Transcriptome Characteristics and Six Alternative
生物学基本名词详细解释(中英文对照)

生物学基本名词详细解释(中英文对照)/english/speciality/5091.html组织相容性Histocompatibility字面上讲是指不同组织共存的能力;严格地讲是指所有移植蛋白的一致性,这是阻止移植和器官排斥的需要。
组织相容性的分子基础是修饰几乎所有人类细胞表面的一套移植蛋白。
这些蛋白是由位于6号染色体上的一段称为主要组织相溶性复合体基因,MHC编码的。
这些蛋白高度多态。
例如,它们在不同的人中显示差异。
尽管很多人会有一些相同的MHC分子,极少数人有完全相同的MHC分子。
微小的差别导致这些蛋白质被移植受体的免疫系统识别为外来的而进行破坏。
对成功的移植来说这些蛋白质应该在供体和受体之间相匹配。
双胞胎相配的几率最高,接下来是兄弟姐妹。
在一般人群中只有10万分之一的比例是MHC匹配的,可以允许移植。
Literally, the ability of different tissues to “get along”; strictly, identity in all of the transplantation proteins, which is a requirement for the prevention of graft or organ rejection. The molecular basis of histocompatibility is a set of transplantation proteins that decorate the surface of nearly all human cells. These proteins are encoded by genes that are grouped on a part of chromosome 6 called the major histocompatibility complex, or MHC. These proteins are highly “polymorphic” i.e., they show variation in different individuals. Although many in dividuals may share some identical MHC molecules, a very low number share all the MHC molecules. The consequence of these minor differences is that these proteins are recognized by the transplant recipient’s immune system as being foreign, and so are targe ted for destruction (since the immune system’s job is to eradicate any foreign proteins or cells that invade the body). For successful transplantation these proteins ideally should be matched between donor and recipient. Twins have the highest rate of matc h, followed by siblings. In the general population only 1 in 100,000 individuals is sufficiently “MHC matched” to another person to allow transplantation.X射线结晶学X-ray Crystallography阐述蛋白质、DNA或其它生物分子的原子水平的三维结构的技术。
转录和转录组学transcriptome PPT课件

1.2.1.组成
• 全酶: 2´ (核心酶 + ) • 核心酶 : 2´
1.2.2.作用
• α亚基: 决定那些基因被转录。 • β亚基: 催化与模板配对的相邻NTP
以3´, 5´-磷酸二酯键相连。
• β´亚基:促进酶与模板链结合,并使
DNA双链打开。 ( 核心酶: 催化RNA链的延长,参与整个 过程。)
• 1.单链小分子; • 2.含有稀有碱基或修饰碱基; • 3. 5′端总是磷酸化, 5′末端往往是pG; • 4. 3′端是CpCpAoH序列; • 5.三叶草结构; • 6.三级结构是倒L型。
三级结构呈倒L形
2.3 rRNA:
• 原核生物:70S--由50S和30S 组成 • 真核生物:80S--由60S和40S 组成
个茎。1~3个环,含13b保守序 列CAAA,AC,AGUC,GUG
核苷酸链断裂点
槌头状结构,最简单的核酶
核酶的意义
• 动摇了酶是蛋白质的传统概念。 • 为地球上生命起源早期可能是先出
现RNA提供证据。
• 为人工合成核酶以破坏某些病原微
生物,消除体内有害基因提供理论 基础。
2.5 核内不均一RNA(hnRNA)
2.7 反义RNA:
• 可与mRNA形成双链,抑制翻译。
2.8 microRNA 调节mRNA的水平
二、RNA的合成----转录(transcription)
指在RNA聚合酶催化下,以DNA为模板, NTP为原料,合成RNA的过程。
转录概述
• DNA为模板合成RNA的过程 • RNA聚合酶 • 原料:ATP,UTP,CTP,GTP (NTP) • Mg2+,Mn2+ • 合成方向:5´→3´ • 连接方式:3´,5´-磷酸二酯键
睡莲品种‘公牛眼’和‘泰国王’授粉后子房的发育差异研究
睡莲品种‘公牛眼’和‘泰国王’授粉后子房的发育差异研究作者:唐毓玮李佳慧毛立彦黄秋伟龙凌云於艳萍苏群来源:《热带作物学报》2022年第04期摘要:睡蓮属(Nymphaea)植物是我国新兴的水生花卉,其中重瓣型睡莲的花态丰满艳丽,一直倍受睡莲爱好者、育种者的喜爱和关注,在水生园林景观中的应用日渐广泛,但在育种的过程中,育种者经常遇到杂交不结实的现象,导致重瓣型睡莲品种难以培育。
为探索重瓣睡莲难以结实的原因,以重瓣睡莲中较具代表性的品种‘泰国王’为实验组,以育性较好的品种‘公牛眼’为对照组,利用石蜡切片技术对其授粉后不同时期的子房进行显微结构比较,采用扫描电镜进一步观察样本的柱头和胚珠,通过RNA-seq测序分析发育与败育子房的基因表达差异。
结果表明:2个睡莲品种的柱头表面均分布着多细胞单列乳突,细胞间的连接处形成一圈圈凹槽,乳突数量庞大、排列紧密,配合自身的结构能更容易捕获外来的花粉。
‘公牛眼’授粉7d后子房中大部分胚珠发育成红色种子,10d后子房膨大成果实,种皮由红色转黑色,形成成熟的种子;重瓣睡莲‘泰国王’授粉7d后无种子形成,子房中有部分胚珠发育,其表皮变为红色,形态特征与‘公牛眼’授粉4 d后的红色胚珠相似,由此推测少量胚珠可能完成了受精,但无法进一步发育,10 d后子房完全败育。
显微结构观察可知,授粉7d后2个品种的胚珠发育产生明显差异,‘公牛眼’的胚珠明显增大,合子开始分裂形成胚,而‘泰国王’胚珠内珠心萎缩,胚珠逐步败育消解,授粉10 d后,‘公牛眼’胚珠的珠被增厚形成种皮,胚开始发育,进一步向种子形态转变。
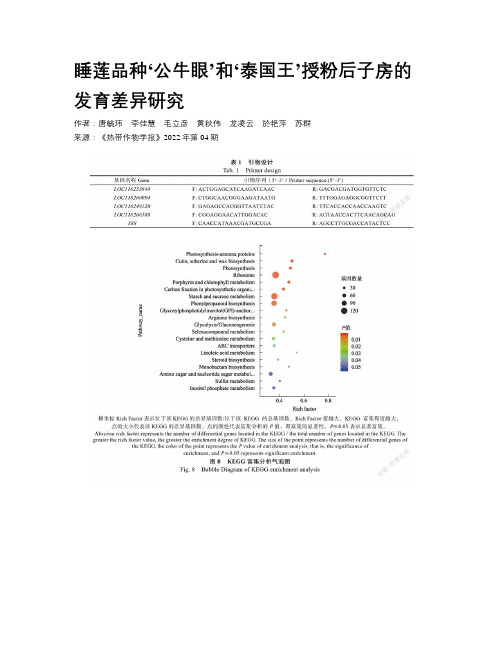
由转录组测序分析可知,差异基因显著富集在光合作用——天线蛋白、光合作用、卟啉和叶绿素代谢、核糖体、淀粉和蔗糖代谢等通路,这些通路可能是影响睡莲子房发育的关键途径,推测‘泰国王’子房败育可能是因为自身光合作用受到抑制,进而影响了蔗糖、淀粉等营养物质的生成。
关键词:睡莲;杂交授粉;子房发育;胚珠;转录组中图分类号:S682.32 文献标识码:ADifferences in Ovary Development Between Nymphaea‘Bull’s Eye’ and Nymphaea‘King of Siam’ after PollinationTANG Yuwei LI Jiahui MAO Liyan HUANG Qiuwei LONG Lingyun YU Yanping SU Qun1. Guangxi Subtropical Crop Research Institute, Nanning, Guangxi 530001, China;2. Flower Research Institute, Guangxi Academy of Agricultural Sciences, Nanning, Guangxi 530007, ChinaAbstract:Nymphaea (water lily) is an aquatic flower that has been introduced in recent years in our country. In particular, double petal water lily are very popular and full of love and attention by water lily lovers and breeders. And it has been increasingly widely used in the field of aquatic garden landscapes. But in the process of breeding, breeders often encounter the situation of failed hybridization. This makes it hard for breeders to breed double flower. In order to study on the reason that double petal water lily is difficult to set fruit, we selected the more representative cultivar Nymphaea‘King of Siam’ fr om double petal water lily as the experimental group, and the better fertile cultivar Nymphaea‘Bull’s Eye’ as the control group. Firstly, the microstructure of ovaries at different stages after pollination was compared by paraffin section technique. Secondly,the stigmas and ovules were further observed by scanning electron microscope. Finally, the differences of gene expression between developing and abortive ovary were analyzed by RNA-Seq. The results showed that multicellular uniserial papillae were distributed on the stigma surface of the two water lily varieties, and a circle of grooves were formed at the junction between single cells. The number of papillae was large and closely arranged. We think this structure can more easilycapture foreign pollen. Meanwhile, after 7 days of N. ‘Bull’s Eye’ pollination, most of the ovules developed into red seeds, 10 days later, the ovary expanded into fruit, and the seed coat changed from red to black to form mature seeds. On the other hand, after 7 days of pollination, no seeds were formed in N. ‘King of Siam’. However, some ovules in its ovary develop, and its epidermis turns red. The morphological characteristics are similar to the red ovules after 4 days of N. ‘Bull’s Eye’ pollination. Therefore, we speculate that a small number of ovules may have completed fertilization, but can not develop further. Than, the ovary was completely aborted after 10 days. Microstructural observation showed that there were significant differences in ovule development between the two varieties after 7 days of pollination. The ovules of N. ‘Bull’s Eye’ increased significantly, and the zygotes began to form embryos, while the nucellus in the ovules of N. ‘King of Siam’ shrank and the ovules gradually disappeared. After 10 days of pollin ation, the embryo of N. ‘Bull’s Eye’ began to develop and further changed to seed morphology. In additional, the enrichment analysis of differential genes showed that photosynthesis antenna protein,photosynthesis, porphyrin and chlorophyll metabolism, ribosome, starch and sucrose metabolism may be the key pathways affecting the development of waterlily ovary. Consequently, we speculate that the abortion of the ovary of N. ‘King of Siam’ may be due to the inhibition of its own photosynthesis, which affects the production of nutrients such as sucrose and starch.Keywords:Nymphaea; cross pollination; ovary development; ovule; transcriptomeDOI:10.3969/j.issn.1000-2561.2022.04.019授粉是種子植物生殖过程中的重要阶段[1],授粉成功的子房会进一步发育成果实,而个体间不亲和的授粉会影响花粉与雌蕊的识别、受精以及胚珠的发育[2-4],进而导致子房败育无法结实。
槟榔不同发育时期果实转录组特征分析

槟榔不同发育时期果实转录组特征分析作者:押辉远陈叶张岩松许启泰来源:《热带作物学报》2020年第07期摘要:檳榔(Areca catechu L.)果实是四大南药之一。
槟榔果的研究主要集中在生理生化、生防菌、有效成分及药理、加工和利用等方面,对槟榔果的发育及其次生物质形成的分子机制尚不清楚。
本研究对不同发育时期的槟榔果皮和果核进行转录组测序,鉴定槟榔果不同发育时期的关键基因,以探讨果实发育相关基因的表达特征及次生物质形成有关的基因调控。
结果显示,槟榔果皮中检测到4491个差异基因,其中617个差异基因共参与了111条KEGG代谢通路,生物过程代谢类有82个通路,共257个差异基因被注释,参与次生代谢途径共有5个,共27个差异基因。
槟榔果核中检测到5443个差异基因,其中898个差异基因共参与了118条通路,466个差异基因被注释在生物代谢类通路上,共涉及89条通路,参与次生代谢相关的基因有53个,参与次生代谢途径共7条。
进一步分析表明:随着果实的发育,果皮中80%次级代谢通路差异相关基因呈下调表达趋势;而果核中71.4%次级代谢通路差异相关基因呈上调表达趋势。
本研究结果在转录组水平揭示了槟榔果发育的生物学过程,发现了不同时期槟榔果皮和果核中次级代谢相关调控基因表达的变化规律,也为槟榔的遗传育种研究奠定了基础。
关键词:槟榔;果实发育时期;转录组中图分类号:S31文献标识码:AAnalysis of Transcriptome Characteristics of Areca atDifferent Developmental StagesYA Huiyuan1, CHEN Ye1, ZHANG Yansong1, XU Qitai1,21. School of Food and Medicine, Luoyang Normal University, Luoyang, Henan 471934,China;2. Hainan Green Areca Science &Technology Development Co., Ltd.,Ding’an, Hainan 571200, ChinaAbstract:Areca(Areca catechu L.)isone of the four primary medicinal plants in south China.In the study, the high-throughput sequencing technology was used to sequence the transcriptome of the peel and kernel in different periods to find differentially expressed genes in different developmental stages. Among the peels, 4491 differential genes were divided, of which 617 differential genes were involved in 111 KEGG metabolic pathways,257 differential genes with 82 pathways in the biological process metabolism class, and a total of 27 genes with 5 genes involved in the secondary metabolic pathway. There were 5443 differential genes in the betel nut kernel, according to the KEGG pathway annotation results, 898 differential genes were involved in 118 pathways, 466 differential genes were annotated on biological metabolic pathways for 89 pathways, 53 genes involved in secondary metabolism with 7 secondary metabolic pathways involved. Further analysis showed that with the development of fruit, 80% of the secondary metabolic pathways in the pericarp showed a down-regulated expression of the genes, while 71.4% of the secondary metabolic pathways in the kernel showed up-regulated expression. The results of the study preliminarily revealed the overall characteristics of the transcriptome of different tissues and different developmental stages of areca nut, and found that the expression of secondary metabolism-related regulatory genes in betel nut and pit were observed in different stages, which was the medicinal development and secondary of betel nut.Keywords:Areca catechu L.;fruit development period; transcriptomeDOI: 10.3969/j.issn.1000-2561.2020.07.001槟榔(Areca catechu L.)是棕榈科槟榔属常绿乔木,广泛分布于南亚和东南亚等国家[1]。
褪黑素对碱性盐胁迫下猴樟呼吸代谢相关酶活性的影响

褪黑素对碱性盐胁迫下猴樟呼吸代谢相关酶活性的影响朱育贤施诗岂子雁吴思诺王秋楠韩浩章*(宿迁学院建筑工程学院,江苏宿迁223800)摘要碱性盐环境影响植物的生长发育,褪黑素能有效缓解盐碱逆境对植物的伤害。
本研究以猴樟幼苗为材料,研究外源褪黑素处理对猴樟幼苗呼吸代谢相关酶活性的影响,以期为褪黑素处理提高植物耐碱性盐胁迫的机理研究提供依据。
结果表明,30~50μmol/L褪黑素处理明显提高碱性盐胁迫下猴樟幼苗中丙酮酸脱氢酶、柠檬酸合成酶、异柠檬酸脱氢酶、琥珀酸脱氢酶和苹果酸脱氢酶的活性,100~150μmol/L褪黑素处理对猴樟幼苗呼吸代谢相关酶活性的促进作用不明显。
适宜浓度褪黑素处理能通过促进呼吸代谢提高猴樟幼苗耐碱性盐胁迫能力。
关键词猴樟;褪黑素;碱性盐胁迫;呼吸代谢;酶活性中图分类号Q945;S792.23文献标识码A文章编号1007-5739(2023)24-0088-04DOI:10.3969/j.issn.1007-5739.2023.24.025开放科学(资源服务)标识码(OSID):Effects of Melatonin on Respiratory Metabolism Related Enzyme Activities ofCinnamomum bodinieri Under Alkaline Salt StressZHU Yuxian SHI Shi QI Ziyan WU Sinuo WANG Qiunan HAN Haozhang*(School of Architecture and Engineering,Suqian University,Suqian Jiangsu223800) Abstract Alkaline salt environment affects plant growth and development,and melatonin can effectively alleviate the damage of saline-alkali stress on plants.This study used Cinnamomum bodinieri seedlings as materials to investigate the effects of exogenous melatonin on respiratory metabolism related enzyme activities of Cinnamomum bodinieri,in order to provide a basis for the research on the mechanism of improving plant resistance to alkaline salt stress by mela-tonin treatment.The results showed that30-50μmol/L melatonin treatment significantly increased the activities of pyru-vate dehydrogenase,citrate synthetase,isocitrate dehydrogenase,succinate dehydrogenase and malate dehydrogenase of Cinnamomum bodinieri under alkaline salt stress.Melatonin treatment with100-150μmol/L didn't significantly promote the activity of respiratory metabolism related enzymes of Cinnamomum bodinieri.The appropriate concentration of mela-tonin can improve the alkaline salt tolerance of Cinnamomum bodinieri seedlings by promoting respiratory metabolism.Keywords Cinnamomum bodinieri;melatonin;alkaline salt stress;respiratory metabolism;enzyme activity土壤盐碱化是影响植物生长发育和限制作物产量的严峻问题,全世界约20%的灌溉农田存在不同程度的盐碱化,已超过地球陆地面积的7%[1]。
真核生物基因表达调控

酸性激活域 (D/E-rich) 谷氨酰胺(Q)富含域 脯氨酸(P)富含域
蛋白质-蛋白质结合域 (dimerization, co-factors)
1) TF最常见的DNA binding domain
Zinc Finger
bZIP
Homeodomain
bHLH
(1) 锌指(zinc finger)
2. The pri5’ capping 3’ formation / polyA splicing
3. Mature transcripts are transported to the cytoplasm for translation
Chromatin
epigenetic control
Protein degradation RNA silencing
一般而言的基因表达调控范畴
二、基因表达的时间性及空间性
(一)时间特异性
按功能需要,某一特定基因的表达严格按 特定的时间顺序发生,称之为基因表达的时间 特异性(temporal specificity)。
Cys-X2-4-Cys-X3-Phe-X5-Leu-X2-His-X3-His C-terminal: α-helix binding DNA
常结合GC box
(2) 碱性亮氨酸拉链 bZIP
(3) 碱性螺旋-环-螺旋bHLH
bHLH蛋白(basic Helix-Loop-Helix)
2) TF常见的trans-activation domain
– usually expressed at high level – the level of their gene expression may vary
基因表达 细胞特异和物种演化
基因表达细胞特异和物种演化English Response:Gene expression is a complex and tightly regulated process that plays a crucial role in determining the characteristics and functions of cells. It involves the transcription of DNA into RNA and the subsequent translation of RNA into proteins. Gene expression can be influenced by a variety of factors, including genetic mutations, environmental stimuli, and cellular signaling pathways.One of the key features of gene expression is its cell specificity. This means that different cells within an organism express different sets of genes. This cell-specific gene expression is essential for the development and function of multicellular organisms, as it allows for the specialization of cells into different tissues and organs. For example, muscle cells express genes that are responsible for muscle contraction, while nerve cellsexpress genes that are involved in transmitting electrical signals.The cell specificity of gene expression is achieved through a variety of mechanisms. One important mechanism is the presence of tissue-specific transcription factors. These transcription factors are proteins that bind to specific DNA sequences and either promote or repress the transcription of genes. For example, the transcriptionfactor MyoD is essential for the expression of muscle-specific genes in muscle cells.Another mechanism that contributes to cell-specificgene expression is epigenetic modifications. These modifications are chemical changes to DNA or histones that can alter the accessibility of DNA to transcription factors. For example, DNA methylation is a type of epigenetic modification that can repress gene expression by blockingthe binding of transcription factors to DNA.In addition to cell specificity, gene expression isalso subject to species evolution. Over time, changes inthe DNA sequence of genes can lead to changes in gene expression. These changes can have a variety of effects, including alterations in the function of cells, the development of new traits, and the evolution of new species. For example, changes in the expression of genes involved in beak morphology have been linked to the evolution ofdifferent beak shapes in Darwin's finches.The study of gene expression is a rapidly growing field of research. Advances in molecular biology techniques have made it possible to identify and characterize the genesthat are expressed in different cells and tissues. This research has provided valuable insights into the development and function of multicellular organisms and has also helped to elucidate the mechanisms of evolution.Chinese Response:基因表达是一个复杂且受严格调控的过程,在决定细胞的特征和功能方面起着至关重要的作用。
转录组学的英文
转录组学的英文Transcriptomics, the study of the transcriptome, is an emerging field of research in biology. It aims to understand the complete set of RNA molecules produced by a cell or a population of cells under certain conditions. At its core, transcriptomics seeks to reveal the complex mechanisms of gene expression and its regulation.The transcriptome is the set of all RNA molecules, including coding and non-coding RNAs, that are produced by a cell or an organism. Transcriptomics investigates the messenger RNA (mRNA) that directs the synthesis of proteins, as well as other RNA molecules that are not translated into proteins, such as ribosomal RNA (rRNA), transfer RNA (tRNA), and small RNA molecules, such as microRNA (miRNA) and small interfering RNA (siRNA).Advances in high-throughput sequencing technologies have enabled transcriptomics to become a powerful tool in understanding the genetic basis of various biological phenomena. Transcriptomicdata can provide insights into the molecular mechanisms of developmental processes, cellular responses to environmental stimuli, and disease pathogenesis.One of the key applications of transcriptomics is in molecular diagnostics. By comparing the transcriptomes of healthy and diseased tissues, researchers can identify disease-specific biomarkers that may be used for early diagnosis or in the development of targeted therapies.Transcriptomics has also been used in drug discovery to identify new drug targets andpotential drug candidates. By analyzing the expression levels of genes in different tissues and cell types, researchers can identify pathways that are specific to certain diseases and develop drugs that target those pathways.Another emerging application of transcriptomics is in the field of synthetic biology. By engineering gene expression patterns, researchers can create custom metabolic pathways or functional cellular systems. Transcriptomic data can informthe design of these systems and enable researchers to optimize their function.Despite its many potential applications, transcriptomics faces several challenges. One major challenge is the huge amount of data that is generated by high-throughput sequencing technologies. Analyzing and interpreting this data requires advanced computational algorithms and infrastructure. Additionally, interpretation of transcriptomic data is complex, involving comparisons of gene expression across multiple samples, tissues, and conditions.In conclusion, transcriptomics is a rapidly evolving field with many potential applications in biology, medicine, and synthetic biology. By understanding the transcriptome, researchers can gain insight into the complex mechanisms of gene expression and regulation, identify biomarkers for early diagnosis of disease, develop new drugs and therapies, and engineer new cellular systems. Despite the challenges, transcriptomics holds greatpromise as a powerful tool for biological research and discovery.。
伯杰氏细菌鉴定手册最新版弧菌科部分
GENUS XIII.SHEWANELLA491M NaCl.Growth occurs at4–30ЊC.Optimum growth tem-perature is25ЊC.Grows between pH6–8.Optimum pH for growth is7.0.Chemoheterotrophic facultative anaerobe.Can grow an-aerobically using nitrate,fumarate,iron,manganese, TMAO,thiosulfate,and elemental sulfur as alternative elec-tron acceptors with lactate acting as the carbon source.Cat-alase,oxidase,and lipase positive.Amylase and gelatinase negative.Glucose,galactose,lactate,acetate,pyruvate,cit-rate,succinate,glutamate,Casamino acids,yeast extract, and peptone are used aerobically as energy sources.Fruc-tose,glycerol,sorbitol,arabinose,formate,and ethanol are not utilized.Isolated from the accessory nidamental glands of female adults of the squid species Loligo pealei.The mol%GםC of the DNA is:45.0(HPLC).Type strain:ANG-SQ1,ATCC700345.GenBank accession number(16S rRNA):AF011335.12.Shewanella violacea Nogi,Kato and Horikoshi1999,341VP(Effective publication:Nogi,Kato and Horikoshi1998b, 337.)Јce.a.L.gen.n.violacea of violet.Cells are straight or slightly curved rods,0.8–1.0ן2–4l m.Colonies on marine agar are circular,smooth,convex with entire edges,and butyrous in consistency.After2–3d, colonies are nonpigmented;after more than7d,colonies appear violet.NaCl is required for growth;optimal levels for growth are2–3%.No growth with6%NaCl.Psychrophilic.Grows optimally between4–10ЊC.Baro-philic.Optimal pressure for growth is30MPa.Other characteristics are as given in the genus descrip-tion and in Table BXII.c.148.Acid is produced from cel-lobiose and d-galactose,but not from dl-arabinose,d-fruc-tose,glycerol,inositol,lactose,maltose,d-mannitol,d-man-nose,d-raffinose,l-rhamnose,d-sorbitol,sucrose,d-treha-lose,or d-xylose.Isolated from the Ryukyu Trench,northwest Pacific Ocean,at a depth of5110m.The mol%GםC of the DNA is:47(T m).Type strain:DSS12,JCM10179.GenBank accession number(16S rRNA):D21225.13.Shewanella woodyi Makemson,Fulayfil,Landry,Van Ert,Wimpee,Widder and Case1997,1039VPwoodЈy.i.M.L.gen.n.woodyi of Woody,in honor of the American biologist J.Woodland Hastings.Cells are rod-shaped,0.4–1.0ן1.4–2.0l m.Colonies on marine agar are pink-orange due to the accumulation of cytochromes.NaCl is required for growth.Growth factors are not required.Psychrophilic.Growth occurs between4and25ЊC;op-timum temperature,20–25ЊC;no growth at30ЊC.Other characteristics are as given in the genus descrip-tion and in Tables BXII.c.148and BXII.c.151.Isolated from squid ink,seawater and marine snow(col-lected from the Alboran Sea).The mol%GםC of the DNA is:39(by measurement of the relative binding of DNA-binding-fluorescent dyes bis-benzimide and chromomycin A3).Type strain:MS32,ATCC51908,DSM12036.GenBank accession number(16S rRNA):AF003549.Order XI.“Vibrionales”Vib.ri.o.naЈles.M.L.masc.n.Vibrio type genus of the order;-ales ending to denote order;M.L.fem.pl.n.Vibrionales the order of bacteria whose circumscription is based on thegenus Vibrio.Description is the same as for the family Vibrionaceae.Type genus:Vibrio Pacini1854,411.Family I.Vibrionaceae Ve´ron1965,5245ALJ.J.F ARMER III AND J.M ICHAEL J ANDAVib.ri.o.naЈce.ae.M.L.masc.n.Vibrio type genus of the family;-aceae ending to denote family;M.L.fem.pl.n.Vibrionaceae the family of bacteria whose circumscription is based on the genusVibrio.Gram-negative straight or curved rods.Motile by means of polar flagella.Additional lateralflagella may be produced when grown on solid media;these differ in wavelength and antigenicity from the polarflagellum and may number from a few to over100flagella/cell.Do not form endospores or microcysts.Chemoor-ganotrophs.Facultative anaerobes,having both a respiratory and a fermentative metabolism.Oxygen is a universal electron ac-ceptor.Do not denitrify.Most strains:are oxidase positive,reduce nitrate to nitrite,ferment d-glucose and utilize it as a sole or principal source of carbon and energy,grow in minimal media with d-glucose or other compounds as the sole source of carbon and energy and use NH4םas the sole nitrogen source.A few species require vitamins and amino acids.Ferment and utilize a wide variety of simple and complex carbohydrates and utilize a wide variety of other carbon sources.Most species require Naםor a seawater base for growth and require0.5–3%NaCl for op-timum growth.Several species are bioluminescent;other species include a few bioluminescent strains.Primarily aquatic.Found in fresh,brackish,and sea water,often in association with aquatic animals and plants.Several species are pathogenic for humans.FAMILY I.VIBRIONACEAE 492Other species are pathogenic forfish,eels,and other aquatic animals.The mol%GםC of the DNA is38–51%.The family is classified in the phylum Proteobacteria in the class Gammapro-teobacteria.Type genus:Vibrio Pacini1854,411.Historical overview A history of the family Vibrionaceae as it has appeared in Bergey’s Manual is given in Table BXII.c.152. Related families include Enterobacteriaceae,Aeromonadaceae,and Pasteurellaceae.The family Vibrionaceae has undergone intense study since thefirst edition of Bergey’s Manual of Systematic Bac-teriology(Krieg and Holt,1984)was published in1984.In their chapter on the family Vibrionaceae in that edition,Baumann and Schubert(1984)included the genera Vibrio,Photobacterium,Ae-romonas,and Plesiomonas.These are the same four genera in-cluded in the original classification of the family proposed by Ve´ron almost twenty years earlier(Ve´ron,1965).In this Manual Aeromonas and Plesiomonas are classified in other families(Table BXII.c.152).For practical identification schemes,it is still useful to consider Aeromonas and Plesiomonas together with other oxidase positive genera of fermentative bacteria such as Vibrio and Pho-tobacterium(Table BXII.c.153).A detailed history of changes in the classification of Vibrio and related genera that occurred as new methods were introduced has been given by Farmer(1992). These methods include examinations of the structure,function, and regulation of proteins;comparison of mol%GםC content; DNA–DNA hybridization;rRNA–DNA hybridization;5S rRNA cataloging and sequence comparisons;and16S rRNA gene se-quence comparisons(Fig.BXII.c.157).The family Vibrionaceae presently includes three genera:Genus1.Vibrio(the type genus)Genus II.PhotobacteriumGenus III.SalinivibrioThe type strain of the type and only species of the genus Allomonas,Allomonas enterica,is very closely related to Vibrioflu-vialis by DNA–DNA hybridization studies and phenotypic analysis (Kalina et al.,1984).Allomonas and Allomonas enterica are not described separately in this edition of the Manual;the reader is referred to the description of V.fluvialis.The two species of the genus Listonella,Listonella anguillarum(the type species)and Lis-tonella pelagia,are included in the genus Vibrio in this edition of the Manual as Vibrio anguillarum and Vibrio pelagius.*F URTHER DESCRIPTIVE INFORMATIONHabitats The ecological niches of members of the family Vibrionaceae have been described by Campbell(1957);Baumann and Baumann(1981a);Sakazaki and Balows(1981);and Simidu and Tsukamoto(1985)).In humans,some vibrios cause diarrhea, wound infections,and occasionally other extraintestinal infec-tions.In aquatic animals,vibrios cause wound and generalized infections.Many vibrios and related organisms are also widely distributed in aquatic environments.Many factors govern the distribution of these organisms,but the most important probably include:particular human,animal or plant hosts;inorganic nu-trients and carbon sources available;temperature;salinity;dis-solved oxygen;and depth below the surface for the species that are found in the ocean(Simidu and Tsukamoto,1985).A few species are adapted to particular hosts.For example,Vibrio chol-erae serogroup O1is adapted to humans and is the cause of cholera,a life-threatening diarrheal disease.Recent studies have shown that the ecology of this organism is more complex than originally thought.Photobacterium leiognathi is usually isolated fromfish in shallow tropical water,and P.phosphoreum is usually found in the luminous organs offish that live at depths of200–1200meters(Hastings and Nealson,1981).Isolation Most members of the Vibrionaceae grow well on ordinary complex media.Samples are spread onto solid medium or diluted in an enrichment broth.NaCl concentrations of0.5–0.85%satisfy the requirements of most species,although a few require greater concentrations of NaCl.Incubation temperatures are also important.A few species grow only at temperatures Ͻ25ЊC;others grow at25ЊC but not at35–37ЊC.General and selective media for Vibrionaceae are described in the chapter on the genus Vibrio.Identification Methods for the isolation and identification of Vibrio spp.from clinical specimens and non-clinical samples are discussed in detail in the chapter describing the genus Vibrio. Assignment of non-clinical isolates to a species can be problem-atic because over50species of Vibrio and Photobacterium must be considered and because comparative data for these organisms are sparse relative to data available for clinically important spe-cies.The US Centers for Disease Control and Prevention maintain computer programs and databases for the identification of iso-lates subjected to a battery of45–60phenotypic tests;for details contact the Vibrio Laboratory at the CDC.These alternatives to phenotypic methods are now being used routinely and have proven extremely useful in a research setting. It will be important to evaluate the sensitivity and specificity,and to understand the advantages and disadvantages of these meth-ods.In the United States,the reporting of cultures from human specimens is subject to specific government regulations(the Clin-ical Laboratory Improvement Amendments of1988),which has limited the application of these approaches in clinical and public health laboratories.A CKNOWLEDGMENTSWe dedicate this chapter to M.Ve´ron for giving us the name Vibrionaceae and for all his contributions to our understanding of the family,its or-ganisms,and their close and distant relatives.F URTHER R EADINGBaumann,P.and L.Baumann.1977.Biology of the marine enterobac-teria:genera Beneckea and Photobacterium.Ann.Rev.Microbiol.31:39–61.Baumann,P.and L.Baumann.1981.The marine Gram-negative eubac-teria.In Starr,Stolp,Tru¨per,Balows and Schlegel(Editors),The Pro-karyotes,a Handbook on Habitats,Isolation and Identification of Bacteria,1st Ed.,Springer-Verlag,New York.pp.1352–1394. Baumann,P.,L.Baumann,S.S.Bang and M.J.Woolkalis.1980.Reeval-uation of the taxonomy of Vibrio,Beneckea,and Photobacterium:aboli-tion of the genus Beneckea.Curr.Microbiol.4:127–132. Baumann,P.,L.Baumann and M.Mandel.1971.Taxonomy of marine bacteria:the genus Beneckea.J.Bacteriol.107:268–294. Baumann,P.,A.L.Furniss and J.V.Lee.1984.Genus I.Vibrio.In Krieg and Holt(Editors),Bergey’s Manual of Systematic Bacteriology,1st Ed.,Vol.1,The Williams&Wilkins Co.,Baltimore.pp.518–538. Baumann,P.and R.H.W.Schubert.1984.Family II.Vibrionaceae.In Krieg and Holt(Editors),Bergey’s Manual of Systematic Bacteriology,1st Ed.,Vol.1,The Williams&Wilkins Co.,Baltimore.pp.516–517. Brenner,D.J.,G.R.Fanning,F.W.Hickmann-Brenner,J.V.Lee,A.G.Stei-FAMILY VIBRIONACEAE493gerwalt,B.R.Davis and J.J.Farmer.1983a.DNA relatedness among Vibrionaceae,with emphasis on the Vibrio species associated with human infection.INSERM Colloq.114:175–184.Chakraborty,S.,G.B.Nair and S.Shinoda.1997.Pathogenic vibrios in the natural aquatic environment.Rev.Environ.Health.12:63–80. Farmer,J.J.1992.The family Vibrionaceae.In Balows,Tru¨per,Dworkin,Harder and Schleifer(Editors),The Prokaryotes.A Handbook on the Biology of Bacteria:Ecophysiology,Isolation,Identification,Ap-plications,2nd Ed.,Vol.3,Springer-Verlag,New York.pp.2938–2951. Farmer,J.J.,M.J.Arduino and F.W.Hickman-Brenner.1992.Aeromonas and Plesiomonas.In Balows,Tru¨per,Dworkin,Harder and Schleifer (Editors),The Prokaryotes.A Handbook on the Biology of Bacteria:FAMILY I.VIBRIONACEAE 494GENUS I.VIBRIO495FIGURE BXII.c.157.Relationship of most of the species of Vibrio and relatives based on16S rRNA gene sequences.* Only the type strain of each species was included.The distances in the tree were calculated using1101positions (the least-squares method,Jukes-Cantor model).(Courtesy T.Lilburn of the Ribosomal Database Project.)*Editorial Note:Photobacterium damselae subsp.damselae is a junior objective synonym of Vibrio damsela.Vibrio pelagius and Vibrio anguillarum are synonyms of Listonella pelagia and Listonella anguillarum,respectively.and volume(15–67%)and a conversion of rods into coccal forms called spherical ultramicrocells(Holmquist and Kjelleberg,1993; Kondo et al.,1994;Nelson et al.,1997).As the length of nutrient starvation increases,cytoplasmic inclusions and granules disap-pear,cell cultivability decreases,and the nuclear region becomes compressed(Hood et al.,1986).There are also noticeable dif-ferences in the integrity of the outer membrane and cell wall. Some changes may be linked to specific nutrient starvation(for example,nitrogen starvation produces longfilaments and phos-phorus starvation produces swollen large rods),whereas others occur regardless of the type of nutritional stress(Holmquist and Kjelleberg,1993).“Non-culturable”V.cholerae O1strains pro-duced in response to nutrient deprivation display a number of ultrastructural changes,which include an undulating outer mem-brane,a surface layer offinefibers,and a thicker peptidoglycan layer(Kondo et al.,1994).FAMILY I.VIBRIONACEAE 496Poly-b-hydroxybutyrate granules(PHB)can be found in a number of Vibrio species,including V.cholerae O1and O139and V.harveyi(Hood et al.,1986;Sun et al.,1994;Finkelstein et al., 1997).In V.cholerae,accumulation of PHB appears to be related to colonial opacity and growth on glycerol-containing media(Fin-kelstein et al.,1997).In V.harveyi,PHB accumulation is de-pendent on cell density and is controlled by the autoinducer,N-(3-hydroxybutanoyl)homoserine lactone(Sun et al.,1994). Other kinds of granules can be found in vibrios,including elec-tron dense lipoid particles and electron translucent inclusions of unknown composition(Sun et al.,1994;Finkelstein et al., 1997).Cell wall composition Vibrios contain the same three lipo-polysaccharide(LPS)elements found in other Gram-negative bacteria:lipid A,core polysaccharide,and an O polysaccharide side chain that determines serological specificity.The most ex-tensive work on biochemical characterization of Vibrio LPS has been done on V.cholerae.The lipid A portion consists of a b(1Ј-6)-linked glucosamine disaccharide backbone with two phos-phoryl groups(Janda,1998).Pyrophosphorylethanolamine is linked to one of these phosphoryl groups at the C-1position of the reducing sugar,and a phosphate group ester is bound to the nonreducing glucosamine residue(Manning et al.,1994).Three fatty acids are ester linked at hydroxyl positions to this disaccha-ride backbone:tetradecanoic acid(C14:0),hexadecanoic acid (C16:0),and3-hydroxydodecanoic acid(C12:03OH).A fourth,3-hydroxytetradecanoic acid(C14:03OH),is connected to the back-bone by an amide bond.The core oligosaccharide region of V.cholerae contains KDO (keto-3-deoxy-d-mannose-octulosonic acid),d-glucose,heptose (l-glycero-d-manno-heptose),d-fructose,and ethanolamine phosphate(Manning et al.,1994).KDO,a normal constituent of the core oligosaccharide of enteric LPS,was originally thought to be absent in Vibrio species.However,when conventional per-iodate-thiobarbituric acid tests were replaced by strong acid hy-drolysates,KDO was detected in Vibrio(Janda,1998).The KDO molecule of V.cholerae differs in several aspects from those of enteric bacteria such as Escherichia coli and the genus Salmonella: only a single KDO molecule has been detected in the core oli-gosaccharide of V.cholerae,and the KDO moiety is phosphory-lated at the C4position(Kondo et al.,1990;Manning et al., 1994).The C5position binds to a distal portion of the core region (heptose)similar to the KDO-C5binding of l-glycero-d-manno-heptose(Janda,1998).The other sugars form the remaining portion of the core oligosaccharide region and often contain additional sugar substitutions at various positions.The O-polysaccharide side chain of V.cholerae O1is a homo-polymer of d-perosamine(4,6-dideoxy-d-mannose)approxi-mately17–18units in length(Manning et al.,1994;Knirel et al., 1997).The amino groups of perosamine units are commonly acetylated with3-deoxy-l-glycero-tetronic acid.Another com-pound,quinovosamine,is thought to be a“capping sugar”on either the distal or the proximal end of the O antigen(Manning et al.,1994).An unusual sugar,4-amino-4,6-dideoxy-2-O-methyl-mannose is present only in the LPS of serogroup Ogawa and may have a role in serological specificity(Itoh et al.,1994).The LPS composition of V.cholerae O139—a second serotype capable of causing pandemic cholera—is remarkably similar to that of O1(Hisatsune et al.,1993;Isshiki et al.,1996).The lipid A moieties of O1and O139,including fatty acid substitutions, appear to be identical(Hisatsune et al.,1993).The core oligo-saccharide region contains two subtle differences:the presence of2-aminoethyl phosphate,which is the O-acetyl group,and the presence of a second fructose molecule(Knirel et al.,1997).The most profound differences between O-groups1and139occur in the O-polysaccharide side chain.Unlike serogroup O1,which has long O-polysaccharide side chains,V.cholerae O139has a short chain LPS similar to“SR strains”(Knirel et al.,1997).These truncated side chains migrate with the core oligosaccharide-lipid A fraction in LPS SDS-PAGE gels(Waldor et al.,1994).Classic “ladder-like”profiles of silver stained LPS side chains in SDS-PAGE gels are absent in O139strains(Hisatsune et al.,1993; Nandy et al.,1995).Perosamine,the main component of the O1 side chain,is also absent in O139strains(Hisatsune et al.,1993); instead,the unique sugar colitose(3,6-dideoxy-l-galactose)—which is not found in any other Vibrio species—is the main side chain subunit in O139strains(Hisatsune et al.,1993).The ab-breviated O-polysaccharide side chain of V.cholerae O139appears to be a hexasaccharide containing colitose residues and a cyclic phosphate group(Knirel et al.,1997).The LPS of other Vibrio species is similar in many aspects to that of O139.KDO-phosphate has been detected in V.parahaemolyticus by gas chromatography-mass spectrometry analysis(Janda,1998).The O-polysaccharide side chains of Vibrio species produce only a single fast-migrating band on silver-stained SDS-PAGE gels(Amaro et al.,1992;Iguchi et al.,1995).This result suggests that the side chains are short;a chain length ofՅ10monosaccharides has been proposed for V.parahaemolyticus(Iguchi et al.,1995).Some species however (e.g.,V.vulnificus)may exhibit ladder-like patterns by immu-noblotting with whole cell antisera;this result suggests that the LPS O-polysaccharide side chains are mbert et al.(1983) studied the cellular fatty acids of most of the Vibrionaceae and postulated that differences among the Vibrio species might prove useful for identification.Flagella Two types offlagella are synthesized by vibrios in different environments.In liquid culture,swimmer cells predom-inate due to production of a single sheathed polarflagellum in most species(Figs.BXII.c.158and BXII.c.159).The sheath is an extension of the outer membrane(Fig.BXII.c.160).The polar flagella are24–30nm in diameter with a central core14–16nm in thickness with a wavelength of1.4–1.8l m(Baumann et al., 1984b;Janda,1998).Some Vibrio species(e.g.,V.harveyi,V.fischeri, V.logei,and V.salmonicida)produce tufts(3–12)of polarflagella (Fig.BXII.c.161)with a wavelength of approximately3.6l m (Baumann et al.,1984b;Ishimaru et al.,1996).Polarflagella provide chemotactic motility in liquid media and derive their energy from the sodium membrane potential(McCarter,1995). In some marine vibrios(e.g.,V.anguillarum),the polarflagellum appears critical for disease production in estuarinefish(Milton et al.,1996;O’Toole et al.,1996).When vibrios come into contact with solid surfaces,a series of morphogenetic changes are ini-tiated that result in the conversion of swimmer cells into swarmer cells in some marine species such as V.parahaemolyticus,V.algi-nolyticus,V.diabolicus,and V.pectenicida(Rague´ne`s et al.,1997a; Lambert et al.,1998).During this process,cell septation ceases, the cells elongate from1to30l m,and numerous lateralflagella are formed(Fig.BXII.c.162)(McCarter and Silverman,1990). These lateralflagella,14–15nm in diameter with a wavelength of0.9l m,are distinct from polarflagella.They are unsheathed, have a different protein subunit composition,and are internally driven by the protonmotive force(Baumann et al.,1984b; McCarter,1995).Formation of lateralflagella permits swarmerGENUS I.VIBRIO497FIGURE BXII.c.158.Leifsonflagella stain of Vibrio cholerae.(Source:CDC archive,courtesy of EdEwing.)FIGURE BXII.c.159.Electron micrograph of Vibrio alginolyticus grown in liquid medium.Note the sheathed polarflagellum and absence of pe-ritrichousflagella.Shadowed preparation.ן13,000.(Reproduced with permission from C.Golten and W.A.Scheffers,Netherlands Journal of Sea Research9:351–364,1975,᭧Netherlands Institute for SeaResearch.)FIGURE BXII.c.160.Electron micrograph of a polarflagellum of Vibrio alginolyticus.Note that the sheath has partially disintegrated exposing the inner core.Negatively stained preparation.ן30,000.(Courtesy of R.D. Allen.)migration across solid surfaces and results in progressive spread-ing of the bacterial colony(McCarter and Silverman,1990),a phenomenon called swarming.Swarming in many vibrio species is dependent upon a number of factors including agar concen-tration,media composition,iron availability,temperature,and relative viscosity(Baumann et al.,1984b;McCarter and Silver-man,1990).The microscopic morphology of vibrio cells removed from different concentric zones of swarming has been studied in some Vibrio strains(Sar and Rosenberg,1989).Innermost zones consist of irregular cells including bent rods that pro-gressively evolve into short rods and then into large rods with bundles of detachedflagella(Fig.BXII.c.163),whereas cells inFAMILY I.VIBRIONACEAE498FIGURE BXII.c.161.Electron micrograph of Vibriofischeri.Note the tufts of sheathed polarflagella.Negatively stained preparation.ן23,000. (Reproduced with permission from:J.L.Reichelt and P.Baumann,Ar-chives of Mikrobiology94:283–330,1973,᭧Springer-Verlag,Berlin.)FIGURE BXII.c.162.Electron micrograph of Vibrio alginolyticus grown on solid medium.Note the thick,sheathedflagellum and numerous un-sheathed lateralflagella.Shadowed preparation.ן18,000.(Reproduced with permission from W.E.de Boer et al.,Netherlands Journal of Sea Research9:197–213,1975,᭧Netherlands Institute for Sea Research.)the outermost circles of swarming colonies consist of longfila-mentous forms.Fimbriae Fimbriae are produced by a number of pathogenic vibrios such as V.cholerae O1and non-O1,V.parahaemolyticus, and V.vulnificus(Hall et al.,1988;Honda et al.,1988;Gander and LaRocco,1989;Nakasone and Iwanaga,1990).Several dif-ferent morphologic types offimbriae have been described in V. cholerae O1.These include both wavy pili3nm in diameter and rigidfilaments5–6nm wide and180–800nm in length(Hall et al.,1988).The most important of these pili is composed of the protein TcpA;these pili are5–6nm wide and form bundles of parallel undulatingfilaments up to15l m long(Hall et al.,1988). TcpA formation is coregulated with cholera toxin expression and is a key determinant of in vivo colonization.The gene encoding TcpA appears to reside on a pathogenicity island.Capsules Capsules have been detected surrounding cells of strains of V.cholerae O139and V.vulnificus strains with a variety of staining techniques such as uranyl acetate,polycationic fer-ritin,and ruthenium red(Janda,1998).The V.vulnificus poly-saccharide capsule is60nm thick and has a low electron density (Amako et al.,1984;Hayat et al.,1993).The carbohydrate com-position of the capsule of V.vulnificus varies from strain to strain. Sugars detected in different isolates include␣-N-acetyl quino-vosamine,␣-N-acetyl galactosamine uronic acid,rhamnosamine, and fucosamine(Hayat et al.,1993).Colonial morphology Most vibrios grow well on a variety of media,including protein-based agars and marine and seawater media,if sufficient Naםis present(Baumann et al.,1984b; Farmer and Hickman-Brenner,1992).On most selective media, vibrios appear as smooth,buff-to-cream-colored colonies2–5mm in diameter,with an entire margin after overnight incubation (Baumann et al.,1984b;Janda,1998).Some species tend to pro-duce grayish colonies,particularly on blood agar.Considerable variation in colonial morphology has been reported for some species,and this is best demonstrated by observing colonies with a dissecting microscope at10–25magnification with oblique lighting.V.cholerae strains can have several different colonial mor-phologies(smooth,rough,and rugose forms)in response to different growth conditions.Rugose colonies are often chlorine-resistant.They are usually found in older cultures and are com-posed of an amorphous intercellular matrix of aggregated bac-teria and exopolysaccharide material(Morris et al.,1996).For-mation of rugose colonies can be enhanced by growth in en-richment broths such as alkaline peptone water(APW)or by picking the growth that has migrated up the sides of a culture tube.They can largely be avoided by picking a smooth colony and freezing it.In addition to these colony types,several path-GENUS I.VIBRIO499FIGURE rge bundle offlagella in a culture of Vibrio harveyi. These bundles are frequently observed in cultures grown on solid me-dium.Negatively stained preparation.ן13,000.(Courtesy of R.D.Allen.) ogenic vibrios,including V.cholerae and V.vulnificus,produce opaque and translucent varieties of smooth colonies on common media such as heart infusion and meat extract agars(Simpson et al.,1987;Finkelstein et al.,1992,1997).Cells from colonies of these different morphologies differ from each other in a num-ber of characteristics,including encapsulation,cell surface com-position,cellular metabolism,and ability to survive under adverse conditions.Pigmentation Several Vibrio species produce pigmented col-onies.V.nigripulchritudo produces an insoluble blue-black pig-ment that accumulates in a crystalline form within the colonies (Baumann et al.,1984b).Other Vibrio species also produce blue-black crystals under various growth conditions,but typically do not produce the characteristic blue-black colonies of V.nigripul-chritudo.Similar blue-black colonies are produced by a few strains of Kluyvera(Farmer et al.,1981a).Other pigmented species in-clude V.gazogenes(red)and V.fischeri and V.logei(yellow-orange).A few strains of V.cholerae produce a brown diffusible melanin-like pigment(Ivins and Holmes,1980).Life cycles The marine environment is the natural habitat of vibrios,and the life cycle of these organisms is probably quite complex.A model for the life of V.cholerae in Gulf Coast estuaries has been proposed(Hood et al.,1984).Depending on nutrient scarcity and the density/availability of particulate matter,V.chol-erae can exist in several states.An epibiotic form attached to plankton predominates during periods of relatively high nu-trient/particulate matter concentrations;this form changes to a microvibrio form(small rounded cells)during times of nutrient and particulate deprivation(Hood et al.,1984;Janda,1998).This latter form may be analogous to the“viable but nonculturable state”(Colwell,1984)that has been described for many Vibrio species including V.cholerae,V.parahaemolyticus,V.vulnificus,V. anguillarum,V.campbellii,V.harveyi,and V.fischeri during colder seasons of the year(Oliver,1995).The microvibrio and“non-culturable”stages may be dormant phases for vibrios in winter from which subsequent blooms are triggered in response to in-creasing temperatures in spring and summer.However,it is still unclear whether any dormant state actually exists and whether blooms are due to the growth of a small number of cultivable cells(present at all times)or the actual resuscitation of dormant cells(Ravel et al.,1995;Bogosian et al.,1998).Nutrition and growth conditions Vibrio species vary in their nutrition and growth requirements.The most important feature is that Naםis required for or stimulates growth.Minimum con-centrations of Naםrequired for optimal growth(Fig.BXII.c.164) range from5–15mM(0.029–0.087%)for V.cholerae and V.metsch-nikovii to600–700mM(3.5–4.1%)for Salinivibrio costicola(Bau-mann et al.,1984b).Most species grow well in solid or liquid media containing0.5–2%NaCl.Some species(Photobacterium ili-opiscarium)form bacterial aggregates in broth culture containing 2%NaCl(Onarheim et al.,1994).The“salt requirement”of a strain will often depend on the test conditions.The main variables are temperature,the growth medium used prior to testing,the suspending medium,and the testing medium.All Vibrio species except V.cholerae and V.mimicus have an absolute requirement for Naם(Fig.BXII.c.164).In some instances,this requirement may be partially offset by concentra-tions of Mg2םor Ca2םsimilar to those normally present in sea-water(Baumann et al.,1984b).However,most species exhibit a specific requirement for Naם(Pujalte and Garay,1986;Borrego et al.,1996).The range and optimum concentrations of NaCl supporting growth of some of the more recently described Vibrio species are listed in Table BXII.c.154.No single medium or NaCl concentration is optimal for the recovery or growth of all Vibrio species.Many vibrios will grow in mildly alkaline conditions.Al-though most species prefer a pH range of7–8(Rague´ne`s et al., 1997a),some species,including V.cholerae and V.metschnikovii, will even grow at a pH of10(Baumann et al.,1984b).Vibrios also vary in their temperature requirements for growth.Almost all Vibrio species grow well at18–22ЊC.Some will grow at0–4ЊC,whereas others can grow at temperatures up to 45ЊC.The temperature at which vibrios can grow is also de-pendent upon other factors including the composition of the medium and the NaCl concentration(Onarheim et al.,1994).Most Vibrio species do not require specific organic growth factors such as vitamins or amino acids,although amino acid supplementation may be required to revive some strains stored for prolonged periods(Baumann et al.,1984b).Complex nu-trients are required to induce growth of some species(Baumann et al.,1984b;Rague´ne`s et al.,1997a).Such required supplements include yeast extract(for V.anguillarum,Moritella marina,and V. logei)and a seawater base(for V.diabolicus).Vibrios use a variety of compounds as carbon and energy。
双低剂量肺动脉血管造影(CTPA)成像技术在诊断肺动脉栓塞的可行性研究
68·中国CT和MRI杂志 2024年1月 第22卷 第1期 总第171期【通讯作者】成启华,女,主任医师,主要研究方向:临床医学。
E-mail:****************FeasibilityCHINESE JOURNAL OF CT AND MRI, JAN. 2024, Vol.22, No.1 Total No.171含观察两组图像的CT值、信噪比(signal to noise ratio,SNR)、剂量长度乘积(dose length product,DLP)、容积CT剂量指数(computed tomography dose index,CTDIvoI)。
CT值测量的测试点选择在肺动脉主干及左右肺动脉分支取三个敏感区的平均CT 值,肺动脉干层面的胸前空气区左中右三个区域取三个敏感区,大小范围15mm,测量标准差的平均值作为背景噪声。
SNR=CT 值/背景噪声[4]。
DLP和CTDIvoI均由CT机自动测量。
图1 低碘对比剂剂量受检者VR图像;图2 低碘对比剂剂量受检者MIP显示分支;121.4 数据统计 数据采用SPSS 22.0统计学软件分析处理,计数资料采用率(%)表示,行χ2检验,计量资料用(χ-±s)表示,行t 检验,P<0.05为差异有统计学意义。
2 结 果2.1 两种检查的图像质量对比 两种检查方式的图像质量对比差异无统计学意义(P>0.05),见表1。
2.2 两种检查方式的定量参数对比 低剂量组与常规剂量组的CT 值、背景噪声、SNR、DLP、CTDIvoI相比差异具有统计学意义(P<0.01),见表2。
2.3 两种检查对于肺动脉栓塞的检出率 低剂量组30例肺动脉栓塞检出率为93%,常规剂量组30例肺动脉栓塞检出率为90%,低剂量组与常规剂量组肺动脉栓塞检出率对比差异无统计学意义(P>0.05),见表3。
3 讨 论 肺动脉栓塞是一种危机重症,受检者的死亡率非常高,对受检者的生命健康及生命安全有着非常严重的威胁[5]。
- 1、下载文档前请自行甄别文档内容的完整性,平台不提供额外的编辑、内容补充、找答案等附加服务。
- 2、"仅部分预览"的文档,不可在线预览部分如存在完整性等问题,可反馈申请退款(可完整预览的文档不适用该条件!)。
- 3、如文档侵犯您的权益,请联系客服反馈,我们会尽快为您处理(人工客服工作时间:9:00-18:30)。
Transcriptome Characteristics and Six Alternative Expressed Genes Positively Correlated with the Phase Transition of Annual Cambial Activities in Chinese Fir (Cunninghamia lanceolata(Lamb.)Hook)Zhanjun Wang1.¤,Jinhui Chen1.,Weidong Liu1,Zhanshou Luo1,Pengkai Wang1,Yanjuan Zhang1, Renhua Zheng2,Jisen Shi1*1Key Laboratory of Forest Genetics and Biotechnology,Ministry of Education of China,Nanjing Forestry University,Nanjing,China,2Fujian Academies of Forestry, Southern Mountain Timber Forest Cultivation Lab,the Ministry of Forestry,Fuzhou,ChinaAbstractBackground:The molecular mechanisms that govern cambial activity in angiosperms are well established,but little is known about these molecular mechanisms in gymnosperms.Chinese fir(Cunninghamia lanceolata(Lamb.)Hook),a diploid (2n=2x=22)gymnosperm,is one of the most important industrial and commercial timber species in China.Here,we performed transcriptome sequencing to identify the repertoire of genes expressed in cambium tissue of Chinese fir.Methodology/Principal Findings:Based on previous studies,the four stage-specific cambial tissues of Chinese fir were defined using transmission electron microscopy(TEM).In total,20million sequencing reads(3.6Gb)were obtained using Illumina sequencing from Chinese fir cambium tissue collected at active growth stage,with a mean length of131bp and a N50of90bp.SOAPdenovo software was used to assemble62,895unigenes.These unigenes were further functionally annotated by comparing their sequences to public protein databases.Expression analysis revealed that the altered expression of six homologous genes(ClWOX1,ClWOX4,ClCLV1-like,ClCLV-like,ClCLE12,and ClPIN1-like)correlated positively with changes in cambial activities;moreover,these six genes might be directly involved in cambial function in Chinese fir.Further,the full-length cDNAs and DNAs for ClWOX1and ClWOX4were cloned and analyzed.Conclusions:In this study,a large number of tissue/stage-specific unigene sequences were generated from the active growth stage of Chinese fir cambium.Transcriptome sequencing of Chinese fir not only provides extensive genetic resources for understanding the molecular mechanisms underlying cambial activities in Chinese fir,but also is expected to be an important foundation for future genetic studies of Chinese fir.This study indicates that ClWOX1and ClWOX4could be possible reverse genetic target genes for revealing the molecular mechanisms of cambial activities in Chinese fir.Citation:Wang Z,Chen J,Liu W,Luo Z,Wang P,et al.(2013)Transcriptome Characteristics and Six Alternative Expressed Genes Positively Correlated with the Phase Transition of Annual Cambial Activities in Chinese Fir(Cunninghamia lanceolata(Lamb.)Hook).PLoS ONE8(8):e71562.doi:10.1371/journal.pone.0071562 Editor:Nicholas S.Foulkes,Karlsruhe Institute of Technology,GermanyReceived December22,2012;Accepted July1,2013;Published August12,2013Copyright:ß2013Wang et al.This is an open-access article distributed under the terms of the Creative Commons Attribution License,which permits unrestricted use,distribution,and reproduction in any medium,provided the original author and source are credited.Funding:This work was supported by grants from National Science Foundation of China grant30930077,30901156,31170619;‘Pandeng’Scholar grant of Jiangsu Province BK2008051;Public welfare projects of Chinese National Forest Bureau(201004049);Forest Tree Seeds and Seedlings Science and Technique Foundation of Fujian Province grants Fujian Forestry(2009-4);Jiangsu Provincial Graduate Student Innovation Project grant CXZZ11_0507;Doctorate Fellowship Foundation of Nanjing Forestry University grant2011YB017;and Priority Academic Program Development of Jiangsu Higher Education Institutions fund(PAPD).The funders had no role in study design,data collection and analysis,decision to publish,or preparation of the manuscript.Competing Interests:The authors have declared that no competing interests exist.*E-mail:jshi@¤Current address:Department of Life Science,Hefei Normal University,Hefei,China.These authors contributed equally to this work.IntroductionWood formation involves the cambium cell activities of division and differentiation,including cell expansion,cell wall thickening, lignification,and programmed cell death[1–3].Cambium cells can maintain themselves periclinally,and they give rise to xylem tissue(wood)centripetally and phloem tissue(bast)centrifugally [4–6].Therefore,an understanding of the regulation of cambial activities could facilitate the improvement of wood yield and quality.However,cambial activity is a complex process,which is regulated by both genetic and environmental signals[7–9].While previous studies have focused mainly on anatomical,biochemical, and cytological aspects[4,10–14],the molecular mechanisms of cambial activities are not well understood.Fortunately,with the completion of genome sequences for model plants such as Arabidopsis[15]and Populus[16],many molecular mechanisms and genes involved in cambial activity,such as TDIF/CLE41/ CLE44-TDR/PXY-WOX4[17–22],Class III HD-Zip/KANADI[23–26],have been identified in angiosperms[27].Unfortunately,little is known about the molecular mechanisms and genes in gymnosperms.There has been a rapid development of genomicand molecular tools,especially next-generation sequencing(NGS) technologies,such as the Illumina Genome Analyzer,Roche 454GS FLX Titanium,and ABI SOLiD[28–30],which have been widely applied to transcriptome sequencing in cambium and other vascular tissues,such as Eucalyptus[31],Liriodendron[32], Acacia[33],and so on.In addition,the altered expression of many regulatory genes correlates strongly with changes in cambial activity[34],and indeed many such genes are highly expressed when the cambium is active.Hence,the molecular mechanisms of cambial activity in gymnosperms can be revealed using NGS in the active growth stage of the cambium zone. Nonetheless,as a result of the structural characteristics and number of features of cambium cells[35,36],cambium tissue is difficult to identify accurately and sample successfully[37,38].For instance,actively growing cambial cells are characterized by large central vacuoles,thin tangential walls,and more cell layers.By contrast,dormant cambial cells are characterized by numerous subdivided vacuoles,thick tangential walls,and fewer cell layers [35,36].Laser microdissection(LMD),a technique for collecting cell-or tissue-specific material[39],has been successfully applied in sampling of cambium in Populus[40–42]and Picea glauca[43]. However,the amount of RNA collected using this method often does not meet the requirements of NGS;moreover,LMD has not been wildly used with woody plants[43].Thus,currently cambium tissues are currently collected primarily using the following methods:(i)scraping the inner surface of fresh bark [44,45];(ii)scraping the debarked surface of immature xylem of living trees[46,47];(iii)simultaneously scraping cambium tissue using both methods(i)and(ii)[48,49].With the return of cambial activity in early springtime,the bark of Chinese fir is easily removed from the tree stem,with the separation occurring at the cambial layer[46].The cambium tissues can then be scraped from the exposed surface of the xylem[46].Chinese fir(Cunninghamia lanceolata(Lamb.)Hook)is an evergreen coniferous tree that primarily distributed in southern China and northern Vietnam[50].It is one of the most important coniferous species[51–53],with high yield,excellent wood quality, versatile uses,and pest resistant[53,54].However,as of February 2012,only220nucleotides,445expressed sequence tags(ESTs) [52],and85proteins from Chinese fir had been deposited in the US National Center for Biotechnology Information(NCBI) GenBank database.In conclusion,the complex genetic back-ground and limited genomic information available in this species are obstacles to understanding the molecular mechanisms on cambial activity.In this study,we used TEM to define four stage-specific cambium tissues and used transcriptome sequencing to identify genes that are activated during cambial growth.Based on a bioinformatics analysis of assembled transcriptome data,house-keeping gene selection for qRT-PCR and expression analysis of17 orthologous genes were performed in four stage-specific cambium tissues.Two genes that showed positive correlations with changes in cambial activity were further cloned and analyzed.ResultsAnatomical Observations of the CambiumThe four stages of cambial activity were defined by comparing the results of TEM analysis between our study and Peng’s[55]as follows:reactivation(February28,S1),active growth(May26,S2), transition to dormancy(October12,S3),and dormancy(January 17,S4)(Figures1A–D).The structure of the cambium in the four stages can be described as follows:during S1,the cambial zone consists of three layers of cells containing dense cytoplasm and many small vacuoles(Figure1A);during S2,the cambial zone has nine layers of cells containing a large central vacuole,an organelle-rich cytoplasm,and a thin cell wall(Figure1B);during S3,the cambial zone has three layers of cells containing dense cytoplasm and a large central vacuole and some small vacuoles(Figure1C); during S4,the cambial zone has three to four layers of cells containing dense cytoplasm(Figure1D).These results provided a cytological foundation for further study of the molecular aspects underlying cambial activity.Assembly of SequencesTo identify key genes involved in cambial development in Chinese fir,transcriptome sequencing was performed using Illumina sequencing technology.Since the expression of related genes of cambial development is strongly correlated with the activity of cambium cells(which are vigorous during activated stage of cambial development),activated stage Chinese fir cambium tissue was used to construct the transcriptome sequenc-ing library(Figure S1).The library was sequenced with Illumina HiSeq2000.A total of20million sequencing reads(3.6Gb)were generated.After filtering out low quality reads,all reads were assembled by SOAPdenovo[56].The longest assembled sequenc-es containing no Ns were labeled contigs(Figure S2).Mapping reads back to contigs and combining paired-end information linked contigs into scaffolds.Scaffold length was estimated according to average segment length of each pair of reads. Unknown bases were filled with Ns.After filling gaps in scaffolds using paired-end reads,we obtained sequences called unigenes. Using information analysis of mapping reads back to contigs and paired-ends,we obtained637,458contigs,with a mean length of 131bp and N50of90bp,the length distributions of these contigs are shown in Table1;96.46%of the contigs displayed a length from75to400bp.Contig joining and gap filling were used to assemble118,391scaffolds with an average length of336bp and total length of11.84Mb(Table1).Finally,62,895unigenes were generated from the cambium transcriptome,with an average length of505bp,an N50of613bp,and a total length of 31.75Mb(Table1).There were37,499unigenes(59.62%)with lengths varying from200to400bp,19,188unigenes(30.51%) with lengths ranging from401to1000bp,and6256unigenes (9.87%)with lengths of more than1000bp(Table1).Quality Analysis of Assembled UnigenesTo assess the quality and coverage of assembled unigenes,we analyzed the sequencing depth range.The sequencing depth ranged from0.19to2517folds,in which79.71%of the unigenes were more than10reads,40.50%of the unigenes were more than 100reads,and20.29%of the unigenes varied from1to10reads (Figure2).To further analyze sequencing bias,random distribution of reads was detected in unigenes.Interestingly,comparison of the location of reads among the59ends and other positions of all unigenes showed that the reads seldom appeared near the3’ends of all unigenes(Figure3).In addition,the length of unigenes was compared between hit and no-hit in protein databases by BLAST matches.Consequently,70.63%of unigenes over500bp in length had BLAST matches,45.74%of unigenes between300bp and 500bp had BLAST matches,whereas only29.78%of unigenes shorter than300bp had BLAST matches(Figure4).These results were similar to previous reports of transcriptome research on Ipomoea batatas[57]and Camellia sinensis[58].To detect the sequence similarity in gene level between Chinese fir and Pinus taeda,TBLASTX alignment was performed on Chinese fir transcriptomes against a draft genome sequence of Pinus taeda.21,281unigenes (,33.84%of all the 62,895unigenes)of Chinese fir had significant matches in the Pinus taeda genome sequence (Table S1).Taken together,these results demonstrated that the quality of the Chinese fir unigenes was indeed high.Functional AnnotationTo predict putative functions of the assembled unigenes,we compared all unigenes with public protein databases,namely NCBI non-redundant protein (Nr),Swiss-Prot protein,Clusters of Orthologous Groups (COG),Gene Ontology (GO),and the Kyoto Encyclopedia of Genes and Genomes (KEGG)(Figure S2)[59].Among the 62,895unigenes,27,629and 18,157were identified in Nr and Swiss-Prot (Table 2),respectively.The E value,similarity,and species distributions of the annotated results were compared with Nr and Swiss-Prot.Nr and Swiss-Prot results respectively showed that 74.59%(20,609)and 79.74%(14,478)of unigenes had high homology (E value range 10–5to 10–50),and 60.67%(16,761)and 54.05%(9815)showed 100%similarity (Figures 5A–D ).The species distribution analysis showed that the Chinese fir unigenes were more closely related to sequences of Arabidopsis (8754,28.11%)and Oryza (7838,25.17%)than to Populus (1349,4.33%),Vitis (1104, 3.55%),and Pinus or Picea (824, 2.65%)(Figure 5E ).These results underscore the paucity of referencegenomes and incompletely defined genetic backgrounds for gymnosperms [60].All unigenes were compared with the COG database,and 15,662unigenes of the Chinese fir transcriptome had COG classifications (Figure 6,Tables 2,S2).Among the 25COG categories,the cluster for ‘general function prediction only’(2605,16.63%)represented the largest group,followed by ‘replication,recombination and repair’(1495,9.55%),‘transcription’(1308,8.35%),‘posttranslational modification,protein turnover,chaper-ones’(1238,7.90%),and ‘signal transduction mechanisms’(1015,6.48%).There were only a few annotated unigenes for the categories ‘extracellular structures’(4,0.03%)and ‘nuclear structure’(5,0.03%)(Table S2).Blast2GO (Nr annotation)was applied to assign GO categories [61],and WEGO software [62]showed that 24,791unigenes of the Chinese fir transcriptome had GO classifications (Table 2),including ‘cellular component’(11,819)was a more dominant GO functional classification of the annotated unigenes than the ‘biological process’(7051)and ‘molecular function’(5921).The cellular component category was mostly composed of ‘cell’(3923,63.75%),‘cell part’(3922,63.73%),and ‘organelle’(2902,47.16%).The ‘biological process’consisted mostly of the ‘metabolic process’(2220,36.07%)and ‘cellular process’(2147,34.89%),and ‘molecular function’was comprised mostlyofFigure 1.Analysis of cambium ultrastructure.The photographs display the transverse section of second vascular tissue that contains phloem (Ph)and xylem (Xy).CZ,Cambial Zone.The following stages are shown:(A)Reactivation (February 28,stage 1),(B)active growth (May 26,stage 2),(C)transition to dormancy (October 12,stage 3),and (D)dormancy (January 17,stage 4).Bars =2.0m m (A),10.0m m (B),and 5.0m m (C,D).doi:10.1371/journal.pone.0071562.g001‘binding’(2659,43.21%)and ‘catalytic’(2457,39.93%)(Figure 7,Table S3).These results were similar to those published for non-model plant transcriptomes [57,58].A total of 14,402annotated sequences of the cambium transcriptome were assigned to 125KEGG reference canonical pathways (Tables 2,S4).The pathways most represented were ‘secondary metabolites’(5277),‘amino acid metabolism’(1693),‘plant-pathogen interaction’(1197),‘carbohydrate metabolism’(1183),and ‘transcription’(1093)(Table 3).These results partially explained why a large number of secondary metabolites exist in Chinese fir.Coding Sequence Prediction and Expression AnalysisTo further infer unigene function for Chinese fir,we identified unigene coding sequences and searched protein databases using BLASTX (E value ,1025)in the following order:Nr,Swiss-Prot,KEGG,and COG.If unigenes were matched in one database,they were not further analyzed against another database.These BLAST results were applied to collect coding DNA sequence information from unigenes and then translate them into peptide sequences.Unigenes with no matches in BLASTX (using ESTScan)[63]were employed to predict coding DNA sequences and then translate them into peptide sequences.Finally,30,818and 4037unigenes had their coding DNA sequences identified by BLASTX and ESTScan,respectively.Expression of all unigenes was denoted by reads per kilobase of exon model per million mapped reads (RPKM)values [64].Table 4lists the top 10most frequently expressed unigenes in the cambial transcriptome.The average RPKM value for all unigenes was 30.16.eIF-3was a Stable Housekeeping Gene for qRT-PCR in Four Stage-specific CambiumsTo obtain reliable results from qRT-PCR analysis,the ideal reference gene (internal control gene)should be selected in samples regardless of different tissues,developmental stages,and/or sample treatment [65–67],and the combination of geNorm [68],NormFinder [69],and BestKeeper [67,70,71]software that had been applied to identify reference genes from the transcrip-Table1.Length distribution of assembled contigs and unigenes.Nucleotide length (bp)Contigs Unigenes 75–100500,2450100–20077,620409201–30026,11525,464301–40010,93911,626401–50063006673501–60040694329601–70028243048701–80019962154801–90014651666901–1000115413181001–1200161418631201–1400103212321401–16006718661601–18004656291801–2000332466.20006171152Total 637,45862,895N50(bp)90613Mean length (bp)131505Total nucleotides length (bp)83,701,04631,748,167doi:10.1371/journal.pone.0071562.t001Figure 2.Distribution of uniquely mapped reads of assembled unigenes.The horizontal axis indicates the number of reads.The vertical axis indicates the number of assembled unigenes.doi:10.1371/journal.pone.0071562.g002tome data [71,72].We selected nine housekeeping genes from the transcriptome data on Chinese fir cambium,and identified their expression by RT-PCR before qRT-PCR analysis (Table S5).The analysis of expressional stability in the nine housekeeping genes was carried out by qRT-PCR.geNorm,NormFinder,and BestKeeper were used to analyze the results.geNorm evaluatedhousekeeping gene results from most stable to least stable:ClUBQ ,CleIF-4A ,CleIF-3,ClActin ,ClEF-1a ,ClGAPDH,Cl40S ,Cl b -TU ,Cl a -TU (Figure S3,Table S6).NormFinder analyzed housekeeping genes from most stable to least stable:ClActin ,CleIF-4A ,ClUBQ ,CleIF-3,ClEF-1a ,ClGAPDH ,Cl40S ,Cl b -TU ,Cl a -TU (Figure S4,Table S6).BestKeeper described the housekeeping genes from most stable to least stable:ClGAPDH ,CleIF-3,ClEF-1a ,ClUBQ ,CleIF-4A ,ClActin ,Cl40S ,Cl a -TU ,Cl b -TU (Figure S5,Table S6).Considering that the difference found with BestKeeper is larger than the other results,we chose the CleIF-3as the most appropriate reference genes for qRT-PCR analysis of cambial development in Chinese fir.The Altered Expression of Six Genes was Correlated Positively with Changes in Cambial ActivityTo identify genes in the Chinese fir transcriptome database that regulate cambial activity,17highly expressed genes (RPKM value .30.16)(Tables 5,S7)that are known homologs of model-plant regulatory genes were selected as candidates to perform expression analysis of stages S1–S4in cambium tissues.In angiosperms,these regulatory genes play important roles in controlling cambium initiation and development [20–22,34,73,74],vascular develop-ment and differentiation [6,20,40,75–77],and plant stem cell fate [20–22,40,77].Therefore,it is interesting to examine the functional conservation of these regulatory genes between angiosperms and gymnosperms.The 17candidate genes could be categorized as follows:(i)WOX and CLV genes:ClWOX1,ClWOX4,ClWOX8,ClWOX9,ClCLV1,ClCLV1-like ,ClCLV2,CLV-like ,and ClCLE12;(ii)ClassIII HD-Zip genes:ClREV ,ClPHB1,andFigure 3.Random distribution of HiSeq 2000sequencing reads in the assembled unigenes.The horizontal axis indicates relative position in gene (5’R 3’).The vertical axis indicates the number of assembled reads.doi:10.1371/journal.pone.0071562.g003Figure parison of length distribution between hit and no-hit unigenes.The horizontal axis indicates length distribution of hit and no-hit assembled unigenes.The vertical axis indicates the percentage of hit and no-hit assembled unigenes.doi:10.1371/journal.pone.0071562.g004ClATHB15;(iii)hormone-related genes:ClPIN1-like ,ClAUX ,and ClARR7;and (iv)ClSHR and ClSCR .Comparison of the results of the gene expression analysis (Figure 8)and the histological analysis (Figure 1)revealed that the observed altered expression of the 17candidates could be classified into three groups.The first group (ClWOX1,ClWOX4,ClCLV1-like ,ClCLV-like ,ClCLE12,ClPIN1-like ,ClWOX8,ClATHB15,and ClWOX9)was expressed most highly during S2.Six genes (ClWOX1,ClWOX4,ClCLV1-like ,ClCLV-like ,ClCLE12,and ClPIN1-like )were upregulated during S1and S2and downregulated during S3and S4.Interestingly,ClWOX4expres-sion increased 415-fold between S2and S4.The second group (ClCLV1,ClREV ,and ClSCR )was expressed most highly during S3.In the third group,the expression of ClSHR graduallydecreasedFigure 5.Assigned distribution of unigenes.E value (A,B),similarity (C,D),and species (E)distributions using results from the Nr (A,C,E)and Swiss-Prot (B,D,E)databases.All unigenes were blasted with a cut-off E value of 1E 25.doi:10.1371/journal.pone.0071562.g005Table 2.Summary of the percentage of unigenes annotated for Chinese fir.Database Number of unigenes Percent of unigenes annotated Nr 27,62943.93Swiss-Prot 18,15728.87COG 15,66224.90GO 24,79139.42KEGG14,40222.90doi:10.1371/journal.pone.0071562.t002doi:10.1371/journal.pone.0071562.g006Figure7.GO classification of the transcriptome.GO classification of the Chinese fir transcriptome.The horizontal axis indicates the three main categories that include molecular function,cellular component,and biological process,and certain categories that are self evident.The horizontal axis indicates the three main categories that include molecular function,cellular component,and biological process,and certain categories that are self evident.The right vertical axis indicates the number of assembled unigenes in each category,and the left vertical axis indicates the percentage of a certain type of subcategory in that main category.doi:10.1371/journal.pone.0071562.g007from S1to S4,and that of ClAUX correlated negatively withcambial activity.These results revealed that the altered expression of six candidate genes,namely ClWOX1,ClWOX4,ClCLV1-like ,ClCLV-like ,ClCLE12,and ClPIN1-like ,correlated positively with changes in cambial activity;moreover,these six genes might be directly involved in cambial function in Chinese fir.Molecular Characterization of ClWOX1and ClWOX4The altered expression of six genes (ClWOX1,ClWOX4,ClCLV1-like ,ClCLV-like ,ClCLE12,and ClPIN1-like )was correlated positively with changes in cambial activity.WOX genes perform similar regulation functions in the plant embryo development of both gymnosperms and angiosperms [78–80],and it has been revealed that WOX4can regulate the cambial activity in angiosperms [21].In addition,WOX1has also been identified with regulating lateral organ development in angiosperms [81,82].ClWOX1and ClWOX4were selected to clone from among the six genes mentioned in the preceding paragraph.The gene structure of ClWOX1and ClWOX4are displayed in Figure S6.These results will facilitate phylogenetic analyses and structural comparison of the WOX gene family in gymnosperms and angiosperms.As shown in Figure S7,WUS homodomain and WUS box sequences are similar to the results of the previous study showing that the multiple alignment of amino acid sequences predicted WOX proteins [82,83](Figure S7).Based on the full amino acid sequences of 108WOX homologs,a phylogenetic tree could be resolved into the following three subgroups:WUS ,WOX9,WOX13lineage [82–84](Figure S8).ClWOX1and ClWOX4were assigned to the WUS lineage.ClWOX1was closely related to PSWUS (Pinus )and AtWOX6(Arabidopsis ).ClWOX4was more closely related to PsitWOX4(Pinus sitchensis ).DiscussionFor conifers with very large genomes (18to 35Gb)[85],studies of wood formation and vascular tissue activity have focused on transcript profile investigations in conifer trees to date [86].Moreover,recent development of NGS technologies provides revolutionary tools for detecting transcriptome especially with the advent of RNA-sequencing (RNA-seq)[87].For example,Illumina,454,and SOLiD have been widely applied on NSG platforms [28–30,87–89].However,each sequencing technology has its own advantage and disadvantage [87–89].For example,Table 3.Mapping of assembled unigenes of the Chinese fir transcriptome to KEGG pathways.KEGG pathways KEGG sub-pathways Number of unigenes MetabolismCarbohydrate metabolism 1183Lipid metabolism 824Amino acid metabolism 1693Nucleotide metabolism605Glycan biosynthesis and degradation 191Metabolism of cofactors and vitamins 232Secondary metabolites5277Genetic information processing Replication and repair 660Transcription1093Folding,sorting and degradation 206Translation210Plant-pathogen interaction1197doi:10.1371/journal.pone.0071562.t003Table 4.The top 10most frequently expressed unigenes in the Chinese fir transcriptome.Rank Unigene ID RPKM value Function annotation 1Unigene 42991005.0917Ribosomal protein S72Unigene 19270731.2727Mtn21-like protein 3Unigene 26045687.10274-coumarate:CoA ligase 4Unigene 5166595.8491Unknown5Unigene 11581583.4117Plasma intrinsic protein 2,16Unigene 33274583.1480Malate dehydrogenase 7Unigene 23196576.9831Lactoylglutathione lyase 8Unigene 13199570.8247Actin 9Unigene 18523566.9721Ubiquitin10Unigene 15174561.1256UPF0727protein WS02710_H03doi:10.1371/journal.pone.0071562.t004Figure 8.Expression analysis of 17homologous genes in cambium tissues during the four growth stages,S1–S4,using qRT-PCR.Error bars represent the standard error of three biological replicates.doi:10.1371/journal.pone.0071562.g008SOLiD increases the accuracy of sequencing results against double error correction,but with the longest run times[88,89].454has the advantage of longer reads,fast run times,good choice for de novo assembly,but existing higher reagent costs and error rates in homopolymeric tracts[88,89].The Illumina technology has high coverage,cost-effectiveness,and high-throughput,but inherent shorter read-lengths,less feasible for de novo assembly[88,89]. Specially,Illumina HiSeq2000has been shown that it is currently the most widely used NGS platform[89].In the present study,we used Illumina HiSeq2000to perform transcriptome sequencing for activated stage cambium of Chinese fir.20million sequencing reads(3.6Gb)were generated.The quality of transcriptome data is a decisive factor for various downstream analyses[90].However,the quality of the data may be affected by several factors,including contaminant of adapter/ primer sequences,poor quality reads,and assembly errors[90].In this study,a total of62,895unigenes were assembled using SOAPdenovo software,with an average length of505bp and a N50of613bp.For non-model plants,owing to absence of homology unigenes,the average length of unigenes after assembly from Illumina transcriptome sequencing is usually less than 500bp,such as is the case with Camellia sinensis(mean=355bp) [58],Dendrocalamus latiflorus(mean=461bp)[91],Hevea brasiliensis (mean=485bp)[92].Surprisingly,Chinese fir unigenes have an average length of505bp.Moreover,comparison analysis of Chinese fir and Pinus taeda in gene sequence similarity also suggested that the quality of our results was credible.However, each assembly strategy has an effect on the accuracy of transcriptome data[87,93].To further improve the accuracy of assembled Chinese fir transcriptome sequences,comparative study of different assembly software tools for NGS technologies are worthy of special attention in future analysis.QRT-PCR is a common means of confirming the expression pattern and the quality of transcriptome data[94].The ideal reference gene is essential for the accurate measurement in qRT-PCR[67].Our transcriptome database presumably provides a great potential source of candidate reference genes.Through comprehensive analysis of geNorm,NormFinder,and BestKeeper, CleIF-3was identified as the most appropriate reference gene for qRT-PCR analysis during cambial development in Chinese fir. CleIF-3not only supplies a housekeeping gene for gene expression analysis in cambium,but also considers as the potential reference gene for gene expression analysis in samples regardless of different tissues,developmental stages,and/or sample treatment.In2012, Hossain et al.indicated that eEF-1B b1(locus Atlg30230)is involved in plant growth and cell elongation,and plays an important role in plant development and cell wall biosynthesis in Arabidopsis[95].In the future,we should examine the functional conservation of housekeeping genes between Chinese fir and Arabidopsis.WUSCHEL(WUS)-related homeobox(WOX)genes,with15 members in Arabidopsis,play an important role in controlling plant stem cell fate and organogenesis[20–22,80,81].In Arabidopsis, WOX4can promote the proliferation of procambial/cambial stem cells[20–22].In petunia(Petunia6hybrida),an ortholog of Arabidopsis WOX1(MAEWEST,MAW)and CHORIPETALA SUZANNE(CHSU)regulates petal fusion and leaf development, and WOX1and PRESSED FLOWER(PRS)redundantly show a similar function in Arabidopsis[81].Although the functions of ClWOX1and ClWOX4need to be more accurately determined to verify whether they indeed regulate cambial activity in Chinese fir, our results suggest that these two genes may regulate cambial activity.These results also demonstrate that the quality of the transcriptome data is sufficient for gene cloning,and provide insight for further research on stage-specific cambial activity.In addition,previous studies showed that WOX2,WOX8and WOX9 have similar function in embryonic growth both in gymnosperms (Picea abies)and angiosperms(Arabidopsis)[78,79].Thus,we have initiated transgenic analysis of the ClWOX1and ClWOX4in Arabidopsis.The functional verification of ClWOX1and ClWOX4 genes,which correlated positively with changes in cambial activity, can help to further understand the mechanisms of cambial activityTable5.The17homologous genes used for qRT-PCR.Gene Unigene ID RPKM value Putative annotation ID E value ClWOX1Unigene5635990.27WUSCHEL-related homeobox1gi|61217290| 2.00E-06 ClWOX4Unigene30112144.58WUSCHEL-related homeobox4gi|30693997| 4.00E-29 ClWOX8Unigene58521145.76WUSCHEL-related homeobox8gi|75286325| 2.00E-53 ClWOX9Unigene372938.05WUSCHEL-related homeobox9gi|61217281|7.00E-13 ClCLV1Unigene2021934.14CLAVATA1gi|290796119| 3.00E-91 ClCLV1-like Unigene2606535.74CLAVATA1-like gi|290882856| 1.00E-22 ClCLV2Unigene6021539.28CLAVATA2gi|6049567| 1.00E-92 ClCLV-like Unigene5788632.16CLAVATA-like gi|104642235| 5.00E-56 ClCLE12Unigene5977836.69CLE12(CLAVATA3/ESR-RELATED12)gi|18409123| 1.00E-06 ClREV Unigene2293957.94REVOLUTA gi|75203823| 1.00E-14 ClPHB1Unigene56464133.88PHB1gi|71370257| 1.00E-98 ClATHB15Unigene38884111.27ATHB-15gi|75216693| 5.00E-45 ClSHR Unigene6057030.89SHR gi|75213595| 2.00E-90 ClSCR Unigene4826137.52SCARECROW gi|112012486| 2.00E-54 ClPIN1-like Unigene38615123.70PIN1-like auxin transport protein gi|25956262| 1.00E-39 ClAUX Unigene13481412.34Auxin influx transport protein gi|151564283| 4.00E-42 ClARR7Unigene6076265.09Two-component response regulator ARR7gi|51316125| 2.00E-39 doi:10.1371/journal.pone.0071562.t005。
