subunit 4 delta, 44kD a (EIF3S4), mRNA 1085NM_003757 Homo sapiens eukaryotic translation initiation factor 3, subunit 2 beta, 36kD a (EIF3S2), mRNA 622NM_001360 Homo sapiens 7-dehydrocholesterol reductase (DHCR7), mRNA 653。
生化中英对照单词

Chapter 14 Protein Biosynthesis
第十四章 蛋白质的生物合成
Antibiotics 抗生素 Cap-site binding protein 帽子结合蛋白 Chloromycetin 氯霉素 Diphtheria toxin 白喉毒素 Eukaryote 真核生物 Genetic code 遗传密码 Insulin 胰岛素 Interferon 干扰素 Molecular chaperone 分子伴侣 Parathyroid hormone 甲状旁腺激素 Streptomycin 链霉素 Translational initiation complex 翻译起始复合物 Transpeptidase 转肽酶
丝氨酸/苏氨酸蛋白磷酸酶
Chapter 12 DNA Biosynthesis 第十二章 DNA生物合成
Bidirectional replication 双向复制 Endonuclease 内切核酸酶 Exonuclease 外切核酸酶 Gene expression 基因表达 Polymerases 聚合酶类 Primase 引发酶 Primosome 引发体 Proliferating cell nuclear antigen 增殖细胞核抗原 Recombination repairing 重组修复 Replicon 复制子 Reverse transcriptase 逆转录酶 Semiconservative replication 半保留复制 Single stranded DNA binding protein 单链DNA结合蛋白 Telomerase 端粒酶 Telomere 端粒 DNA topoisomerase DNA拓扑异构酶
Chapter 1 Nucleic Acid 第一章 核酸
生物化学原理课件(英文):Chapter32 Nucleotide Metabolism

Why does UMP Cure Orotic Aciduria?
Carbamoyl Phosphate
X Orotate UMP Synthetase
Disease (-UMP)
– No UMP/excess orotate
Feedback Inhibition
☺Disease (+UMP)
Purines are synthesized on the Ribose ring
The metabolic origin of the nine atoms in the purine ring system
Many Steps Require an Activated Ribose Sugar (PRPP)
Catalyzes conversion of NDP to dNDP Highly regulated enzyme Regulates the level of cellular dNTPs Activated prior to DNA synthesis Controlled by feedback inhibition
The synthesis of AMP and GMP from IMP
Salvage Pathway for Purines
Hypoxanthine
or
+ PRPP = IMP or GMP + PPi
Guanine
(HGPRTase)
Adenine + PRPP = AMP + PPi
(APRTase)
de novo Pathway
Salvage Pathway
De novo Synthesis Committed step: This is the point of no return
中山大学生科院 细胞生物学试卷 (9)

中山大学生科院细胞生物学期末试卷(生物科学、生物技术专业、进修教师、交换生,140人)姓名∶专业∶一、填空题(每空1分,共10分)1. 人细胞中DNA总长度为米2. 一个既含有输入细胞核又含有输出细胞核信号的蛋白质分子合成后将会。
3. 如果在一个正处于缩短状态的微管溶液中加入不能被水解的GTP类似物, 微管将。
4. 细胞分裂的最后一步, 即产生两个子细胞的过程称为。
5. 2003年的诺贝尔生理学或医学奖奖给了英国科学家曼斯菲尔德和美国科学家劳特布尔, 表彰他们在方面作出的杰出贡献。
6.同源染色体配对和联会的部位形成,进而发生重组。
7.染色体上有主缢痕和次缢痕,着丝粒位于。
8.内质网中BiP蛋白的功能是,以阻止它们的输出。
9. AP1和AP2都是披网格蛋白小泡外被装配必需的辅助蛋白, 但是二者的作用部位不同:AP1 的披网格蛋白小泡的装配。
10. 通过起动子交换重组实验, 证明了U1 snRNA输出细胞核的信号与相关。
二、判断以下各题是否正确, 若正确, 用T表示, 不正确用F表示, 不需说明。
(每题1分,共10分)1. 一个含有输入过氧化物酶体和输入ER两种信号的蛋白质将定位于ER。
( )2. 在多次跨膜蛋白中,奇数跨膜片段(从N-端起始)只能作为起始跨膜信号,而偶数片段只能作为停止跨膜信号。
( )3. 细胞质膜将细胞分割成功能各异、不通透的区室。
( )4. “在多数真核细胞中质膜的含量最少、功能最大”, 此话正确吗? ( )5. Ras基因是一种癌基因。
( )6. 虽然细胞周期各时相的长短都有不同程度的变化,但变化最大的是G1期。
( )7. 一个同时含有输入线粒体信号和ER驻留信号的蛋白质最终定位于线粒体而不能驻留ER。
( )8. 脊椎动物和芽殖酵母都是使用一种 Cdk控制细胞周期的时相转换。
( )9. 核纤层是由核纤层蛋白A、核纤层蛋白B和核纤层蛋白C构成的,其中只有核纤层蛋白A与内核膜相连,核纤层蛋白B和C则与染色质相连。
卤键、碳氢活化偶联机理的量子化学计算及药物结合口袋的识别与应用研究

第一部分包括第二章及第三章。第二章为药物-靶标间卤键作用的量子化学 计算研究。卤键是一种非常重要的非共价作用,其强度和氢键相当,但具有更强 的方向性。卤素原子头部存在正电势区域,称为 σ-hole 区域,通过该区域和其它 原子的负电势区域发生吸引作用。我们先前的研究发现带有羧基的含卤小分子在 去质子化后带有负电荷,σ-hole 区域的表面静电势显示为负值,但仍然存在 VS,max。在真空中,该类型卤键的相互作用能很弱为排斥作用,但仍具有能阱。 在溶剂环境中,这种排斥作用转变为吸引作用。我们将这种类型的卤键称之为负 电荷卤键。在本论文中,我们研究了负电荷到卤素原子的距离在不同溶剂环境下 对负电荷卤键的影响。发现随着距离的增加,卤键强度逐渐变强,在真空中尤其 明显。带负电荷的卤键强度可以通过所处的环境和距离来调节,在材料和药物设 计中可利用这些特性来改善材料的功能和提高药物活性。第三章为铑催化的碳氢 (C(sp2)-H/ C(sp3)-H)活化偶联机理的量子化学计算研究。过渡金属配合物催化 碳氢键活化构建碳碳、碳氧键等,被广泛应用于药物分子、天然产物、材料分子 的合成。反应机理的理论研究可以了解反应过程,指导并设计反应。合作课题组 Zhou 等人 利用 萘 胺 化合物和 重氮 化合物 在三价铑 的催 化下进 行 环化 反应 (annulation reaction)获得了 1-氢-苯并[g]吲哚啉。在第三章中,我们利用量子
《细胞生物学》——细胞8章 细胞骨架1
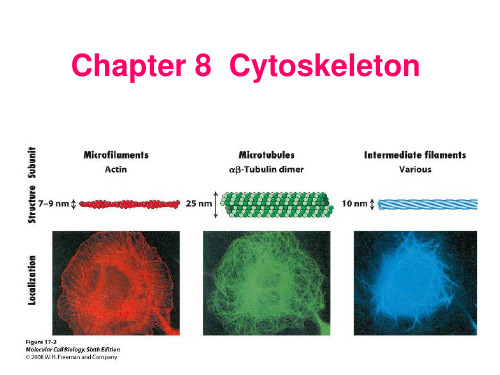
A 球状肌动蛋白 B 纤维状肌动蛋白
G蛋 C白 D Arp2/3
提交
3. 微丝的组装不包括下列哪个阶段() A. 成核反应 B. 纤维的延长 C. 踏车行为 D. 解离阶段
单选题 10分
3. 微丝的组装不包括下列哪个阶段()
A 成核反应 B 纤维的延长 C 踏车行为 D 解离阶段
即正极与负极之别
(2)肌动蛋白单体组装成微丝的过程
① 缓慢成核期:肌动蛋白单体与起始复合物结合→形成寡聚体(至少2-3
个单体)。
包括2种肌动蛋白相关蛋白(Arp2/3)和5种
其它蛋白。
② 快速延长期:肌动蛋白单体具有ATP酶活性,可利用水解ATP释放的能
量来快速组装单体。当微丝的组装速度快于肌动蛋白水解ATP的速度时,在
提交
本章主要内容
微丝与细胞运动 微管及其功能 中间丝
第一节 微丝与细胞运动
微丝(microfilament, MF) 肌动蛋白丝(actin filament) 纤维状肌动蛋白
(fibrous, F-actin)
由肌动蛋白单体组装而成的 直径为7 nm的纤维状结构 存在于所有真核细胞中 微丝的组装/去组装 微丝结合蛋白
(MF binding protein)
一、微丝的组成及其组装 二、微丝网络结构的调节与细胞运动
一、微丝的组成及其组装
(一)微丝的结构与成分 (二)微丝的组装及其动力学特性 (三)影响微丝组装的特异性药物
(一)微丝的结构与成分
• 微丝的主要结构成分:肌动蛋白(actin)。
• 肌动蛋白在细胞内有2种存在形式:①球状肌动蛋白(G-actin): 肌动蛋白单体;②纤维状肌动蛋白(F-actin):由多个单体组装 而成。
HRAC-Classification of Herbicides According to Mod
Classification of Herbicides According to Mode of ActionFarmers, advisors and researchers should know which herbicides are best suited to combat specific resistant weeds. To support the use of herbicides suitable for resistance management the enclosed classification of herbicides is proposed.The herbicides are classified alphabetically according to their target sites, modes of action, similarity of induced symptoms or chemical classes.If different herbicide groups share the same mode or site of action only one letter is used. In the case of photosynthesis inhibitors subclasses C1, C2 and C3 indicate different binding behaviour at the binding protein D1 or different classes. Bleaching can be caused by different ways. Accordingly subgroups F1, F2 and F3 are introduced. Growth inhibition can be induced by herbicides from subgroups K1, K2 and K3. Herbicides with unknown modes or sites of action are classified in group Z as "unknown" until they can be grouped exactly.Classification of HerbicidesIn order to avoid confusion with I and O categories J and Q are omitted. New herbicides will be classified in the respective groups or in new groups (R, S, T...).Since the system was in part developed in co-operation with the "Weed Science Society of America (WSSA)" new herbicides should be categorised jointly by HRAC and WSSA.For reference the numerical system of the WSSA is listed, too.The aim of HRAC is to create a uniform classification of herbicide modes of action in as many countries as possible.Such a classification system can be useful for many instances but there are cases where weeds exhibit multiple resistance across many of the groups listed and in these cases the key may be of limited value.The system itself is not based on resistance risk assessment but can be used by the farmer or advisor as a tool to choose herbicides in different mode of action groups, so that mixtures or rotations of active ingredients can be planned.For a figure of the chemical groups involved each HRAC Group, click on the letter in the table below. A synoptic graphic of the HRAC groups is also available.The WSSA and HRAC systems differ in minor ways. Herbicides in italics are listed on the HRAC classification system but are not listed on the WSSA classification.January 2005 HRAC: Herbicide classificationRemarks:According to information and comments following herbicides are classified in the January 2005 version in HRAC (WSSA) groups:B (2): cancelled: #9; #9; procarbazone Approved ISO name: propoxycarbazone E (14): cancelled: pyrazogyl Approved name: pyraclonil。
BTB-1_DataSheet_MedChemExpress
Inhibitors, Agonists, Screening Libraries Data SheetBIOLOGICAL ACTIVITY:BTB–1 is a potent, selective and reversible mitotic motor protein Kif18A inhibitor with an IC 50 of 1.69 μM.IC50 & Target: IC50: 1.69 μM (Kif18A)[1]In Vitro: BTB–1 blocks the motility of Kif18A in a reversible manner. BTB–1 inhibits Kif18A in an adenosine triphosphate(ATP)–competitive but microtubule–uncompetitive manner and slows down the progression of cells through mitosis. 100 μM BTB–1does not significantly inhibit any of the other tested mitotic kinesins. BTB–1 competes with ATP for Kif18A binding only when the motor–protein is associated with its pseudosubstrate microtubules. HeLa cells treated with BTB–1 accumulate in mitosis in a dose–dependent manner [1]. BTB–1 shows cell toxicity with an EC 50 values of 35.8 μM. HeLa cells treated with 50 μM BTB–1 reveals severe defects in spindle morphology and chromosome alignment. Treatment with high concentrations of BTB–1 does not result in elongated spindles [2].PROTOCOL (Extracted from published papers and Only for reference)Kinase Assay:[1] BTB–1 is prepared in DMSO. The activity of His–Kif18A motor at increasing concentrations of ATP is monitored in the presence of 3 μM Mts and increasing concentrations of BTB–1 (0.21 μM, 0.42 μM, 0.85 μM, 1.7 μM) or DMSO as control [1].References:[1]. Catarinella M, et al. BTB–1: a small molecule inhibitor of the mitotic motor protein Kif18A. Angew Chem Int Ed Engl. 2009;48(48):9072–6.[2]. Braun J, et al. Synthesis and biological evaluation of optimized inhibitors of the mitotic kinesin Kif18A. ACS Chem Biol. 2015 Feb 20;10(2):554–60.Product Name:BTB–1Cat. No.:HY-101770CAS No.:86030-08-2Molecular Formula:C 12H 8ClNO 4S Molecular Weight:297.71Target:Microtubule/Tubulin; Microtubule/Tubulin Pathway:Cell Cycle/DNA Damage; Cytoskeleton Solubility:10 mM in DMSOCaution: Product has not been fully validated for medical applications. For research use only.Tel: 609-228-6898 Fax: 609-228-5909 E-mail: tech@ Address: 1 Deer Park Dr, Suite Q, Monmouth Junction, NJ 08852, USA。
tubulin乙酰化微管位点_解释说明以及概述
tubulin乙酰化微管位点解释说明以及概述1. 引言1.1 概述在细胞生物学领域中,微管是一种重要的细胞骨架蛋白质结构,参与了多种重要的细胞过程,如细胞分裂、细胞运输和形态维持等。
微管由α-和β- tubulin二聚体组成,这些二聚体会通过不同的修饰方式来调节其功能。
其中,乙酰化是一种常见的修饰方式,可以发生在微管上的特定位点上。
1.2 文章结构本文将首先对tubulin乙酰化的基本概念进行介绍,并解释其在细胞中扮演的生理作用。
接着,我们将详细阐述tubulin乙酰化微管位点的解释和说明。
随后,我们将阐述我们所采取的研究方法以及实验设计、材料、方法步骤和数据分析等内容。
在结果及讨论部分,我们将呈现实验结果并对其进行详尽讨论。
最后,我们将总结文章中的主要观点,并对研究结果进行解读和意义分析。
同时,我们还会展望未来以及可能推进该领域研究方向。
1.3 目的本文的主要目的是深入探讨tubulin乙酰化微管位点的解释和说明。
通过了解乙酰化修饰在微管上的发生位置,我们可以更好地理解这种修饰方式对细胞功能和调节过程的影响。
此外,我们还希望通过研究方法和实验设计等部分,为后续相关研究提供有价值的借鉴和指导。
最终,我们希望通过本文的撰写能够促进对tubulin乙酰化微管位点的深入研究,并为细胞生物学领域的发展做出一定贡献。
参考文献:(根据引用格式添加所需参考文献)2. 正文:2.1 tubulin乙酰化的基本概念tubulin乙酰化是一种重要的细胞生物学过程,指的是在微管蛋白tubulin上发生乙酰化修饰作用。
tubulin是微管的主要组成单位,它包含α-tubulin和β-tubulin两个亚单位。
乙酰化通常发生在α-tubulin的Lys40位点上。
这个修饰过程由乙酰转移酶(acetyltransferase)催化,而去乙酰过程则由脱乙酰酶(deacetylase)催化。
2.2 tubulin乙酰化的生理作用tubulin乙酰化在细胞分裂、细胞运动以及细胞形态维持等方面起着关键作用。
自噬_检测指南
Autophagosome formation
Class III phosphatidylinositol 3-kinase (PtdIns3K) complex:
PtdIns3K Vps34 (vacuolar protein sorting 34), a myristoylated serine/threonine kinase Vps15, Atg14 ; Beclin 1 ;
Vesicle fusion and autophagosome breakdown
In mammalian cells, the fusion event requires the lysosomal membrane protein LAMP-2 and the small GTPase Rab7.
Maturation,Degradation: Vesicle fusion and autophagosome breakdown
Induction
Normal conditions Basal-level autophagy is very low; Autophagy inhibitor: serine/threonine protein kinase TOR(target of rapamycin) input information from multiple upstream signal transduction pathways (discussed below) and negatively regulates another serine/threonine kinase, Atg1, in nutrient-rich conditions
- 1、下载文档前请自行甄别文档内容的完整性,平台不提供额外的编辑、内容补充、找答案等附加服务。
- 2、"仅部分预览"的文档,不可在线预览部分如存在完整性等问题,可反馈申请退款(可完整预览的文档不适用该条件!)。
- 3、如文档侵犯您的权益,请联系客服反馈,我们会尽快为您处理(人工客服工作时间:9:00-18:30)。
Synthesis and Microtubule Binding of Fluorescent PaclitaxelDerivativesErkan Baloglu,a David G.I.Kingston,a Pruthvi Patel,b Sabarni K.Chatterjee band Susan L.Bane b,*aDepartment of Chemistry,M/C 0212,Virginia Polytechnic Institute and State University,Blacksburg,VA 24061,USAbDepartment of Chemistry,State University of New York at Binghamton,Binghamton,NY 13902-6016,USAReceived 8February 2001;accepted 18May 2001Abstract—The preparation of two new fluorescent derivatives of paclitaxel in which the fluorophore is bonded to paclitaxel at the C-10position is reported.Both analogues,10-deacetyl-10-(m -aminobenzoyl)paclitaxel (1,BTax)and 10-deacetyl-10-[7-(diethyla-mino)coumarin-3-carbonyl]paclitaxel (2,CTax)retain good activity as promoters of in vitro tubulin assembly.Microtubule bind-ing enhances the emission intensity of both probes.#2001Elsevier Science Ltd.All rights reserved.The complex natural product paclitaxel (Taxol 1)(3),first isolated from Taxus brevifolia ,1is a member of a large family of taxane diterpenoids.2Paclitaxel is widely used to treat solid tumors,particularly those of the breast and ovaries.The drug exerts its pharmacological effects by interacting with cellular microtubules.3Microtubules are primarily composed of tubulin,a 100kDal heterodimer that reversibly assembles to form the core of the microtubule.Proper spatial and temporal control of microtubule dynamics is essential for cellular homeostasis.Paclitaxel disrupts the dynamic processes of microtubules by binding to the polymer at a single site on b -tubulin.4Fluorescence spectroscopy is a powerful and versatile tool for the study of biological systems and has been extensively applied to the study of paclitaxel–micro-tubule interactions.Small,environmentally sensitive probes based on the aminobenzoate fluorophore have been used to explore the local environment of the pacli-taxel binding site.5À8This fluorophore,however,must be excited with ultraviolet light,which limits its useful-ness.Probes that are exited by visible light are more desirable.They are best suited for visualization of microtubule-bound paclitaxel using fluorescence micro-scopy and can be easily detected in plate readers.These structures tend to be large (rhodamine,fluorescein,andBODIPY)and their fluorescence emission spectra are relatively insensitive to their environment.These fluorophores are most commonly attached to paclitaxel through the C-7position,9À13the most synthetically accessible site on the ‘northern hemisphere’of paclitaxel (C-6–C-12positions).Structural changes in this portion of the molecule appear to have less impact on its anti-microtubule activity than modifications in the ‘southern hemisphere’.14We have been probing the nature of the paclitaxel–tubulin interaction through synthesis and in vitro bio-logical investigation of fluorescent paclitaxel derivatives.In our previous studies,we prepared and investigated paclitaxels in which the fluorophore is contained in the ‘southern hemisphere’(i.e.,at C-2and C-30).7,8We chose the less utilized C-10position to initiate our investigations into the ‘northern hemisphere’of pacli-taxel.Moreover,we chose to use 7-(diethylamino)cou-marin-3-carboxylic acid as one of the labels.This fluorophore has not yet been used as a probe for the paclitaxel–tubulin interaction.Other coumarin deriva-tives have been used to label paclitaxel,but the resulting probes had absorption maxima at high energy ($370nm).10,13Inclusion of an electron withdrawing group at the 3-position of 7-(diethylamino)coumarin yields a probe that is excited by visible light (410–430nm)and is also environmentally sensitive.15Repor-ted herein are the synthesis of two C-10modified fluor-escent paclitaxels and their in vitro tubulin binding properties.0960-894X/01/$-see front matter #2001Elsevier Science Ltd.All rights reserved.P I I :S 0960-894X(01)00453-XBioorganic&Medic inal Chemistry Letters 11(2001)2249–2252*Corresponding author.Tel.:+1-607-777-2927;fax:+1-607-777-4478;e-mail:sbane@ResultsSynthesis of fluorescent paclitaxels modified at the C-10positionThe sequence leading to the required fluorescent pacli-taxels is shown in Scheme 1.The known paclitaxel derivative (4)16was esterified at the C-10position with 3-nitrobenzoic acid,employing the carbodiimide-based coupling protocol to yield the paclitaxel analogue 5.Concomitant removal of both silyl protecting groups,followed by hydrogenation,gave the desired amine (1,BTax)in good yield.CTax (2)was prepared from the common intermediate 4by acylation at C-10as before utilizing 7-(diethylamino)coumarin-3-carboxylic acid,followed by simultaneous removal of the protecting groups.17Microtubule activityThe activities of the ligands were assessed by their abil-ities to induce purified tubulin to assemble in vitro.Both Btax and CTax were nearly equipotent to paclitaxel in promoting tubulin assembly,although CTax did not promote assembly quite as rapidly as paclitaxel (Fig.1).The absorption,excitation and emission spectra of the two fluorescent ligands were obtained in a variety of solvents and bound to microtubules.The effect of microtubule binding on the emission spectra of the compounds is illustrated in Figure 2.The emission intensity of BTax increased and underwent a small blue shift upon microtubule binding (Fig.2A).Since the absorption maximum of the ligand in buffer is near 320nm,ultraviolet radiation is required for excitation of this fluorophore.CTax possesses photochemical properties that will make it more useful than BTax as a fluorescent probe.The absorption and emission maximaare both in the visible region of the electromagnetic spectrum.The emission intensity is very strongly affec-ted by environmental polarity,while the emission energy is less so (Table 1).Microtubule binding by CTax is accompanied by a significant increase in emis-sion intensity (Fig.2B).The appearance of fluorescence coincides with assembly of the protein (Fig.3),which further demonstrates that the fluorescence changes that occur in CTax in the presence of tubulin are due to microtubulebinding.Scheme 1.Synthesis of fluorescent paclitaxelderivatives.Figure 1.Promotion of tubulin assembly by paclitaxel,BTax and CTax.Tubulin in PMEG buffer (0.1M Pipes,1mM MgSO 4,2mM EGTA and 0.1mM GTP,pH 6.9)was equilibrated to 37 C prior to addition of the ligand.Assembly was monitored by apparent light scattering (absorption at 350nm).Solid curve:10m M BTax;dashed curve:10m M paclitaxel.Dash dot dot curve:5m M paclitaxel;dotted curve:5m M CTax.2250 E.Baloglu et al./Bioorg.Med.Chem.Lett.11(2001)2249–2252The small change in emission energy observed when BTax and CTax bind to microtubules indicates that the substituent is solvent accessible.This observation is consistent with the structure–activity data for C-10analogues of paclitaxel.18Based on their spectroscopic observations,Evangelio et al.13and Sengupta et al.6hypothesized that the C-7and C-10position of pacli-taxel may be in a cationic microenvironment.These predictions were made before the electron crystal-lographicstruc ture of tubulin was published.19It is nowknown that tubulin has at least two arginine residues located on the exterior of the paclitaxel binding site,19and at least one of these is in close proximity to the C-7position of the tubulin-bound ligand.20The spectro-scopic data for microtubule-bound Btax and CTax are also consistent with the presence of a charged amino acid in close proximity to the fluorophore.ConclusionThe C-10position can be used to prepare fluorescent derivatives of paclitaxel that retain good in vitro assembly-promoting activity.The small change in emis-sion intensity when BTax binds to microtubules limits its utility as a probe.The 7-(diethylamino)coumarin-3-carbonyl fluorophore,which has not been previously used to fluorescently label paclitaxel,produces a probe with many possible utilities.Since the probe can be excited with visible light,the fluorescent paclitaxel-microtubule association can be readily observed in standard plate readers and possibly in live cells.Unlike the rhodamine and fluorescein paclitaxel derivatives,13the emission intensity of the diethylaminocoumarin fluorophore in CTax undergoes a large change in emis-sion intensity in upon microtubule binding.Thus,background fluorescence due to unbound ligand will be low when CTax is used as a probe.AcknowledgementsWe thank Sergey Osipov for technical assistance,Dr.Barbara Poliks for performing the molecular modeling and the Nebraska Center for Mass Spectrometry at the University of Nebraska for mass spectrometric assis-tance.References and Notes1.Wani,M.C.;Taylor,H.L.;Wall,M.E.;Coggon,P.;McPhail,A.T.J.Am.Chem.Soc.1971,93,2325.2.Baloglu,E.;Kingston,D.G.I.J.Nat.Prod.1999,62,1448.3.Horwitz,S.B.Ann.Oncol.1994,5,S3.Figure 2.Emission spectra of BTax and CTax in the presence and absence of tubulin.(A)Solid curve:BTax (10m M)in PMEG buffer was incubated with 10m M tubulin at 37 C prior to collection of the emission spectrum.Dashed curve:Emission spectrum of 10m M BTax in PMEG buffer.The excitation wavelength was 320nm.(B)Solid curve:CTax (0.4m M)in PMEG buffer was incubated with 5m M tubulin at 37 C prior to collection of the emission spectrum.Dashed curve:Emission spectrum of 0.4m M CTax in PMEG buffer.The excitation wavelength was 420nm.Table 1.Absorption and emission maxima and relative emission intensity of CTax in solvent and bound to microtubules SolventAbsorption maximum (nm)Emission maximum (nm)Relative fluorescence intensity aDioxane 41344742Ethyl acetate41545240Dimethylsulfoxide 42746918Acetonitrile 422464 1.0Ethanol 4234610.11Methanol4244620.272%DMSO/water 428b 462b 0.06Microtubules430c4684.8daFor solvents:intensity at emission maximum,normalized to acetoni-trile value.The absorptivity of the solvent samples at the excitation wavelength (420nm)was equal.For microtubules:intensity of bound drug relative to free drug.bWithout DMSO cosolvent and at higher concentrations the ligand undergoes self-association.The absorption maximum of aggregated samples is 434nm and the emission maximum is 530nm.cApproximate value due to turbidity of microtubule-containing solu-tion.dConcentration of free drug at 0.4m M,concentration of bound drug at 0.4m M in presence of 5m Mtubulin.Figure 3.Promotion of tubulin assembly by CTax.Tubulin (10m M)in PMEG buffer was equilibrated to 37 C prior to addition of 10m M CTax.Assembly was monitored by apparent light scattering (dotted curve)or by emission at 465nm (solid curve;excitation at 420nm).E.Baloglu et al./Bioorg.Med.Chem.Lett.11(2001)2249–225222514.Jordan,M.A.;Wilson,L.Curr.Opin.Cell Biol.1998,10,123.5.Sengupta,S.;Boge,T.C.;Georg,G.I.;Himes,R.H.Bio-chemistry1995,34,11889.6.Sengupta,S.;Boge,T.C.;Liu,Y.;Hepperle,M.;Georg,G.I.;Himes,R.H.Biochemistry1997,36,5179.7.Han,Y.;Chaudhary,A.G.;Chordia,M.D.;Sackett,D.L.; Perez-Ramirez,B.;Kingston,D.G.;Bane,S.Biochemistry 1996,35,14173.8.Li,Y.;Poliks,B.;Cegelski,L.;Poliks,M.;Gryczynski,Z.; Piszczek,G.;Jagtap,P.G.;Studelska, D.R.;Kingston,D.G.I.;Schaefer,J.;Bane,S.Biochemistry2000,39,281.9.Dubois,J.;Le,G.M.T.;Gueritte-Voegelein,F.;Guenard,D.;Tollon,Y.;Wright,M.Bioorg.Med.Chem.1995,3,1357.10.Souto,A.A.;Acuna,A.U.;Andreu,J.M.;Barasoain,I.; Abal,M.;Amat-Guerri,F.Angew.Chem.,Int.Ed.Engl.1995, 34,2710.11.Guy,R.;Scott,Z.;Sloboda,R.;Nicolaou,K.Chem.Biol. 1996,3,1021.12.Rao,C.S.;Chu,J.J.;Liu,R.S.;Lai,Y.K.Bioorg.Med. Chem.1998,6,2193.13.Evangelio,J.A.;Abal,M.;Barasoain,I.;Souto,A.A.; Lillo,M.P.;Acuna,A.U.;Amat-Guerri,F.;Andreu,J.M. Cell Motil.Cytoskeleton1998,39,73.14.Kingston,D.G.I.J.Nat.Prod.2000,63,726.15.Bangar Raju,B.;Varadarajan,T.S.J.Phys.Chem.1994, 98,8903.16.Paclitaxel derivative4was prepared by protecting the known10-deacetyl-20-(tert-butyldimethylsilyl)paclitaxel as its triethylsilyl ether at C-7position.Datta, A.;Hepperle, .Chem.1995,60,761.17.General procedure for esterification at the C-10position:1 equivalent of the carboxylic acid was stirred in toluene at room temperature with1equivalent of dicyclohexyl carbodi-imide(DCC)or1-[3-(dimethylamino)propyl-3-ethylcarbodi-imide hydrochloride(EDC)for15min.0.01equivalent of4-(dimethylamino)pyridine(DMAP)was added and stirred for 5min.0.1equivalent of10-deacetyl-20-(tert-butyldimethylsi-lyl)-7-(triethylsilyl)paclitaxel was then introduced and stirred at rt for24–48h.In order to overcome the solubility problem of7-(diethylamino)coumarin-3-carboxylic acid,CH2Cl2/DMF (10:1)solvent system was used for2.The progress of the reaction was monitored by thin layer chromatography and when the reaction was complete,the reaction mixture was diluted with EtOAc.The organic layer washed with water (Â2),NaHCO3(Â2),brine and dried over sodium sulfate.The crude product was applied on a preparative thin layer chro-matography plate,developed with solvent system of40% EtOAc/hexane to give the desired products.1H NMR of1 (400MHz,CDCl3)d8.12–8.14(d,2H),7.72–7.48(d,2H),7.61 (t,1H),7.34–7.53(m,12H),7.24(t,1H),7.02–7.05(d,1H), 6.88–6.90(dd,1H),6.49(s,1H),6.24(t,1H),5.77–5.80(dd, 1H),5.70–5.72(d,1H),4.95–4.97(d,1H),4.80(s,1H),4.47 (m,1H),4.30–4.32(d,1H),4.19–4.21(d,1H),3.85–3.87(d, 1H),3.82(bs,2H),3.62(bs,1H),2.66–2.67(d,1H),2.57(m, 1H),2.39(s,3H),2.34(m,1H),1.89(m,1H),1.81(s,3H),1.69 (s,3H),1.66(s,3H),1.30(s,3H),1.24(s,3H);13C NMR of1 (100MHz)d203.45,172.66,170.38,167.05,166.97,166.57, 146.56,142.07,137.89,133.72,133.59,133.24,131.96,130.19, 129.97,129.41,129.13,129.01,128.72,128.69,128.37,127.02, 120.17,120.08,115.99,84.44,81.18,79.04,76.49,75.85,74.95, 73.17,72.38,72.25,58.66,55.03,45.72,43.22,35.72,35.68, 29.68,27.04,22.63,22.07,14.88,9.57.HR-FABMS of1m/z found953.343872(M+Na)+,calcd953.3473(M+Na)+;LR-FABMS m/z found953.5(M+Na)+.1H NMR of2 (400MHz,CDCl3)d8.48(s,1H),8.12–8.14(d,2H),7.74–7.76 (d,2H),7.61(t,1H),7.34–7.53(m,11H),7.05–7.07(d,1H), 6.60–6.63(dd,1H),6.52(s,1H),6.45–6.46(d,1H),6.25(t, 1H),5.78–5.80(dd,1H),5.70–5.72(d,1H),4.95–4.97(d,1H), 4.801–4.807(d,1H),4.40–4.48(m,1H),4.30–4.32(d,1H), 4.20–4.22(d,1H),3.86–3.88(d,1H),3.43–3.49(q,4H),2.57 (m,1H),2.40(s,3H),2.30–2.34(m,3H),1.87–1.94(m,2H), 1.82(s,3H),1.71(s,3H),1.32(s,3H),1.23–1.28(m,15H);13C NMR of2(100MHz)d203.65,172.56,170.40,166.98,164.32, 158.80,153.23,150.22,141.79,137.89,133.68,133.63,133.18, 131.92,131.47,130.19,129.16,128.99,128.68,128.34,127.04, 109.73,107.84,107.37,69.77,84.44,81.18,79.04,76.49,75.73, 74.96,73.20,72.38,72.19,58.69,55.01,45.79,45.19,43.27, 35.74,35.66,26.57,22.63,21.66,14.92,12.43,9.58LR-FABMS(M+H)+m/z found1055.6.18.Kant,J.;O’Keeffe,W.S.;Chen,S.-H.;Farina,V.;Fair-child,C.;Johnson,K.;Kadow,J.F.;Long,B.H.;Vyas,D. Tetrahedron Lett.1994,35,5543.19.Nogales,E.;Wolf,S.G.;Downing,K.H.Nature1998, 391,199.20.Rao,S.;He,L.F.;Chakravarty,S.;Ojima,I;Orr,G.A.; Horwitz,S.B.J.Biol.Chem.1999,274,37990.2252 E.Baloglu et al./Bioorg.Med.Chem.Lett.11(2001)2249–2252。
